Clinical Relevance of FOXP3, PD-L1, PD-1, and miR-155 Gene Expression and Genetic Variants in HPV-Negative Oral Carcinomas
Abstract
1. Introduction
1.1. Pathogenesis and HPV Status
1.2. Immune Response and Checkpoint Mechanisms
1.3. Regulatory T Cells and FOXP3 as Prognostic Modulators
1.4. MicroRNA-155: Dual Role in Tumorigenesis and Immune Regulation
1.5. Immune Gene Polymorphisms in OSCC: Clinical Implications
1.6. Immunotherapy Context and Study Rationale
2. Results
2.1. Association of Gene Polymorphisms with Demographic and Clinicopathological Features
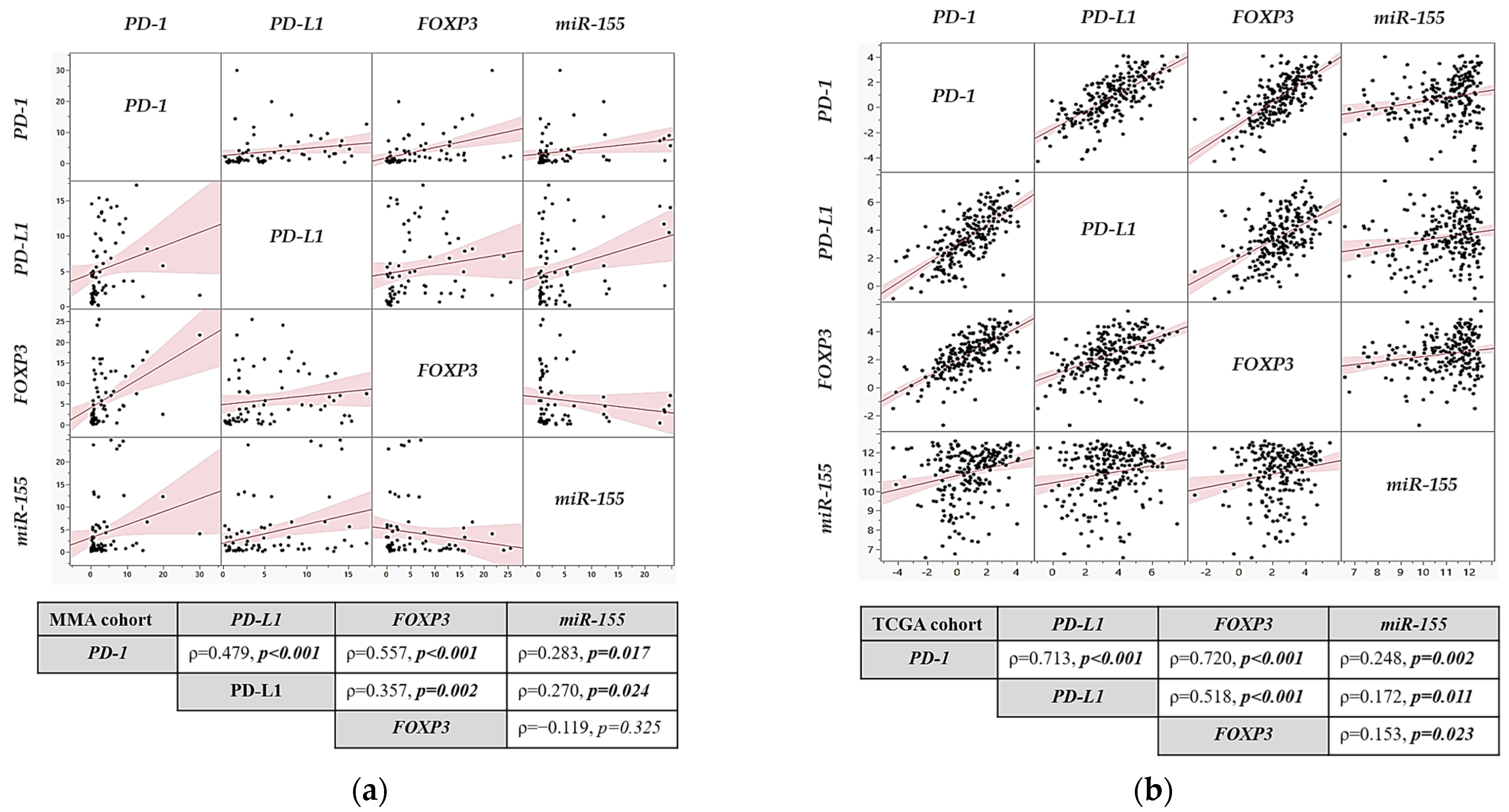
2.2. Association of Gene Expression Profiles with Clinicopathological Features in MMA and TCGA Cohorts
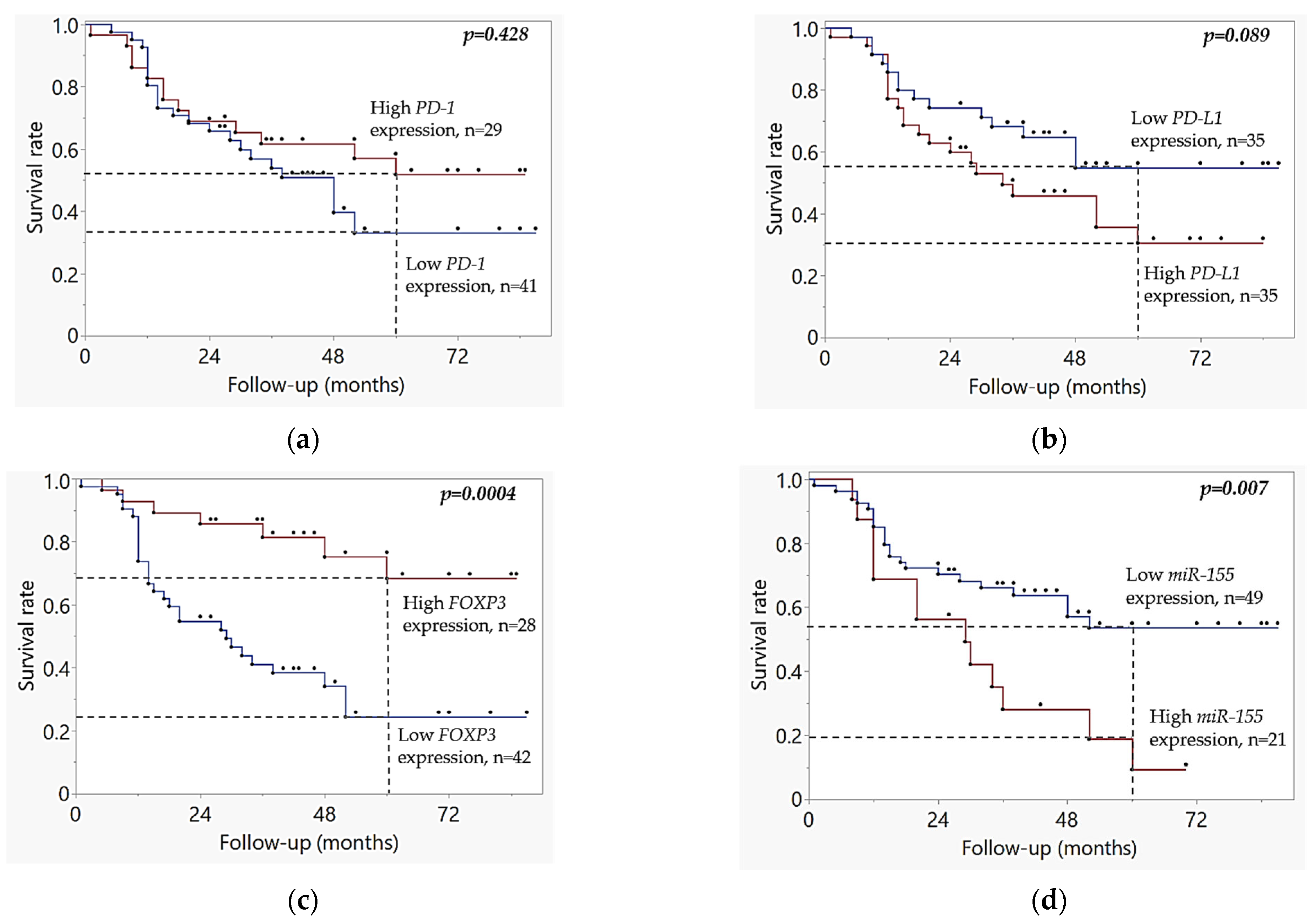
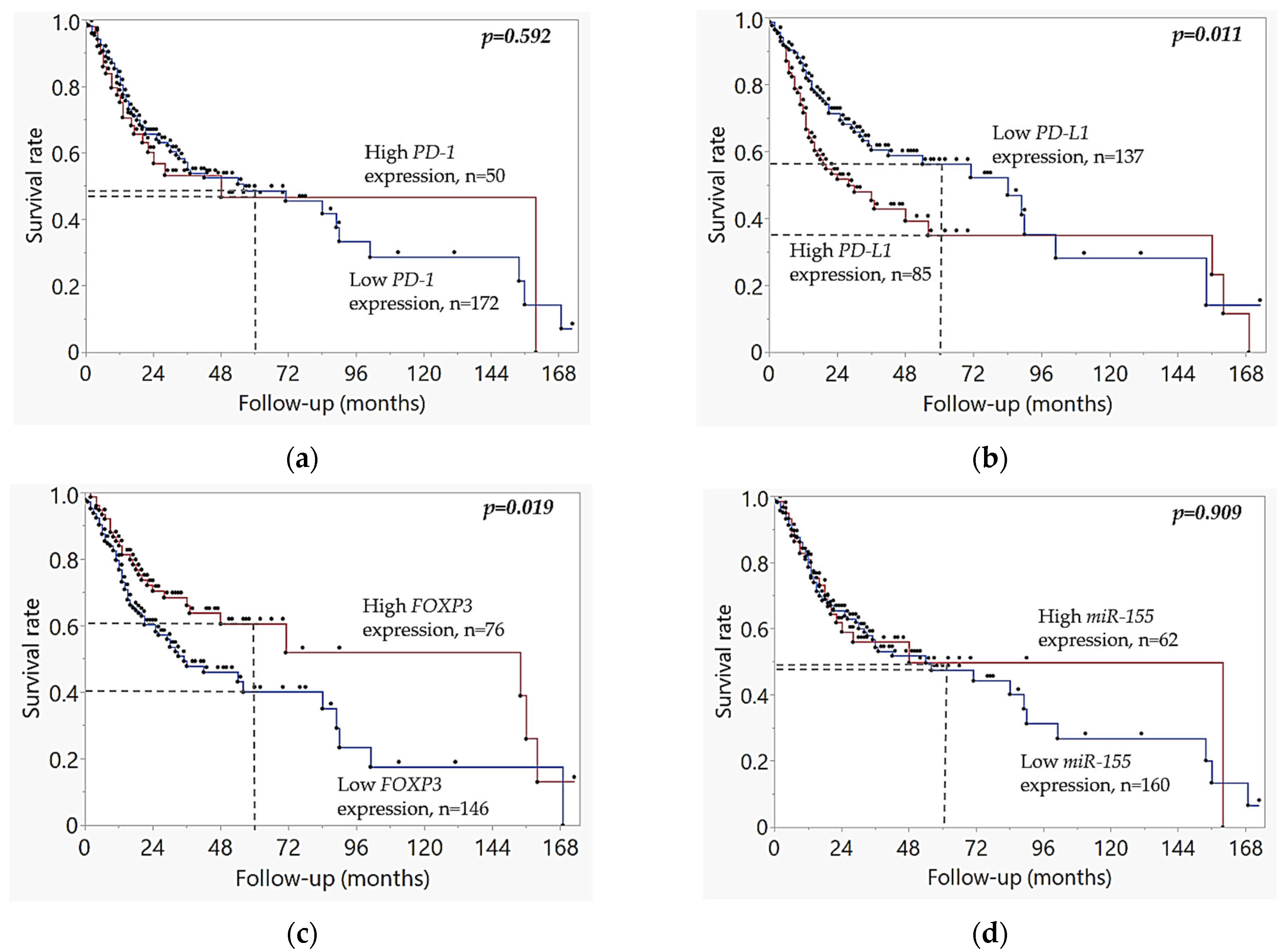
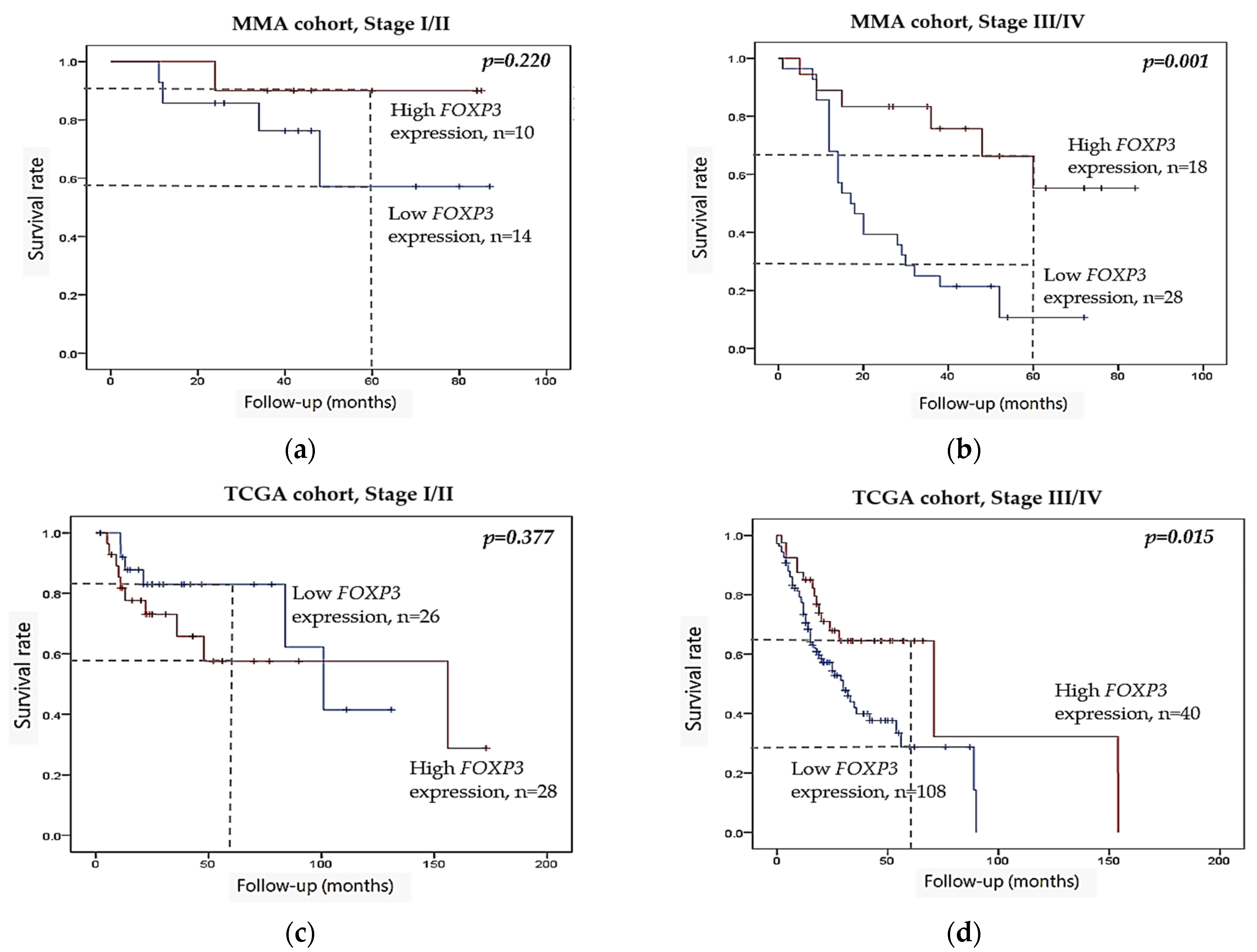
2.3. Kaplan–Meier Survival Analysis
2.4. Cox Regression Analysis
3. Discussion
3.1. Clinical Implications
3.2. Study Limitations
3.3. Conclusions and Future Directions
4. Materials and Methods
4.1. MMA Patient Cohort and Ethical Approvals
4.2. Sample Processing: HPV Status Determination, SNP Genotyping, CNV Analysis, and Gene and miRNA Expression Analysis
4.3. TCGA Validation Cohort
4.4. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
Abbreviations
| AJCC | American Joint Committee on Cancer |
| APCs | Antigen-Presenting Cells |
| BIC | B-cell Integration Cluster |
| CI | Confidence Interval |
| CNV | Copy Number Variation |
| CTLA-4 | Cytotoxic T-Lymphocyte-Associated Protein 4 |
| CTLs | Cytotoxic T Lymphocytes |
| EMT | Epithelial-Mesenchymal Transition |
| FFPE | Formalin-Fixed Paraffin-Embedded |
| FOXP3 | Forkhead Box P3 |
| FoxP3FL | Forkhead Box P3 Full-Length Isoform |
| FOXP3ΔE2 | FOXP3 Isoform Lacking Exon 2 |
| FOXP3ΔE2ΔE7 | FOXP3 Isoform Lacking Exons 2 and 7 |
| GAPDH | Glyceraldehyde-3-Phosphate Dehydrogenase |
| HER2 | Human Epidermal Growth Factor Receptor 2 |
| HNSCC | Head and Neck Squamous Cell Carcinoma |
| HPV | Human Papillomavirus |
| HR | Hazard Ratio |
| ICIs | Immune Checkpoint Inhibitors |
| IFN-γ | Interferon Gamma |
| IHC | Immunohistochemistry |
| IL-2 | Interleukin 2 |
| JAK/STAT | Janus Kinase/Signal Transducer and Activator of Transcription |
| miRNA (miR) | MicroRNA |
| MMA | Military Medical Academy |
| NF-κB | Nuclear Factor Kappa-Light-Chain-Enhancer of Activated B Cells |
| NSCLC | Non-Small Cell Lung Cancer |
| OSCC | Oral Squamous Cell Carcinoma |
| PD-1 | Programmed Cell Death Protein 1 |
| PD-L1/2 | Programmed Death-Ligand 1/2 |
| QPCR | Quantitative Polymerase Chain Reaction |
| RNA-seq | RNA Sequencing |
| ROC/AUC | Receiver Operating Characteristic/Area Under the Curve |
| RT | Reverse Transcription |
| SNVs | Single Nucleotide Variants |
| SOCS1 | Suppressor of Cytokine Signaling 1 |
| STAT | Signal Transducer and Activator of Transcription |
| TCGA | The Cancer Genome Atlas |
| Th | T helper cell |
| TILs | Tumor-Infiltrating Lymphocytes |
| TME | Tumor Microenvironment |
| TNFα | Tumor Necrosis Factor Alpha |
| Treg | Regulatory T Cell |
| UNG | Uracil-N-Glycosylase |
| UTR | Untranslated Region |
References
- Tan, Y.; Wang, Z.; Xu, M.; Li, B.; Huang, Z.; Qin, S.; Nice, E.C.; Tang, J.; Huang, C. Oral squamous cell carcinomas: State of the field and emerging directions. Int. J. Oral. Sci. 2023, 15, 44. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Irimie, A.I.; Ciocan, C.; Gulei, D.; Mehterov, N.; Atanasov, A.G.; Dudea, D.; Berindan-Neagoe, I. Current Insights into Oral Cancer Epigenetics. Int. J. Mol. Sci. 2018, 19, e670. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Li, X. An analysis of influencing factors of oral frailty in the elderly in the community. BMC Oral. Health 2024, 24, 260. [Google Scholar] [CrossRef] [PubMed]
- Vijayvargiya, P.; Trivedi, S.; Rupji, M.; Song, H.; Liu, Y.; Jiang, R.; Kaka, A.S.; Chen, G.Z.; Stokes, W.; Steuer, C.; et al. Comparison of the Seventh and Eighth Edition of American Joint Committee on Cancer (AJCC) Staging for Selected and Nonselected Oropharyngeal Squamous Cell Carcinomas. Oncologist 2022, 27, 48–56. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef]
- Topalian, S.L.; Drake, C.G.; Pardoll, D.M. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 2015, 27, 450–461. [Google Scholar] [CrossRef]
- Jiang, X.; Wang, J.; Deng, X.; Xiong, F.; Ge, J.; Xiang, B.; Wu, X.; Ma, J.; Zhou, M.; Li, X.; et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol. Cancer 2019, 18, 10. [Google Scholar] [CrossRef]
- Gonzalez, H.; Hagerling, C.; Werb, Z. Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes. Dev. 2018, 32, 1267–1284. [Google Scholar] [CrossRef]
- Kim, S.K.; Cho, S.W. The Evasion Mechanisms of Cancer Immunity and Drug Intervention in the Tumor Microenvironment. Front. Pharmacol. 2022, 13, 868695. [Google Scholar] [CrossRef]
- Lin, X.; Kang, K.; Chen, P.; Zeng, Z.; Li, G.; Xiong, W.; Yi, M.; Xiang, B. Regulatory mechanisms of PD-1/PD-L1 in cancers. Mol. Cancer 2024, 23, 108. [Google Scholar] [CrossRef]
- Burtness, B.; Harrington, K.J.; Greil, R.; Soulières, D.; Tahara, M.; de Castro, G.; Psyrri, A.; Basté, N.; Neupane, P.; Bratland, Å.; et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): A randomised, open-label, phase 3 study. Lancet 2019, 394, 1915–1928. [Google Scholar] [CrossRef]
- Al-Azzawi, H.M.A.; Hamza, S.A.; Paolini, R.; Lim, M.; Patini, R.; Celentano, A. PD-L1/PD-1 Expression in the Treatment of Oral Squamous Cell Carcinoma and Oral Potentially Malignant Disorders: An Overview of Reviews. J. Pers. Med. 2025, 15, 126. [Google Scholar] [CrossRef]
- Yamashita, M.; Yano, H.; Komohara, Y.; Yamada, R.; Fujiwara, Y.; Hirayama, M.; Seki, Y.; Yoshida, R.; Nakayama, H. Cytotoxic T Lymphocyte Density and PD-L1 Expression Predict the Response to Anti-PD1 Therapy in Recurrent Oral Squamous Cell Carcinoma. Microbiol. Immunol. 2025, 69, 350–358. [Google Scholar] [CrossRef]
- Mortezaee, K. FOXP3 (in)stability and cancer immunotherapy. Cytokine 2024, 178, 156589. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gong, R.; Zhao, C.; Lei, K.; Sun, X.; Ren, H. Human FOXP3 and tumour microenvironment. Immunology 2023, 168, 248–255. [Google Scholar] [CrossRef]
- Yagi, H.; Nomura, T.; Nakamura, K.; Yamazaki, S.; Kitawaki, T.; Hori, S.; Maeda, M.; Onodera, M.; Uchiyama, T.; Fujii, S.; et al. Crucial role of FOXP3 in the development and function of human CD25+CD4+ regulatory T cells. Int. Immunol. 2004, 16, 1643–1656. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Dong, P.; Ren, M.; Song, Y.; Qian, X.; Yang, Y.; Li, S.; Zhang, X.; Liu, F. PD-L1 Expression Is Associated with Tumor FOXP3(+) Regulatory T-Cell Infiltration of Breast Cancer and Poor Prognosis of Patient. J. Cancer 2016, 7, 784–793. [Google Scholar] [CrossRef]
- Que, Y.; Xiao, W.; Guan, Y.; Liang, Y.; Yan, S.; Chen, H.; Li, Q.; Xu, B.; Zhou, Z.; Zhang, X. PD-L1 Expression Is Associated with FOXP3+ Regulatory T-Cell Infiltration of Soft Tissue Sarcoma and Poor Patient Prognosis. J. Cancer 2017, 8, 2018–2025. [Google Scholar] [CrossRef]
- Song, J.-J.; Zhao, S.-J.; Fang, J.; Ma, D.; Liu, X.-Q.; Chen, X.-B.; Wang, Y.; Cheng, B.; Wang, Z. Foxp3 overexpression in tumor cells predicts poor survival in oral squamous cell carcinoma. BMC Cancer 2016, 16, 530. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Liu, H.; Su, Y.; Zhang, T.; Chu, M.; Liang, L.; Liao, G. Foxp3 expressed by tongue squamous cell carcinoma cells correlates with clinicopathologic features and overall survival in tongue squamous cell carcinoma patients. Oral. Oncol. 2011, 47, 566–570. [Google Scholar] [CrossRef]
- Seminerio, I.; Descamps, G.; Dupont, S.; de Marrez, L.; Laigle, J.-A.; Lechien, J.R.; Kindt, N.; Journe, F.; Saussez, S. Infiltration of FoxP3+ Regulatory T Cells is a Strong and Independent Prognostic Factor in Head and Neck Squamous Cell Carcinoma. Cancers 2019, 11, 227. [Google Scholar] [CrossRef]
- Shang, B.; Liu, Y.; Jiang, S.; Liu, Y. Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: A systematic review and meta-analysis. Sci. Rep. 2015, 5, 15179. [Google Scholar] [CrossRef]
- Badoual, C.; Hans, S.; Rodriguez, J.; Peyrard, S.; Klein, C.; Agueznay, N.E.H.; Mosseri, V.; Laccourreye, O.; Bruneval, P.; Fridman, W.H.; et al. Prognostic value of tumor-infiltrating CD4+ T-cell subpopulations in head and neck cancers. Clin. Cancer Res. 2006, 12, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.-J.; Zhang, C.-Y.; Zhou, Z.-T.; Ma, J.-Y.; Liu, Y.; Bao, Z.-X.; Jiang, W.-W. MicroRNA-155 in oral squamous cell carcinoma: Overexpression, localization, and prognostic potential. Head Neck 2015, 37, 970–976. [Google Scholar] [CrossRef]
- Kim, H.; Yang, J.M.; Ahn, S.-H.; Jeong, W.-J.; Chung, J.-H.; Paik, J.H. Potential Oncogenic Role and Prognostic Implication of MicroRNA-155-5p in Oral Squamous Cell Carcinoma. Anticancer Res. 2018, 38, 5193–5200. [Google Scholar] [CrossRef] [PubMed]
- Kirave, P.; Gondaliya, P.; Kulkarni, B.; Rawal, R.; Garg, R.; Jain, A.; Kalia, K. Exosome mediated miR-155 delivery confers cisplatin chemoresistance in oral cancer cells via epithelial-mesenchymal transition. Oncotarget 2020, 11, 1157–1171. [Google Scholar] [CrossRef]
- Litak, J.; Grajkowska, W.; Bogucki, J.; Kowalczyk, P.; Petniak, A.; Podkowiński, A.; Szumiło, J.; Kocki, J.; Roliński, J.; Rahnama-Hezavah, M.; et al. PD-L1/miR-155 Interplay in Pediatric High-Grade Glioma. Brain Sci. 2022, 12, 324. [Google Scholar] [CrossRef]
- Zhou, R.-M.; Li, Y.; Wang, N.; Huang, X.; Cao, S.-R.; Shan, B.-E. Association of programmed death-1 polymorphisms with the risk and prognosis of esophageal squamous cell carcinoma. Cancer Genet. 2016, 209, 365–375. [Google Scholar] [CrossRef]
- Polcaro, G.; Liguori, L.; Manzo, V.; Chianese, A.; Donadio, G.; Caputo, A.; Scognamiglio, G.; Dell’Annunziata, F.; Langella, M.; Corbi, G.; et al. rs822336 binding to C/EBPβ and NFIC modulates induction of PD-L1 expression and predicts anti-PD-1/PD-L1 therapy in advanced NSCLC. Mol. Cancer 2024, 23, 63. [Google Scholar] [CrossRef]
- Ohhara, Y.; Tomaru, U.; Kinoshita, I.; Hatanaka, K.C.; Noguchi, T.; Hatanaka, Y.; Amono, T.; Matsuno, Y.; Dosaka-Akita, H. Polymorphisms of the PD-L1 gene 3’-untranslated region are associated with the expression of PD-L1 in non-small cell lung cancer. Genes. Chromosomes Cancer 2024, 63, e23216. [Google Scholar] [CrossRef]
- Grenda, A.; Krawczyk, P.; Kucharczyk, T.; Błach, J.; Reszka, K.; Chmielewska, I.; Buczkowski, J.; Kieszko, R.; Siwiec, J.; Kubiatowski, T.; et al. Impact of copy number variant and single nucleotide polymorphism of the programmed death-ligand 1 gene, programmed death-ligand 1 protein expression and therapy regimens on overall survival in a large group of Caucasian patients with non-small cell lung carcinoma. Oncol. Lett. 2021, 21, 449. [Google Scholar] [CrossRef]
- Chen, P.-J.; Lin, C.-W.; Lu, H.-J.; Chuang, C.-Y.; Yang, S.-F.; Chou, Y.-E. The impact of FOXP3 polymorphisms on oral cancer progression and clinicopathological characteristics. J. Cancer 2023, 14, 1195–1201. [Google Scholar] [CrossRef]
- Xie, K.; Ma, H.; Liang, C.; Wang, C.; Qin, N.; Shen, W.; Gu, Y.; Yan, C.; Zhang, K.; Dai, N.; et al. A functional variant in miR-155 regulation region contributes to lung cancer risk and survival. Oncotarget 2015, 6, 42781–42792. [Google Scholar] [CrossRef]
- Lin, C.-W.; Lu, J.-W.; Chuang, C.-Y.; Hsieh, W.-Y.; Tsai, Y.-J.; Yang, S.-F.; Lin, S.-H. Clinical significance of long non-coding RNA MIR155HG genetic variants and susceptibility to oral cancer. Sci. Rep. 2025, 15, 9956. [Google Scholar] [CrossRef]
- Ferris, R.L.; Blumenschein, G.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2016, 375, 1856–1867. [Google Scholar] [CrossRef]
- Lenouvel, D.; González-Moles, M.Á.; Ruiz-Ávila, I.; Gonzalez-Ruiz, L.; Gonzalez-Ruiz, I.; Ramos-García, P. Prognostic and clinicopathological significance of PD-L1 overexpression in oral squamous cell carcinoma: A systematic review and comprehensive meta-analysis. Oral. Oncol. 2020, 106, 104722. [Google Scholar] [CrossRef]
- Nocini, R.; Vianini, M.; Girolami, I.; Calabrese, L.; Scarpa, A.; Martini, M.; Morbini, P.; Marletta, S.; Brunelli, M.; Molteni, G.; et al. PD-L1 in oral squamous cell carcinoma: A key biomarker from the laboratory to the bedside. Clin. Exp. Dent. Res. 2022, 8, 690–698. [Google Scholar] [CrossRef]
- Yee, D.; Shah, K.M.; Coles, M.C.; Sharp, T.V.; Lagos, D. MicroRNA-155 induction via TNF-α and IFN-γ suppresses expression of programmed death ligand-1 (PD-L1) in human primary cells. J. Biol. Chem. 2017, 292, 20683–20693. [Google Scholar] [CrossRef]
- Mhawech-Fauceglia, P.; Wang, D.; Ali, L.; Lele, S.; Huba, M.A.; Liu, S.; Odunsi, K. Intraepithelial T cells and tumor-associated macrophages in ovarian cancer patients. Cancer Immun. 2013, 13, 1. [Google Scholar]
- Frey, D.M.; Droeser, R.A.; Viehl, C.T.; Zlobec, I.; Lugli, A.; Zingg, U.; Oertli, D.; Kettelhack, C.; Terracciano, L.; Tornillo, L. High frequency of tumor-infiltrating FOXP3(+) regulatory T cells predicts improved survival in mismatch repair-proficient colorectal cancer patients. Int. J. Cancer 2010, 126, 2635–2643. [Google Scholar] [CrossRef]
- Saito, T.; Nishikawa, H.; Wada, H.; Nagano, Y.; Sugiyama, D.; Atarashi, K.; Maeda, Y.; Hamaguchi, M.; Ohkura, N.; Sato, E.; et al. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat. Med. 2016, 22, 679–684. [Google Scholar] [CrossRef]
- Kindt, N.; Descamps, G.; Seminerio, I.; Bellier, J.; Lechien, J.R.; Mat, Q.; Pottier, C.; Delvenne, P.; Journé, F.; Saussez, S. High stromal Foxp3-positive T cell number combined to tumor stage improved prognosis in head and neck squamous cell carcinoma. Oral. Oncol. 2017, 67, 183–191. [Google Scholar] [CrossRef]
- Perrone, G.; Ruffini, P.A.; Catalano, V.; Spino, C.; Santini, D.; Muretto, P.; Spoto, C.; Zingaretti, C.; Sisti, V.; Alessandroni, P.; et al. Intratumoural FOXP3-positive regulatory T cells are associated with adverse prognosis in radically resected gastric cancer. Eur. J. Cancer 2008, 44, 1875–1882. [Google Scholar] [CrossRef]
- Shah, W.; Yan, X.; Jing, L.; Zhou, Y.; Chen, H.; Wang, Y. A reversed CD4/CD8 ratio of tumor-infiltrating lymphocytes and a high percentage of CD4(+)FOXP3(+) regulatory T cells are significantly associated with clinical outcome in squamous cell carcinoma of the cervix. Cell. Mol. Immunol. 2011, 8, 59–66. [Google Scholar] [CrossRef]
- Dong, Y.; Yang, C.; Pan, F. Post-Translational Regulations of Foxp3 in Treg Cells and Their Therapeutic Applications. Front. Immunol. 2021, 12, 626172. [Google Scholar] [CrossRef]
- Li, C.; Jiang, P.; Wei, S.; Xu, X.; Wang, J. Regulatory T cells in tumor microenvironment: New mechanisms, potential therapeutic strategies and future prospects. Mol. Cancer 2020, 19, 116. [Google Scholar] [CrossRef]
- Lafont, V.; Sanchez, F.; Laprevotte, E.; Michaud, H.-A.; Gros, L.; Eliaou, J.-F.; Bonnefoy, N. Plasticity of gamma delta T cells: Impact on the anti-tumor response. Front. Immunol. 2014, 5, 622. [Google Scholar] [CrossRef]
- Contreras-Castillo, E.; García-Rasilla, V.Y.; García-Patiño, M.G.; Licona-Limón, P. Stability and plasticity of regulatory T cells in health and disease. J. Leukoc. Biol. 2024, 116, 33–53. [Google Scholar] [CrossRef]
- Liu, H.; Chen, M.; Hong, B.; Xiao, Y.; Chen, Q.; Qian, Y. Single-nucleus RNA sequencing and spatial transcriptomics reveal an immunosuppressive tumor microenvironment related to metastatic dissemination during pancreatic cancer liver metastasis. Theranostics 2025, 15, 5337–5357. [Google Scholar] [CrossRef]
- Sato, Y.; Liu, J.; Lee, E.; Perriman, R.; Roncarolo, M.G.; Bacchetta, R. Co-Expression of FOXP3FL and FOXP3Δ2 Isoforms Is Required for Optimal Treg-Like Cell Phenotypes and Suppressive Function. Front. Immunol. 2021, 12, 752394. [Google Scholar] [CrossRef]
- Mailer, R.K.W. Alternative Splicing of FOXP3-Virtue and Vice. Front. Immunol. 2018, 9, 530. [Google Scholar] [CrossRef]
- Szylberg, Ł.; Karbownik, D.; Marszałek, A. The Role of FOXP3 in Human Cancers. Anticancer Res. 2016, 36, 3789–3794. [Google Scholar]
- Incorvaia, L.; Fanale, D.; Badalamenti, G.; Brando, C.; Bono, M.; De Luca, I.; Algeri, L.; Bonasera, A.; Corsini, L.R.; Scurria, S.; et al. A “Lymphocyte MicroRNA Signature” as Predictive Biomarker of Immunotherapy Response and Plasma PD-1/PD-L1 Expression Levels in Patients with Metastatic Renal Cell Carcinoma: Pointing towards Epigenetic Reprogramming. Cancers 2020, 12, 3396. [Google Scholar] [CrossRef]
- Song, T.-Y.; Long, M.; Zhao, H.-X.; Zou, M.-W.; Fan, H.-J.; Liu, Y.; Geng, C.-L.; Song, M.-F.; Liu, Y.-F.; Chen, J.-Y.; et al. Tumor evolution selectively inactivates the core microRNA machinery for immune evasion. Nat. Commun. 2021, 12, 7003. [Google Scholar] [CrossRef]
- Mohan, S.; Hakami, M.A.; Dailah, H.G.; Khalid, A.; Najmi, A.; Zoghebi, K.; Halawi, M.A.; Alotaibi, T.M. From inflammation to metastasis: The central role of miR-155 in modulating NF-κB in cancer. Pathol. Res. Pract. 2024, 253, 154962. [Google Scholar] [CrossRef]
- Lu, L.-F.; Thai, T.-H.; Calado, D.P.; Chaudhry, A.; Kubo, M.; Tanaka, K.; Loeb, G.B.; Lee, H.; Yoshimura, A.; Rajewsky, K.; et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 2009, 30, 80–91. [Google Scholar] [CrossRef]
- Bala, S.; Marcos, M.; Kodys, K.; Csak, T.; Catalano, D.; Mandrekar, P.; Szabo, G. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor {alpha} (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease. J. Biol. Chem. 2011, 286, 1436–1444. [Google Scholar] [CrossRef]
- Janović, A.; Bracanović, Đ.; Antić, S.; Marković-Vasiljković, B. Demographic and imaging features of oral squamous cell cancer in Serbia: A retrospective cross-sectional study. BMC Oral. Health 2024, 24, 141. [Google Scholar] [CrossRef]
- Tsimafeyeu, I.; Imyanitov, E.; Zavalishina, L.; Raskin, G.; Povilaitite, P.; Savelov, N.; Kharitonova, E.; Rumyantsev, A.; Pugach, I.; Andreeva, Y.; et al. Agreement between PDL1 immunohistochemistry assays and polymerase chain reaction in non-small cell lung cancer: CLOVER comparison study. Sci. Rep. 2020, 10, 3928. [Google Scholar] [CrossRef]
- Venina, A.R.; Ivantsov, A.O.; Iyevleva, A.G.; Kuligina, E.S.; Preobrazhenskaya, E.V.; Yurlov, D.O.; Rawlinson, K.E.; Kosmin, A.V.; Savelov, N.A.; Raskin, G.A.; et al. PCR-based analysis of PD-L1 RNA expression in lung cancer: Comparison with commonly used immunohistochemical assays. Ann. Diagn. Pathol. 2022, 59, 151968. [Google Scholar] [CrossRef]
- Supic, G.; Stefik, D.; Ivkovic, N.; Sami, A.; Zeljic, K.; Jovic, S.; Kozomara, R.; Vojvodic, D.; Stosic, S. Prognostic impact of miR-34b/c DNA methylation, gene expression, and promoter polymorphism in HPV-negative oral squamous cell carcinomas. Sci. Rep. 2022, 12, 1296. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Chen, Y.; Lun, A.T.L.; Smyth, G.K. From reads to genes to pathways: Differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 2016, 5, 1438. [Google Scholar] [CrossRef]
- Liu, R.; Holik, A.Z.; Su, S.; Jansz, N.; Chen, K.; Leong, H.S.; Blewitt, M.E.; Asselin-Labat, M.-L.; Smyth, G.K.; Ritchie, M.E. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 2015, 43, e97. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome. Biol. 2010, 11, R25. [Google Scholar] [CrossRef]
- Budczies, J.; Klauschen, F.; Sinn, B.V.; Győrffy, B.; Schmitt, W.D.; Darb-Esfahani, S.; Denkert, C. Cutoff Finder: A comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS ONE 2012, 7, e51862. [Google Scholar] [CrossRef]
| Variables | Total MMA (n = 134) | PD-1 rs36084323 | PD-L1 rs822336 | PD-L1 rs4143815 | PD-L1 CNV 1 | FOXP3 rs3761548 | FOXP3 rs2232365 | miR-155 rs767649 | |
|---|---|---|---|---|---|---|---|---|---|
| wt/ht/hom 2 | wt/ht/hom | wt/ht/hom | No/Yes | wt/ht/hom | wt/ht/hom | wt/ht/hom | |||
| Sex | Male | 107 | 95/11/1 | 42/46/19 | 43/52/12 | 78/29 | 54/3/50 | 64/4/39 | 53/54/0 |
| Female | 27 | 21/6/0 | 10/10/7 | 14/10/3 | 19/8 | 12/3/12 | 17/4/6 | 14/13/0 | |
| p/p 3 | 0.226/0.202 | 0.621/1.000 | 0.519/0.386 | 0.812 | 0.173/0.668 | 0.056/0.828 | 0.829 | ||
| Age (median) | <58 | 63 | 55/8/0 | 30/22/11 | 25/27/11 | 45/18 | 31/3/29 | 39/5/19 | 30/33/0 |
| ≥58 | 71 | 61/9/1 | 22/34/15 | 33/34/4 | 52/19 | 35/3/33 | 42/3/26 | 37/34/0 | |
| p/p 3 | 0.639/1.000 | 0.138/0.053 | 0.095/0.486 | 0.848 | 0.989/1.000 | 0.542/0.860 | 0.729 | ||
| Smoking | Never | 33 | 28/5/0 | 13/14/6 | 17/11/5 | 25/8 | 15/3/15 | 20/3/10 | 19/14/0 |
| Ever | 101 | 88/12/1 | 39/42/20 | 40/51/10 | 72/29 | 51/3/47 | 61/5/35 | 48/53/0 | |
| p/p 3 | 0.759/0.771 | 0.979/1.000 | 0.220/0.314 | 0.662 | 0.330/0.690 | 0.652/1.000 | 0.316 | ||
| Alcohol | No | 21 | 15/6/0 | 6/11/4 | 9/9/3 | 17/4 | 8/1/12 | 13/0/8 | 16/5/0 |
| Yes | 113 | 101/11/1 | 46/45/22 | 49/52/12 | 80/33 | 58/5/50 | 68/8/37 | 51/62/0 | |
| p/p 3 | 0.055/0.038 | 0.511/0.339 | 0.881/1.000 | 0.432 | 0.530/0.343 | 0.439/0.624 | 0.018 | ||
| Tumor size | T 1/2 | 93 | 80/12/1 | 41/34/18 | 37/44/12 | 64/29 | 47/4/42 | 55/6/32 | 47/46/0 |
| T 3/4 | 41 | 36/5/0 | 11/22/8 | 20/18/3 | 33/8 | 19/2/20 | 26/2/13 | 20/21/0 | |
| p/p 3 | 0.794/1.000 | 0.124/0.083 | 0.495/0.451 | 0.210 | 0.903/0.710 | 0.876/0.704 | 0.851 | ||
| Nodal status | N− | 50 | 43/7/0 | 15/24/11 | 21/23/6 | 37/13 | 26/4/20 | 28/3/19 | 23/27/0 |
| N+ | 84 | 73/10/1 | 37/32/15 | 36/39/9 | 60/24 | 40/2/42 | 53/5/26 | 44/40/0 | |
| p/p 3 | 0.702/1.000 | 0.272/0.142 | 0.974/0.858 | 0.843 | 0.222/0.721 | 0.696/0.467 | 0.296 | ||
| Tumor stage | I | 8 | 7/1/0 | 3/3/2 | 5/3/0 | 8/0 | 4/0/4 | 5/0/3 | 2/6/0 |
| II | 35 | 30/5/0 | 15/14/6 | 13/17/5 | 25/10 | 16/3/16 | 20/3/12 | 18/17/0 | |
| III | 63 | 55/7/1 | 27/23/13 | 28/28/7 | 43/20 | 33/3/27 | 37/3/23 | 34/29/0 | |
| IV | 28 | 24/4/0 | 7/16/5 | 11/14/3 | 21/7 | 13/0/15 | 19/2/7 | 13/15/0 | |
| p/p 3 | 0.967/0.995 | 0.655/0.402 | 0.855/0.548 | 0.296 | 0.717/0.916 | 0.894/0.827 | 0.463 | ||
| Recurrence | No | 58 | 47/11/0 | 22/25/11 | 30/23/5 | 43/15 | 28/4/26 | 34/5/19 | 25/33/0 |
| Yes | 76 | 69/6/1 | 30/31/15 | 27/39/10 | 54/32 | 38/2/36 | 47/3/26 | 42/34/0 | |
| p/p 3 | 0.116/0.127 | 0.964/1.000 | 0.165/0.053 | 0.846 | 0.496/0.863 | 0.527/0.725 | 0.162 | ||
| Variables | Total MMA (n = 70) | PD-1 Expression Median (25–75%) | PD-L1 Expression Median (25–75%) | FOXP3 Expression Median (25–75%) | miR-155 Expression Median (25–75%) | |
|---|---|---|---|---|---|---|
| Sex | Male | 58 | 1.892 (0.718–4.133) | 3.905 (1.689–7.933) | 3.631 (0.806–10.999) | 1.422 (0.568–4.76) |
| Female | 12 | 1.531 (0.925–4.335) | 2.655 (1.544–10.467) | 3.548 (0.982–11.470) | 5.465 (0.485–6.689) | |
| p | 0.803 | 0.876 | 0.827 | 0.399 | ||
| Age (median) | <58 | 35 | 2.445 (0.979–5.349) | 4.897 (1.965–9.334) | 4.582 (1.132–11.977) | 1.422 (0.502–9.004) |
| ≥58 | 35 | 1.057 (0.599–3.862) | 2.655 (1.347–7.201) | 2.600 (0.735–10.999) | 1.489 (0595–4.959) | |
| p | 0.072 | 0.115 | 0.279 | 0.742 | ||
| Smoking | Never | 12 | 1.531 (0.783–3.599) | 6.027 (2.324–11.726) | 5.312 (3.046–8.624) | 0.871 (0.432–4.556) |
| Ever | 58 | 1.891 (0.761–4.790) | 3.215 (1.604–7.812) | 2.590 (0.806–11.546) | 1.538 (0.606–5.71) | |
| p | 0.779 | 0.236 | 0.454 | 0.289 | ||
| Alcohol | No | 19 | 1.845 (0.823–5.603) | 4.857 (2.974–7.036) | 3.748 (0.903–11.722) | 1.417 (0.463–5.468) |
| Yes | 51 | 1.588 (0.709–4.106) | 2.772 (1.198–9.669) | 2.657 (0.822–10.912) | 1.641(0.616–16.62) | |
| p | 0.602 | 0.306 | 0.697 | 0.627 | ||
| Tumor size | T 1/2 | 40 | 2.032 (0.806–5.332) | 3.652 (1.735–8.701) | 4.683 (1.659–12.976) | 1.344 (0.513–5.239) |
| T 3/4 | 30 | 1.634 (0.657–3.481) | 3.631 (1.074–8.501) | 1.230 (0.687–6.125) | 1.708 (0.558–6.499) | |
| p | 0.419 | 0.652 | 0.007 | 0.307 | ||
| Nodal status | N− | 34 | 1.369 (0.630–6.182) | 3.652 (1.373–6.914) | 3.121 (0.882–11.377) | 1.454 (0.410–5.467) |
| N+ | 36 | 2.137 (0.872–3.509) | 3.803 (1.899–9.614) | 3.769 (0.778–8.041) | 1.565 (0.647–5.945) | |
| p | 0.742 | 0.180 | 0.962 | 0.417 | ||
| Tumor stage | I | 7 | 1.032 (0.621–5.603) | 1.891 (1.431–4.735) | 3.748 (1.779–12.904) | 0.595 (0.315–4.959) |
| II | 17 | 1.845 (0.715–6.839) | 3.645 (1.353–8.739 | 4.507 (1.183–13.239) | 2.336 (0.469–5.561) | |
| III | 20 | 1.951 (0.889–3.176) | 4.199 (2.243–9.614) | 7.111 (1.705–13.121) | 1.144 (0.398–6.473) | |
| IV | 26 | 1.763 (0.736–4.456) | 4.459 (1.682–8.768) | 1.163 (0.637–4.696) | 1.641 (0.981–6.654) | |
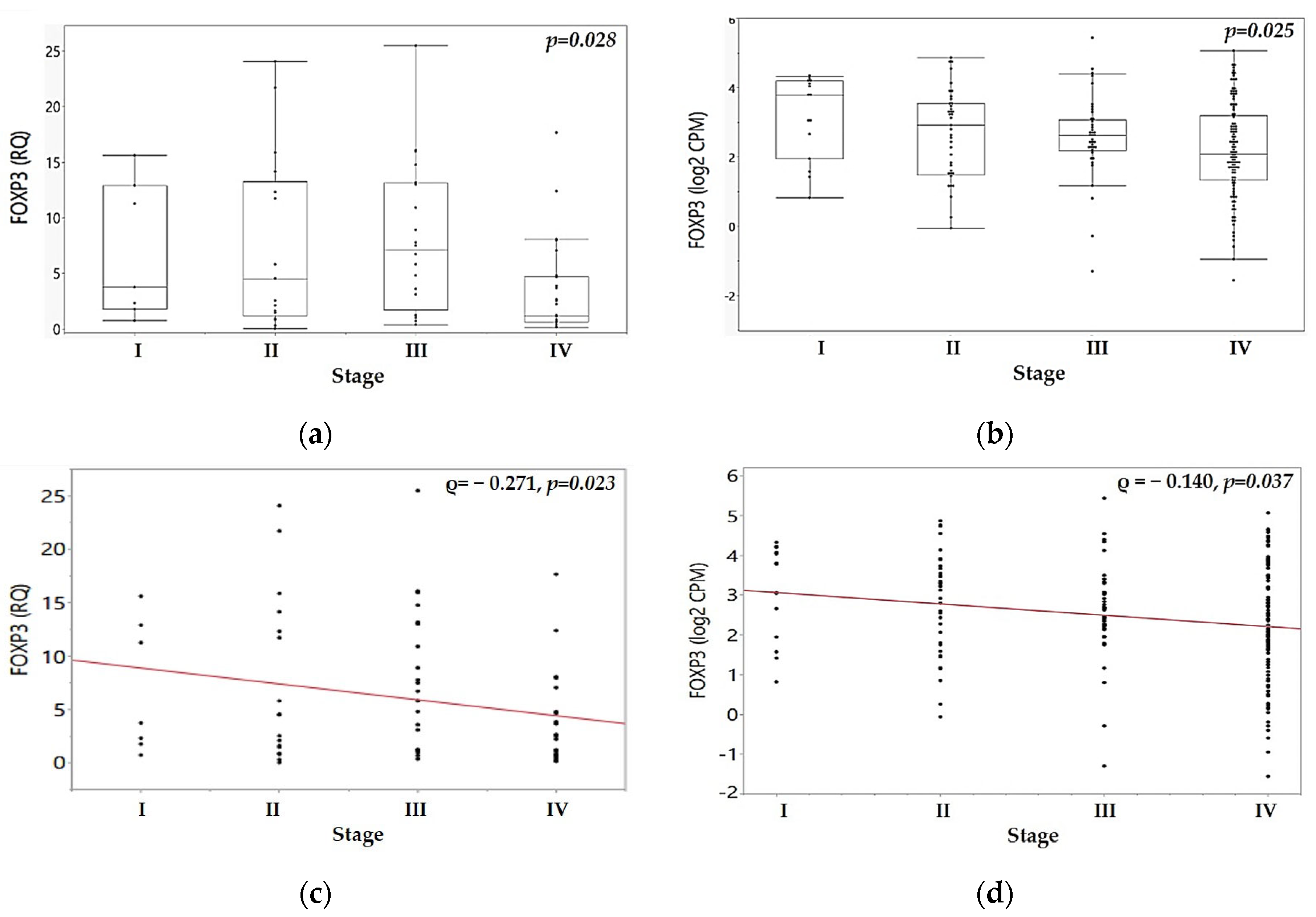
| p | 0.973 | 0.390 | 0.028 | 0.303 | ||
| Recurrence | No | 31 | 1.845 (0.823–6.351) | 3.015 (1.509–7.157) | 7.777 (1.463–13.161) | 0.733 (0.338–3.561) |
| Yes | 39 | 1.588 (0.771–3.466) | 4.151 (1.821–10.321) | 2.107 (0.733–4.657) | 2.593 (0.879–12.51) | |
| p | 0.365 | 0.566 | 0.002 | 0.002 | ||
| Variables | Total TCGA (n = 222) | PD-1 Expression Median (25–75%) | PD-L1 Expression Median (25–75%) | FOXP3 Expression Median (25–75%) | miR-155 Expression Median (25–75%) | |
|---|---|---|---|---|---|---|
| Sex | Male | 143 | 0.761 (−0.090–2.135) | 3.907 (2.729–4.906) | 2.733 (1.770–3.676) | 11.474 (10.755–11.894) |
| Female | 79 | 0.718 (−0.402–1.974) | 3.379 (2.286–3.379) | 2.275 (1.483–3.285) | 11.076 (10.220–11.690) | |
| p | 0.390 | 0.087 | 0.098 | 0.032 | ||
| Age (median) | <58 | 72 | 0.505 (−0.527–1.565) | 3.623 (2.691–4.857) | 2.392 (1.545–3.283) | 10.995 (10.279–11.543) |
| ≥58 | 150 | 0.978 (−0.077–2.358) | 3.106 (2.268–4.295) | 2.467 (1.635–3.498) | 11.360 (10.560–11.988) | |
| p | 0.015 | 0.077 | 0.479 | 0.043 | ||
| Smoking | Never | 101 | 0.761 (−0.424–2.099) | 3.337 (2.408–4.309) | 2.477 (1.591–3.440) | 11.127 (10.386–11.911) |
| Ever | 121 | 0.735 (−0.100–2.010) | 3.465 (2.596–4.817) | 2.400 (1.545–3.449) | 11.263 (10.414–11.754) | |
| p | 0.718 | 0.381 | 0.706 | 0.770 | ||
| Alcohol | No | 75 | 0.950 (−0.253–2.373) | 3.739 (2.646–4.700) | 2.433 (1.944–3.478) | 11.312 (10.621–11.880) |
| Yes | 142 | 0.675 (−0.241–2.011) | 3.365 (2.502–4.766) | 2.447 (1.470–3.440) | 11.153 (10.304–11.889) | |
| p | 0.623 | 0.626 | 0.499 | 0.480 | ||
| Tumor size | T 1/2 | 93 | 0.0977 (−0.060–2.434) | 3.465 (2.255–4.967) | 2.679 (1.626–3.708) | 11.318 (10.587–12.070) |
| T 3/4 | 115 | 0.496 (−0.396–1.724) | 3.339 (2.485–4.334) | 2.266 (1.520–3.159) | 11.065 (10.231–11.690) | |
| p | 0.036 | 0.558 | 0.048 | 0.060 | ||
| Nodal status | N− | 87 | 0.665 (−0.376–2.123) | 3.302 (2.301–4.455) | 2.435 (1.467–3.497) | 11.105 (10.219–11.950) |
| N+ | 102 | 0.617 (−0.241–1.992) | 3.407 (2.458–4.532) | 2.396 (1.678–3.409) | 11.128 (10.342–11.707) | |
| p | 0.716 | 0.494 | 0.854 | 0.837 | ||
| Tumor stage | I | 15 | 1.609 (0.183–2.613) | 4.455 (2.866–5.176) | 3.781 (1.947–4.200) | 11.498 (10.816–12.070) |
| II | 39 | 0.977 (0.051–2.549) | 3.462 (2.646–4.964) | 2.918 (1.483–3.547) | 11.446 (10.636–12.153) | |
| III | 40 | 1.017 (0.048–2.018) | 3.926 (2.697–4.633) | 2.629 (2.182–3.069) | 11.467 (10.382–11.915) | |
| IV | 108 | 0.371 (−0.454–1.677) | 3.140 (2.344–4.287) | 2.078 (1.334–3.199) | 10.941 (10.157–11.597) | |
| p | 0.038 | 0.081 | 0.025 | 0.019 | ||
| Cox Regression Analysis | Variables | OS | |
|---|---|---|---|
| HR [95% CI] | p | ||
| Univariate | Sex | 0.612 (0.312–1.201) | 0.153 |
| Age (≥median) | 0.672 (0.403–1.122) | 0.121 | |
| Smoking | 1.848 (0.942–3.625) | 0.074 | |
| Alcohol | 1.478 (0.996–2.194) | 0.052 | |
| T 1/2 vs. 3/4 | 1.349 (1.147–1.586) | 0.0003 | |
| Nodal status | 2.168 (1.236–3.804) | 0.007 | |
| Tumor stage | 2.892 (2.008–4.167) | 0.0000001 | |
| Recurrence | 17.245 (7.812–38.071) | 0.0000001 | |
| PD-1 rs36084323 | 0.614 (0.291–1.299) | 0.202 | |
| PD-L1 rs822336 | 1.017 (0.735–1.406) | 0.920 | |
| PD-L1 rs4143815 | 1.281 (0.909–1.805) | 0.158 | |
| PD-L1 CNV | 1.148 (0.679–1.943) | 0.606 | |
| FOXP3 rs3761548 | 0.939 (0.732–1.204) | 0.621 | |
| FOXP3 rs2232365 | 0.923 (0.714–1.194) | 0.544 | |
| miR-155 rs767649 | 0.629 (0.387–1.022) | 0.061 | |
| PD-1 expression | 0.691 (0.344–1.388) | 0.299 | |
| PD-L1 expression | 1.746 (0.886–3.442) | 0.107 | |
| FOXP3 expression | 0.252 (0.109–0.583) | 0.001 | |
| miR-155 expression | 2.388 (1.246–4.573) | 0.009 | |
| Multivariate | Recurrence | 32.126 (7.446–138.608) | 0.000003 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivkovic, N.; Misic, D.; Kozomara, R.; Jovic, S.; Sami, A.; Velikic, G.; Stosic, S.; Supic, G. Clinical Relevance of FOXP3, PD-L1, PD-1, and miR-155 Gene Expression and Genetic Variants in HPV-Negative Oral Carcinomas. Int. J. Mol. Sci. 2025, 26, 7218. https://doi.org/10.3390/ijms26157218
Ivkovic N, Misic D, Kozomara R, Jovic S, Sami A, Velikic G, Stosic S, Supic G. Clinical Relevance of FOXP3, PD-L1, PD-1, and miR-155 Gene Expression and Genetic Variants in HPV-Negative Oral Carcinomas. International Journal of Molecular Sciences. 2025; 26(15):7218. https://doi.org/10.3390/ijms26157218
Chicago/Turabian StyleIvkovic, Nemanja, Debora Misic, Ruzica Kozomara, Sasa Jovic, Ahmad Sami, Gordana Velikic, Srboljub Stosic, and Gordana Supic. 2025. "Clinical Relevance of FOXP3, PD-L1, PD-1, and miR-155 Gene Expression and Genetic Variants in HPV-Negative Oral Carcinomas" International Journal of Molecular Sciences 26, no. 15: 7218. https://doi.org/10.3390/ijms26157218
APA StyleIvkovic, N., Misic, D., Kozomara, R., Jovic, S., Sami, A., Velikic, G., Stosic, S., & Supic, G. (2025). Clinical Relevance of FOXP3, PD-L1, PD-1, and miR-155 Gene Expression and Genetic Variants in HPV-Negative Oral Carcinomas. International Journal of Molecular Sciences, 26(15), 7218. https://doi.org/10.3390/ijms26157218








