Efficient Assessment and Optimisation of Medium Components Influencing Extracellular Xylanase Production by Pediococcus pentosaceus G4 Using Statistical Approaches
Abstract
1. Introduction
2. Results and Discussion
2.1. Assessment of Medium Components by Plackett–Burman Design
2.2. Optimisation of Selected Medium Compositions by Central Composite Design
2.2.1. Extracellular Xylanase Production of P. pentosaceus G4
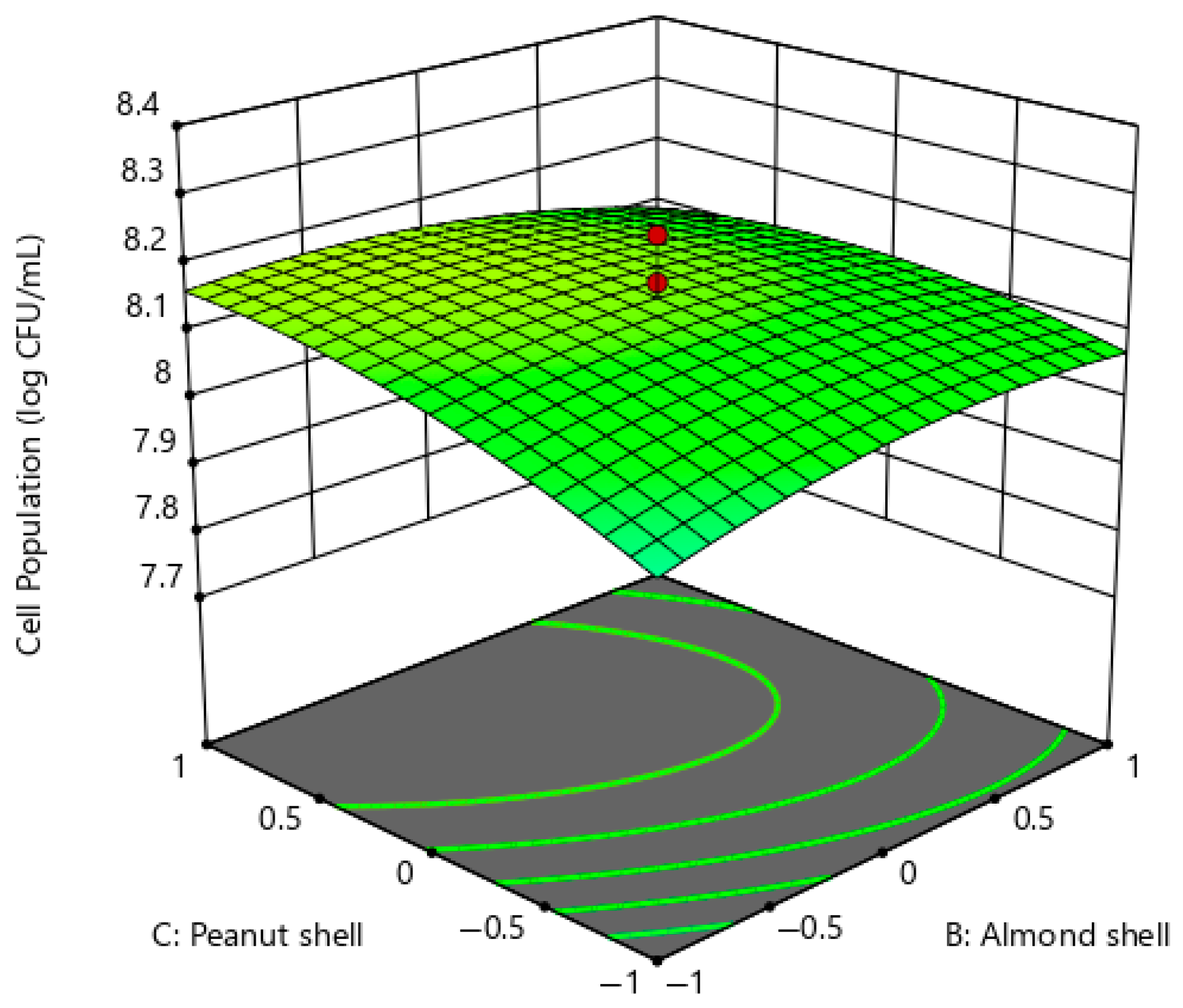
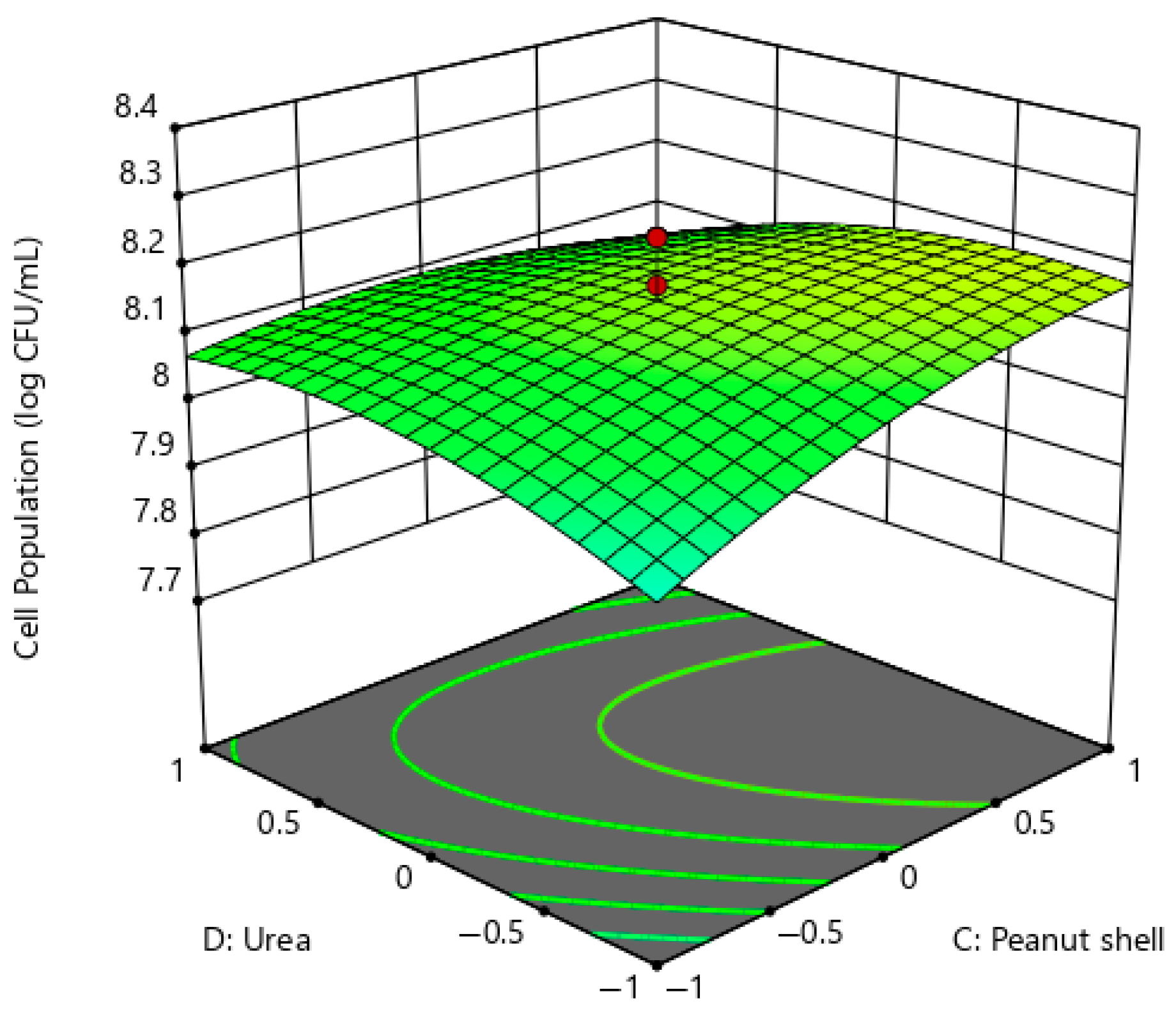
2.2.2. Correlation of Other Factors Associated with Extracellular Xylanase Production of P. pentosaceus G4
Cell Viability of P. pentosaceus G4
Lactic Acid Production and pH
Sugar Utilisation
3. Materials and Methods
3.1. Inoculum Maintenance and Preparation
3.2. Experimental Design of Optimisation of Extracellular Xylanase Production
3.2.1. Plackett–Burman Design
3.2.2. Central Composite Design
3.3. Extracellular Xylanase Production of P. pentosaceus G4
3.4. Determination of Cell Viability
3.5. Determination of Lactic Acid Concentration
3.6. Determination of Sugar Utilisation
3.7. Initial and Final pH Determination
3.8. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Saavedra-Bouza, A.; Escuder-Rodríguez, J.-J.; deCastro, M.-E.; Becerra, M.; González-Siso, M.-I. Xylanases from thermophilic archaea: A hidden treasure. Curr. Res. Biotechnol. 2023, 5, 100116. [Google Scholar] [CrossRef]
- Mendonça, M.; Barroca, M.; Collins, T. Endo-1,4-β-xylanase-containing glycoside hydrolase families: Characteristics, singularities and similarities. Biotechnol. Adv. 2023, 65, 108148. [Google Scholar] [CrossRef]
- Devi, S.; Dwivedi, D.; Bhatt, A.K. Utilization of agroresidues for the production of xylanase by Bacillus safensis XPS7 and optimization of production parameters. Fermentation 2022, 8, 221. [Google Scholar] [CrossRef]
- Chakdar, H.; Kumar, M.; Pandiyan, K.; Singh, A.; Nanjappan, K.; Kashyap, P.L.; Srivastava, A.K. Bacterial xylanases: Biology to biotechnology. 3 Biotech 2016, 6, 150. [Google Scholar] [CrossRef]
- Shrestha, S.; Chio, C.; Khatiwada, J.R.; Kognou, A.L.M.; Qin, W. Formulation of the agro-waste mixture for multi-enzyme (pectinase, xylanase, and cellulase) production by mixture design method exploiting Streptomyces sp. Bioresour. Technol. Rep. 2022, 19, 101142. [Google Scholar] [CrossRef]
- Nabais, J.M.V.; Laginhas, C.E.C.; Carrott, P.J.M.; Ribeiro Carrott, M.M.L. Production of activated carbons from almond shell. Fuel Process. Technol. 2011, 92, 234–240. [Google Scholar] [CrossRef]
- Di Michele, A.; Pagano, C.; Allegrini, A.; Blasi, F.; Cossignani, L.; Raimo, E.D.; Faieta, M.; Oliva, E.; Pittia, P.; Primavilla, S.; et al. Hazelnut shells as source of active ingredients: Extracts preparation and characterization. Molecules 2021, 26, 6607. [Google Scholar] [CrossRef]
- Pączkowski, P.; Puszka, A.; Gawdzik, B. Effect of eco-friendly peanut shell powder on the chemical resistance, physical, thermal, and thermomechanical properties of unsaturated polyester resin Composites. Polymers 2021, 13, 3690. [Google Scholar] [CrossRef]
- Bilal, M.; Iqbal, H.M.N. Recent advancements in the life cycle analysis of lignocellulosic biomass. Curr. Sustain./Renew. Energy Rep. 2020, 7, 100–107. [Google Scholar] [CrossRef]
- Dwivedi, P.; Vivekanand, V.; Ganguly, R.; Singh, R.P. Parthenium sp. as a plant biomass for the production of alkalitolerant xylanase from mutant Penicillium oxalicum SAUE-3.510 in submerged fermentation. Biomass Bioenergy 2009, 33, 581–588. [Google Scholar] [CrossRef]
- Terrone, C.C.; Freitas, C.D.; Terrasan, C.R.F.; Almeida, A.F.D.; Carmona, E.C. Agroindustrial biomass for xylanase production by Penicillium chrysogenum: Purification, biochemical properties and hydrolysis of hemicelluloses. Electron. J. Biotechnol. 2018, 33, 39–45. [Google Scholar] [CrossRef]
- Sadaf, A.; Khare, S.K. Production of Sporotrichum thermophile xylanase by solid state fermentation utilizing deoiled Jatropha curcas seed cake and its application in xylooligosachharide synthesis. Bioresour. Technol. 2014, 153, 126–130. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, Y.-M.; Lebaka, V.R.; Wee, Y.-J. Lactic acid for green chemical industry: Recent advances in and future prospects for production technology, recovery, and applications. Fermentation 2022, 8, 609. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Kumar, B.; Verma, P. A detailed overview of xylanases: An emerging biomolecule for current and future prospective. Bioresour. Bioprocess. 2019, 6, 40. [Google Scholar] [CrossRef]
- Allikian, K.; Edgar, R.; Syed, R.; Zhang, S. Fundamentals of Fermentation Media. In Essentials in Fermentation Technology; Berenjian, A., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 41–84. [Google Scholar]
- Panda, B.P.; Ali, M.; Javed, S. Fermentation process optimization. Res. J. Microbiol. 2007, 2, 201–208. [Google Scholar] [CrossRef]
- Lim, Y.H.; Foo, H.L.; Loh, T.C.; Mohamad, R.; Abdul Rahim, R. Rapid evaluation and optimization of medium components governing tryptophan production by Pediococcus acidilactici TP-6 isolated from Malaysian food via statistical approaches. Molecules 2020, 25, 779. [Google Scholar] [CrossRef]
- Limkar, M.B.; Pawar, S.V.; Rathod, V.K. Statistical optimization of xylanase and alkaline protease co-production by Bacillus spp. using Box-Behnken Design under submerged fermentation using wheat bran as a substrate. Biocatal. Agric. Biotechnol. 2019, 17, 455–464. [Google Scholar] [CrossRef]
- Singh, V.; Haque, S.; Niwas, R.; Srivastava, A.; Pasupuleti, M.; Tripathi, C.K.M. Strategies for fermentation medium optimization: An in-depth review. Front. Microbiol. 2017, 7, 2087. [Google Scholar] [CrossRef]
- Topakas, E.; Panagiotou, G.; Christakopoulos, P. Xylanases: Characteristics, sources, production, and applications. In Bioprocessing Technologies in Biorefinery for Sustainable Production of Fuels, Chemicals, and Polymers; Yang, S.-T., El-Enshasy, H.A., Thongchul, N., Eds.; Wiley: New York, NY, USA, 2013; Chapter 9; pp. 147–166. [Google Scholar]
- Jacyna, J.; Kordalewska, M.; Markuszewski, M.J. Design of experiments in metabolomics-related studies: An overview. J. Pharm. Biomed. Anal. 2019, 164, 598–606. [Google Scholar] [CrossRef]
- Dhaver, P.; Pletschke, B.; Sithole, B.; Govinden, R. Optimization, purification, and characterization of xylanase production by a newly isolated Trichoderma harzianum strain by a two-step statistical experimental design strategy. Sci. Rep. 2022, 12, 17791. [Google Scholar] [CrossRef]
- Walia, A.; Guleria, S.; Mehta, P.; Chauhan, A.; Parkash, J. Microbial xylanases and their industrial application in pulp and paper biobleaching: A review. 3 Biotech 2017, 7, 11. [Google Scholar] [CrossRef]
- Coman, G.; Bahrim, G. Optimization of xylanase production by Streptomyces sp. P12-137 using response surface methodology and central composite design. Ann. Microbiol. 2011, 61, 773–779. [Google Scholar] [CrossRef]
- Bibra, M.; Kunreddy, V.R.; Sani, R.K. Thermostable xylanase production by Geobacillus sp. strain DUSELR13, and its application in ethanol production with lignocellulosic biomass. Microorganisms 2018, 6, 93. [Google Scholar] [CrossRef]
- Thite, V.S.; Nerurkar, A.S.; Baxi, N.N. Optimization of concurrent production of xylanolytic and pectinolytic enzymes by Bacillus safensis M35 and Bacillus altitudinis J208 using agro-industrial biomass through response surface methodology. Sci. Rep. 2020, 10, 3824. [Google Scholar] [CrossRef] [PubMed]
- Fernandes de Souza, H.; Aguiar Borges, L.; Dédalo Di Próspero Gonçalves, V.; Vitor dos Santos, J.; Sousa Bessa, M.; Fronja Carosia, M.; Vieira de Carvalho, M.; Viana Brandi, I.; Setsuko Kamimura, E. Recent advances in the application of xylanases in the food industry and production by actinobacteria: A review. Food Res. Int. 2022, 162, 112103. [Google Scholar] [CrossRef] [PubMed]
- Pariza, M.W.; Johnson, E.A. Evaluating the safety of microbial enzyme preparations used in food processing: Update for a new century. Regul. Toxicol. Pharmacol. 2001, 33, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Lee, F.H.; Wan, S.Y.; Foo, H.L.; Loh, T.C.; Mohamad, R.; Abdul Rahim, R.; Idrus, Z. Comparative study of extracellular proteolytic, cellulolytic, and hemicellulolytic enzyme activities and biotransformation of palm kernel cake biomass by lactic acid bacteria isolated from Malaysian foods. Int. J. Mol. Sci. 2019, 20, 4979. [Google Scholar] [CrossRef]
- Zabidi, N.A.M.; Foo, H.L.; Loh, T.C.; Mohamad, R.; Abdul Rahim, R. Enhancement of versatile extracellular cellulolytic and hemicellulolytic enzyme productions by Lactobacillus plantarum RI 11 isolated from Malaysian food using renewable natural polymers. Molecules 2020, 25, 2607. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Z.; Zhao, H.; Xu, Y.; Cui, F. Statistical optimization of xylanase production from new isolated Penicillium oxalicum ZH-30 in submerged fermentation. Biochem. Eng. J. 2007, 34, 82–86. [Google Scholar] [CrossRef]
- Bedade, D.; Berezina, O.; Singhal, R.; Deska, J.; Shamekh, S. Extracellular xylanase production from a new xylanase producer Tuber maculatum mycelium under submerged fermentation and its characterization. Biocatal. Agric. Biotechnol. 2017, 11, 288–293. [Google Scholar] [CrossRef]
- Nagar, S.; Gupta, V.K.; Kumar, D.; Kumar, L.; Kuhad, R.C. Production and optimization of cellulase-free, alkali-stable xylanase by Bacillus pumilus SV-85S in submerged fermentation. J. Ind. Microbiol. Biotechnol. 2010, 37, 71–83. [Google Scholar] [CrossRef]
- Ikram-ul-Haq, M.M.J.; Khan, T.S. An innovative approach for hyperproduction of cellulolytic and hemicellulolytic enzymes by consortium of Aspergillus niger MSK-7 and Trichoderma viride MSK-10. Afr. J. Biotechnol. 2006, 5, 609–614. [Google Scholar]
- Jecu, L. Solid state fermentation of agricultural wastes for endoglucanase production. Ind. Crops Prod. 2000, 11, 1–5. [Google Scholar] [CrossRef]
- Gowdhaman, D.; Manaswini, V.S.; Jayanthi, V.; Dhanasri, M.; Jeyalakshmi, G.; Gunasekar, V.; Sugumaran, K.R.; Ponnusami, V. Xylanase production from Bacillus aerophilus KGJ2 and its application in xylooligosaccharides preparation. Int. J. Biol. Macromol. 2014, 64, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Velkova, Z.; Gocheva, V.; Kostov, G.; Atev, A. Optimization of nutritive media composition for hylanase production be Aspergillus awamori. Bulg. J. Agric. Sci. 2007, 13, 651–656. [Google Scholar]
- Lama, L.; Calandrelli, V.; Gambacorta, A.; Nicolaus, B. Purification and characterization of thermostable xylanase and β-xylosidase by the thermophilic bacterium Bacillus thermantarcticus. Res. Microbiol. 2004, 155, 283–289. [Google Scholar] [CrossRef]
- Mmango-Kaseke, Z.; Okaiyeto, K.; Nwodo, U.U.; Mabinya, L.V.; Okoh, A.I. Optimization of cellulase and xylanase production by Micrococcus species under submerged fermentation. Sustainability 2016, 8, 1168. [Google Scholar] [CrossRef]
- Duarte, M.C.T.; Portugal, E.P.; Ponezi, A.N.; Bim, M.A.; Tagliari, C.V.; Franco, T.T. Production and purification of alkaline xylanases. Bioresour. Technol. 1999, 68, 49–53. [Google Scholar] [CrossRef]
- Bhalla, A.; Bischoff, K.M.; Sani, R.K. Highly thermostable xylanase production from a thermophilic Geobacillus sp. strain WSUCF1 utilizing lignocellulosic biomass. Front. Bioeng. Biotechnol. 2015, 3, 84. [Google Scholar] [CrossRef]
- Blanco, A.; Vidal, T.; Colom, J.F.; Pastor, F. Purification and properties of xylanase A from alkali-tolerant Bacillus sp. strain BP-23. Appl. Environ. Microbiol. 1995, 61, 4468–4470. [Google Scholar] [CrossRef]
- Tseng, M.-J.; Yap, M.-N.; Ratanakhanokchai, K.; Kyu, K.L.; Chen, S.-T. Purification and characterization of two cellulase free xylanases from an alkaliphilic Bacillus firmus. Enzym. Microb. Technol. 2002, 30, 590–595. [Google Scholar] [CrossRef]
- Poosarla, V.G.; Chandra, T. Purification and characterization of novel halo-acid-alkali-thermo-stable xylanase from Gracilibacillus sp. TSCPVG. Appl. Biochem. Biotechnol. 2014, 173, 1375–1390. [Google Scholar] [CrossRef] [PubMed]
- Giridhar, P.V.; Chandra, T.S. Production of novel halo-alkali-thermo-stable xylanase by a newly isolated moderately halophilic and alkali-tolerant Gracilibacillus sp. TSCPVG. Process Biochem. 2010, 45, 1730–1737. [Google Scholar] [CrossRef]
- Boucherba, N.; Gagaoua, M.; Copinet, E.; Bettache, A.; Duchiron, F.; Benallaoua, S. Purification and characterization of the xylanase produced by Jonesia denitrificans BN-13. Appl. Biochem. Biotechnol. 2014, 172, 2694–2705. [Google Scholar] [CrossRef]
- Dahlberg, L.; Holst, O.; Kristjansson, J.K. Thermostable xylanolytic enzymes from Rhodothermus marinus grown on xylan. Appl. Microbiol. Biotechnol. 1993, 40, 63–68. [Google Scholar] [CrossRef]
- Xin, F.; He, J. Characterization of a thermostable xylanase from a newly isolated Kluyvera species and its application for biobutanol production. Bioresour. Technol. 2013, 135, 309–315. [Google Scholar] [CrossRef]
- Dheeran, P.; Nandhagopal, N.; Kumar, S.; Jaiswal, Y.K.; Adhikari, D.K. A novel thermostable xylanase of Paenibacillus macerans IIPSP3 isolated from the termite gut. J. Ind. Microbiol. Biotechnol. 2012, 39, 851–860. [Google Scholar] [CrossRef]
- Seyis, I.; Aksoz, N. Effect of carbon and nitrogen sources on xylanase production by Trichoderma harzianum 1073 D3. Int. Biodeterior. Biodegrad. 2005, 55, 115–119. [Google Scholar] [CrossRef]
- Pasalari, A.; Homaei, A. Isolation and molecular identification of xylanase-producing bacteria from Ulva flexuosa of the Persian Gulf. Processes 2022, 10, 1834. [Google Scholar] [CrossRef]
- Ravanal, M.-C.; Rosa, L.; Polanco, R.; Eyzaguirre, J.; Espinosa, Y.; Levicán, G.; Chávez, R.; Vaca, I. Glucose-induced production of a Penicillium purpurogenum xylanase by Aspergillus nidulans. Mycoscience 2012, 53, 152–155. [Google Scholar] [CrossRef]
- Knob, A.; Fortkamp, D.; Prolo, T.; Izidoro, S.C.; Almeida, J.M. Agro-residues as alternative for xylanase production by filamentous fungi. BioResources 2014, 9, 5738–5773. [Google Scholar]
- Wang, Y.; Wu, J.; Lv, M.; Shao, Z.; Hungwe, M.; Wang, J.; Bai, X.; Xie, J.; Wang, Y.; Geng, W. Metabolism characteristics of lactic acid bacteria and the expanding applications in food industry. Front. Bioeng. Biotechnol. 2021, 9, 612285. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, E.H.M.; Avanci, N.C.; Romano, L.H.; Branco, D.L.; de Pádua, A.X.; Ward, R.J.; Baptista Neto, Á.D.; Lourenzoni, M.R. Recombinant xylanase production by Escherichia coli using a non-induced expression system with different nutrient sources. Braz. J. Chem. Eng. 2020, 37, 29–39. [Google Scholar] [CrossRef]
- Shi, X.; Xie, J.; Liao, S.; Wu, T.; Zhao, L.-G.; Ding, G.; Wang, Z.; Xiao, W. High-level expression of recombinant thermostable β-glucosidase in Escherichia coli by regulating acetic acid. Bioresour. Technol. 2017, 241, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.D.; Nadar, C.G.; Muir, J.; Arora, A. Green and clean process to obtain low degree of polymerisation xylooligosaccharides from almond shell. J. Clean. Prod. 2019, 241, 118237. [Google Scholar] [CrossRef]
- Ebringerová, A.; Hromádková, Z.; Košt’álová, Z.; Sasinková, V. Chemical valorization of agricultural by-products: Isolation and characterization of xylan-based antioxidants from almond shell biomass. BioResources 2008, 3, 60–70. [Google Scholar] [CrossRef]
- Queirós, C.S.G.P.; Cardoso, S.; Lourenço, A.; Ferreira, J.; Miranda, I.; Lourenço, M.J.V.; Pereira, H. Characterization of walnut, almond, and pine nut shells regarding chemical composition and extract composition. Biomass Convers. Biorefinery 2020, 10, 175–188. [Google Scholar] [CrossRef]
- Cho, C.H.; Hatsu, M.; Takamizawa, K. The production of D-xylose by enzymatic hydrolysis of agricultural wastes. Water Sci. Technol. 2002, 45, 97–102. [Google Scholar] [CrossRef]
- Barbieri, G.S.; Bento, H.B.S.; de Oliveira, F.; Picheli, F.P.; Dias, L.M.; Masarin, F.; Santos-Ebinuma, V.C. Xylanase production by Talaromyces amestolkiae valuing agroindustrial byproducts. BioTech 2022, 11, 15. [Google Scholar] [CrossRef]
- Do, T.T.; Quyen, D.T.; Dam, T.H. Purification and characterization of an acid-stable and organic solvent-tolerant xylanase from Aspergillus awamori VTCC-F312. ScienceAsia 2012, 38, 157–165. [Google Scholar] [CrossRef]
- Yang, C.-H.; Yang, S.-F.; Liu, W.-H. Production of xylooligosaccharides from xylans by extracellular xylanases from Thermobifida fusca. J. Agric. Food Chem. 2007, 55, 3955–3959. [Google Scholar] [CrossRef] [PubMed]
- Raju, G.; Kumarappa, S.; Gaitonde, V. Mechanical and physical characterization of agricultural waste reinforced polymer composites. J. Mater. Environ. Sci. 2012, 3, 907–916. [Google Scholar]
- Arumugam, N.; Biely, P.; Puchart, V.; Singh, S.; Pillai, S. Structure of peanut shell xylan and its conversion to oligosaccharides. Process Biochem. 2018, 72, 124–129. [Google Scholar] [CrossRef]
- Kamble, R.D.; Jadhav, A.R. Isolation, purification, and characterization of xylanase produced by a new species of Bacillus in solid state fermentation. Int. J. Microbiol. 2012, 2012, 683193. [Google Scholar] [CrossRef]
- Paul, M.; Nayak, D.P.; Thatoi, H. Optimization of xylanase from Pseudomonas mohnii isolated from Simlipal Biosphere Reserve, Odisha, using response surface methodology. J. Genet. Eng. Biotechnol. 2020, 18, 81. [Google Scholar] [CrossRef]
- Kumar, A.R.; Hegde, S.S.; Ganesh, K.N.; Khan, M.I. Structural changes enhance the activity of Chainia xylanase in low urea concentrations. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2003, 1645, 164–171. [Google Scholar] [CrossRef]
- Yin, T.; Miao, L.L.; Guan, F.F.; Wang, G.L.; Peng, Q.; Li, B.X.; Guan, G.H.; Li, Y. Optimized medium improves expression and secretion of extremely thermostable bacterial xylanase, XynB, in Kluyveromyces lactis. J. Microbiol. Biotechnol. 2010, 20, 1471–1480. [Google Scholar] [CrossRef]
- Sá-Pereira, P.; Mesquita, A.; Duarte, J.C.; Barros, M.R.A.; Costa-Ferreira, M. Rapid production of thermostable cellulase-free xylanase by a strain of Bacillus subtilis and its properties. Enzym. Microb. Technol. 2002, 30, 924–933. [Google Scholar] [CrossRef]
- Marimuthu, M.; Sorimuthu, A.; Muruganantham, S. Production and optimization of xylanase enzyme from Bacillus subtilis using agricultural wastes by solid state fermentation. Int. J. Pharm. Investig. 2019, 9, 169–173. [Google Scholar] [CrossRef]
- Ellatif, S.A.; Abdel Razik, E.S.; AL-surhanee, A.A.; Al-Sarraj, F.; Daigham, G.E.; Mahfouz, A.Y. Enhanced production, cloning, and expression of a xylanase gene from endophytic fungal strain Trichoderma harzianum kj831197.1: Unveiling the in vitro anti-fungal activity against phytopathogenic fungi. J. Fungi 2022, 8, 447. [Google Scholar] [CrossRef]
- Battan, B.; Sharma, J.; Kuhad, R. High-level xylanase production by alkaliphilic Bacillus pumilus ASH under solid-state fermentation. World J. Microbiol. Biotechnol. 2006, 22, 1281–1287. [Google Scholar] [CrossRef]
- Sanghi, A.; Garg, N.; Sharma, J.; Kuhar, K.; Kuhad, R.C.; Gupta, V.K. Optimization of xylanase production using inexpensive agro-residues by alkalophilic Bacillus subtilis ASH in solid-state fermentation. World J. Microbiol. Biotechnol. 2008, 24, 633–640. [Google Scholar] [CrossRef]
- Adhyaru, D.N.; Bhatt, N.S.; Modi, H.A. Enhanced production of cellulase-free, thermo-alkali-solvent-stable xylanase from Bacillus altitudinis DHN8, its characterization and application in sorghum straw saccharification. Biocatal. Agric. Biotechnol. 2014, 3, 182–190. [Google Scholar] [CrossRef]
- Palaniswamy, M.; Pradeep, B.V.; Sathya, R.; Angayarkanni, J. Isolation, identification and screening of potential xylanolytic enzyme from litter degrading fungi. Afr. J. Biotechnol. 2008, 7, 1978–1982. [Google Scholar] [CrossRef]
- Amezaga, M.R.; Booth, I.R. Osmoprotection of Escherichia coli by peptone is mediated by the uptake and accumulation of free proline but not of proline-containing peptides. Appl. Environ. Microbiol. 1999, 65, 5272–5278. [Google Scholar] [CrossRef]
- Davami, F.; Eghbalpour, F.; Nematollahi, L.; Barkhordari, F.; Mahboudi, F. Effects of peptone supplementation in different culture media on growth, metabolic pathway and productivity of CHO DG44 Cells; a new insight into amino acid profiles. Iran. Biomed. J. 2015, 19, 194–205. [Google Scholar] [CrossRef]
- Sepahy, A.A.; Ghazi, S.; Sepahy, M.A. Cost-effective production and optimization of alkaline xylanase by indigenous Bacillus mojavensis AG137 fermented on agricultural waste. Enzym. Res. 2011, 2011, 593624. [Google Scholar] [CrossRef]
- Bocchini, D.A.; Alves-Prado, H.F.; Baida, L.C.; Roberto, I.C.; Gomes, E.; Da Silva, R. Optimization of xylanase production by Bacillus circulans D1 in submerged fermentation using response surface methodology. Process Biochem. 2002, 38, 727–731. [Google Scholar] [CrossRef]
- Atalla, S.M.M.; Ahmed, N.E.; Awad, H.M.; El Gamal, N.G.; El Shamy, A.R. Statistical optimization of xylanase production, using different agricultural wastes by Aspergillus oryzae MN894021, as a biological control of faba bean root diseases. Egypt. J. Biol. Pest Control 2020, 30, 125. [Google Scholar] [CrossRef]
- Ravindran, R.; Williams, G.A.; Jaiswal, A.K. Spent coffee waste as a potential media component for xylanase production and potential application in juice enrichment. Foods 2019, 8, 585. [Google Scholar] [CrossRef]
- Geetha, K.; Gunasekaran, P. Optimization of nutrient medium containing agricultural waste for xylanase production by Bacillus pumilus B20. Biotechnol. Bioprocess. Eng. 2010, 15, 882–889. [Google Scholar] [CrossRef]
- Long, C.; Liu, J.; Gan, L.; Zeng, B.; Long, M. Optimization of xylanase production by Trichoderma orientalis using corn cobs and wheat bran via statistical strategy. Waste Biomass Valorization 2019, 10, 1277–1284. [Google Scholar] [CrossRef]
- Senthilkumar, S.R.; Ashokkumar, B.; Chandra Raj, K.; Gunasekaran, P. Optimization of medium composition for alkali-stable xylanase production by Aspergillus fischeri Fxn 1 in solid-state fermentation using central composite rotary design. Bioresour. Technol. 2005, 96, 1380–1386. [Google Scholar] [CrossRef] [PubMed]
- Jahnen-Dechent, W.; Ketteler, M. Magnesium basics. Clin. Kidney J. 2012, 5, i3–i14. [Google Scholar] [CrossRef]
- Sissi, C.; Palumbo, M. Effects of magnesium and related divalent metal ions in topoisomerase structure and function. Nucleic Acids Res. 2009, 37, 702–711. [Google Scholar] [CrossRef]
- Lee, D.-Y.D.; Galera-Laporta, L.; Bialecka-Fornal, M.; Moon, E.C.; Shen, Z.; Briggs, S.P.; Garcia-Ojalvo, J.; Süel, G.M. Magnesium flux modulates ribosomes to increase bacterial survival. Cell 2019, 177, 352–360.e313. [Google Scholar] [CrossRef]
- Lusk, J.E.; Williams, R.J.P.; Kennedy, E.P. Magnesium and the Growth of Escherichia coli. J. Biol. Chem. 1968, 243, 2618–2624. [Google Scholar] [CrossRef]
- Watcharawipas, A.; Watanabe, D.; Takagi, H. Sodium acetate responses in Saccharomyces cerevisiae and the Ubiquitin ligase Rsp5. Front. Microbiol. 2018, 9, 2495. [Google Scholar] [CrossRef]
- Lim, Y.H.; Foo, H.L.; Loh, T.C.; Mohamad, R.; Abdul Rahim, R.; Idrus, Z. Optimized medium via statistical approach enhanced threonine production by Pediococcus pentosaceus TL-3 isolated from Malaysian food. Microb. Cell Factories 2019, 18, 125. [Google Scholar] [CrossRef]
- Kaushal, R.; Sharma, N.; Dogra, V. Optimization of the production and molecular characterization of cellulase-free xylanase from an alkalophillic Bacillus subtilis SD8 isolated from paper mill effluent. Appl. Biochem. Microbiol. 2015, 51, 551–559. [Google Scholar] [CrossRef]
- Patel, K.; Dudhagara, P. Optimization of xylanase production by Bacillus tequilensis strain UD-3 using economical agricultural substrate and its application in rice straw pulp bleaching. Biocatal. Agric. Biotechnol. 2020, 30, 101846. [Google Scholar] [CrossRef]
- Sharma, D.; Sharma, G.; Mahajan, R. Development of strategy for simultaneous enhanced production of alkaline xylanase-pectinase enzymes by a bacterial isolate in short submerged fermentation cycle. Enzym. Microb. Technol. 2019, 122, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Cubas-Cano, E.; González-Fernández, C.; Ballesteros, M.; Tomás-Pejó, E. Biotechnological advances in lactic acid production by lactic acid bacteria: Lignocellulose as novel substrate. Biofuels Bioprod. Biorefining 2018, 12, 290–303. [Google Scholar] [CrossRef]
- Jiang, J.; Yang, B.; Ross, R.P.; Stanton, C.; Zhao, J.; Zhang, H.; Chen, W. Comparative genomics of Pediococcus pentosaceus isolated from different niches reveals genetic diversity in carbohydrate metabolism and immune system. Front. Microbiol. 2020, 11, 253. [Google Scholar] [CrossRef]
- Raman, J.; Kim, J.-S.; Choi, K.R.; Eun, H.; Yang, D.; Ko, Y.-J.; Kim, S.-J. Application of lactic acid bacteria (LAB) in sustainable agriculture: Advantages and limitations. Int. J. Mol. Sci. 2022, 23, 7784. [Google Scholar] [CrossRef]
- Nwamba, M.C.; Sun, F.; Mukasekuru, M.R.; Song, G.; Harindintwali, J.D.; Boyi, S.A.; Sun, H. Trends and hassles in the microbial production of lactic acid from lignocellulosic biomass. Environ. Technol. Innov. 2021, 21, 101337. [Google Scholar] [CrossRef]
- Carvalho, I.P.C.D.; Detmann, E.; Mantovani, H.C.; Paulino, M.F.; Valadares Filho, S.D.C.; Costa, V.A.C.; Gomes, D.I. Growth and antimicrobial activity of lactic acid bacteria from rumen fluid according to energy or nitrogen source. Rev. Bras. Zootec. 2011, 40, 1260–1265. [Google Scholar] [CrossRef]
- Nel, H.A.; Bauer, R.; Vandamme, E.J.; Dicks, L.M. Growth optimization of Pediococcus damnosus NCFB 1832 and the influence of pH and nutrients on the production of pediocin PD-1. J. Appl. Microbiol. 2001, 91, 1131–1138. [Google Scholar] [CrossRef]
- Karne, H.; Moharir, S. Optimization of lactic acid production from different substrates using Rhizopus oryzae. Mater. Today Proc. 2023. [Google Scholar] [CrossRef]
- Brown, D.M.; Grunden, A.M.; Pawlak, J.J. Statistical optimization of black liquor-containing media for growth and lactic acid production by Paenibacillus glucanolyticus SLM1. Bioresour. Technol. Rep. 2021, 13, 100629. [Google Scholar] [CrossRef]
- Altaf, M.; Naveena, B.J.; Reddy, G. Use of inexpensive nitrogen sources and starch for L(+) lactic acid production in anaerobic submerged fermentation. Bioresour. Technol. 2007, 98, 498–503. [Google Scholar] [CrossRef]
- Katepogu, H.; Wee, Y.J.; Anu Appaiah, K.; Chinni, S.V.; Gopinath, S.C.B.; Syed, A.; Verma, M.; Lebaka, V.R. Lactic acid production by Pediococcus pentosaceus HLV1 from banana crop residue: An economic and renewable resource. Biomass Convers. Biorefinery 2023. [Google Scholar] [CrossRef]
- Coelho, L.; De Lima, C.; Rodovalho, C.; Bernardo, M.; Contiero, J. Lactic acid production by new Lactobacillus plantarum LMISM6 grown in molasses: Optimization of medium composition. Braz. J. Chem. Eng. 2011, 28, 27–36. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, C.; Cai, D.; Wang, Z.; Qin, P.; Tan, T. The optimization of L-lactic acid production from sweet sorghum juice by mixed fermentation of Bacillus coagulans and Lactobacillus rhamnosus under unsterile conditions. Bioresour. Technol. 2016, 218, 1098–1105. [Google Scholar] [CrossRef]
- Heenan, C.N.; Adams, M.C.; Hosken, R.W.; Fleet, G.H. Growth medium for culturing probiotic bacteria for applications in vegetarian food products. LWT—Food Sci. Technol. 2002, 35, 171–176. [Google Scholar] [CrossRef]
- Watson, S.P.; Clements, M.O.; Foster, S.J. Characterization of the starvation-survival response of Staphylococcus aureus. J. Bacteriol. 1998, 180, 1750–1758. [Google Scholar] [CrossRef]
- Cui, Y.; Qu, X. Genetic mechanisms of prebiotic carbohydrate metabolism in lactic acid bacteria: Emphasis on Lacticaseibacillus casei and Lacticaseibacillus paracasei as flexible, diverse and outstanding prebiotic carbohydrate starters. Trends Food Sci. Technol. 2021, 115, 486–499. [Google Scholar] [CrossRef]
- Pajak, B.; Siwiak, E.; Sołtyka, M.; Priebe, A.; Zieliński, R.; Fokt, I.; Ziemniak, M.; Jaśkiewicz, A.; Borowski, R.; Domoradzki, T.; et al. 2-Deoxy-d-Glucose and its analogs: From diagnostic to therapeutic agents. Int. J. Mol. Sci. 2020, 21, 234. [Google Scholar] [CrossRef]
- Krisnawati, R.; Cahyanto, M.N.; Sardjono, S.; Suroto, D.A.; Widada, J. RNA-seq data of Aspergillus tubingensis NBRC 31125 in carbon catabolite repressor related to xylanase production. Data Brief 2022, 45, 108700. [Google Scholar] [CrossRef]
- Farliahati, M.R.; Ramanan, R.N.; Mohamad, R.; Puspaningsih, N.N.T.; Ariff, A.B. Enhanced production of xylanase by recombinant Escherichia coli DH5α through optimization of medium composition using response surface methodology. Ann. Microbiol. 2010, 60, 279–285. [Google Scholar] [CrossRef]
- Alokika; Singh, B. Enhanced production of bacterial xylanase and its utility in saccharification of sugarcane bagasse. Bioprocess Biosyst. Eng. 2020, 43, 1081–1091. [Google Scholar] [CrossRef]
- Foo, H.; Loh, T.; Lai, P.; Lim, Y.; Kufli, C.; Rusul, G. Effects of adding Lactobacillus plantarum I-UL4 metabolites in drinking water of rats. Pak. J. Nutr. 2003, 2, 283–288. [Google Scholar] [CrossRef]
- Alshelmani, M.I.; Loh, T.C.; Foo, H.L.; Lau, W.H.; Sazili, A.Q. Biodegradation of palm kernel cake by cellulolytic and hemicellulolytic bacterial cultures through solid state fermentation. Sci. World J. 2014, 2014, 729852. [Google Scholar] [CrossRef]
- Nakkarach, A.; Foo, H.L.; Song, A.A.-L.; Nitisinprasert, S.; Withayagiat, U. Promising discovery of beneficial Escherichia coli in the human gut. 3 Biotech 2020, 10, 296. [Google Scholar] [CrossRef]
- Borshchevskaya, L.; Gordeeva, T.; Kalinina, A.; Sineokii, S. Spectrophotometric determination of lactic acid. J. Anal. Chem. 2016, 71, 755–758. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]

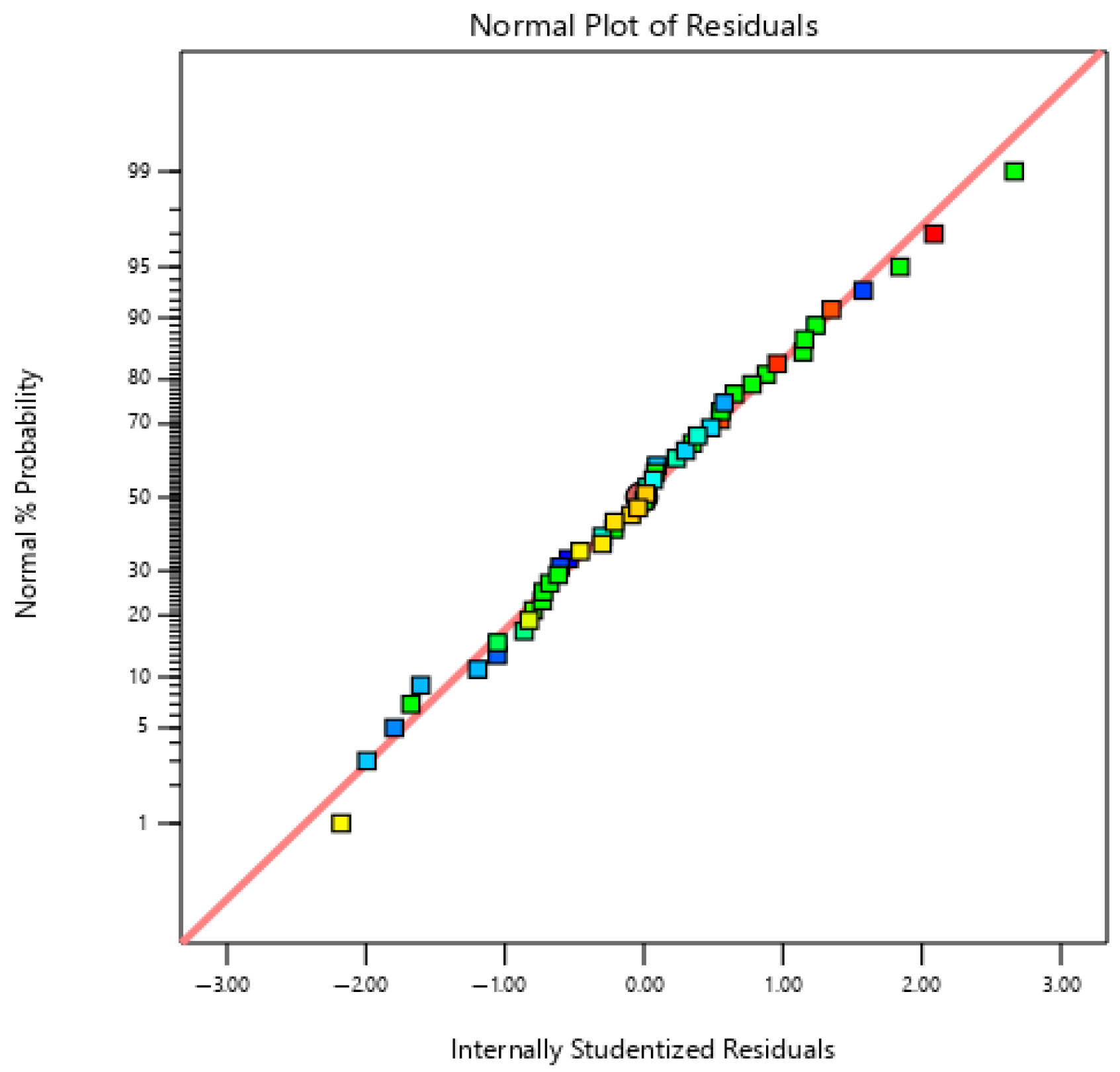
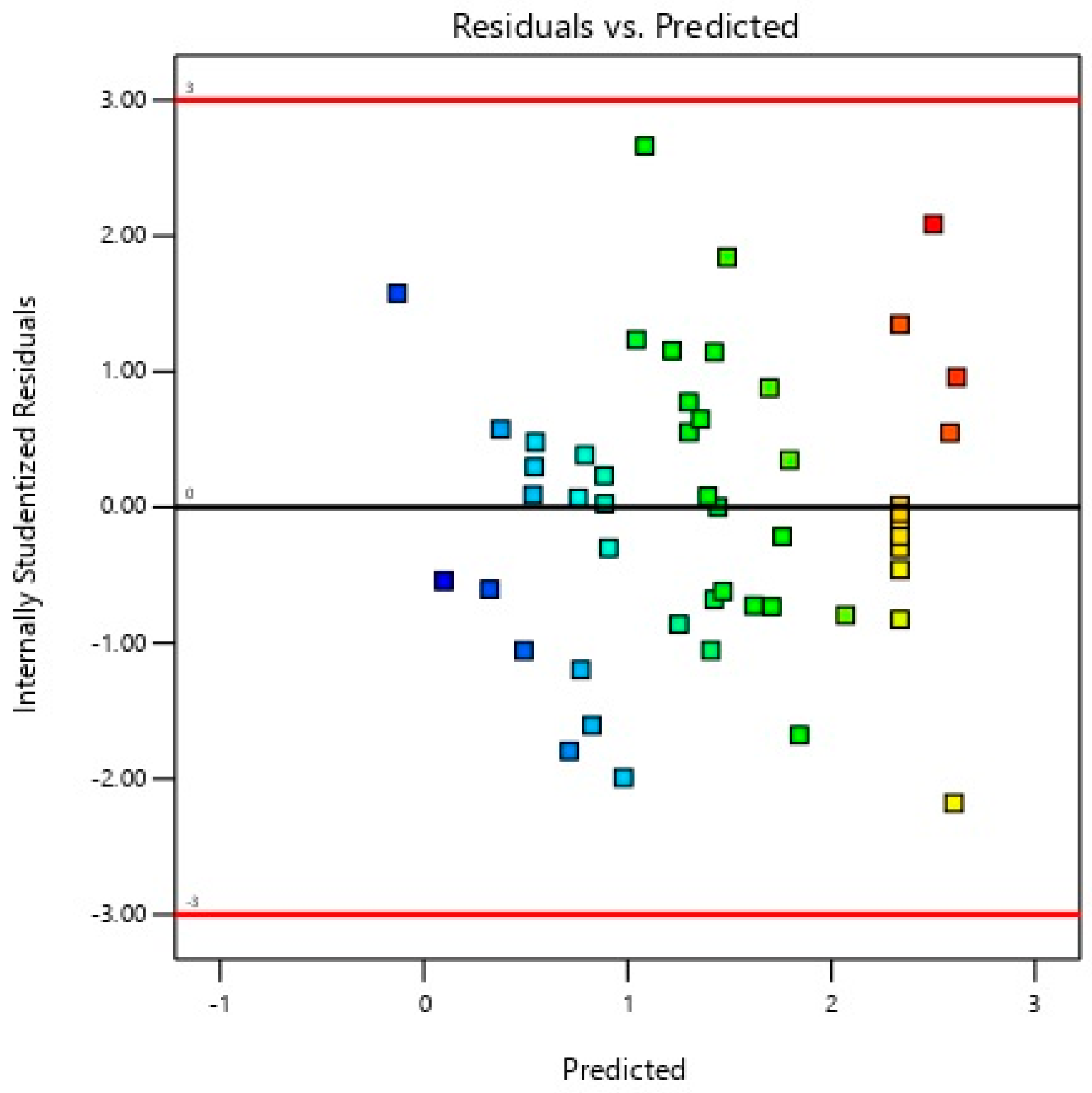

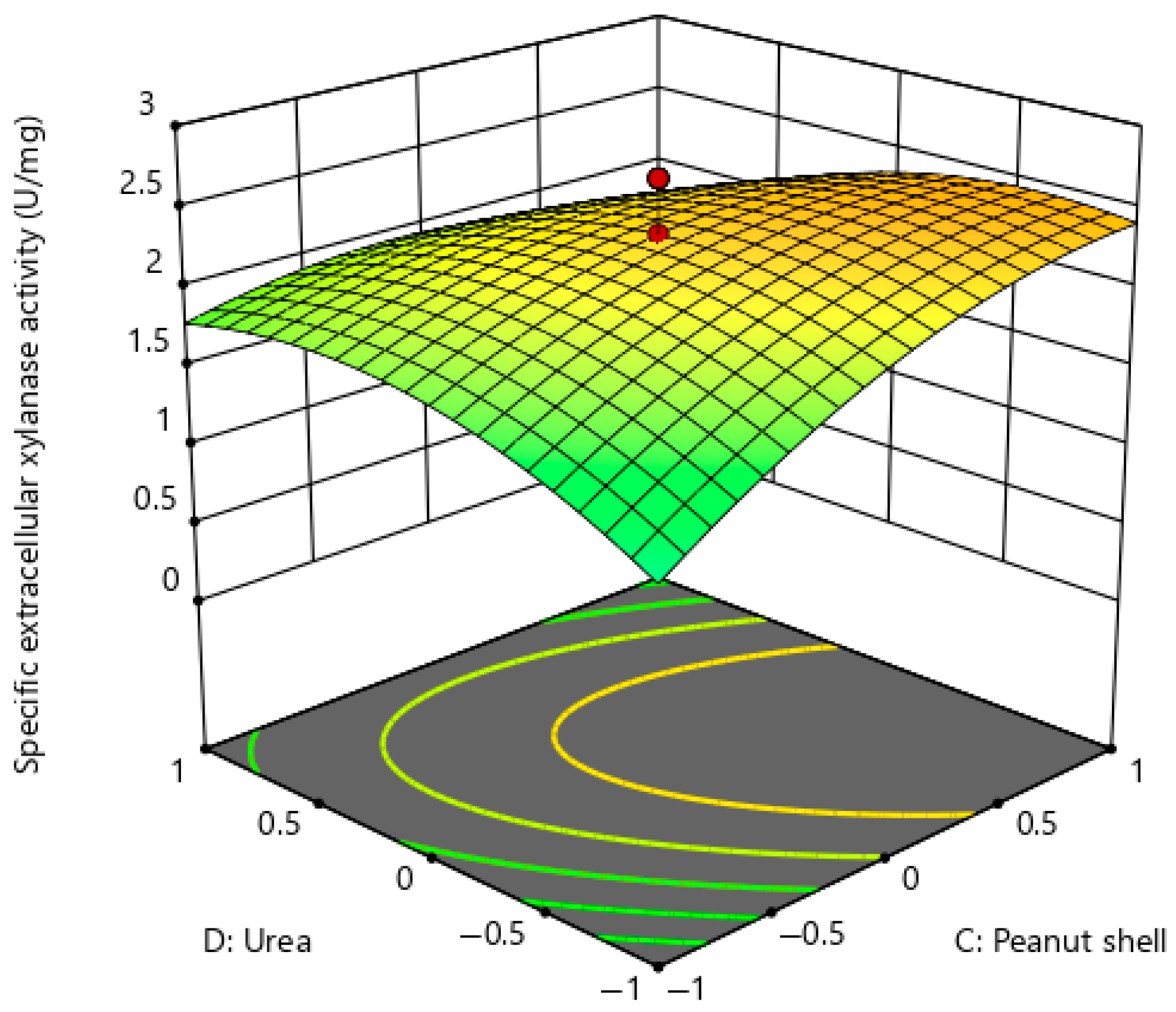
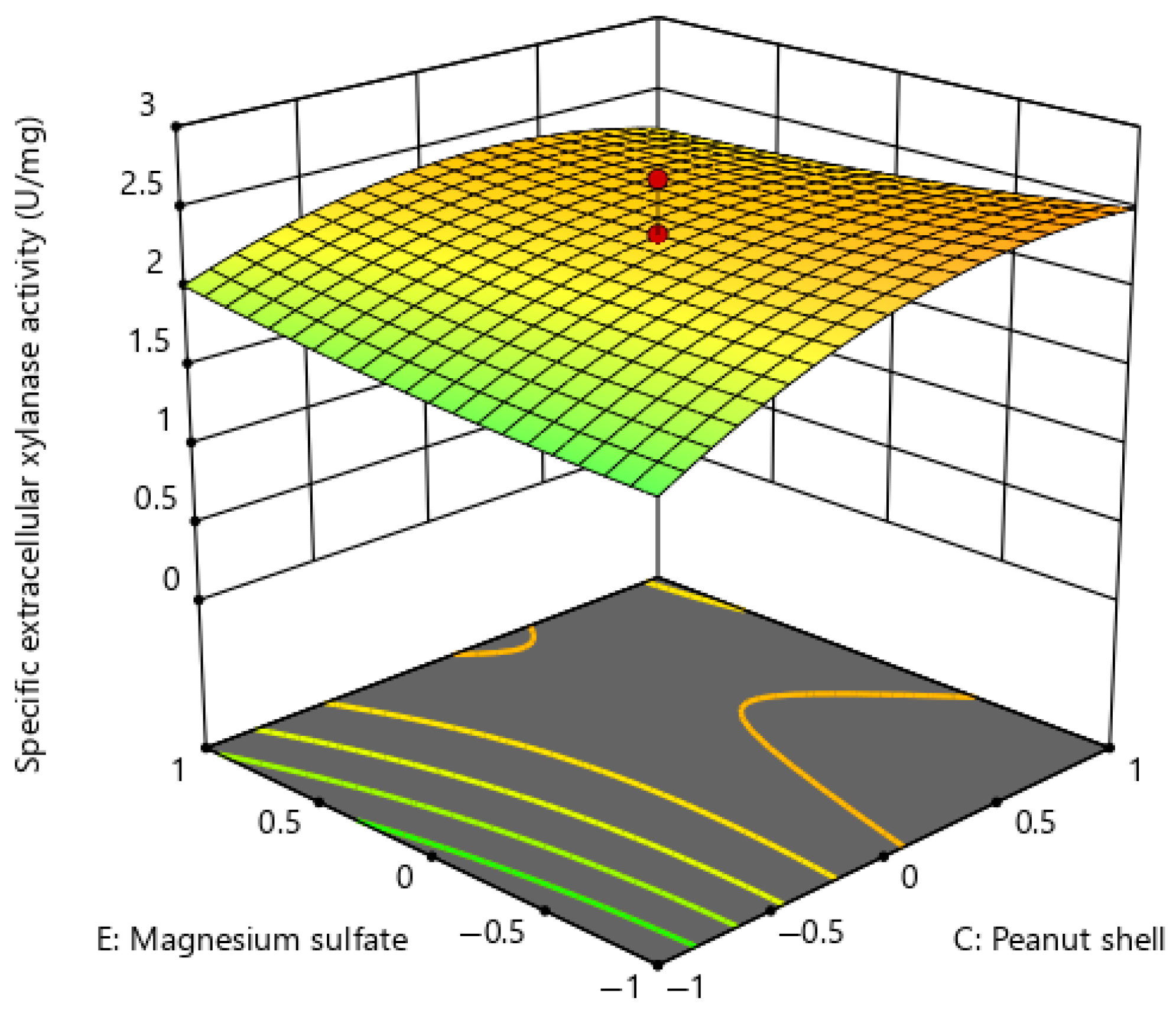
| Experimental Run | A | B | C | D | E | F | G | H | J | K | L | M | N | O | P | Q | R | S | T | Specific Extracellular Xylanase Activity (U/mg) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 0.2913 cd ± 0.0295 |
| 2 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | 0.6062 b ± 0.0497 |
| 3 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 0.2570 d ± 0.0442 |
| 4 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 0.1289 efg ± 0.0381 |
| 5 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | 0.0135 i ± 0.0024 |
| 6 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | 0.0121 i ± 0.0037 |
| 7 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | 0.1552 ef ± 0.0133 |
| 8 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | 0.1071 efg ± 0.0086 |
| 9 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 0.1207 efg ± 0.0050 |
| 10 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | 0.0989 fgh ± 0.0620 |
| 11 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 0.3486 c ± 0.0098 |
| 12 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | 0.0000 i ± 0.0000 |
| 13 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 0.0741 ghi ± 0.0058 |
| 14 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 0.1817 e ± 0.0046 |
| 15 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 0.6453 b ± 0.0382 |
| 16 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1.0329 a ± 0.0307 |
| 17 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | 0.0278 hi ± 0.0039 |
| 18 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | 0.0000 i ± 0.0000 |
| 19 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 0.0000 i ± 0.0000 |
| 20 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | 0.0000 i ± 0.0000 |
| Source | Sum of Squares | df | Mean Square | F-Value | p-Value | |
|---|---|---|---|---|---|---|
| Model | 1.40 | 15 | 0.0931 | 159.83 | <0.0001 | Significant |
| A—Glucose | 0.2121 | 1 | 0.2121 | 364.30 | <0.0001 | |
| B—Almond shell | 0.1905 | 1 | 0.1905 | 327.10 | <0.0001 | |
| C—Peanut shell | 0.1870 | 1 | 0.1870 | 321.10 | <0.0001 | |
| D—Hazelnut shell | 0.0130 | 1 | 0.0130 | 22.34 | 0.0091 | |
| E—Pistachio shell | 0.0738 | 1 | 0.0738 | 126.75 | 0.0004 | |
| F—Walnut shell | 0.0198 | 1 | 0.0198 | 34.05 | 0.0043 | |
| G—Malt extract | 0.0054 | 1 | 0.0054 | 9.22 | 0.0385 | |
| H—Xylan | 0.1903 | 1 | 0.1903 | 326.83 | <0.0001 | |
| J—Peptone | 0.0389 | 1 | 0.0389 | 66.76 | 0.0012 | |
| K—Yeast extract | 0.2222 | 1 | 0.2222 | 381.70 | <0.0001 | |
| L—Meat extract | 0.0372 | 1 | 0.0372 | 63.94 | 0.0013 | |
| N—Urea | 0.1384 | 1 | 0.1384 | 237.69 | 0.0001 | |
| P—Sodium acetate | 0.0288 | 1 | 0.0288 | 49.48 | 0.0022 | |
| Q—Magnesium sulphate | 0.0266 | 1 | 0.0266 | 45.62 | 0.0025 | |
| S—Dipotassium hydrogen phosphate | 0.0120 | 1 | 0.0120 | 20.63 | 0.0105 | |
| Residual | 0.0023 | 4 | 0.0006 | |||
| Cor Total | 1.40 | 19 |
| Experimental Run | Specific Extracellular Xylanase Activity (U/mg) | Cell Population (Log CFU/mL) | Lactic Acid (g/mL) | Utilised Sugar (g/L) | Initial pH | Final pH | ||
|---|---|---|---|---|---|---|---|---|
| Experimental | Predicted * | Experimental | Predicted * | Experimental | Experimental | Experimental | Experimental | |
| 1 | 0.1888 no ± 0.0456 | −0.1308 | 7.8030 klm ± 0.0906 | 7.73844 | 11.0105 j ± 0.0101 | 4.6196 m ± 0.0984 | 6.21 | 4.85 |
| 2 | 0.5277 ijklmno ± 0.0655 | 0.7697 | 7.8667 ijklm ± 0.1191 | 7.90031 | 13.1940 ghij ± 0.0253 | 6.6145 ijklm ± 0.1873 | 6.23 | 4.66 |
| 3 | 0.2773 lmno ± 0.0644 | 0.4908 | 7.8127 jklm ± 0.0737 | 7.84304 | 11.2079 j ± 0.0209 | 5.0546 lm ± 0.1171 | 6.25 | 4.77 |
| 4 | 1.6239 bcdefghij ± 0.0951 | 1.0844 | 8.0794 bcdefgh ± 0.0919 | 7.96835 | 14.7503 abcdefghi ± 0.4851 | 8.9243 cdefghij ± 0.1050 | 6.22 | 4.51 |
| 5 | 1.8737 abcdefgh ± 0.0439 | 1.6952 | 8.0961 bcdefgh ± 0.0653 | 8.05298 | 15.0929 abcdefghi ± 0.0419 | 9.3443 cdefghi ± 0.1281 | 6.26 | 4.45 |
| 6 | 2.6942 abc ± 0.0751 | 2.5828 | 8.2407 abcd ± 0.0668 | 8.21065 | 16.8351 abc ± 0.0253 | 11.5041 abc ± 1.4970 | 6.21 | 4.30 |
| 7 | 1.8624 abcdefgh ± 0.0551 | 1.4892 | 8.0918 bcdefgh ± 0.0730 | 8.01477 | 15.0830 abcdefghi ± 0.0041 | 9.3892 cdefghi ± 0.0794 | 6.21 | 4.45 |
| 8 | 1.9088 abcdefgh ± 0.1221 | 2.0696 | 8.0948 bcdefgh ± 0.0201 | 8.13588 | 15.5981 abcdefgh ± 0.0290 | 9.4492 cdefghi ± 0.0937 | 6.28 | 4.44 |
| 9 | 0.5541 ijklmno ± 0.0778 | 0.5354 | 7.8934 hijklm ± 0.1061 | 7.87604 | 13.3914 efghij ± 0.0116 | 6.9894 ghijklm ± 0.1333 | 6.4 | 4.66 |
| 10 | 1.4579 efghijkl ± 0.0715 | 1.3004 | 8.0546 cdefghi ± 0.0527 | 7.9939 | 14.5354 abcdefghi ± 0.0838 | 8.5493 cdefghijk ± 0.0520 | 6.4 | 4.56 |
| 11 | 1.4135 efghijkl ± 0.0661 | 1.3013 | 8.0212 efghi ± 0.1247 | 8.0169 | 14.3902 bcdefghi ± 0.0402 | 8.3993 cdefghijkl ± 0.1171 | 6.42 | 4.57 |
| 12 | 1.7159 bcdefghi ± 0.0225 | 1.7591 | 8.0833 bcdefgh ± 0.0757 | 8.0982 | 14.9129 abcdefghi ± 0.0363 | 9.0893 cdefghij ± 0.2810 | 6.44 | 4.47 |
| 13 | 1.2931 efghijklmn ± 0.0665 | 1.0425 | 7.9972 efghijk ± 0.0725 | 7.9646 | 14.2102 cdefghi ± 0.0307 | 8.2793 cdefghijkl ± 0.0260 | 6.41 | 4.57 |
| 14 | 1.8652 abcdefgh ± 0.0233 | 1.7945 | 8.0835 bcdefgh ± 0.0464 | 8.0782 | 15.0174 abcdefghi ± 0.0101 | 9.4942 bcdefghi ± 0.2637 | 6.42 | 4.47 |
| 15 | 0.5771 ijklmno ± 0.0400 | 0.9806 | 7.9263 ghijklm ± 0.0397 | 7.9627 | 13.2346 fghij ± 0.0323 | 6.7945 hijklm ± 0.0937 | 6.41 | 4.66 |
| 16 | 1.6574 bcdefghij ± 0.0700 | 1.42542 | 8.0794 bcdefgh ± 0.0934 | 8.0397 | 14.8548 abcdefghi ± 0.0058 | 9.3743 cdefghi ± 0.1308 | 6.43 | 4.47 |
| 17 | 0.6448 ijklmno ± 0.0069 | 0.5471 | 7.9284 ghijklm ± 0.0243 | 7.8885 | 13.6005 efghij ± 0.0209 | 8.0244 defghijklm ± 0.1308 | 6.28 | 4.60 |
| 18 | 1.4518 efghijkl ± 0.0958 | 1.2178 | 8.0101 efghij ± 0.0744 | 7.9987 | 14.4483 bcdefghi ± 0.1057 | 8.4143 cdefghijkl ± 0.0937 | 6.21 | 4.56 |
| 19 | 0.8933 hijklmno ± 0.0420 | 0.8879 | 7.9512 fghijklm ± 0.0697 | 7.9305 | 13.7398 defghij ± 0.1424 | 8.0544 cdefghijklm ± 0.1580 | 6.29 | 4.58 |
| 20 | 1.0771 fghijklmno ± 0.0838 | 1.2514 | 7.9894 efghijkl ± 0.0521 | 8.0041 | 14.1696 cdefghi ± 0.0154 | 8.1443 cdefghijkl ± 0.1873 | 6.22 | 4.58 |
| 21 | 1.5047 cdefghijk ± 0.0476 | 1.8441 | 8.0687 bcdefghi ± 0.0746 | 8.1307 | 14.7677 abcdefghi ± 0.1108 | 8.9693 cdefghij ± 0.0794 | 6.27 | 4.53 |
| 22 | 2.9243 a ± 0.0919 | 2.5016 | 8.3531 a ± 0.1033 | 8.2366 | 17.3171 a ± 0.0201 | 13.1090 a ± 0.9382 | 6.23 | 4.21 |
| 23 | 1.4889 defghijk ± 0.0116 | 1.3571 | 8.0390 defghi ± 0.0447 | 8.0299 | 14.5470 abcdefghi ± 0.1220 | 8.6093 cdefghijk ± 0.1950 | 6.22 | 4.56 |
| 24 | 1.5597 cdefghijk ± 0.0219 | 1.7075 | 8.0665 bcdefghi ± 0.1074 | 8.0992 | 14.6516 abcdefghi ± 0.0363 | 8.8943 cdefghij ± 0.1670 | 6.21 | 4.55 |
| 25 | 0.8473 hijklmno ± 0.0317 | 0.9080 | 7.9674 fghijklm ± 0.0685 | 7.9561 | 13.5482 efghij ± 0.0058 | 7.4439 efghijklm ± 0.1043 | 6.43 | 4.64 |
| 26 | 1.4436 efghijkl ± 0.0991 | 1.4430 | 8.0187 efghi ± 0.0379 | 8.0222 | 14.5122 abcdefghi ± 0.0266 | 8.4743 cdefghijkl ± 0.1281 | 6.41 | 4.56 |
| 27 | 1.4097 efghijklm ± 0.0324 | 1.3928 | 8.0269 efghi ± 0.0413 | 8.0343 | 14.3438 bcdefghi ± 0.0209 | 8.3483 cdefghijkl ± 0.0323 | 6.42 | 4.57 |
| 28 | 1.4742 efghijkl ± 0.0663 | 1.6207 | 8.0526 cdefghi ± 0.0898 | 8.0639 | 14.6748 abcdefghi ± 0.1108 | 8.8643 cdefghij ± 0.2505 | 6.4 | 4.53 |
| 29 | 0.9333 ghijklmno ± 0.0631 | 0.8859 | 7.9867 efghijkl ± 0.0522 | 7.9723 | 13.7224 defghij ± 0.0253 | 8.0799 cdefghijklm ± 0.7327 | 6.44 | 4.58 |
| 30 | 1.1946 efghijklmno ± 0.0547 | 1.4078 | 7.9935 efghijk ± 0.1328 | 8.0342 | 14.2044 cdefghi ± 0.0154 | 8.3093 cdefghijkl ± 0.2602 | 6.43 | 4.58 |
| 31 | 0.6038 ijklmno ± 0.0120 | 0.54299 | 7.9212 ghijklm ± 0.0323 | 7.9077 | 13.5017 efghij ± 0.0266 | 7.0899 fghijklm ± 0.0437 | 6.41 | 4.65 |
| 32 | 0.7716 hijklmno ± 0.0313 | 0.75780 | 7.9324 ghijklm ± 0.0556 | 7.9330 | 13.5947 efghij ± 0.0154 | 8.0244 defghijklm ± 0.1587 | 6.43 | 4.59 |
| 33 | 0.0000 o ± 0.0000 | 0.09803 | 7.7740 m ± 0.1687 | 7.8150 | 6.2253 k ± 0.1192 | 0.0000 n ± 0.0000 | 6.37 | 5.36 |
| 34 | 1.3018 efghijklmn ± 0.0442 | 1.42451 | 8.0094 efghij ± 0.0462 | 8.0377 | 14.2218 cdefghi ± 0.0768 | 8.2943 cdefghijkl ± 0.0541 | 6.31 | 4.57 |
| 35 | 0.5331 ijklmno ± 0.0906 | 0.82385 | 7.8749 ijklm ± 0.0681 | 7.9561 | 13.2462 fghij ± 0.1018 | 6.8994 hijklm ± 0.1050 | 6.31 | 4.68 |
| 36 | 0.8602 hijklmno ± 0.0678 | 0.7901 | 7.9722 efghijklm ± 0.0231 | 7.9603 | 13.6760 efghij ± 0.0612 | 8.0844 cdefghijkl ± 0.1522 | 6.35 | 4.58 |
| 37 | 0.2138 mno ± 0.0929 | 0.3228 | 7.7893 lm ± 0.0812 | 7.8361 | 12.3461 ij ± 0.0058 | 5.2796 klm ± 0.0150 | 6.32 | 4.77 |
| 38 | 1.3565 efghijklmn ± 0.0600 | 1.4682 | 8.0321 efghi ± 0.0947 | 8.0546 | 13.8211 defghij ± 0.0307 | 8.1593 cdefghijkl ± 0.0600 | 6.38 | 4.58 |
| 39 | 0.3886 klmno ± 0.0556 | 0.7137 | 7.8075 jklm ± 0.0280 | 7.8894 | 12.6829 ij ± 0.0402 | 5.6545 jklm ± 0.0150 | 6.11 | 4.73 |
| 40 | 0.4811 jklmno ± 0.0642 | 0.3766 | 7.8680 ijklm ± 0.0557 | 7.8554 | 12.8571 hij ± 0.0201 | 6.5995 ijklm ± 0.0912 | 6.49 | 4.68 |
| 41 | 2.2082 abcdef ± 0.0705 | 2.6027 | 8.1059 bcdefg ± 0.1303 | 8.1965 | 15.6794 abcdefgh ± 0.0302 | 9.7192 abcdefghi ± 0.0260 | 6.32 | 4.43 |
| 42 | 2.7889 ab ± 0.0787 | 2.6151 | 8.2694 ab ± 0.0686 | 8.2481 | 17.1719 ab ± 0.0058 | 12.9440 ab ± 0.0750 | 6.3 | 4.22 |
| 43 | 2.2604 abcdef ± 0.0929 | 2.3373 | 8.1396 bcdef ± 0.1079 | 8.1515 | 16.0221 abcdefg ± 0.0154 | 10.4992 abcdef ± 0.1200 | 6.3 | 4.43 |
| 44 | 2.2200 abcdef ± 0.0986 | 2.3373 | 8.1022 bcdefg ± 0.0786 | 8.1515 | 16.0046 abcdefg ± 0.1366 | 10.1392 abcdefgh ± 0.0541 | 6.3 | 4.41 |
| 45 | 2.3411 abcde ± 0.1555 | 2.3373 | 8.1743 abcde ± 0.0568 | 8.1515 | 16.5621 abcd ± 0.0253 | 10.8441 abcde ± 0.1800 | 6.3 | 4.37 |
| 46 | 2.3140 abcde ± 0.1454 | 2.3373 | 8.1436 bcdef ± 0.0500 | 8.1515 | 16.1847 abcde ± 0.0101 | 10.5892 abcde ± 0.1050 | 6.3 | 4.38 |
| 47 | 2.1264 abcdefg ± 0.0970 | 2.3373 | 8.1004 bcdefg ± 0.0371 | 8.1515 | 15.8885 abcdefg ± 0.0266 | 10.1242 abcdefgh ± 0.0937 | 6.3 | 4.43 |
| 48 | 2.6822 abcd ± 0.0323 | 2.3373 | 8.2436 abc ± 0.0510 | 8.1515 | 15.7607 abcdefg ± 0.0116 | 9.9592 abcdefghi ± 0.2163 | 6.3 | 4.44 |
| 49 | 2.2831 abcde ± 0.1130 | 2.3373 | 8.1186 bcdefg ± 0.0257 | 8.1515 | 16.0859 abcdef ± 0.2125 | 10.3792 abcdefg ± 0.2116 | 6.3 | 4.41 |
| 50 | 2.3265 abcde ± 0.0481 | 2.3373 | 8.1442 bcdef ± 0.0614 | 8.1515 | 16.5854 abcd ± 0.0402 | 11.1141 abcd ± 0.1819 | 6.3 | 4.31 |
| Regression Model | Sequential p-Value | Lack of Fit p-Value | Adjusted R2 | Predicted R2 | |
|---|---|---|---|---|---|
| Linear | 0.0517 | 0.0001 | 0.1254 | 0.0078 | |
| 2FI | 0.2764 | 0.0001 | 0.1790 | 0.1708 | |
| Quadratic | <0.0001 | 0.0502 | 0.8714 | 0.7323 | Suggested |
| Cubic | 0.6845 | 0.0202 | 0.8545 | −0.6719 | Aliased |
| Source | Sum of Squares | df | Mean Square | F-Value | p-Value | |
|---|---|---|---|---|---|---|
| Model | 26.25 | 20 | 1.31 | 17.60 | <0.0001 | Significant |
| A—Glucose | 3.37 | 1 | 3.37 | 45.15 | <0.0001 | Significant |
| B—Almond shell | 0.0022 | 1 | 0.0022 | 0.0291 | 0.8658 | |
| C—Peanut shell | 2.51 | 1 | 2.51 | 33.67 | <0.0001 | Significant |
| D—Urea | 0.2175 | 1 | 0.2175 | 2.92 | 0.0984 | |
| E—Magnesium sulphate | 0.0003 | 1 | 0.0003 | 0.0039 | 0.9504 | |
| AB | 0.1886 | 1 | 0.1886 | 2.53 | 0.1226 | |
| AC | 0.0003 | 1 | 0.0003 | 0.0046 | 0.9465 | |
| AD | 0.0368 | 1 | 0.0368 | 0.4933 | 0.4881 | |
| AE | 0.1058 | 1 | 0.1058 | 1.42 | 0.2434 | |
| BC | 1.37 | 1 | 1.37 | 18.37 | 0.0002 | Significant |
| BD | 0.0415 | 1 | 0.0415 | 0.5566 | 0.4616 | |
| BE | 0.1579 | 1 | 0.1579 | 2.12 | 0.1565 | |
| CD | 3.48 | 1 | 3.48 | 46.65 | <0.0001 | Significant |
| CE | 0.5601 | 1 | 0.5601 | 7.51 | 0.0104 | Significant |
| DE | 0.1866 | 1 | 0.1866 | 2.50 | 0.1245 | |
| A2 | 4.31 | 1 | 4.31 | 57.82 | <0.0001 | Significant |
| B2 | 4.07 | 1 | 4.07 | 54.52 | <0.0001 | Significant |
| C2 | 3.61 | 1 | 3.61 | 48.39 | <0.0001 | Significant |
| D2 | 5.58 | 1 | 5.58 | 74.76 | <0.0001 | Significant |
| E2 | 0.1281 | 1 | 0.1281 | 1.72 | 0.2004 | |
| Residual | 2.16 | 29 | 0.0746 | |||
| Lack of Fit | 1.98 | 22 | 0.0900 | 3.42 | 0.0502 | Not significant |
| Pure Error | 0.1841 | 7 | 0.0263 | |||
| Cor Total | 28.42 | 49 |
| Media | Specific Extracellular Xylanase Activity (U/mg) | Medium Composition (g/L) | Medium Cost (USD/L) | Increase Specific Extracellular Xylanase Activity (Fold) | Medium Cost Reduction (Fold) | |
|---|---|---|---|---|---|---|
| Composition | Total | |||||
| MRS | 0.8809 | - | 11.11 | Baseline (1.00) | Baseline (1.00) | |
| Optimised Medium | 2.7646 | Glucose | 0.96 | 1.391 | 3.13 | 7.99 |
| Almond Shell | Free | |||||
| Peanut Shell | Free | |||||
| Urea | 0.40 | |||||
| Magnesium Sulphate | 0.031 | |||||
| Source | Sequential p-Value | Lack of Fit p-Value | Adjusted R2 | Predicted R2 | |
|---|---|---|---|---|---|
| Linear | 0.0255 | 0.0054 | 0.1594 | 0.0271 | |
| 2FI | 0.1688 | 0.0070 | 0.2509 | 0.2003 | |
| Quadratic | <0.0001 | 0.2045 | 0.7790 | 0.5415 | Suggested |
| Cubic | 0.8328 | 0.0759 | 0.7210 | −3.2719 | Aliased |
| Source | Sum of Squares | df | Mean Square | F-Value | p-Value | |
|---|---|---|---|---|---|---|
| Model | 0.6864 | 20 | 0.0343 | 9.64 | <0.0001 | Significant |
| A—Glucose | 0.0949 | 1 | 0.0949 | 26.65 | <0.0001 | Significant |
| B—Almond shell | 0.0000 | 1 | 0.0000 | 0.0092 | 0.9241 | |
| C—Peanut shell | 0.0914 | 1 | 0.0914 | 25.66 | <0.0001 | Significant |
| D—Urea | 0.0022 | 1 | 0.0022 | 0.6206 | 0.4372 | |
| E—Magnesium sulphate | 0.0051 | 1 | 0.0051 | 1.44 | 0.2406 | |
| AB | 0.0027 | 1 | 0.0027 | 0.7507 | 0.3934 | |
| AC | 0.0000 | 1 | 0.0000 | 0.0100 | 0.9212 | |
| AD | 0.0039 | 1 | 0.0039 | 1.09 | 0.3056 | |
| AE | 0.0054 | 1 | 0.0054 | 1.50 | 0.2300 | |
| BC | 0.0408 | 1 | 0.0408 | 11.45 | 0.0021 | Significant |
| BD | 0.0026 | 1 | 0.0026 | 0.7395 | 0.3969 | |
| BE | 0.0078 | 1 | 0.0078 | 2.20 | 0.1485 | |
| CD | 0.1021 | 1 | 0.1021 | 28.67 | <0.0001 | Significant |
| CE | 0.0105 | 1 | 0.0105 | 2.94 | 0.0969 | |
| DE | 0.0098 | 1 | 0.0098 | 2.76 | 0.1075 | |
| A2 | 0.0880 | 1 | 0.0880 | 24.71 | <0.0001 | Significant |
| B2 | 0.0649 | 1 | 0.0649 | 18.21 | 0.0002 | Significant |
| C2 | 0.0738 | 1 | 0.0738 | 20.72 | <0.0001 | Significant |
| D2 | 0.1352 | 1 | 0.1352 | 37.98 | <0.0001 | Significant |
| E2 | 0.0087 | 1 | 0.0087 | 2.45 | 0.1287 | |
| Residual | 0.1033 | 29 | 0.0036 | |||
| Lack of Fit | 0.0882 | 22 | 0.0040 | 1.85 | 0.2045 | Not significant |
| Pure Error | 0.0151 | 7 | 0.0022 | |||
| Cor Total | 0.7897 | 49 |
| No. | Medium Component | Symbol Code | Concentration Unit | Coded Values | |
|---|---|---|---|---|---|
| −1 | +1 | ||||
| 1. | Glucose | A | g/L | 0 | 20 |
| 2. | Almond shell | B | g/L | 0 | 20 |
| 3. | Peanut shell | C | g/L | 0 | 20 |
| 4. | Hazelnut shell | D | g/L | 0 | 20 |
| 5. | Pistachio shell | E | g/L | 0 | 20 |
| 6. | Walnut shell | F | g/L | 0 | 20 |
| 7 | Malt extract | G | g/L | 0 | 5 |
| 8. | Xylan | H | g/L | 0 | 20 |
| 9. | Peptone | J | g/L | 0 | 10 |
| 10. | Yeast extract | K | g/L | 0 | 4 |
| 11. | Meat extract | L | g/L | 0 | 8 |
| 12. | Ammonium citrate | M | g/L | 0 | 2 |
| 13. | Urea | N | g/L | 0 | 4 |
| 14. | Potassium nitrate | O | g/L | 0 | 1 |
| 15. | Sodium acetate | P | g/L | 0 | 5 |
| 16. | Magnesium sulphate | Q | g/L | 0 | 0.2 |
| 18. | Manganese sulphate | R | g/L | 0 | 0.04 |
| 18. | Dipotassium hydrogen phosphate | S | g/L | 0 | 2 |
| 19. | Tween 80 | T | ml/L | 0 | 1 |
| Experimental Run | A | B | C | D | E | F | G | H | J | K | L | M | N | O | P | Q | R | S | T |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 |
| 2 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 |
| 3 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 |
| 4 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 |
| 5 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 |
| 6 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 |
| 7 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 |
| 8 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 |
| 9 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 |
| 10 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 |
| 11 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 |
| 12 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 |
| 13 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 | 1 |
| 14 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 | 1 |
| 15 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 1 |
| 16 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 |
| 17 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 |
| 18 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 |
| 19 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 1 |
| 20 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 |
| Medium Components | Coded Symbol | Coded Values | ||||
|---|---|---|---|---|---|---|
| −α | −1 | 0 | +1 | +α | ||
| Glucose | A | 0 | 10 | 20 | 30 | 40 |
| Almond shell | B | 0 | 10 | 20 | 30 | 40 |
| Peanut shell | C | 0 | 10 | 20 | 30 | 40 |
| Urea | D | 0 | 2 | 4 | 6 | 8 |
| Magnesium sulphate | E | 0 | 0.1 | 0.2 | 0.3 | 0.4 |
| Experimental Run | A | B | C | D | E |
|---|---|---|---|---|---|
| 1 | −1 | −1 | −1 | −1 | −1 |
| 2 | 1 | −1 | −1 | −1 | −1 |
| 3 | −1 | 1 | −1 | −1 | −1 |
| 4 | 1 | 1 | −1 | −1 | −1 |
| 5 | −1 | −1 | 1 | −1 | −1 |
| 6 | 1 | −1 | 1 | −1 | −1 |
| 7 | −1 | 1 | 1 | −1 | −1 |
| 8 | 1 | 1 | 1 | −1 | −1 |
| 9 | −1 | −1 | −1 | 1 | −1 |
| 10 | 1 | −1 | −1 | 1 | −1 |
| 11 | −1 | 1 | −1 | 1 | −1 |
| 12 | 1 | 1 | −1 | 1 | −1 |
| 13 | −1 | −1 | 1 | 1 | −1 |
| 14 | 1 | −1 | 1 | 1 | −1 |
| 15 | −1 | 1 | 1 | 1 | −1 |
| 16 | 1 | 1 | 1 | 1 | −1 |
| 17 | −1 | −1 | −1 | −1 | 1 |
| 18 | 1 | −1 | −1 | −1 | 1 |
| 19 | −1 | 1 | −1 | −1 | 1 |
| 20 | 1 | 1 | −1 | −1 | 1 |
| 21 | −1 | −1 | 1 | −1 | 1 |
| 22 | 1 | −1 | 1 | −1 | 1 |
| 23 | −1 | 1 | 1 | −1 | 1 |
| 24 | 1 | 1 | 1 | −1 | 1 |
| 25 | −1 | −1 | −1 | 1 | 1 |
| 26 | 1 | −1 | −1 | 1 | 1 |
| 27 | −1 | 1 | −1 | 1 | 1 |
| 28 | 1 | 1 | −1 | 1 | 1 |
| 29 | −1 | −1 | 1 | 1 | 1 |
| 30 | 1 | −1 | 1 | 1 | 1 |
| 31 | −1 | 1 | 1 | 1 | 1 |
| 32 | 1 | 1 | 1 | 1 | 1 |
| 33 | −α | 0 | 0 | 0 | 0 |
| 34 | +α | 0 | 0 | 0 | 0 |
| 35 | 0 | −α | 0 | 0 | 0 |
| 36 | 0 | +α | 0 | 0 | 0 |
| 37 | 0 | 0 | −α | 0 | 0 |
| 38 | 0 | 0 | +α | 0 | 0 |
| 39 | 0 | 0 | 0 | −α | 0 |
| 40 | 0 | 0 | 0 | +α | 0 |
| 41 | 0 | 0 | 0 | 0 | −α |
| 42 | 0 | 0 | 0 | 0 | +α |
| 43 | 0 | 0 | 0 | 0 | 0 |
| 44 | 0 | 0 | 0 | 0 | 0 |
| 45 | 0 | 0 | 0 | 0 | 0 |
| 46 | 0 | 0 | 0 | 0 | 0 |
| 47 | 0 | 0 | 0 | 0 | 0 |
| 48 | 0 | 0 | 0 | 0 | 0 |
| 49 | 0 | 0 | 0 | 0 | 0 |
| 50 | 0 | 0 | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, N.L.; Foo, H.L.; Ramli, N.; Halim, M.; Thalij, K.M. Efficient Assessment and Optimisation of Medium Components Influencing Extracellular Xylanase Production by Pediococcus pentosaceus G4 Using Statistical Approaches. Int. J. Mol. Sci. 2025, 26, 7219. https://doi.org/10.3390/ijms26157219
Ali NL, Foo HL, Ramli N, Halim M, Thalij KM. Efficient Assessment and Optimisation of Medium Components Influencing Extracellular Xylanase Production by Pediococcus pentosaceus G4 Using Statistical Approaches. International Journal of Molecular Sciences. 2025; 26(15):7219. https://doi.org/10.3390/ijms26157219
Chicago/Turabian StyleAli, Noor Lutphy, Hooi Ling Foo, Norhayati Ramli, Murni Halim, and Karkaz M. Thalij. 2025. "Efficient Assessment and Optimisation of Medium Components Influencing Extracellular Xylanase Production by Pediococcus pentosaceus G4 Using Statistical Approaches" International Journal of Molecular Sciences 26, no. 15: 7219. https://doi.org/10.3390/ijms26157219
APA StyleAli, N. L., Foo, H. L., Ramli, N., Halim, M., & Thalij, K. M. (2025). Efficient Assessment and Optimisation of Medium Components Influencing Extracellular Xylanase Production by Pediococcus pentosaceus G4 Using Statistical Approaches. International Journal of Molecular Sciences, 26(15), 7219. https://doi.org/10.3390/ijms26157219







