CCR4-NOT Transcription Complex Subunit 7 (CNOT7) Protein and Leukocyte-Associated Immunoglobulin-like Receptor-1 in Breast Cancer Progression: Clinical Mechanistic Insights and In Silico Therapeutic Potential
Abstract
1. Introduction
1.1. Research Problem
1.2. Aim and Objectives
2. Results
2.1. BC Patient Group Participants’ Demographic and Clinical Characteristics
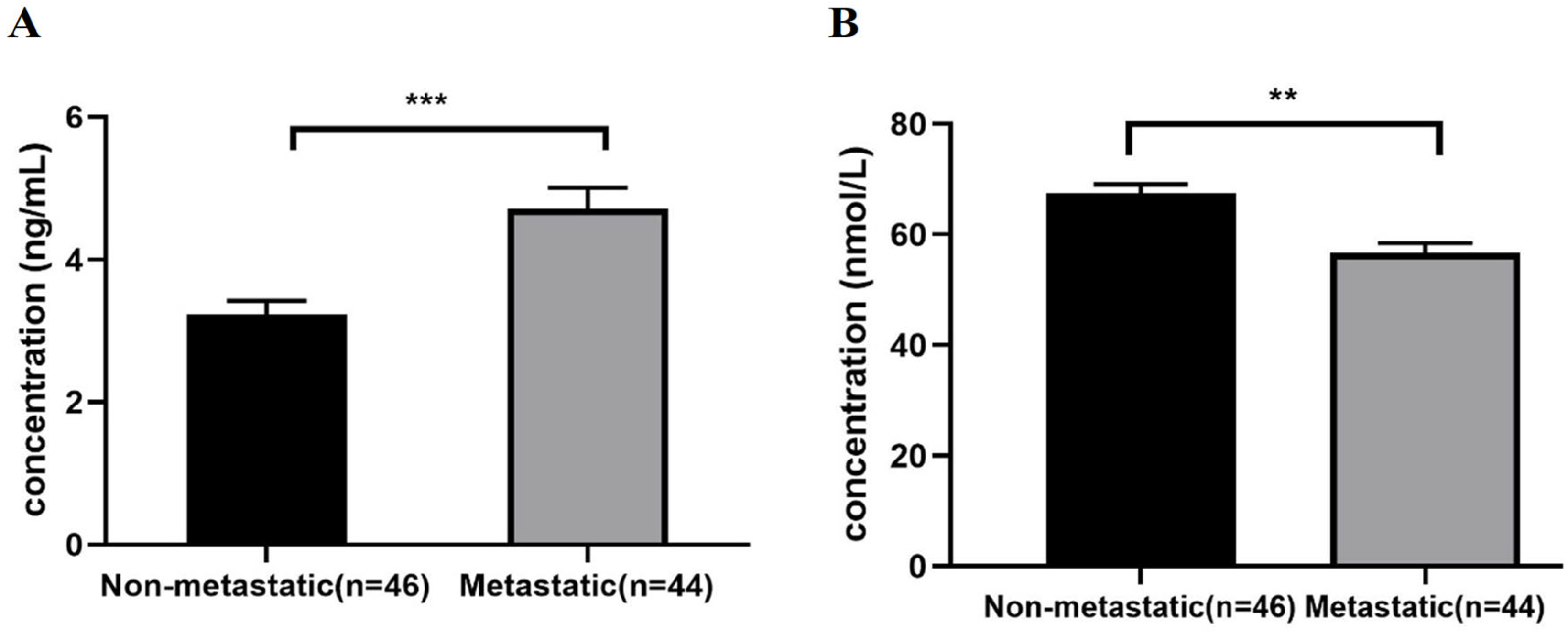
2.2. CNOT7 and LAIR-1 Levels in Peripheral Blood Samples of Metastatic BC Patients Group Compared to the Non-Metastatic Group
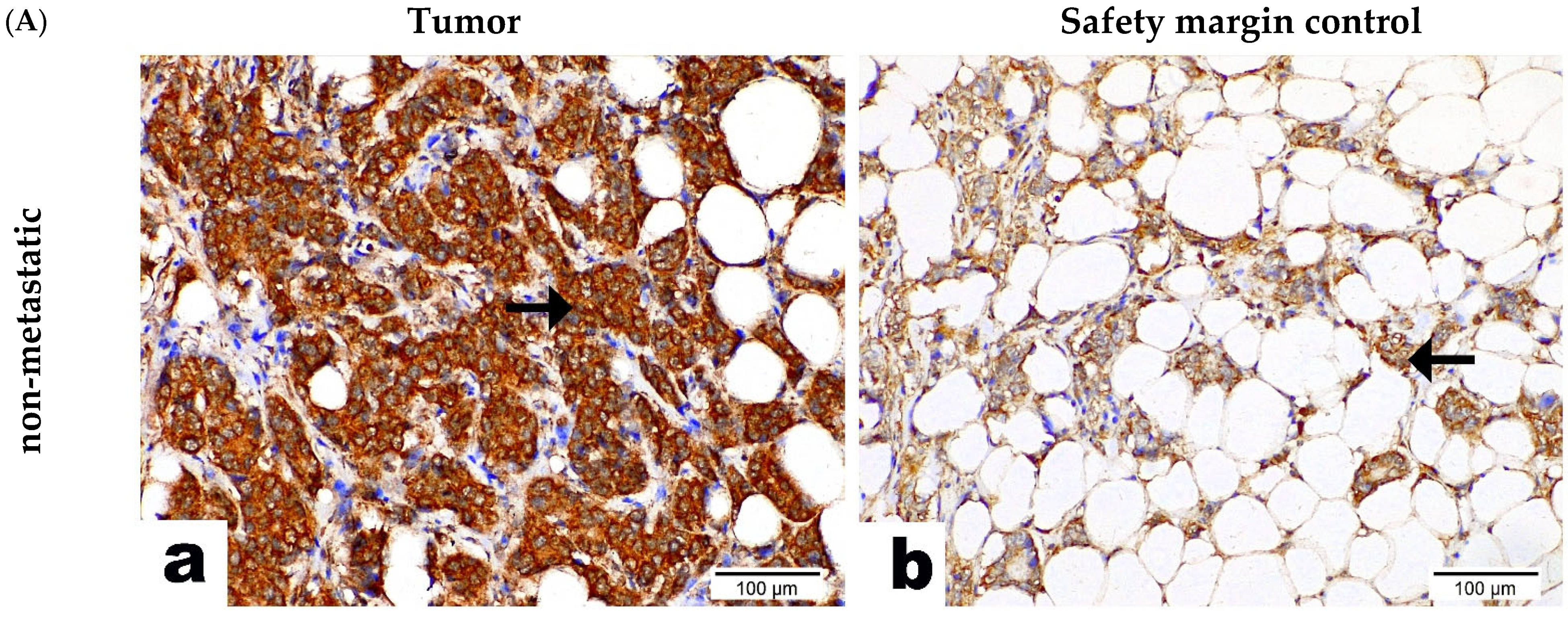
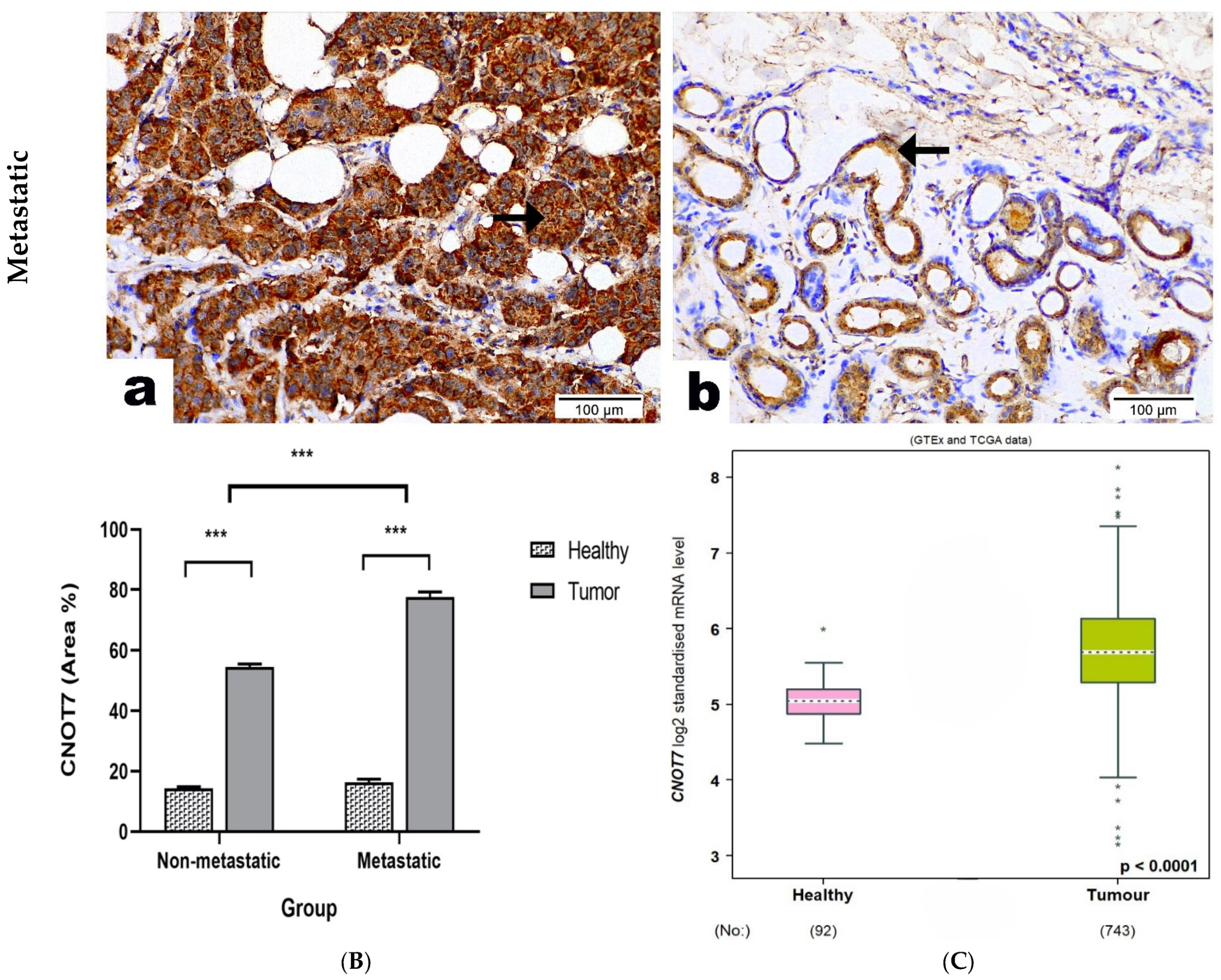
2.3. CNOT7 Protein Expression in BC Patients’ Tissue Samples
2.4. LAIR-1 or CNOT7 Serum Levels’ Association with the Clinicopathological Data of the BC Cohort (n = 90)
2.5. Correlations Results
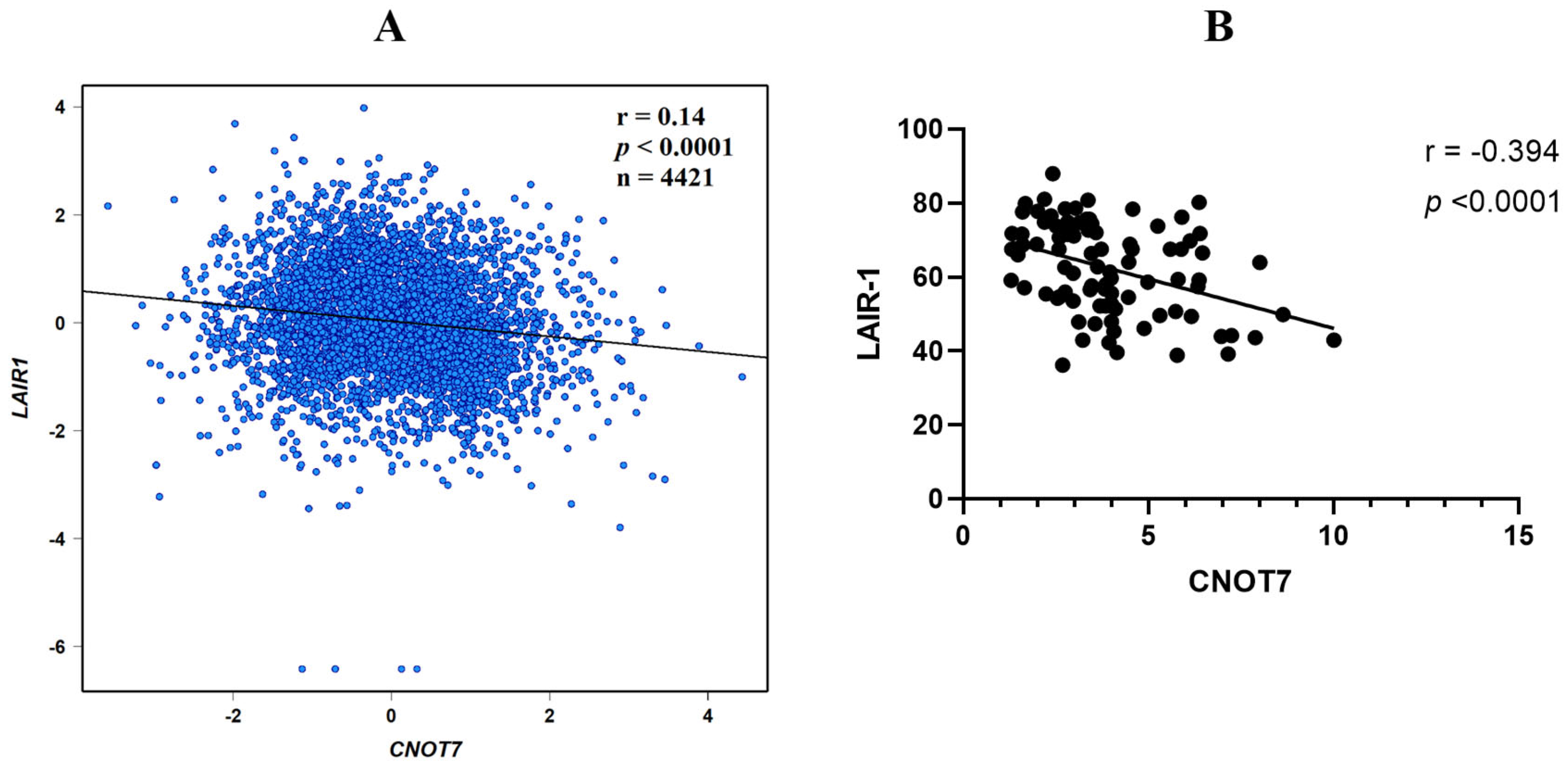
2.6. CNOT7 Correlation with LAIR-1
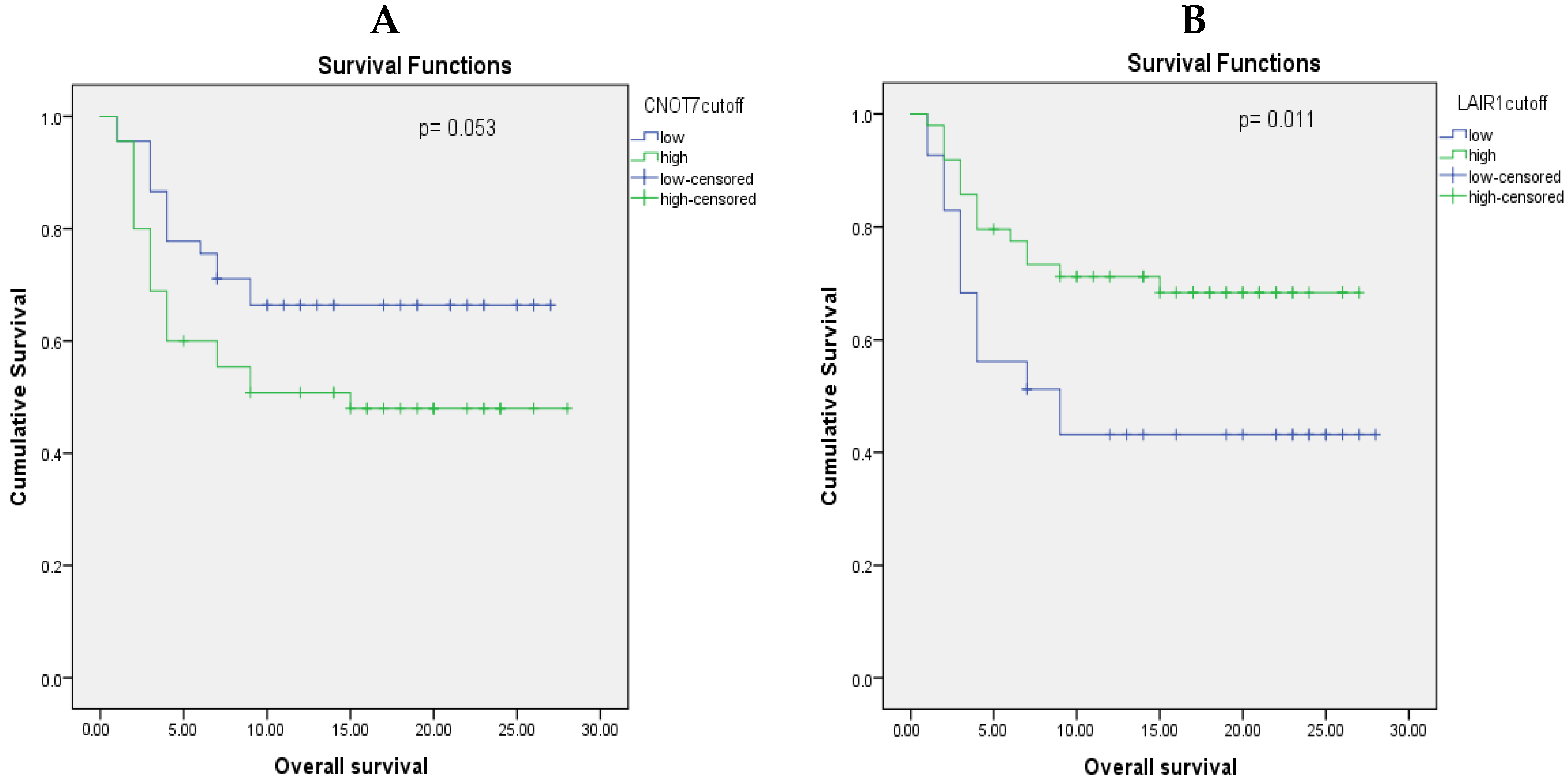
2.7. Prognostic Value of CNOT7 and LAIR-1 Serum Levels
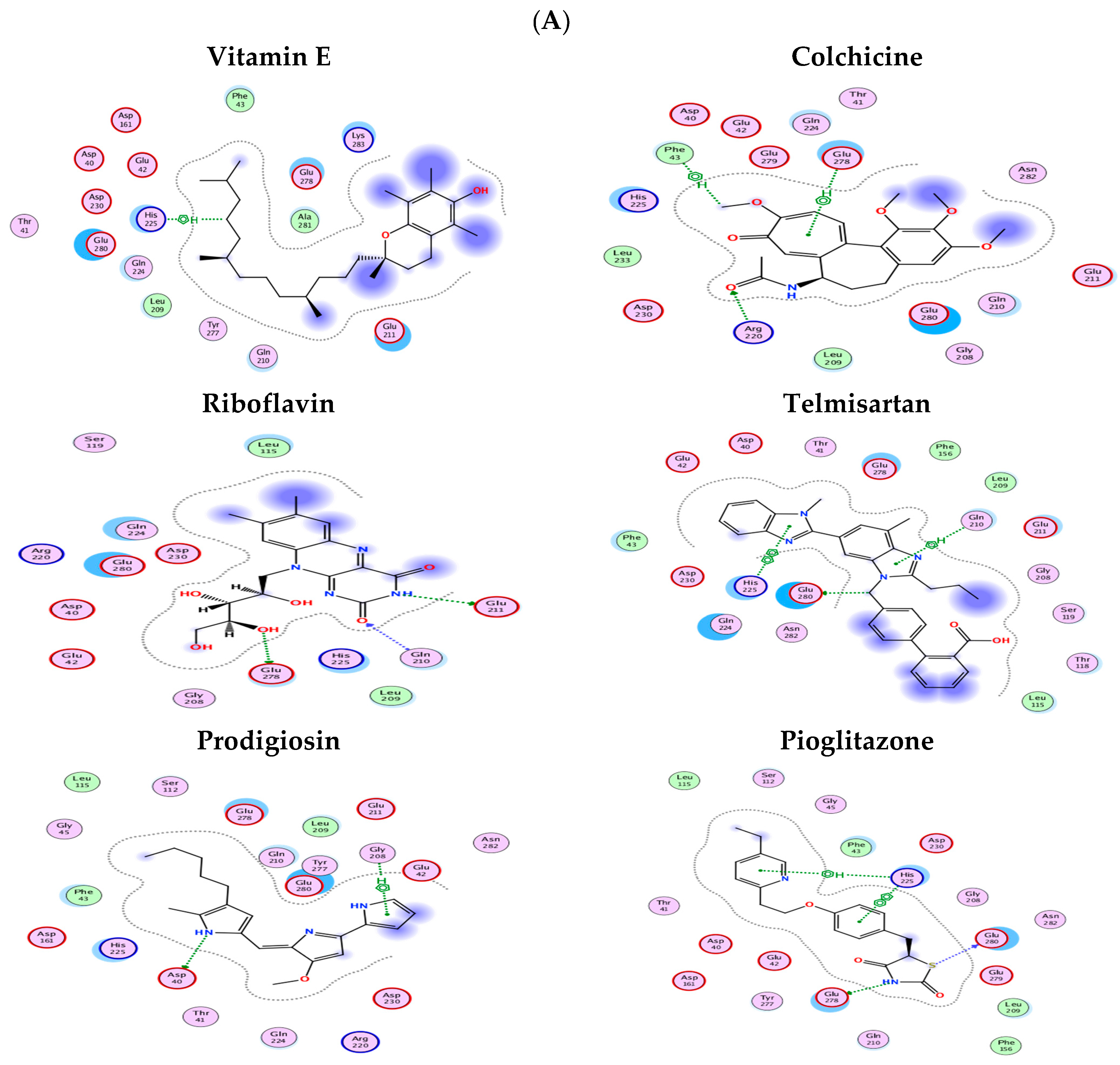
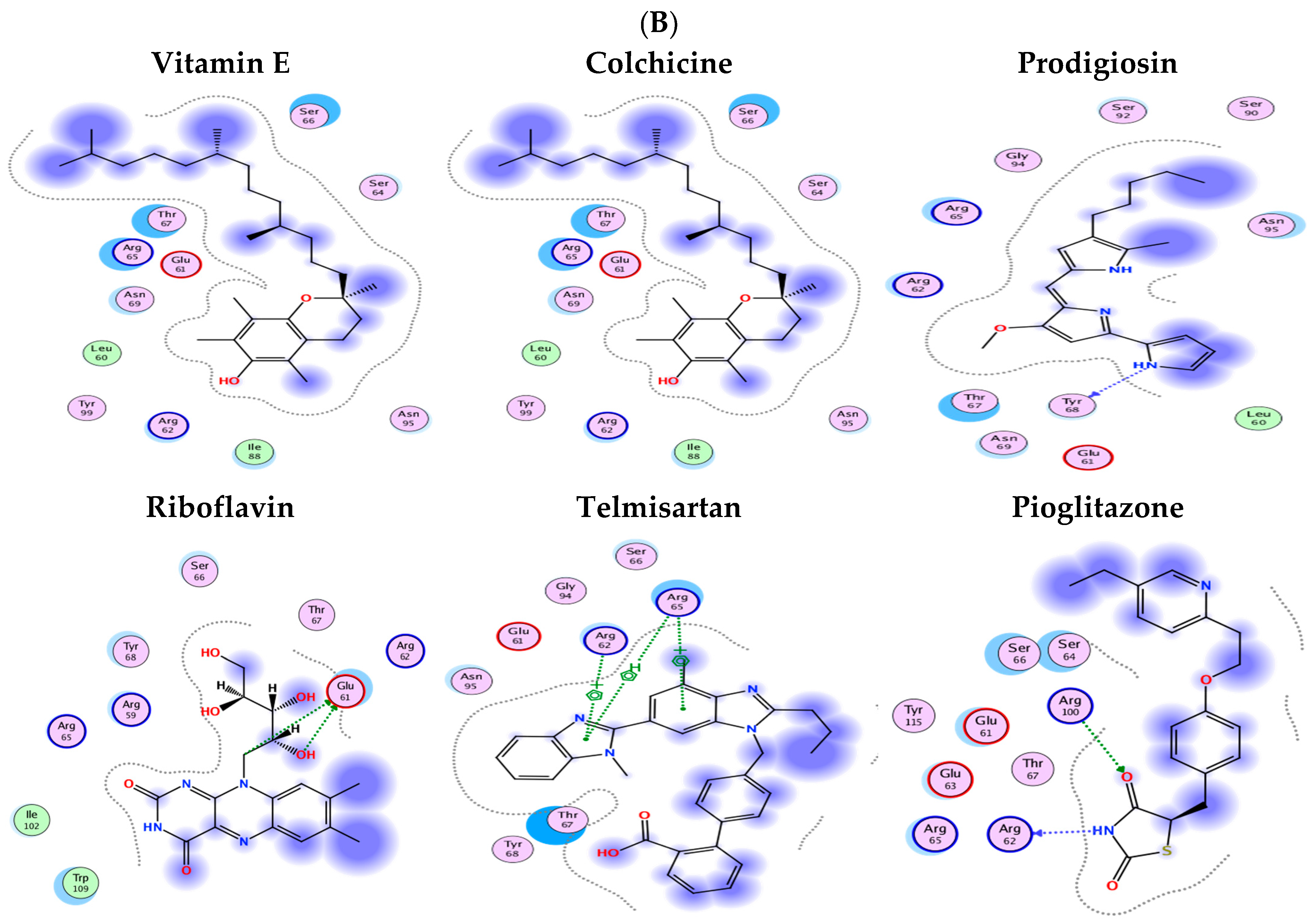
2.8. Molecular Docking Results
3. Discussion
4. Subjects
4.1. Sample Size and the Power Study
4.2. Study Design
Clinical Trial Registration
4.3. BC Patients’ Groups
Patients’ Inclusion Criteria
The Exclusion Criteria
4.4. Patients’ Clinical and Pathological Features
5. Materials and Methods
5.1. Blood Samples
5.2. Paraffin Sections Tissue Samples
5.3. Biochemical Analysis
5.3.1. Human CNOT7 or LAIR-1 ELISA (Mybiosource, San Diego, CA, USA)
5.3.2. Insulin ELISA
5.3.3. CNOT7 IHC Staining Protocol
5.3.4. Quantitative Evaluation of CNOT7 IHC Results “Area %”
5.4. Statistical Analysis
5.5. In Silico Bioinformatics Analysis
5.5.1. Breast Cancer Gene Expression Miner v5.0 (bc-GenExMiner v5.0) Updated on 28 June 2023
5.5.2. Function Module of LinkedOmics
5.5.3. Gene–Gene Interactions and Pathways
5.5.4. Protein–Protein Interactions (PPIs)
5.5.5. Exploring Gene Expression Patterns Across Normal and Tumor Tissues
5.6. Molecular Docking
5.6.1. Preparation of the Protein Structures of LAIR-1 and CNOT7
5.6.2. Ligand Dataset Curation
5.6.3. Docking Simulations
6. Summary and Conclusions
7. Therapeutic Implication(s)
8. Recommendations and Future Perspectives
9. Limitation(s), Possible Research Gap(s)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AFG3L2 | AFG3-like AAA ATPase 2 |
| AGO | Argonaute RISC catalytic component |
| ALDH7A1 | Aldehyde dehydrogenase 7 family, member A1 |
| APP | Amyloid beta (A4) precursor protein |
| BAG3 | BCL2-associated athanogene 3 |
| BC | Breast cancer |
| BTG | B-cell translocation gene |
| CAPZA | Capping protein (actin filament) muscle Z-line, alpha |
| CDK | Cyclin-dependent kinase |
| CKB | Creatine Kinase B |
| EIF | Eukaryotic translation initiation factor |
| EPCAM | Epithelial cell adhesion molecule |
| GO | Gene Ontology |
| GSEA | Gene set enrichment analysis |
| HCC | Hepatocellular carcinoma |
| IFN-γ | Interferon-γ |
| INF | Interferon |
| LAIR-1 | Leukocyte-associated immunoglobulin-like receptor-1 |
| NK | Natural killer |
| NKKB1 | Nuclear Factor Kappa B Subunit 1 |
| PABPC | Poly(A) binding protein cytoplasmic like |
| PI3K/AKT/mTOR | Phosphoinositide 3 kinase (PI3K)/AKT/mammalian (or mechanistic) target of rapamycin (mTOR) |
| PTK | Protein tyrosine kinase |
| PTP | Protein tyrosine phosphatase |
| PTPN | Protein tyrosine phosphatase non-receptor |
| RELA | V-rel avian reticuloendotheliosis viral oncogene homolog A |
| STAT1 | Signal transducer and activator of transcription 1 |
| TGF-β1 | Transforming growth factor-β1 |
| TIME | Tumor immunosuppressive microenvironment |
| TME | Tumor microenvironment |
| UBC | Ubiquitin C |
References
- Hamdy, N.M.; El-Sisi, M.G.; Ibrahim, S.M.; ElNokoudy, H.; Hady, A.A.; Abd-Ellatef, G.E.F.; Sallam, A.-A.M.; Barakat, B.M. In silico analysis and comprehensive review of circular-RNA regulatory roles in breast diseases; a step-toward non-coding RNA precision. Pathol. Res. Pract. 2024, 263, 155651. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.-F.; Huang, K.-C.; Chen, H.-Y.; Hamdy, N.M.; Huang, T.-C.; Chang, H.-Y.; Shieh, T.-M.; Huang, Y.-J.; Hsia, S.-M. Hinokitiol Inhibits Breast Cancer Cells In Vitro Stemness-Progression and Self-Renewal with Apoptosis and Autophagy Modulation via the CD44/Nanog/SOX2/Oct4 Pathway. Int. J. Mol. Sci. 2024, 25, 3904. [Google Scholar] [CrossRef] [PubMed]
- Smyth, M.J.; Dunn, G.P.; Schreiber, R.D. Cancer immunosurveillance and immunoediting: The roles of immunity in suppressing tumor development and shaping tumor immunogenicity. Adv. Immunol. 2006, 90, 1–50. [Google Scholar] [CrossRef]
- Caras, I.; Grigorescu, A.; Stavaru, C.; Radu, D.L.; Mogos, I.; Szegli, G.; Salageanu, A. Evidence for immune defects in breast and lung cancer patients. Cancer Immunol. Immunother. 2004, 53, 1146–1152. [Google Scholar] [CrossRef]
- Alexe, G.; Dalgin, G.S.; Scanfeld, D.; Tamayo, P.; Mesirov, J.P.; DeLisi, C.; Harris, L.; Barnard, N.; Martel, M.; Levine, A.J.; et al. High expression of lymphocyte-associated genes in node-negative HER2+ breast cancers correlates with lower recurrence rates. Cancer Res. 2007, 67, 10669–10676. [Google Scholar] [CrossRef]
- Sabatier, R.; Finetti, P.; Cervera, N.; Lambaudie, E.; Esterni, B.; Mamessier, E.; Tallet, A.; Chabannon, C.; Extra, J.-M.; Jacquemier, J.; et al. A gene expression signature identifies two prognostic subgroups of basal breast cancer. Breast Cancer Res. Treat. 2010, 126, 407–420. [Google Scholar] [CrossRef]
- Moretta, L.; Bottino, C.; Pende, D.; Vitale, M.; Mingari, M.; Moretta, A. Different checkpoints in human NK-cell activation. Trends Immunol. 2004, 25, 670–676. [Google Scholar] [CrossRef]
- Della Chiesa, M.; Carlomagno, S.; Frumento, G.; Balsamo, M.; Cantoni, C.; Conte, R.; Moretta, L.; Moretta, A.; Vitale, M. The tryptophan catabolite L-kynurenine inhibits the surface expression of NKp46- and NKG2D-activating receptors and regulates NK-cell function. Blood 2006, 108, 4118–4125. [Google Scholar] [CrossRef]
- Hammad, R.; Aglan, R.B.; Mohammed, S.A.; Awad, E.A.-E.; Elsaid, M.A.; Bedair, H.M.; Khirala, S.K.; Selim, M.A.; Elqasem, A.A.A.; Rushdi, A.; et al. Cytotoxic T Cell Expression of Leukocyte-Associated Immunoglobulin-Like Receptor-1 (LAIR-1) in Viral Hepatitis C-Mediated Hepatocellular Carcinoma. Int. J. Mol. Sci. 2022, 23, 12541. [Google Scholar] [CrossRef]
- Wang, L.; Simons, D.L.; Lu, X.; Tu, T.Y.; Avalos, C.; Chang, A.Y.; Dirbas, F.M.; Yim, J.H.; Waisman, J.; Lee, P.P. Breast cancer induces systemic immune changes on cytokine signaling in peripheral blood monocytes and lymphocytes. EBioMedicine 2020, 52, 102631. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Jiang, X.; Lin, K.; Zheng, P.; Wu, S.; Zeng, G.; Wei, D. Oncogenic Gene CNOT7 Promotes Progression and Induces Poor Prognosis of Glioma. Mol. Biotechnol. 2025, 67, 2067–2616. [Google Scholar] [CrossRef]
- Ren, C.; Ren, X.; Cao, D.; Zhao, H.; Zhai, Z.; Li, H.; Li, Y.; Fu, X.; He, J.; Zhao, H. CNOT7 depletion reverses natural killer cell resistance by modulating the tumor immune microenvironment of hepatocellular carcinoma. FEBS Open Bio. 2020, 10, 847–860. [Google Scholar] [CrossRef]
- Miao, Y.; Lu, J.; Fan, B.; Sun, L. MicroRNA-126-5p Inhibits the Migration of Breast Cancer Cells by Directly Targeting CNOT7. Technol. Cancer Res. Treat. 2020, 19, 1533033820977545. [Google Scholar] [CrossRef]
- Faraji, F.; Hu, Y.; Yang, H.H.; Lee, M.P.; Winkler, G.S.; Hafner, M.; Hunter, K.W.; Corbo, L. Post-transcriptional Control of Tumor Cell Autonomous Metastatic Potential by CCR4-NOT Deadenylase CNOT7. PLoS Genet. 2016, 12, e1005820. [Google Scholar] [CrossRef]
- Ren, C.; Ma, X.; Zhao, L.; Cao, D.; Zhao, H. CNOT7 is upregulated in hepatocellular carcinoma with hepatitis B virus related cirrhosis and regulates tumorigenesis by targeting signal transducer and activator of transcription 1. Int. J. Clin. Exp. Pathol. 2016, 9, 8886–8898. [Google Scholar]
- Fouët, G.; Bally, I.; Chouquet, A.; Reiser, J.-B.; Thielens, N.M.; Gaboriaud, C.; Rossi, V. Molecular basis of complement c1q collagen-like region interaction with the immunoglobulin-like receptor lair-1. Int. J. Mol. Sci. 2021, 22, 5125. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Narayanan, S.; Peruzzi, G.; Apara, A.; Natarajan, K.; Margulies, D.H.; Coligan, J.E.; Borrego, F. A Single Residue, Arginine 65, Is Critical for the Functional Interaction of Leukocyte-Associated Inhibitory Receptor-1 with Collagens. J. Immunol. 2009, 182, 5446–5452. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, D.; Takahashi, A.; Yanagiya, A.; Yamaguchi, T.; Abe, T.; Kureha, T.; Kuba, K.; Kanegae, Y.; Furuta, Y.; Yamamoto, T.; et al. Essential functions of the CNOT7/8 catalytic subunits of the CCR4-NOT complex in mRNA regulation and cell viability. RNA Biol. 2020, 17, 403–416. [Google Scholar] [CrossRef]
- Chen, C.; Ito, K.; Takahashi, A.; Wang, G.; Suzuki, T.; Nakazawa, T.; Yamamoto, T.; Yokoyama, K. Distinct expression patterns of the subunits of the CCR4–NOT deadenylase complex during neural development. Biochem. Biophys. Res. Commun. 2011, 411, 360–364. [Google Scholar] [CrossRef]
- Yu, J.; Hu, X.; Chen, X.; Zhou, Q.; Jiang, Q.; Shi, Z.; Zhu, H. CNOT7 modulates biological functions of ovarian cancer cells via AKT signaling pathway. Life Sci. 2021, 268, 118996. [Google Scholar] [CrossRef]
- Maragozidis, P.; Papanastasi, E.; Scutelnic, D.; Totomi, A.; Kokkori, I.; Zarogiannis, S.G.; Kerenidi, T.; Gourgoulianis, K.I.; Balatsos, N.A.A. Poly(A)-specific ribonuclease and Nocturnin in squamous cell lung cancer: Prognostic value and impact on gene expression. Mol. Cancer 2015, 14, 187. [Google Scholar] [CrossRef]
- Chapat, C.; Kolytcheff, C.; Le Romancer, M.; Auboeuf, D.; De La Grange, P.; Chettab, K.; Sentis, S.; Corbo, L. hCAF1/CNOT7 regulates interferon signalling by targeting STAT1. EMBO J. 2013, 32, 688–700. [Google Scholar] [CrossRef]
- Aslam, A.; Mittal, S.; Koch, F.; Andrau, J.-C.; Winkler, G.S.; Wickens, M. The Ccr4–NOT deadenylase subunits CNOT7 and CNOT8 have overlapping roles and modulate cell proliferation. Mol. Biol. Cell 2009, 20, 3840–3850. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Adachi, S.; Morita, M.; Tokumasu, M.; Natsume, T.; Suzuki, T.; Yamamoto, T. Post-transcriptional Stabilization of Ucp1 mRNA Protects Mice from Diet-Induced Obesity. Cell Rep. 2015, 13, 2756–2767. [Google Scholar] [CrossRef] [PubMed]
- Elanany, M.M.; Mostafa, D.; Hamdy, N.M. Remodeled tumor immune microenvironment (TIME) parade via natural killer cells reprogramming in breast cancer. Life Sci. 2023, 330, 121997. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gingold, J.A.; Su, X. Immunomodulatory TGF-β Signaling in Hepatocellular Carcinoma. Trends Mol. Med. 2019, 25, 1010–1023. [Google Scholar] [CrossRef]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef]
- Piaszyk-Borychowska, A.; Széles, L.; Csermely, A.; Chiang, H.-C.; Wesoły, J.; Lee, C.-K.; Nagy, L.; Bluyssen, H.A.R. Signal integration of IFN-I and IFN-II with TLR4 involves sequential recruitment of STAT1-Complexes and NFκB to enhance pro-inflammatory transcription. Front. Immunol. 2019, 10, 1253. [Google Scholar] [CrossRef]
- Hammad, R.; Kandeel, E.Z.; Lambert, C.; Sack, U.; Kujumdshiev, S.; Kamhawy, A.; Abo-Elkheir, O.I.; El Hakam, F.E.-Z.A.; Mashaal, A.; Ramadan, M.; et al. Leukemic B cells expression of CD200 and Leukocyte-associated immunoglobulin-like receptor-1 (LAIR-1, CD305) in Chronic Lymphocytic Leukemia patients in relation to Treg frequency. Pathol. Res. Pract. 2024, 263, 155669. [Google Scholar] [CrossRef]
- Joseph, C.; Alsaleem, M.A.; Toss, M.S.; Kariri, Y.A.; Althobiti, M.; Alsaeed, S.; Aljohani, A.I.; Narasimha, P.L.; Mongan, N.P.; Green, A.R.; et al. The ITIM-Containing Receptor: Leukocyte-Associated Immunoglobulin-Like Receptor-1 (LAIR-1) Modulates Immune Response and Confers Poor Prognosis in Invasive Breast Carcinoma. Cancers 2021, 13, 80. [Google Scholar] [CrossRef]
- Lebbink, R.J.; de Ruiter, T.; Adelmeijer, J.; Brenkman, A.B.; van Helvoort, J.M.; Koch, M.; Farndale, R.W.; Lisman, T.; Sonnenberg, A.; Lenting, P.J.; et al. Collagens are functional, high affinity ligands for the inhibitory immune receptor LAIR-1. J. Exp. Med. 2006, 203, 1419–1425. [Google Scholar] [CrossRef]
- Poggi, A.; Pellegatta, F.; Leone, B.E.; Moretta, L.; Zocchi, M.R. Engagement of the leukocyte-associated Ig-like receptor-1 induces programmed cell death and prevents NF-κB nuclear translocation in human myeloid leukemias. Eur. J. Immunol. 2000, 30, 2751–2758. [Google Scholar] [CrossRef] [PubMed]
- Zocchi, M.R.; Pellegatta, F.; Pierri, I.; Gobbi, M.; Poggi, A. Leukocyte-associated Ig-like receptor-1 prevents granulocyte-monocyte colony stimulating factor-dependent proliferation and Akt1/PKB alpha activation in primary acute myeloid leukemia cells. Eur. J. Immunol. 2001, 31, 3667–3675. [Google Scholar] [CrossRef] [PubMed]
- Poggi, A.; Catellani, S.; Bruzzone, A.; Caligaris-Cappio, F.; Gobbi, M.; Zocchi, M.R. Lack of the leukocyte-associated Ig-like receptor-1 expression in high-risk chronic lymphocytic leukaemia results in the absence of a negative signal regulating kinase activation and cell division. Leukemia 2008, 22, 980–988. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, M.; Wu, L.; Mao, L.; Chen, L.; Yu, G.; Deng, W.; Zhang, W.; Liu, B.; Sun, W.; et al. LAIR-1 overexpression and correlation with advanced pathological grade and immune suppressive status in oral squamous cell carcinoma. Head Neck 2019, 41, 1080–1086. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, L.; Zhou, J.; Liu, L.; Fu, Q.; Fu, A.; Feng, X.; Xin, R.; Liu, H.; Gao, Y.; et al. Clinicopathologic significance of LAIR-1 expression in hepatocellular carcinoma. Curr. Probl. Cancer 2019, 43, 18–26. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Miao, F.; Cao, Y.; Xue, J.; Cao, Q.; Zhang, X. Clinical significance of leukocyte-associated immunoglobulin-like receptor-1 expression in human cervical cancer. Exp. Ther. Med. 2016, 12, 3699–3705. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, S.; Zhang, H.; Long, Q.-X.; Liu, X.-B.; Zhu, D.-M. Correlation of the quantitative parameters of ultrasonic elastography in breast cancer with the abnormal apoptosis and invasion of cancer cells in lesions. J. Hainan Med. Univ. 2018, 24, 119–122. [Google Scholar]
- Fan, G.; Aleem, S.; Yang, M.; Miller, W.T.; Tonks, N.K. Protein-tyrosine phosphatase and kinase specificity in regulation of SRC and breast tumor kinase. J. Biol. Chem. 2015, 290, 15934–15947. [Google Scholar] [CrossRef]
- Sonis, S.T.; Hashemi, S.; Epstein, J.B.; Nair, R.G.; Raber-Durlacher, J.E. Could the biological robustness of low level laser therapy (Photobiomodulation) impact its use in the management of mucositis in head and neck cancer patients. Oral Oncol. 2016, 54, 7–14. [Google Scholar] [CrossRef]
- Hamdy, N.M.; Zaki, M.B.; Rizk, N.I.; Abdelmaksoud, N.M.; Abd-Elmawla, M.A.; Ismail, R.A.; Abulsoud, A.I. Unraveling the ncRNA landscape that governs colorectal cancer: A roadmap to personalized therapeutics. Life Sci. 2024, 354, 122946. [Google Scholar] [CrossRef]
- Wander, S.A.; Zhao, D.; Besser, A.H.; Hong, F.; Wei, J.; Ince, T.A.; Milikowski, C.; Bishopric, N.H.; Minn, A.J.; Creighton, C.J.; et al. PI3K/mTOR inhibition can impair tumor invasion and metastasis in vivo despite a lack of anti-proliferative action in vitro: Implications for targeted therapy. Breast Cancer Res. Treat. 2013, 138, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Qian, J.; Li, J.; Zhu, C. Knockdown of lncRNA-HOTAIR downregulates the drug-resistance of breast cancer cells to doxorubicin via the PI3K/AKT/mTOR signaling pathway. Exp. Ther. Med. 2019, 18, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Anwar, M.M.; Albanese, C.; Hamdy, N.M.; Sultan, A.S. Rise of the natural red pigment ‘prodigiosin’ as an immunomodulator in cancer. Cancer Cell Int. 2022, 22, 419. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, V.A.; Pereira, I.C.; Nogueira, T.R.; Martins, J.A.; Péres-Rodrigues, G.; de Jesus e Silva de Almendra, B.; Silva, V.C.; Júnior, D.D.; Leal, F.L.T.; de Castro e Sousa, J.M.; et al. The Role of Vitamin E in Breast Cancer Treatment and Prevention: Current Perspectives. Curr. Nutr. Food Sci. 2020, 17, 134–143. [Google Scholar] [CrossRef]
- Singh, A.; Maheshwari, S.; Kumar, R.; Yadav, J.P.; Kumari, R. Telmisartan-loaded liposomes: An innovative weapon against breast cancer. Intell. Pharm. 2024, 2, 565–570. [Google Scholar] [CrossRef]
- Shen, H.; Huo, R.; Zhang, Y.; Wang, L.; Tong, N.; Chen, W.; Paris, A.J.; Mensah, K.; Chen, M.; Xue, Y.; et al. A Pilot Study To Assess the Suitability of Riboflavin As a Surrogate Marker of Breast Cancer Resistance Protein in Healthy Participants. J. Pharmacol. Exp. Ther. 2024, 390, 162–173. [Google Scholar] [CrossRef]
- Krutilina, R.I.; Hartman, K.L.; Oluwalana, D.; Playa, H.C.; Parke, D.N.; Chen, H.; Miller, D.D.; Li, W.; Seagroves, T.N. Sabizabulin, a Potent Orally Bioavailable Colchicine Binding Site Agent, Suppresses HER2+ Breast Cancer and Metastasis. Cancers 2022, 14, 5336. [Google Scholar] [CrossRef]
- Jiao, X.X.; Lin, S.Y.; Lian, S.X.; Qiu, Y.R.; Li, Z.H.; Chen, Z.H.; Lu, W.Q.; Zhang, Y.; Deng, L.; Jiang, Y.; et al. Inhibition of the breast cancer by pparγ agonist pioglitazone through jak2/stat3 pathway. Neoplasma 2020, 67, 834–842. [Google Scholar] [CrossRef]
- Hamdy, N.M.; Sanad, E.F.; Kassab, S.E.; Essam, M.; Guirguis, M.A.; Basalious, E.B.; Sultan, A.S. Treatment of non-small cell lung cancer using chem-bioinformatics-driven engineering of exosomal cargo-vehicle for telmisartan and pioglitazone targeted-delivery. Sci. Rep. 2025, 15, 25166. [Google Scholar] [CrossRef]
- Xue, J.-N.; Wang, C.-Y.; Zhang, X.-S.; Cao, Q.-Z.; Su, Z.-G. The up-regulated expression of LAIR-1 in tumor patients PBMC. J. Cell. Mol. Immunol. 2008, 24, 373–374+377. [Google Scholar]
- Bloom, H.J.G.; Richardson, W.W. Histological Grading and Prognosis in Breast Cancer. Br. J. Cancer 1957, 11, 359–377. [Google Scholar] [CrossRef] [PubMed]
- Nuttall, F.Q. Body Mass Index: Obesity, BMI, and Health: A Critical Review. Nutr. Today 2015, 50, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Ekta Ghosh, D. Intensive training in breast cancer imaging with article intelligence and deep learning—A review article. J. Adv. Radiol. Med. Imaging 2022, 1, 18–26. [Google Scholar]
- Vajpeyi, R. WHO Classification of Tumours: Pathology and Genetics of Tumours of the Breast and Female Genital Organs. J. Clin. Pathol. 2005, 58, 671–672. [Google Scholar]
- Makki, J. Diversity of breast carcinoma: Histological subtypes and clinical relevance. Clin. Med. Insights Pathol. 2015, 8, 23–31. [Google Scholar] [CrossRef]
- Hsu, S.M.; Raine, L.; Fanger, H. Use of avidin-biotin-peroxidase complex (ABC) in immunoperoxidase techniques: A comparison between ABC and unlabeled antibody (PAP) procedures. J. Histochem. Cytochem. 1981, 29, 577–580. [Google Scholar] [CrossRef]
- Jézéquel, P.; Frénel, J.-S.; Campion, L.; Guérin-Charbonnel, C.; Gouraud, W.; Ricolleau, G.; Campone, M. bc-GenExMiner 3.0: New mining module computes breast cancer gene expression correlation analyses. Database 2013, 2013, bas060. [Google Scholar] [CrossRef]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018, 46, D956–D963. [Google Scholar] [CrossRef]
- Fernandes, J.D.; Hinrichs, A.S.; Clawson, H.; Gonzalez, J.N.; Lee, B.T.; Nassar, L.R.; Raney, B.J.; Rosenbloom, K.R.; Nerli, S.; Rao, A.A.; et al. The UCSC SARS-CoV-2 Genome Browser. Nat. Genet. 2020, 52, 991–998. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Jadhav, G.P.; Kaur, I.; Maryati, M.; Airhihen, B.; Fischer, P.M.; Winkler, G.S. Discovery, synthesis and biochemical profiling of purine-2,6-dione derivatives as inhibitors of the human poly(A)-selective ribonuclease Caf1. Bioorganic Med. Chem. Lett. 2015, 25, 4219–4224. [Google Scholar] [CrossRef]
- Chapat, C.; Chettab, K.; Simonet, P.; Wang, P.; De La Grange, P.; Le Romancer, M.; Corbo, L. Alternative splicing of CNOT7 diversifies CCR4–NOT functions. Nucleic Acids Res. 2017, 45, 8508–8523. [Google Scholar] [CrossRef] [PubMed]
- Sanad, E.F.; Hamdy, N.M.; El-Etriby, A.K.; Sebak, S.A.; El-Mesallamy, H.O. Peripheral leucocytes and tissue gene expression of granzyme B/perforin system and serpinB9: Impact on inflammation and insulin resistance in coronary atherosclerosis. Diabetes Res. Clin. Pract. 2017, 131, 132–141. [Google Scholar] [CrossRef] [PubMed]
- El-Mesallamy, H.O.; Hamdy, N.M.; Zaghloul, A.S.; Sallam, A.M.M. Clinical value of circulating lipocalins and insulin-like growth factor axis in pancreatic cancer diagnosis. Pancreas 2013, 42, 149–154. [Google Scholar] [CrossRef] [PubMed]
- El-Mesallamy, H.O.; Hamdy, N.M.; Rizk, H.H.; El-Zayadi, A.-R. Apelin serum level in Egyptian patients with chronic hepatitis C. Mediat. Inflamm. 2011, 2011, 703031. [Google Scholar] [CrossRef]
- El-Mesallamy, H.O.; Hamdy, N.M.; Zaghloul, A.S.; Sallam, A.M. Serum retinol binding protein-4 and neutrophil gelatinase-associated lipocalin are interrelated in pancreatic cancer patients. Scand J. Clin Lab. Invest. 2012, 72, 602–607. [Google Scholar] [CrossRef]
- El-Mesallamy, H.O.; Hamdy, N.M.; Sallam, A.A. Effect of obesity and glycemic control on serum lipocalins and insulin-like growth factor axis in type 2 diabetic patients. Acta Diabetol. 2013, 50, 679–685. [Google Scholar] [CrossRef]
- Mostafa, A.M.; Hamdy, N.M.; El-Mesallamy, H.O.; Abdel-Rahman, S.Z. Glucagon-like peptide 1 (GLP-1)-based therapy upregulates LXR-ABCA1/ABCG1 cascade in adipocytes. Biochem. Biophys. Res. Commun. 2015, 468, 900–905. [Google Scholar] [CrossRef]
- Hamdy, N.M. Relationship between pro-anti-inflammatory cytokines, T-cell activation and CA 125 in obese patients with heart failure. Med. Sci. Monit. 2011, 17, CR174-9. [Google Scholar] [CrossRef]
- Radwan, S.M.; Hamdy, N.M.; Hegab, H.M.; El-Mesallamy, H.O. Beclin-1 and hypoxia-inducible factor-1α genes expression: Potential biomarkers in acute leukemia patients. Cancer Biomark. 2016, 16, 619–626. [Google Scholar] [CrossRef]
- Kamal, A.M.; Hamdy, N.M.; Hegab, H.M.; El-Mesallamy, H.O. Expression of thioredoxin-1 (TXN) and its relation with oxidative DNA damage and treatment outcome in adult AML and ALL: A comparative study. Hematology 2016, 21, 567–575. [Google Scholar] [CrossRef]
- Bhattacharya, K.; Shamkh, I.M.; Khan, M.S.; Lotfy, M.M.; Nzeyimana, J.B.; Abutayeh, R.F.; Hamdy, N.M.; Hamza, D.; Chanu, N.R.; Khanal, P.; et al. Multi-Epitope Vaccine Design against Monkeypox Virus via Reverse Vaccinology Method Exploiting Immunoinformatic and Bioinformatic Approaches. Vaccines 2022, 10, 2010. [Google Scholar] [CrossRef]
| Group (n) | Statistics | ||
|---|---|---|---|
| Characteristic (Unit) | Non-Metastatic (46) | Metastatic (44) | p |
| Age (years) (n,%) | ꭓ2 = 2.820 NS | ||
| ≤50 | (18, 39.1%) | (25, 56.8%) | |
| >50 | (28, 60.9%) | (19, 43.2%) | |
| BMI (kg/m2) | ꭓ2 = 6.673 p = 0.036 * | ||
| Normal (18.5–24.9) (n, %) | (1, 2.2%) | (2, 4.5%) | |
| Overweight (25–29.9) (n, %) | (5, 10.9%) | (14, 31.8%) | |
| Obese (≥30) (n, %) | (40, 87.0%) | (28, 63.6%) | |
| Diabetic status | ꭓ2 = 2.883 NS | ||
| Yes (n, %) | (20, 43.5%) | (27, 61.4%) | |
| No (n, %) | (26, 56.5%) | (17, 38.6%) | |
| No. of pregnancies | ꭓ2 = 1.558 NS | ||
| ≤2 (n, %) | (17, 37.0%) | (22, 50.0%) | |
| >2 (n, %) | (29, 63.0%) | (22, 50.0%) | |
| Menopausal status | ꭓ2 = 0.552 NS | ||
| Pre (n, %) | (21, 45.7%) | (24, 54.5%) | |
| Post (n, %) | (24, 52.2%) | (20, 45.5%) | |
| BC Family History | ꭓ2 = 0.829 NS | ||
| Yes (n, %) | (17, 37.0%) | (12, 27.3%) | |
| No (n, %) | (29, 63.0%) | (31, 70.5%) | |
| Histological BC subtype | ꭓ2 = 2.007 NS | ||
| IDC (n, %) | (43, 93.5%) | (37, 84.1%) | |
| Other subtypes (n, %) | (3, 6.5%) | (7, 15.9%) | |
| BC molecular subtype | |||
| Luminal A (n, %) | (34, 73.9%) | (17, 38.6%) | ꭓ2 = 15.830 p = 0.001 * |
| Luminal B (n, %) | (9, 19.6%) | (18, 40.9%) | |
| HER-2 overexpression (n, %) | (3, 6.5%) | (2, 4.5%) | |
| TNBC (n, %) | (0, 0%) | (7, 15.9%) | |
| ER status | ꭓ2 = 4.779 p = 0.029 * | ||
| Positive (n, %) | (43, 93.5%) | (34, 77.3%) | |
| Negative (n, %) | (3, 6.5%) | (10, 22.7%) | |
| PR status | ꭓ2 = 1.213 NS | ||
| Positive (n, %) | (37, 80.4%) | (31, 70.5%) | |
| Negative (n, %) | (9, 19.6%) | (13, 29.5%) | |
| HER-2/neu status | ꭓ2 = 1.176 NS | ||
| Positive (n, %) High | (12, 26.1%) | (9, 20.5%) | |
| Negative (n, %) Low | (12, 26.1%) | (16, 36.4%) | |
| None | (22, 47.8%) | (19, 43.2%) | |
| Tumor size (cm) | ꭓ2 = 2.402 NS | ||
| <2 (n, %) | (6, 13.0%) | (2, 4.5%) | |
| 2–5 (n, %) | (31, 67.4%) | (30, 68.2%) | |
| >5 (n, %) | (9, 19.6%) | (12, 27.3%) | |
| LN involvement | ꭓ2 = 13.074 p = 0.004 * | ||
| None (n, %) | (19, 41.3%) | (7, 15.9%) | |
| 1–3 (n, %) | (13, 28.3%) | (11, 25.0%) | |
| 4–9 (n, %) | (5, 10.9%) | (18, 40.9%) | |
| ≥10 (n, %) | (9, 19.6%) | (8, 18.2%) | |
| TNM stage | ꭓ2 = 40.927 p = 0.001 * | ||
| I-II (early stage) (n, %) | (29, 63.0%) | (0, 20.0%) | |
| III-IV (late stage) (n, %) | (17, 37.0%) | (44, 100.0%) | |
| Grade (Bloom–Richardson scale) | ꭓ2 =7.396 p =0.025 * | ||
| I (n, %) | (2, 4.3%) | (0, 0%) | |
| II (n, %) | (30, 65.2%) | (39, 88.6%) | |
| III (n,%) | (14, 30.4%) | (5, 11.4%) | |
| Ki-67 status | ꭓ2 = 1.472 NS | ||
| High (>14%) (n, %) | (21, 45.7%) | (24, 54.5%) | |
| Low (≤14%) (n, %) | (12, 26.1%) | (7, 15.9%) | |
| NA (n, %) | (13, 28.3%) | (13, 29.5%) | |
| Therapy | |||
| Neoadjuvant chemotherapy | ꭓ2 = 26.058 p < 0.001 * | ||
| Yes (n, %) | (7, 15.2%) | (30, 68.2%) | |
| No (n, %) | (39, 84.8%) | (14, 31.8%) | |
| Adjuvant chemotherapy | ꭓ2 = 1.574 NS | ||
| Yes (n, %) | (32, 69.6%) | (25, 56.8%) | |
| No (n, %) | (14, 30.4%) | (19, 43.2%) | |
| Endocrine therapy | ꭓ2 = 0.005 NS | ||
| Yes (n, %) | (30, 65.2%) | (29, 65.9%) | |
| No (n, %) | (16, 34.8%) | (15, 34.1%) | |
| Mastectomy | ꭓ2 = 0.356 NS | ||
| Yes (n, %) | (32, 69.6%) | (28, 63.6%) | |
| No (n, %) | (14, 30.4%) | (16, 36.4%) | |
| Insulin (uIU/mL) # | 30.865 (24.837–49.484) | 30.578 (23.027–45.919) | NS |
| Characteristic (Unit) | LAIR-1 Serum Levels (≤61.01) | Statistics p | CNOT7 Serum Levels (≤3.62) | Statistics p | ||
|---|---|---|---|---|---|---|
| Low | High | Low | High | |||
| Age (years) | ꭓ2 = 0.062 NS | ꭓ2 = 3.607 NS | ||||
| ≤50 (n, %) | (19, 46.3%) | (24, 49.0%) | (26, 57.8%) | (17, 37.8%) | ||
| >50 (n, %) | (22, 53.7%) | (25, 51.0%) | (19, 42.2%) | (28, 62.2%) | ||
| BMI (kg/m2) | ꭓ2 = 0.630 NS | ꭓ2 = 6.067 p = 0.048 * | ||||
| Normal (18.5–24.9) (n, %) | (2, 4.9%) | (1, 2.0%) | (1, 2.2%) | (2, 4.4%) | ||
| Overweight (25–29.9) (n, %) | (8, 19.5%) | (11, 22.4%) | (5, 11.1%) | (14, 31.1%) | ||
| Obese (≥30) (n, %) | (31, 75.6%) | (37, 75.5%) | (39, 86.7%) | (29, 64.4%) | ||
| Diabetic status | ꭓ2 = 7.795 p = 0.005 * | ꭓ2 = 3.607 NS | ||||
| Yes (n, %) | (28, 68.3%) | (19, 38.8%) | (19, 42.2%) | (28, 62.2%) | ||
| No (n, %) | (13, 31.7%) | (30, 61.2%) | (26, 57.8%) | (17, 37.8%) | ||
| No. of pregnancies | ꭓ2 = 0.010 NS | ꭓ2 = 0.045 NS | ||||
| ≤2 (n, %) | (18, 43.9%) | (21, 42.9%) | (20, 44.4%) | (19, 42.2%) | ||
| >2 (n, %) | (23, 56.1%) | (28, 57.1%) | (25, 55.6%) | (26, 57.8%) | ||
| Menopausal status | ꭓ2= 0.109 NS | ꭓ2= 12.231 p = 0.0005 * | ||||
| Pre (n, %) | (21, 51.2%) | (24, 49.0%) | (31, 68.9%) | (14, 31.1%) | ||
| Post (n, %) | (19, 46.3%) | (25, 51.0%) | (14, 31.1%) | (30, 66.7%) | ||
| BC Family History | ꭓ2 = 1.902 NS | ꭓ2 = 0.023 NS | ||||
| Yes (n, %) | (10, 24.4%) | (19, 38.8%) | (15, 33.3%) | (14, 31.1%) | ||
| No (n, %) | (30, 73.2%) | (30, 61.2%) | (30, 66.7%) | (30, 66.7%) | ||
| Histological BC subtype | ꭓ2 = 0.090 NS | ꭓ2 = 1.80 NS | ||||
| IDC (n, %) | (36, 87.8%) | (44, 89.9%) | (38, 84.4%) | (42, 93.3%) | ||
| Other subtypes (n, %) | (5, 12.2%) | (5, 10.2%) | (7, 15.6%) | (3, 6.7%) | ||
| BC molecular subtype | ꭓ2 = 5.812 NS | ꭓ2 = 8.077 p = 0.044 * | ||||
| Luminal A (n, %) | (20, 48.8%) | (31, 63.3%) | (31, 68.9%) | (20, 44.4%) | ||
| Luminal B (n, %) | (12, 29.3%) | (15, 30.6%) | (12, 26.7%) | (15, 33.3%) | ||
| HER-2 overexpression (n, %) | (3, 7.3%) | (2, 4.1%) | (1, 2.2%) | (4, 8.9%) | ||
| TNBC (n, %) | (6, 14.6%) | (1, 2.0%) | (1, 2.2%) | (6, 13.3%) | ||
| BC molecular subtype combined | ꭓ2 = 5.618 NS | ꭓ2 = 6.192 p = 0.045 * | ||||
| Luminal (n, %) | (32, 78.0%) | (46, 93.9%) | (43, 95.6%) | (35, 77.8%) | ||
| HER-2 overexpression (n, %) | (3, 7.3%) | (2, 4.1%) | (1, 2.2%) | (4, 8.9%) | ||
| TNBC (n, %) | (6, 14.6%) | (1, 2.0%) | (1, 2.2%) | (6, 13.3%) | ||
| ER status | ꭓ2 = 6.028 p = 0.014 * | ꭓ2 = 7.283 p = 0.007 * | ||||
| Positive (n, %) | (31, 75.6%) | (46, 93.9%) | (43, 95.6%) | (34, 75.6%) | ||
| Negative (n, %) | (10, 24.4%) | (3, 6.1%) | (2, 4.4%) | (11, 24.4%) | ||
| PR status | ꭓ2 = 3.838 p = 0.05 * | ꭓ2 = 6.016 p = 0.014 * | ||||
| Positive (n, %) | (27, 65.9%) | (41, 83.7%) | (39, 86.7%) | (29, 64.4%) | ||
| Negative (n, %) | (14, 34.1%) | (8, 16.3%) | (6, 13.3%) | (16, 35.6%) | ||
| HER-2/neu status | ꭓ2 = 0.652 NS | ꭓ2 = 1.553 NS | ||||
| Positive (n, %) | (8, 19.5%) | (13, 26.5%) | (8, 17.8%) | (13, 28.9%) | ||
| Negative (n, %) | (33, 80.5%) | (36, 73.5%) | (37, 82.2%) | (32, 71.1%) | ||
| Tumor size (cm) | ꭓ2 = 0.645 NS | ꭓ2 = 0.064 NS | ||||
| <2 (n, %) | (3, 7.3%) | (5, 10.2%) | (4, 8.9%) | (4, 8.9%) | ||
| 2–5 (n, %) | (27, 65.9%) | (34, 69.4%) | (31, 68.9%) | (30, 66.7%) | ||
| >5 (n, %) | (11, 26.8%) | (10, 20.4%) | (10, 22.2%) | (11, 24.4%) | ||
| LN involvement | ꭓ2 = 2.980, NS | ꭓ2 = 9.795 p = 0.020 * | ||||
| None (n, %) | (10, 24.4%) | (16, 32.7%) | (15, 33.3%) | (11, 24.4%) | ||
| 1–3 (n, %) | (10, 24.4%) | (14, 28.6%) | (17, 37.8%) | (7, 15.6%) | ||
| 4–9 (n, %) | (14, 34.1%) | (9, 18.4%) | (8, 17.8%) | (15, 33.3%) | ||
| ≥10 (n, %) | (7, 17.1%) | (10, 20.4%) | (5, 11.1%) | (12, 26.7%) | ||
| TNM stage | ꭓ2 = 10.667 p = 0.001 * | ꭓ2 = 6.156 p = 0.013 * | ||||
| I-II (early stage) (n, %) | (6, 14.6%) | (23, 46.9%) | (20, 44.4%) | (9, 20.0%) | ||
| III-IV (late stage) (n, %) | (35, 85.4%) | (26, 53.1%) | (25, 55.6%) | (36, 80.0%) | ||
| Grade (Bloom–Richardson scale) | ꭓ2 = 4.393 NS | ꭓ2 = 2.067 NS | ||||
| I (n, %) | (0, 0%) | (2, 4.1%) | (2, 4.4%) | (0, 0%) | ||
| II (n, %) | (29, 70.7%) | (40, 81.6%) | (34, 75.6%) | (35, 77.8%) | ||
| III (n, %) | (12, 29.3%) | (7, 14.3%) | (9, 20.0%) | (10, 22.2%) | ||
| Ki-67 status | ꭓ2 = 2.327 NS | ꭓ2 = 2.178 NS | ||||
| High (>14%) (n, %) | (19, 46.3%) | (26, 53.1%) | (11, 24.4%) | (8, 17.8%) | ||
| Low (≤14%) (n, %) | (7, 17.1%) | (12, 24.5%) | (19, 42.2%) | (26, 57.8%) | ||
| NA (n, %) | (15, 36.6%) | (11, 22.4%) | (15, 33.3%) | (11, 24.4%) | ||
| Therapy | ||||||
| Neoadjuvant chemotherapy | ꭓ2 = 3.178 NS | ꭓ2 = 5.553 p = 0.018 * | ||||
| Yes (n, %) | (21, 51.2%) | (16, 32.7%) | (13, 28.9%) | (24, 53.3%) | ||
| No (n, %) | (20, 48.8%) | (33, 67.3%) | (32, 71.1%) | (21, 46.7%) | ||
| Adjuvant chemotherapy | ꭓ2 = 0.746 NS | ꭓ2 = 8.086 p = 0.004 * | ||||
| Yes (n, %) | (24, 58.5%) | (33, 67.3%) | (35, 77.8%) | (22, 48.9%) | ||
| No (n, %) | (17, 41.5%) | (16, 32.7%) | (10, 22.2%) | (23, 51.1%) | ||
| Endocrine therapy | ꭓ2 = 0.003 NS | ꭓ2 = 1.23 NS | ||||
| Yes (n, %) | (27, 65.9%) | (32, 65.3%) | (32, 71.1%) | (27, 60.0%) | ||
| No (n, %) | (14, 34.1%) | (17, 34.7%) | (13, 28.9%) | (18, 40.0%) | ||
| Mastectomy | ꭓ2 = 1.434 NS | ꭓ2 = 3.200 NS | ||||
| Yes (n, %) | (30, 73.2%) | (30, 61.2%) | (26, 57.8%) | (34, 75.6%) | ||
| No (n, %) | (11, 26.8) | (11, 38.8%) | (19, 42.2%) | (11, 24.4%) | ||
| Patient Characteristic | CNOT7 r, p |
|---|---|
| Age # | −0.034, NS |
| BMI # | 0.065, NS |
| Diabetic status | 0.147, NS |
| No. of pregnancies | −0.079, NS |
| Menopausal status | 0.292, 0.006 * |
| BC family history | −0.162, NS |
| Histological subtype | −0.198, NS |
| Molecular subtype | −0.063, NS |
| ER status | −0.209, 0.048 * |
| PR status | −0.166, NS |
| HER-2 status | 0.105, NS |
| Tumor size | 0.029, NS |
| LN involvement | 0.226, 0.032 * |
| TNM stage | 0.256, 0.015 * |
| Grade (Bloom–Richardson scale) | −0.063, NS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elanany, M.M.; Mostafa, D.; Hady, A.A.; Abd Allah, M.Y.Y.; Ahmed, N.S.; Elghazawy, N.H.; Sippl, W.; Yamamoto, T.; Hamdy, N.M. CCR4-NOT Transcription Complex Subunit 7 (CNOT7) Protein and Leukocyte-Associated Immunoglobulin-like Receptor-1 in Breast Cancer Progression: Clinical Mechanistic Insights and In Silico Therapeutic Potential. Int. J. Mol. Sci. 2025, 26, 7141. https://doi.org/10.3390/ijms26157141
Elanany MM, Mostafa D, Hady AA, Abd Allah MYY, Ahmed NS, Elghazawy NH, Sippl W, Yamamoto T, Hamdy NM. CCR4-NOT Transcription Complex Subunit 7 (CNOT7) Protein and Leukocyte-Associated Immunoglobulin-like Receptor-1 in Breast Cancer Progression: Clinical Mechanistic Insights and In Silico Therapeutic Potential. International Journal of Molecular Sciences. 2025; 26(15):7141. https://doi.org/10.3390/ijms26157141
Chicago/Turabian StyleElanany, Mona M., Dina Mostafa, Ahmad A. Hady, Mona Y. Y. Abd Allah, Nermin S. Ahmed, Nehal H. Elghazawy, Wolfgang Sippl, Tadashi Yamamoto, and Nadia M. Hamdy. 2025. "CCR4-NOT Transcription Complex Subunit 7 (CNOT7) Protein and Leukocyte-Associated Immunoglobulin-like Receptor-1 in Breast Cancer Progression: Clinical Mechanistic Insights and In Silico Therapeutic Potential" International Journal of Molecular Sciences 26, no. 15: 7141. https://doi.org/10.3390/ijms26157141
APA StyleElanany, M. M., Mostafa, D., Hady, A. A., Abd Allah, M. Y. Y., Ahmed, N. S., Elghazawy, N. H., Sippl, W., Yamamoto, T., & Hamdy, N. M. (2025). CCR4-NOT Transcription Complex Subunit 7 (CNOT7) Protein and Leukocyte-Associated Immunoglobulin-like Receptor-1 in Breast Cancer Progression: Clinical Mechanistic Insights and In Silico Therapeutic Potential. International Journal of Molecular Sciences, 26(15), 7141. https://doi.org/10.3390/ijms26157141







