Prognostic Implications of T Cell Receptor Repertoire Diversity in Cervical Lymph Nodes of Oral Squamous Cell Carcinoma Patients
Abstract
1. Introduction
2. Results
2.1. Patient Grouping and Survival Outcome
2.2. Investigation of Histopathological Prognostic Factors in OSCC Patients
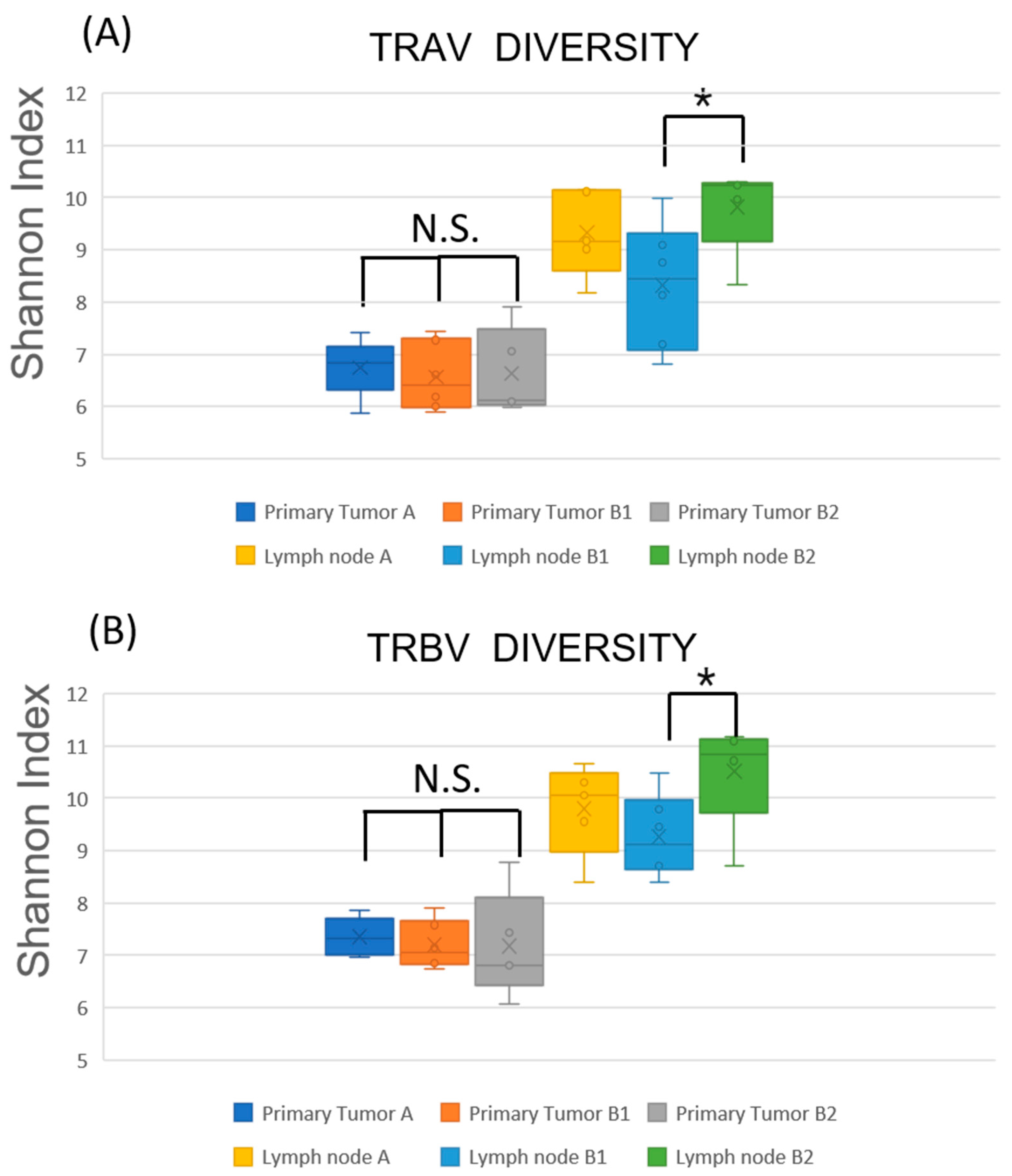
2.3. TCR Repertoire Diversity Reflects Nodal Metastasis and Tumor-Specific Clonal Expansion
2.4. Restricted TCR Diversity in LNs Correlates with Favorable Prognosis
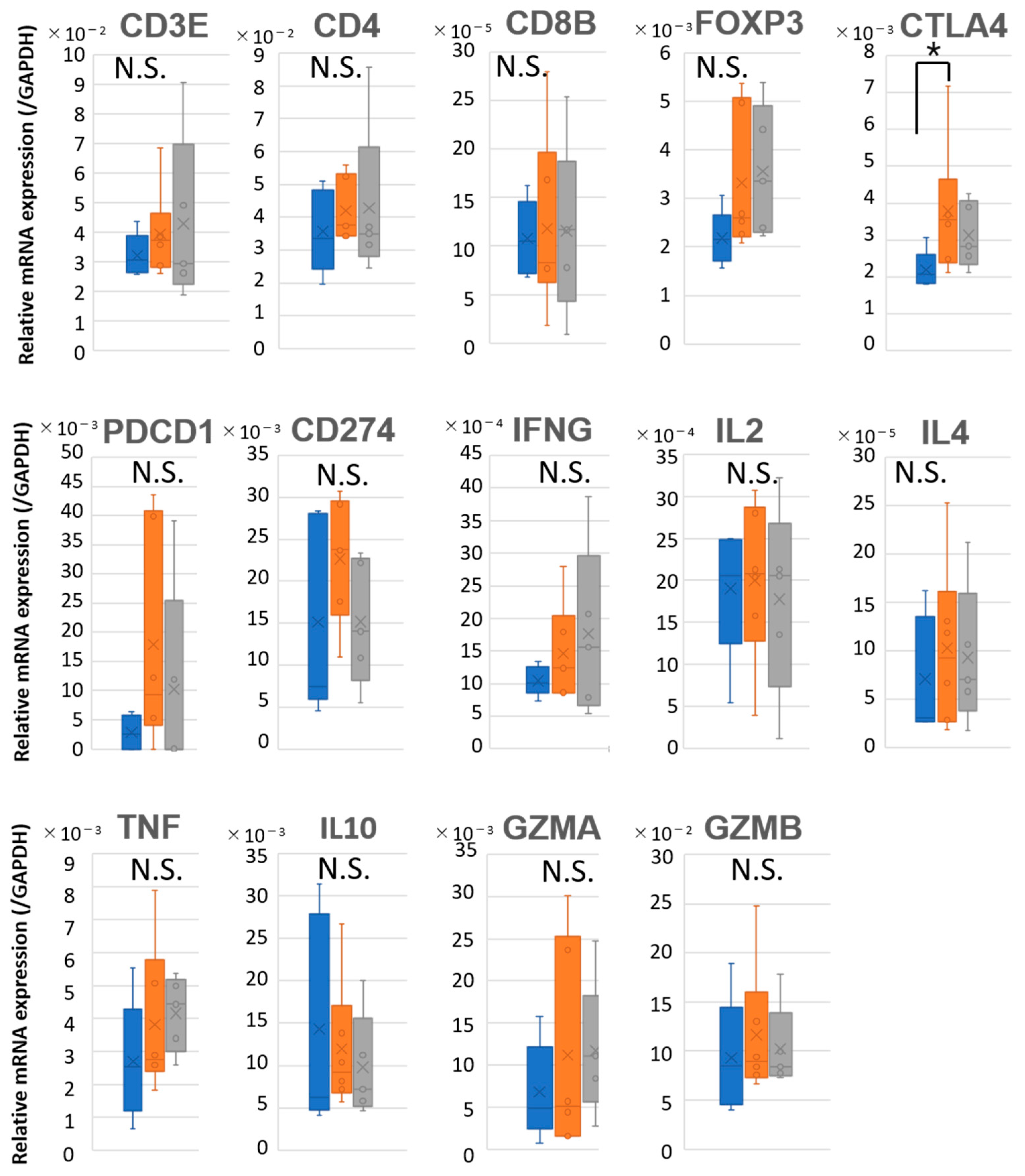
2.5. Prognostic Discrimination Based on Immune Gene Expression in LNs
3. Discussion
4. Materials and Methods
4.1. Sample Collection from OSCC Patients
4.2. Histological Evaluation of OSCC Specimens
4.3. RNA Extraction
4.4. TCR CDR3 Region Sequencing and TCR Repertoire Analysis
4.5. Quantitative Polymerase Chain Reaction
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TDLN | Tumor-draining lymph node |
| OSCC | Oral squamous cell carcinoma |
| LN | Lymph node |
| TCR | T cell receptor |
| CDR3 | Complementarity-determining region 3 |
| qPCR | quantitative polymerase chain reaction |
| GAPDH | Glyceraldehyde-3-Phosphate Dehydrogenase |
| CDLN | Cervical draining lymph node |
| APC | Antigen-presenting cell |
| PFS | Progression-free survival |
| OS | Overall survival |
| Y-K | Yamamoto–Kohama |
| ENE | Extranodal extension |
| TRA | T cell receptor alpha chain |
| TRB | T cell receptor beta chain |
| FOXP3 | Forkhead box P3 |
| CTLA4 | Cytotoxic T-lymphocyte associated protein 4 |
| GZM | Granzyme |
| PDCD1 | Programmed cell death protein 1 (PD-1) |
| CD274 | Cluster of differentiation 274, programmed death-ligand 1 (PD-L1) |
| Treg | Regulatory T cell |
| MDSC | Myeloid-derived suppressor cell |
| ICI | Immune checkpoint inhibitor |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.M.; Ahn, S.J. Prognostic factors of oral squamous cell carcinoma: The importance of recurrence and pTNM stage. Maxillofac. Plast. Reconstr. Surg. 2024, 46, 8. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, P.; Jain, A.; Rajpal, A.; Goyal, M. Cervical lymph node metastasis in squamous cell carcinoma of the buccal mucosa: A retrospective study on pattern of involvement and clinical analysis. J. Oral Maxillofac. Surg. Med. Pathol. 2021, 33, 81–86. [Google Scholar]
- Obayashi, F.; Koizumi, K.; Ito, N.; Higaki, M.; Ishida, Y.; Hamada, A.; Yamasaki, S.; Tani, R.; Yanamoto, S. A Study of the Prognostic Factors for Late Cervical Lymph Node Metastasis and Distant Metastasis in Patients with cT1-2N0 Tongue Cancer. J. Clin. Med. 2024, 13, 976. [Google Scholar] [CrossRef] [PubMed]
- Dolens, E.D.S.; Dourado, M.R.; Almangush, A.; Salo, T.A.; Gurgel Rocha, C.A.; da Silva, S.D.; Brennan, P.A.; Coletta, R.D. The Impact of Histopathological Features on the Prognosis of Oral Squamous Cell Carcinoma: A Comprehensive Review and Meta-Analysis. Front. Oncol. 2021, 11, 784924. [Google Scholar] [CrossRef] [PubMed]
- Delclaux, I.; Ventre, K.S.; Jones, D.; Lund, A.W. The tumor-draining lymph node as a reservoir for systemic immune surveillance. Trends Cancer 2024, 10, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Lei, P.-J.; Padera, T.P. Progression of Metastasis through Lymphatic System. Cells 2021, 10, 627. [Google Scholar] [CrossRef] [PubMed]
- du Bois, H.; Heim, T.A.; Lund, A.W. Tumor-draining lymph nodes: At the crossroads of metastasis and immunity. Sci. Immunol. 2021, 6, eabg3551. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Angelillo, J.; Hugues, S. Lymphatic transport in anti-tumor immunity and metastasis. J. Exp. Med. 2025, 222, e20231954. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, T.; Pignon, J.C.; Wang, B.; Wang, J.; Shukla, S.A.; Dou, R.; Chen, Q.; Hodi, F.S.; Choueiri, T.K.; et al. Landscape of tumor-infiltrating T cell repertoire of human cancers. Nat. Genet. 2016, 48, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Frank, M.L.; Lu, K.; Erdogan, C.; Han, Y.; Hu, J.; Wang, T.; Heymach, J.V.; Zhang, J.; Reuben, A. T-Cell Receptor Repertoire Sequencing in the Era of Cancer Immunotherapy. Clin. Cancer Res. 2023, 29, 994–1008. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Gupta, A.; Sharma, P.; Kumar, M.; Reddy, S. Survival significance of number of positive lymph nodes in oral squamous cell carcinoma. Front. Oncol. 2021, 11, 545433. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Schmidt, J.; Weber, C.; Hoffmann, T.; Wagner, S. Prognostic value of lymph node involvement in oral squamous cell carcinoma: A retrospective cohort study. Clin. Oral Investig. 2022, 26, 1237–1246. [Google Scholar]
- Yamamoto, E.; Kohama, G.; Sunakawa, H.; Iwai, M.; Hiratsuka, H. Mode of invasion, bleomycin sensitivity, and clinical course in squamous cell carcinoma of the oral cavity. Cancer 1983, 51, 2175–2180. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, E.; Miyakawa, A.; Kohama, G. Mode of invasion and lymph node metastasis in squamous cell carcinoma of the oral cavity. Head Neck Surg. 1984, 6, 938–947. [Google Scholar] [CrossRef] [PubMed]
- Abou-Foul, A.K.; Henson, C.; Chernock, R.D.; Huang, S.H.; Lydiatt, W.M.; McDowell, L.; O’Sullivan, B.; Perez-Ordonez, B.; Robinson, M.; Nankivell, P.C.; et al. Standardised definitions and diagnostic criteria for extranodal extension detected on histopathological examination in head and neck cancer: Head and Neck Cancer International Group consensus recommendations. Lancet Oncol. 2024, 25, e286–e296. [Google Scholar] [CrossRef] [PubMed]
- Rahim, M.K.; Okholm, T.L.H.; Jones, K.B.; McCarthy, E.E.; Liu, C.C.; Yee, J.L.; Tamaki, S.J.; Marquez, D.M.; Tenvooren, I.; Wai, K.; et al. Dynamic CD8+ T cell responses to cancer immunotherapy in human regional lymph nodes are disrupted in metastatic lymph nodes. Cell 2023, 186, 1127–1143. [Google Scholar] [CrossRef] [PubMed]
- Sade-Feldman, M.; Yizhak, K.; Bjorgaard, S.L.; Ray, J.P.; de Boer, C.G.; Jenkins, R.W.; Lieb, D.J.; Chen, J.H.; Frederick, D.T.; Barzily-Rokni, M.; et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 2018, 175, 998–1013. [Google Scholar] [CrossRef] [PubMed]
- Reuben, A.; Zhang, J.; Chiou, S.H.; Gittelman, R.M.; Li, J.; Lee, W.C.; Fujimoto, J.; Behrens, C.; Liu, X.; Wang, F.; et al. Comprehensive T Cell Repertoire Characterization of Non-Small Cell Lung Cancer. Nat. Commun. 2020, 11, 603. [Google Scholar] [CrossRef] [PubMed]



| Patient | Age, Year | Gender | Primary Site | pTNM | Clinical Stage | Lymph Node Metastasis | Extranodal Extension | Local Recurrence | Distant Metastasis | Prognosis | PFS (Month) | OS (Month) | Group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 74 | F | Upper Gingiva | T2N0M0 | 2 | — | — | — | — | Alive | 72.3 | 72.3 | A |
| 2 | 79 | M | Lower Gingiva | T4aN0M0 | 4A | — | — | — | — | Alive | 70.8 | 70.8 | A |
| 3 | 71 | M | Tongue, Esophageal | T2N0M0 | 2 | — | — | — | — | Alive | 70.1 | 70.1 | A |
| 4 | 61 | F | Lower Gingiva | T3N0M0 | 3 | — | — | — | — | Alive | 65 | 65 | A |
| 5 | 64 | M | Tongue | T3N0M0 | 3 | — | — | — | — | Alive | 61.1 | 61.1 | A |
| 6 | 71 | F | Tongue | T1N1M0 | 3 | + | — | — | — | Death from other causes | 30.7 | 30.7 | B1 |
| 7 | 64 | M | Lower Gingiva | T4aN2bM0 | 4A | + | + | — | — | Alive | 71.7 | 71.7 | B1 |
| 8 | 67 | M | Tongue | T2N2bM0 | 4A | + | — | — | — | Alive | 71.4 | 71.4 | B1 |
| 9 | 78 | M | Upper Gingiva | T2N1M0 | 3 | + | — | — | — | Alive | 71.4 | 71.4 | B1 |
| 10 | 75 | M | Lower Gingiva | T1N1M0 | 3 | + | — | — | — | Alive | 67.8 | 67.8 | B1 |
| 11 | 69 | F | Tongue | T2N2bM0 | 3 | + | — | — | — | Alive | 64.1 | 64.1 | B1 |
| 12 | 86 | M | Lower Gingiva | T4N1M0 | 4A | + | — | + | — | Death | 10.1 | 11.6 | B2 |
| 13 | 79 | F | Upper Gingiva | T4aN3bM0 | 4B | + | + | + | — | Death | 1.6 | 18.1 | B2 |
| 14 | 68 | M | Tongue | T2N2cM0 | 4A | + | — | + | + | Death | 3.1 | 11.2 | B2 |
| 15 | 50 | F | Tongue | T1N1M0 | 3 | + | — | + | — | Death | 14.6 | 35.6 | B2 |
| 16 | 75 | F | Tongue | T3N2bM0 | 4A | + | + | + | — | Death | 5.4 | 14.3 | B2 |
| Patient | Age Year | Gender | Primary Site | pTNM | Clinical Stage | Histological Type of Primary Tumor (Y-K Classification) | Lymph Node Metastasis | Extranodal Extension | Type of Extranodal Extension ※ | Histological Type of Metastatic Lymph Node | The Number of Metastatic Lymph Nodes | Group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 74 | F | Upper Gingiva | T2N0M0 | 2 | 3 | — | — | — | 0 | A | |
| 2 | 79 | M | Lower Gingiva | T4aN0M0 | 4A | 4C | — | — | — | 0 | A | |
| 3 | 71 | M | Tongue, Esophageal | T2N0M0 | 2 | 2 | — | — | — | 0 | A | |
| 4 | 61 | F | Lower Gingiva | T3N0M0 | 3 | 4C | — | — | — | 0 | A | |
| 5 | 64 | M | Tongue | T3N0M0 | 3 | 2 | — | — | — | 0 | A | |
| 6 | 71 | F | Tongue | T1N1M0 | 3 | 1 | + | — | No ENE Type 1 | Focal | 1 | B1 |
| 7 | 64 | M | Lower Gingiva | T4aN2bM0 | 4A | 4C | + | + | Major ENE Type 4 | Invasive/cystic | 5 | B1 |
| 8 | 67 | M | Tongue | T2N2bM0 | 4A | 3 | + | — | No ENE Type 1 | Focal | 3 | B1 |
| 9 | 78 | M | Upper Gingiva | T2N1M0 | 3 | 4C | + | — | No ENE Type 1 | Focal | 1 | B1 |
| 10 | 75 | M | Lower Gingiva | T1N1M0 | 3 | 3 | + | — | No ENE Type 2 | Invasive/cystic | 1 | B1 |
| 11 | 69 | F | Tongue | T2N2bM0 | 3 | 4C | + | — | No ENE Type 2 | Invasive/cystic | 1 | B1 |
| 12 | 86 | M | Lower Gingiva | T4N1M0 | 4A | 4C | + | — | No ENE Type 1 | Focal | 2 | B2 |
| 13 | 79 | F | Upper Gingiva | T4aN3bM0 | 4B | 4C | + | + | Major ENE Type 4 | Invasive/cystic | 1 | B2 |
| 14 | 68 | M | Tongue | T2N2cM0 | 4A | 4D | + | — | No ENE Type 1 | Focal | 6 | B2 |
| 15 | 50 | F | Tongue | T1N1M0 | 3 | 4D | + | — | No ENE Type 1 | Focal | 1 | B2 |
| 16 | 75 | F | Tongue | T3N2bM0 | 4A | 4C | + | + | Major ENE Type 4 | Invasive/cystic | 2 | B2 |
| CD3E | forward | TGCTGCTGCTGGTTTACTACTGG |
| reverse | TCATAGTCTGGGTTGGGAACAGG | |
| CD4 | forward | AAGTGAACCTGGTGGTGATGAGA |
| reverse | CTCCCGCTTCGAGACCTTTG | |
| CD8B | forward | GCCGGAAGACAGTGGCATCT |
| reverse | TCTCTTCTTGAGGGTGGACTTCTTG | |
| FOXP3 | forward | GGGTAGCCATGGAAACAGCA |
| reverse | TCGCATGTTGTGGAACTTGAAGTA | |
| CTLA4 | forward | ATCTGCAAGGTGGAGCTCATGTA |
| reverse | ATCTGGGCACGGTTCTGGA | |
| PDCD1 | forward | GGTGCCGACTACAAGCGAATTAC |
| reverse | GGAATTGGTGGTGGTGGTCTTAC | |
| CD274 | forward | AAATGGAACCTGGCGAAAGC |
| reverse | GATGAGCCCCTCAGGCATTT | |
| IFNG | forward | CTTTAAAGATGACCAGAGCATCCAA |
| reverse | GGCGACAGTTCAGCCATCAC | |
| IL2 | forward | AGACCCAGGGACTTAATCAGCAATA |
| reverse | TTCTACAATGGTTGCTGTCTCATCA | |
| IL4 | forward | GACTCGCCTACAAAGCCCAGA |
| reverse | AGCTGCTTGTGCCTGTGGAA | |
| TNF | forward | GAATGAGCTTCTGGAGGCTTG |
| reverse | TGGAGTTGATGTCAGTCACTTGG | |
| IL10 | forward | GGCCCAATTGACTGACAGGA |
| reverse | TCAAACTCACTCATGGCTTTGTAGA | |
| GZMA | forward | GACTGGGTGTTGACTGCAGCTC |
| reverse | TGGCTGGGTCATAGCATGG | |
| GZMB | forward | TGACAGCTGCTCACTGTTGG |
| reverse | GTTCTTAGGATTATAGGCTGGATGG | |
| GAPDH | forward | GCACCGTCAAGGCTGAGAAC |
| reverse | TGGTGAAGACGCCAGTGGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumagai, K.; Hamada, Y.; Horie, A.; Shimizu, Y.; Ohashi, Y.; Aoki, R.; Suzuki, T.; Kawaguchi, K.; Kuroda, A.; Tsujikawa, T.; et al. Prognostic Implications of T Cell Receptor Repertoire Diversity in Cervical Lymph Nodes of Oral Squamous Cell Carcinoma Patients. Int. J. Mol. Sci. 2025, 26, 7073. https://doi.org/10.3390/ijms26157073
Kumagai K, Hamada Y, Horie A, Shimizu Y, Ohashi Y, Aoki R, Suzuki T, Kawaguchi K, Kuroda A, Tsujikawa T, et al. Prognostic Implications of T Cell Receptor Repertoire Diversity in Cervical Lymph Nodes of Oral Squamous Cell Carcinoma Patients. International Journal of Molecular Sciences. 2025; 26(15):7073. https://doi.org/10.3390/ijms26157073
Chicago/Turabian StyleKumagai, Kenichi, Yoshiki Hamada, Akihisa Horie, Yudai Shimizu, Yoshihiro Ohashi, Reo Aoki, Taiki Suzuki, Koji Kawaguchi, Akihiro Kuroda, Takahiro Tsujikawa, and et al. 2025. "Prognostic Implications of T Cell Receptor Repertoire Diversity in Cervical Lymph Nodes of Oral Squamous Cell Carcinoma Patients" International Journal of Molecular Sciences 26, no. 15: 7073. https://doi.org/10.3390/ijms26157073
APA StyleKumagai, K., Hamada, Y., Horie, A., Shimizu, Y., Ohashi, Y., Aoki, R., Suzuki, T., Kawaguchi, K., Kuroda, A., Tsujikawa, T., Hoshi, K., & Kakimi, K. (2025). Prognostic Implications of T Cell Receptor Repertoire Diversity in Cervical Lymph Nodes of Oral Squamous Cell Carcinoma Patients. International Journal of Molecular Sciences, 26(15), 7073. https://doi.org/10.3390/ijms26157073








