Expression Profiles of Genes Related to Serotonergic Synaptic Function in Hypothalamus of Hypertensive and Normotensive Rats in Basal and Stressful Conditions
Abstract
1. Introduction
2. Results
2.1. Inter-Strain Differences at Rest
2.1.1. Characteristics of Differentially Expressed Genes
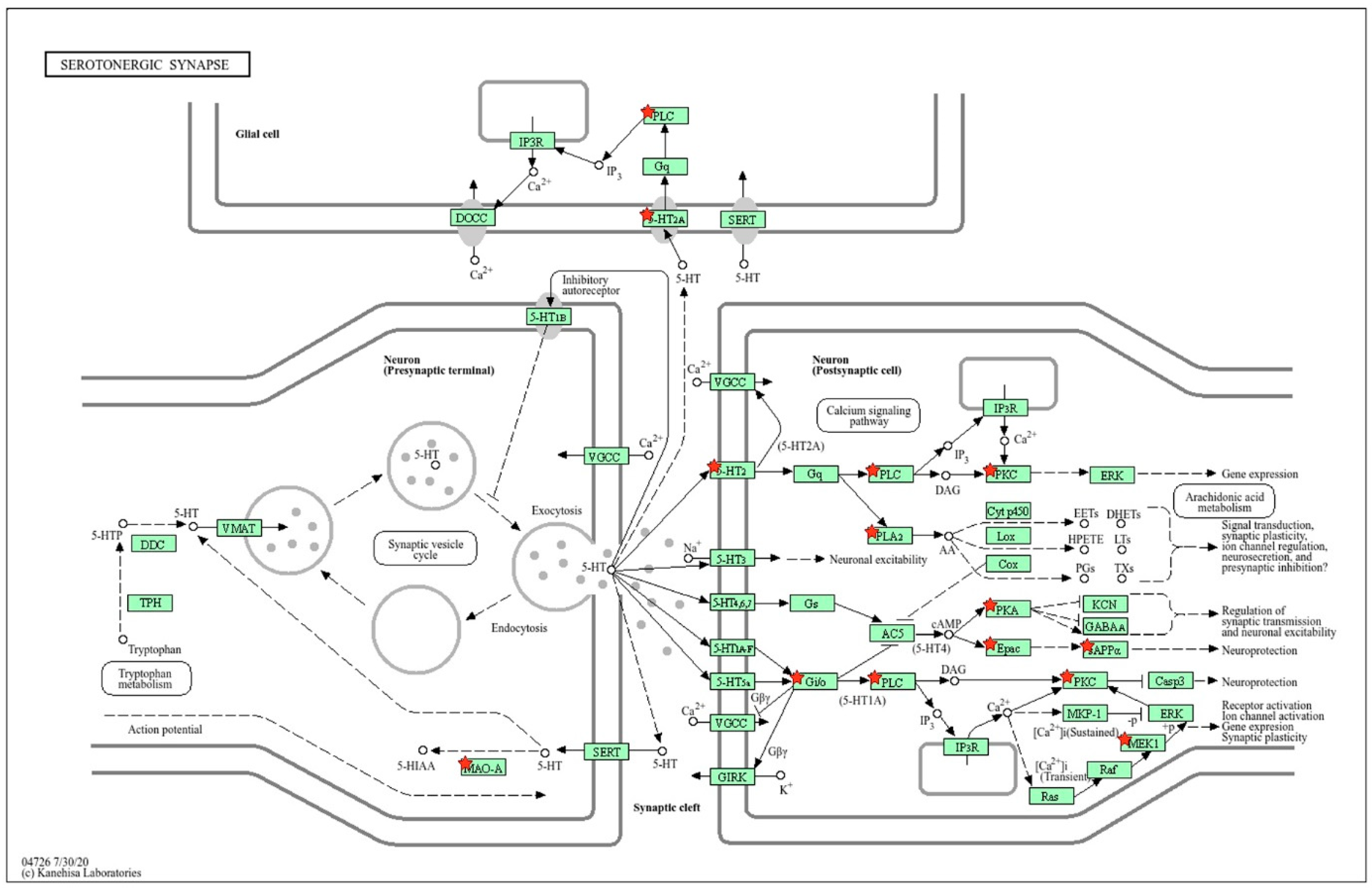
2.1.2. Location of sDEGs Determining Inter-Strain Differences at Rest on the KEGG Map for Serotonergic Synapse
2.2. Stress Response
2.2.1. Genes That Changed Transcription Levels When Exposed to a Single Short-Term Restraint Stress
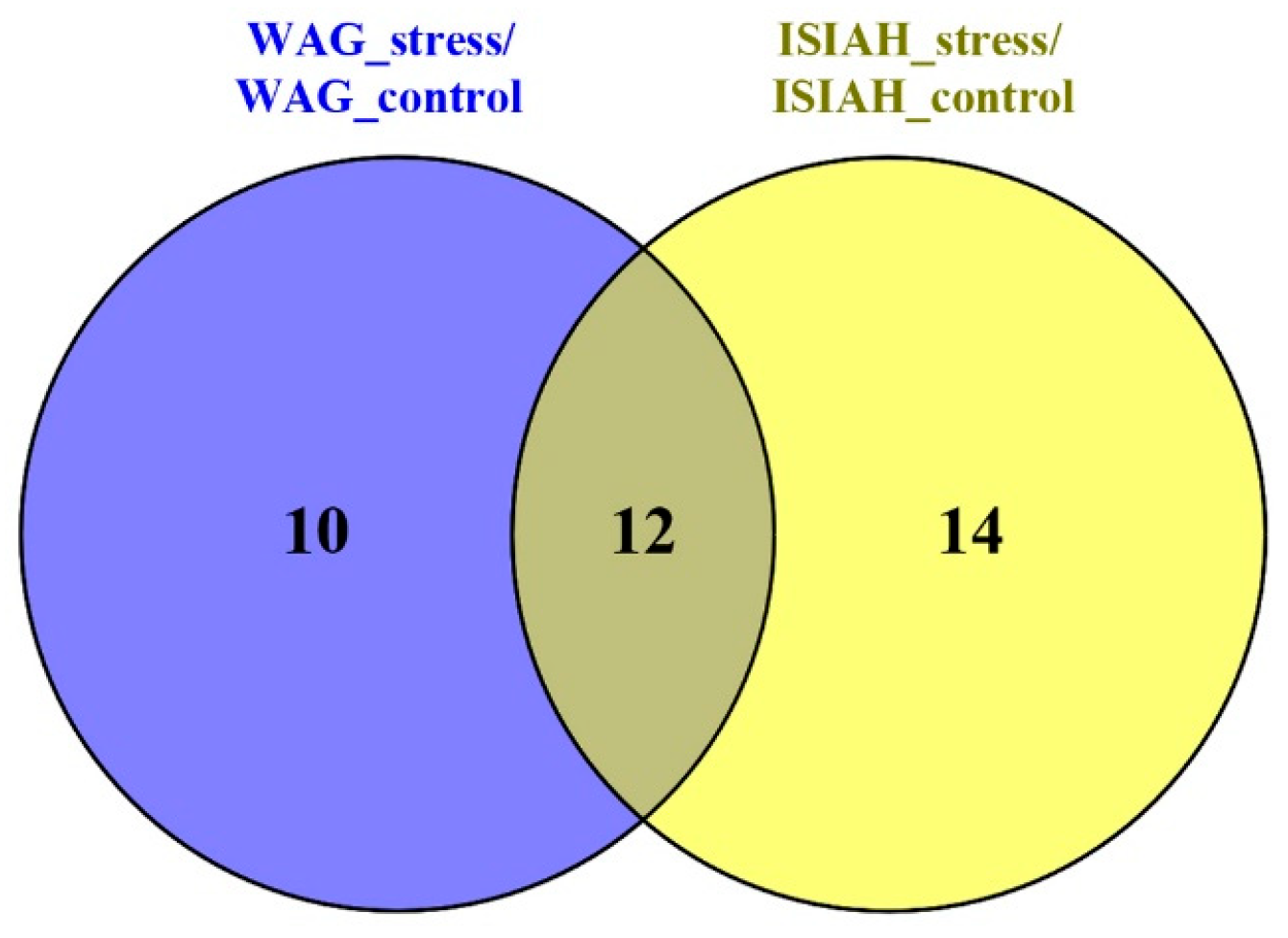
2.2.2. Common Response to Stress
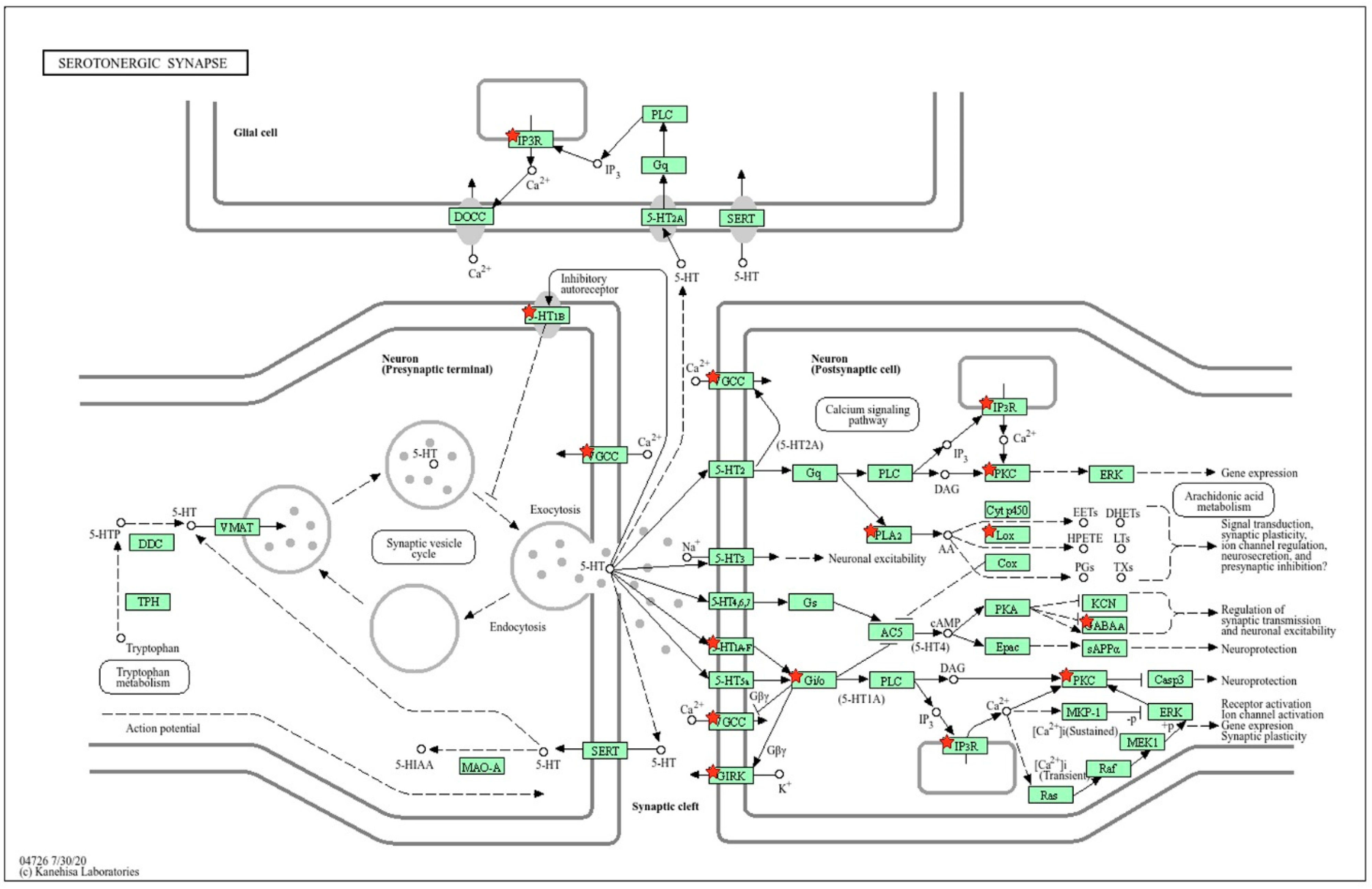
2.2.3. Location of Common sDEGs on the KEGG Map for Serotonergic Synapse
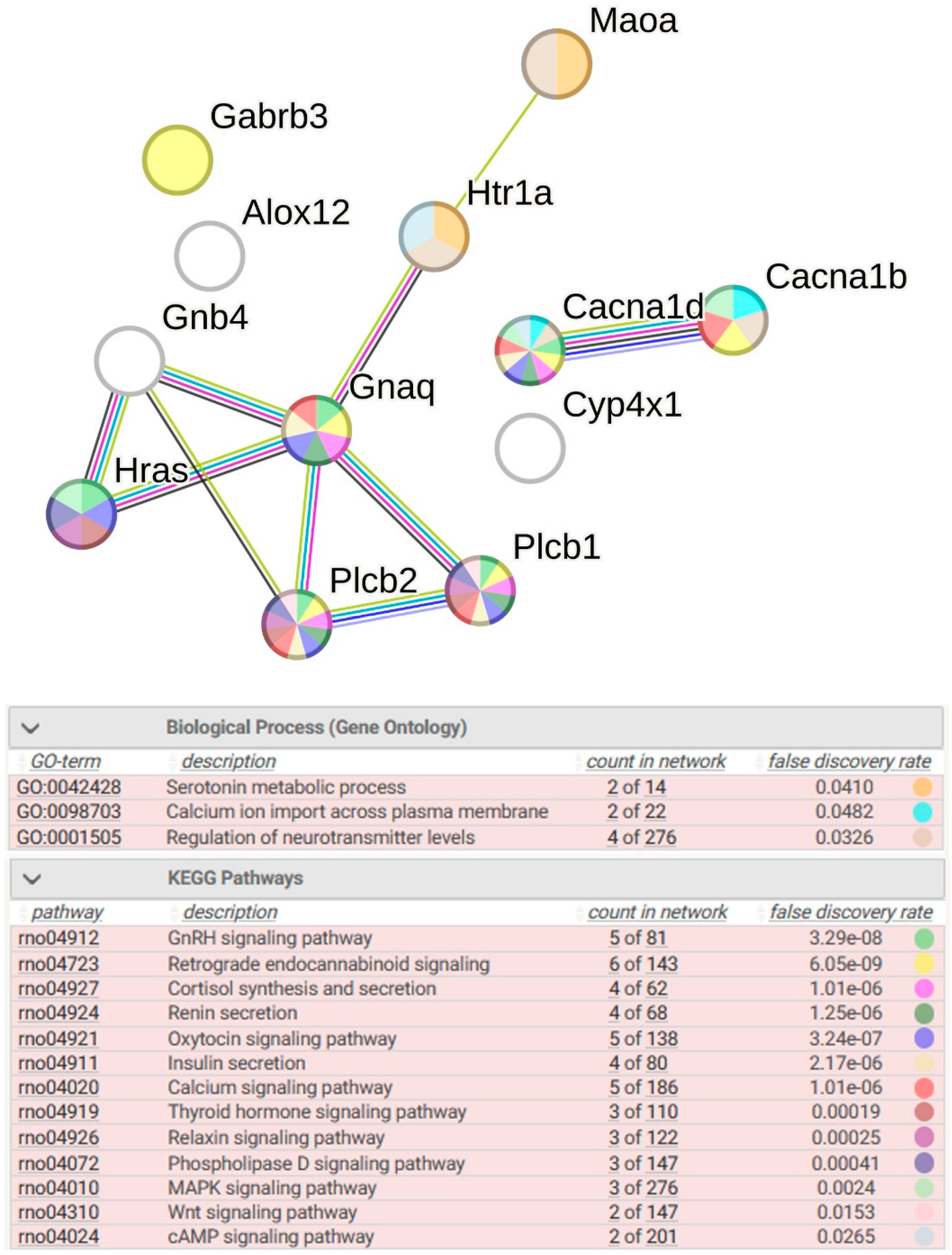
2.2.4. Functional Annotation of Common DEGs
2.2.5. ISIAH-Specific Stress Response
2.2.6. Location of ISIAH-Specific sDEGs on the KEGG Map for Serotonergic Synapse
2.2.7. Functional Annotation of 14 sDEGs Associated with ISIAH-Specific Stress Response
2.2.8. WAG-Specific Stress Response
2.2.9. Location of WAG-Specific sDEGs on the KEGG Map for Serotonergic Synapse
2.2.10. Functional Annotation of 10 sDEGs Associated with WAG-Specific Stress Response
2.2.11. Enrichment Analysis of Promoter Regions of Serotonergic DEGs
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. RNA-Seq
4.3. Functional Annotation of DEGs
4.4. Visualization and Graphing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Watts, S.W. 5-HT in systemic hypertension: Foe, friend or fantasy? Clin. Sci. 2005, 108, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, B.L.; Azmitia, E.C. Structure and function of the brain serotonin system. Physiol. Rev. 1992, 72, 165–229. [Google Scholar] [CrossRef] [PubMed]
- Petrov, T.; Krukoff, T.L.; Jhamandas, J.H. The hypothalamic paraventricular and lateral parabrachial nuclei receive collaterals from raphe nucleus neurons: A combined double retrograde and immunocytochemical study. J. Comp. Neurol. 1992, 318, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Kent, D.L.; Sladek, J.R., Jr. Histochemical, pharmacological and microspectrofluorometric analysis of new sites of serotonin localization in the rat hypothalamus. J. Comp. Neurol. 1978, 180, 221–236. [Google Scholar] [CrossRef]
- Frankfurt, M.; Lauder, J.M.; Azmitia, E.C. The immunocytochemical localization of serotonergic neurons in the rat hypothalamus. Neurosci. Lett. 1981, 24, 227–232. [Google Scholar] [CrossRef]
- Beaudet, A.; Descarries, L. Radioautographic characterization of a serotonin-accumulating nerve cell group in adult rat hypothalamus. Brain Res. 1979, 160, 231–243. [Google Scholar] [CrossRef]
- Moyer, R.W.; Kennaway, D.J. Immunohistochemical localization of serotonin receptors in the rat suprachiasmatic nucleus. Neurosci. Lett. 1999, 271, 147–150. [Google Scholar] [CrossRef]
- Collin, M.; Backberg, M.; Onnestam, K.; Meister, B. 5-HT1A receptor immunoreactivity in hypothalamic neurons involved in body weight control. Neuroreport 2002, 13, 945–951. [Google Scholar] [CrossRef]
- Markel, A.L. Development of a new strain of rats with inherited stress-induced arterial hypertension. In Genetic Hypertension; Sassard, J., Libbey, J., Eds.; Eurotext; Colloque INSERM; John Libbey Eurotext Ltd.: London, UK, 1992; Volume 218, pp. 405–407. [Google Scholar]
- Markel, A.L.; Maslova, L.N.; Shishkina, G.T.; Bulygina, V.V.; Machanova, N.A.; Jacobson, G.S. Developmental influences on blood pressure regulation in ISIAH rats. In Development of the Hypertensive Phenotype: Basic and Clinical Studies; McCarty, R., Blizard, D.A., Chevalier, R.L., Birkenhager, W.H., Reid, J.L., Eds.; Handbook of hypertension; Elsevier: Amsterdam, The Netherlands; Lausanne, Switzerland; New York, NY, USA; Oxford, UK; Shannon, Ireland; Singapore; Tokyo, Japan, 1999; Volume 19, pp. 493–526. [Google Scholar]
- Markel, A.L.; Redina, O.E.; Gilinsky, M.A.; Dymshits, G.M.; Kalashnikova, E.V.; Khvorostova, Y.V.; Fedoseeva, L.A.; Jacobson, G.S. Neuroendocrine profiling in inherited stress-induced arterial hypertension rat strain with stress-sensitive arterial hypertension. J. Endocrinol. 2007, 195, 439–450. [Google Scholar] [CrossRef]
- Makovka, Y.V.; Fedoseeva, L.A.; Oshchepkov, D.Y.; Markel, A.L.; Redina, O.E. Restraint Stress-Induced Expression of Fos and Several Related Genes in the Hypothalamus of Hypertensive ISIAH Rats. Mol. Biol. 2024, 58, 62–70. [Google Scholar] [CrossRef]
- Oshchepkov, D.Y.; Makovka, Y.V.; Fedoseeva, L.A.; Seryapina, A.A.; Markel, A.L.; Redina, O.E. Effect of Short-Term Restraint Stress on the Hypothalamic Transcriptome Profiles of Rats with Inherited Stress-Induced Arterial Hypertension (ISIAH) and Normotensive Wistar Albino Glaxo (WAG) Rats. Int. J. Mol. Sci. 2024, 25, 6680. [Google Scholar] [CrossRef]
- Makovka, Y.V.; Oshchepkov, D.Y.; Fedoseeva, L.A.; Markel, A.L.; Redina, O.E. Effect of Short-Term Restraint Stress on the Expression of Genes Associated with the Response to Oxidative Stress in the Hypothalamus of Hypertensive ISIAH and Normotensive WAG Rats. Antioxidants 2024, 13, 1302. [Google Scholar] [CrossRef]
- Mohammad-Zadeh, L.F.; Moses, L.; Gwaltney-Brant, S.M. Serotonin: A review. J. Vet. Pharmacol. Ther. 2008, 31, 187–199. [Google Scholar] [CrossRef]
- Berger, M.; Gray, J.A.; Roth, B.L. The expanded biology of serotonin. Annu. Rev. Med. 2009, 60, 355–366. [Google Scholar] [CrossRef]
- Watts, S.W.; Morrison, S.F.; Davis, R.P.; Barman, S.M. Serotonin and blood pressure regulation. Pharmacol. Rev. 2012, 64, 359–388. [Google Scholar] [CrossRef] [PubMed]
- Maroteaux, L.; Kilic, F. Frontiers of Serotonin Beyond the Brain. Pharmacol. Res. 2019, 140, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, H.S. Studies on the neuroendocrine role of serotonin. Dan. Med. Bull. 2007, 54, 266–288. [Google Scholar] [PubMed]
- de Wardener, H.E. The hypothalamus and hypertension. Physiol. Rev. 2001, 81, 1599–1658. [Google Scholar] [CrossRef]
- Lawal, H.O.; Krantz, D.E. SLC18: Vesicular neurotransmitter transporters for monoamines and acetylcholine. Mol. Aspects Med. 2013, 34, 360–372. [Google Scholar] [CrossRef]
- Kokubo, Y.; Tomoike, H.; Tanaka, C.; Banno, M.; Okuda, T.; Inamoto, N.; Kamide, K.; Kawano, Y.; Miyata, T. Association of sixty-one non-synonymous polymorphisms in forty-one hypertension candidate genes with blood pressure variation and hypertension. Hypertens. Res. 2006, 29, 611–619. [Google Scholar] [CrossRef]
- Lohoff, F.W.; Lautenschlager, M.; Mohr, J.; Ferraro, T.N.; Sander, T.; Gallinat, J. Association between variation in the vesicular monoamine transporter 1 gene on chromosome 8p and anxiety-related personality traits. Neurosci. Lett. 2008, 434, 41–45. [Google Scholar] [CrossRef]
- Diamant, I.; Clarke, D.J.B.; Evangelista, J.E.; Lingam, N.; Ma’ayan, A. Harmonizome 3.0: Integrated knowledge about genes and proteins from diverse multi-omics resources. Nucleic Acids Res. 2025, 53, D1016–D1028. [Google Scholar] [CrossRef]
- Szymanowicz, O.; Druzdz, A.; Slowikowski, B.; Pawlak, S.; Potocka, E.; Goutor, U.; Konieczny, M.; Ciaston, M.; Lewandowska, A.; Jagodzinski, P.P.; et al. A Review of the CACNA Gene Family: Its Role in Neurological Disorders. Diseases 2024, 12, 90. [Google Scholar] [CrossRef]
- Niesler, B.; Kapeller, J.; Hammer, C.; Rappold, G. Serotonin type 3 receptor genes: HTR3A, B, C, D, E. Pharmacogenomics 2008, 9, 501–504. [Google Scholar] [CrossRef] [PubMed]
- Gatt, J.M.; Williams, L.M.; Schofield, P.R.; Dobson-Stone, C.; Paul, R.H.; Grieve, S.M.; Clark, C.R.; Gordon, E.; Nemeroff, C.B. Impact of the HTR3A gene with early life trauma on emotional brain networks and depressed mood. Depress. Anxiety 2010, 27, 752–759. [Google Scholar] [CrossRef] [PubMed]
- Martin, V.; Riffaud, A.; Marday, T.; Brouillard, C.; Franc, B.; Tassin, J.P.; Sevoz-Couche, C.; Mongeau, R.; Lanfumey, L. Response of Htr3a knockout mice to antidepressant treatment and chronic stress. Br. J. Pharmacol. 2017, 174, 2471–2483. [Google Scholar] [CrossRef]
- Garcia-Garcia, A.L.; Newman-Tancredi, A.; Leonardo, E.D. 5-HT(1A) [corrected] receptors in mood and anxiety: Recent insights into autoreceptor versus heteroreceptor function. Psychopharmacology 2014, 231, 623–636. [Google Scholar] [CrossRef]
- Albert, P.R.; Vahid-Ansari, F. The 5-HT1A receptor: Signaling to behavior. Biochimie 2019, 161, 34–45. [Google Scholar] [CrossRef]
- Ou, X.M.; Storring, J.M.; Kushwaha, N.; Albert, P.R. Heterodimerization of mineralocorticoid and glucocorticoid receptors at a novel negative response element of the 5-HT1A receptor gene. J. Biol. Chem. 2001, 276, 14299–14307. [Google Scholar] [CrossRef]
- Sun, X.; Ming, Q.; Zhong, X.; Dong, D.; Li, C.; Xiong, G.; Cheng, C.; Cao, W.; He, J.; Wang, X.; et al. The MAOA Gene Influences the Neural Response to Psychosocial Stress in the Human Brain. Front. Behav. Neurosci. 2020, 14, 65. [Google Scholar] [CrossRef]
- Jabbi, M.; Korf, J.; Kema, I.P.; Hartman, C.; van der Pompe, G.; Minderaa, R.B.; Ormel, J.; den Boer, J.A. Convergent genetic modulation of the endocrine stress response involves polymorphic variations of 5-HTT, COMT and MAOA. Mol. Psychiatry 2007, 12, 483–490. [Google Scholar] [CrossRef]
- Bouma, E.M.; Riese, H.; Doornbos, B.; Ormel, J.; Oldehinkel, A.J. Genetically based reduced MAOA and COMT functioning is associated with the cortisol stress response: A replication study. Mol. Psychiatry 2012, 17, 119–121. [Google Scholar] [CrossRef]
- Cocco, L.; Martelli, A.M.; Barnabei, O.; Manzoli, F.A. Nuclear inositol lipid signaling. Adv. Enzyme Regul. 2001, 41, 361–384. [Google Scholar] [CrossRef] [PubMed]
- Nagari, A. Functional Genomics Study to Understand the Role of Serotonin in Mouse Embryonic Stem Cells. Indiana University. 2011. Available online: https://scholarworks.indianapolis.iu.edu (accessed on 14 July 2025).
- Duclot, F.; Kabbaj, M. The Role of Early Growth Response 1 (EGR1) in Brain Plasticity and Neuropsychiatric Disorders. Front. Behav. Neurosci. 2017, 11, 35. [Google Scholar] [CrossRef] [PubMed]
- Humblot, N.; Thiriet, N.; Gobaille, S.; Aunis, D.; Zwiller, J. The serotonergic system modulates the cocaine-induced expression of the immediate early genes egr-1 and c-fos in rat brain. Ann. N. Y. Acad. Sci. 1998, 844, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Pagel, J.I.; Deindl, E. Disease progression mediated by egr-1 associated signaling in response to oxidative stress. Int. J. Mol. Sci. 2012, 13, 13104–13117. [Google Scholar] [CrossRef]
- Huang, T.L.; Sung, M.L.; Chen, T.Y. 2D-DIGE proteome analysis on the platelet proteins of patients with major depression. Proteome Sci. 2014, 12, 1. [Google Scholar] [CrossRef]
- Cai, L.; Yan, X.B.; Chen, X.N.; Meng, Q.Y.; Zhou, J.N. Chronic all-trans retinoic acid administration induced hyperactivity of HPA axis and behavioral changes in young rats. Eur. Neuropsychopharmacol. 2010, 20, 839–847. [Google Scholar] [CrossRef]
- Chen, X.N.; Meng, Q.Y.; Bao, A.M.; Swaab, D.F.; Wang, G.H.; Zhou, J.N. The involvement of retinoic acid receptor-alpha in corticotropin-releasing hormone gene expression and affective disorders. Biol. Psychiatry 2009, 66, 832–839. [Google Scholar] [CrossRef]
- Cai, L.; Li, R.; Zhou, J.N. Chronic all-trans retinoic acid administration induces CRF over-expression accompanied by AVP up-regulation and multiple CRF-controlling receptors disturbance in the hypothalamus of rats. Brain Res. 2015, 1601, 1–7. [Google Scholar] [CrossRef]
- Kim, S.; Pajarillo, E.; Digman, A.; Ajayi, I.; Son, D.S.; Aschner, M.; Lee, E. Role of dopaminergic RE1-silencing transcription factor (REST) in manganese-induced behavioral deficits and dysregulating dopaminergic and serotonergic neurotransmission in mice. Neurotoxicology 2025, 108, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Morris-Blanco, K.C.; Kim, T.; Bertogliat, M.J.; Mehta, S.L.; Chokkalla, A.K.; Vemuganti, R. Inhibition of the Epigenetic Regulator REST Ameliorates Ischemic Brain Injury. Mol. Neurobiol. 2019, 56, 2542–2550. [Google Scholar] [CrossRef]
- Cavadas, M.A.; Mesnieres, M.; Crifo, B.; Manresa, M.C.; Selfridge, A.C.; Scholz, C.C.; Cummins, E.P.; Cheong, A.; Taylor, C.T. REST mediates resolution of HIF-dependent gene expression in prolonged hypoxia. Sci. Rep. 2015, 5, 17851. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 30 November 2017).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Klimov, L.O.; Ershov, N.I.; Efimov, V.M.; Markel, A.L.; Redina, O.E. Genome-wide transcriptome analysis of hypothalamus in rats with inherited stress-induced arterial hypertension. BMC Genet. 2016, 17 (Suppl. 1), 13. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Vedi, M.; Smith, J.R.; Thomas Hayman, G.; Tutaj, M.; Brodie, K.C.; De Pons, J.L.; Demos, W.M.; Gibson, A.C.; Kaldunski, M.L.; Lamers, L.; et al. 2022 updates to the Rat Genome Database: A Findable, Accessible, Interoperable, and Reusable (FAIR) resource. Genetics 2023, 224, iyad042. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Chadaeva, I.V.; Filonov, S.V.; Zolotareva, K.A.; Khandaev, B.M.; Ershov, N.I.; Podkolodnyy, N.L.; Kozhemyakina, R.V.; Rasskazov, D.A.; Bogomolov, A.G.; Kondratyuk, E.Y.; et al. RatDEGdb: A knowledge base of differentially expressed genes in the rat as a model object in biomedical research. Vavilovskii Zhurnal Genet. Selektsii 2023, 27, 794–806. [Google Scholar] [CrossRef]
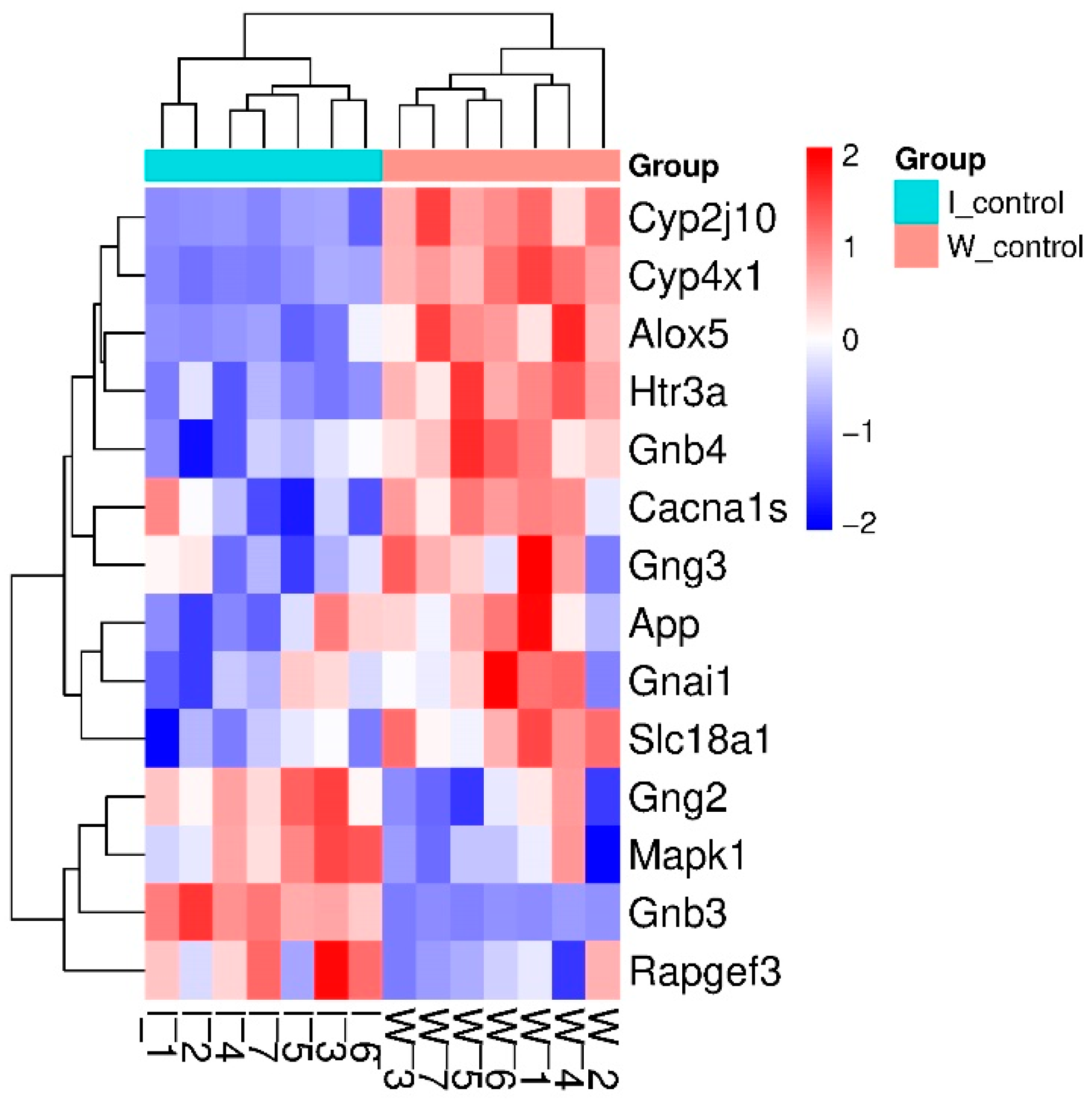
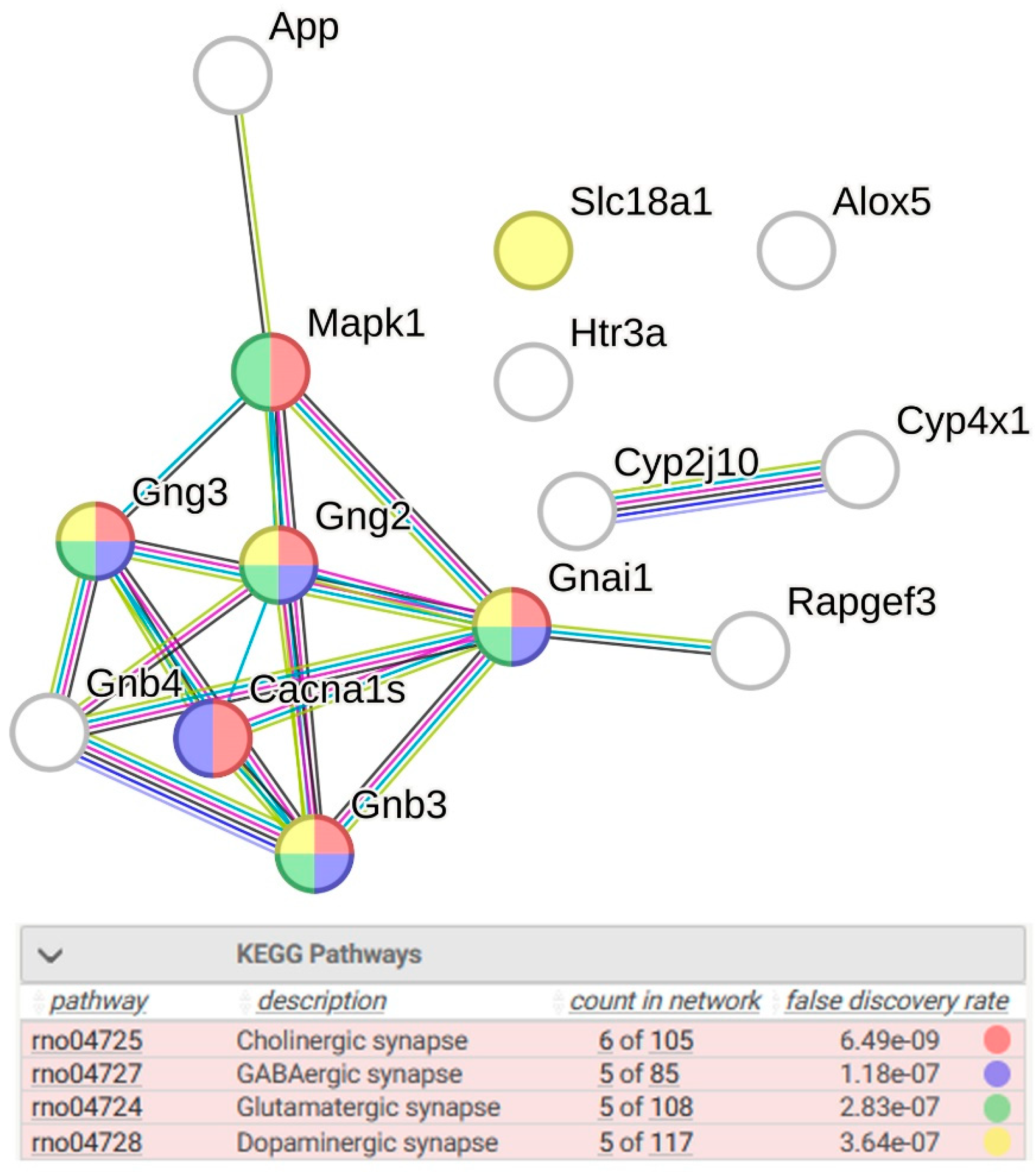
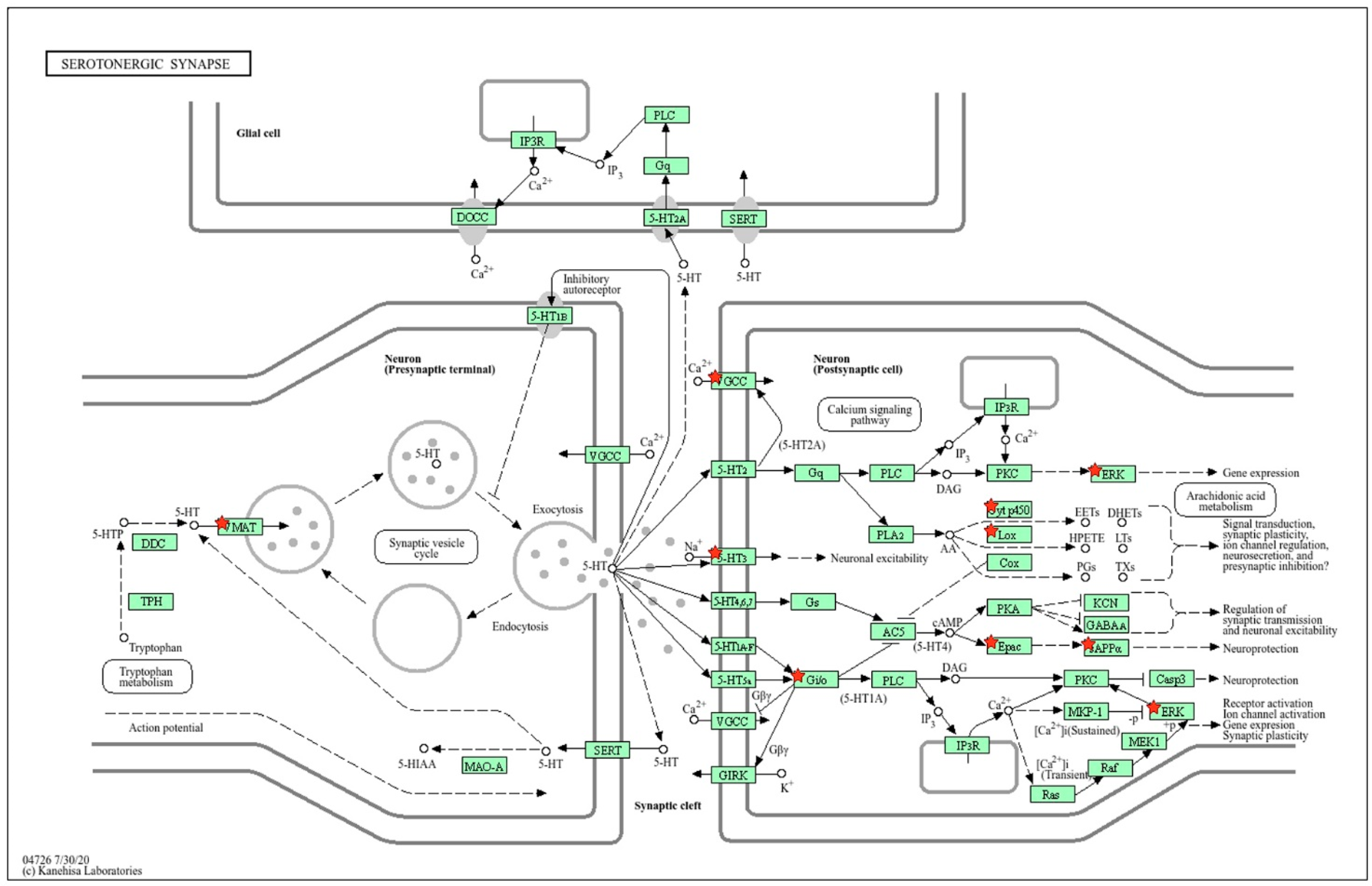
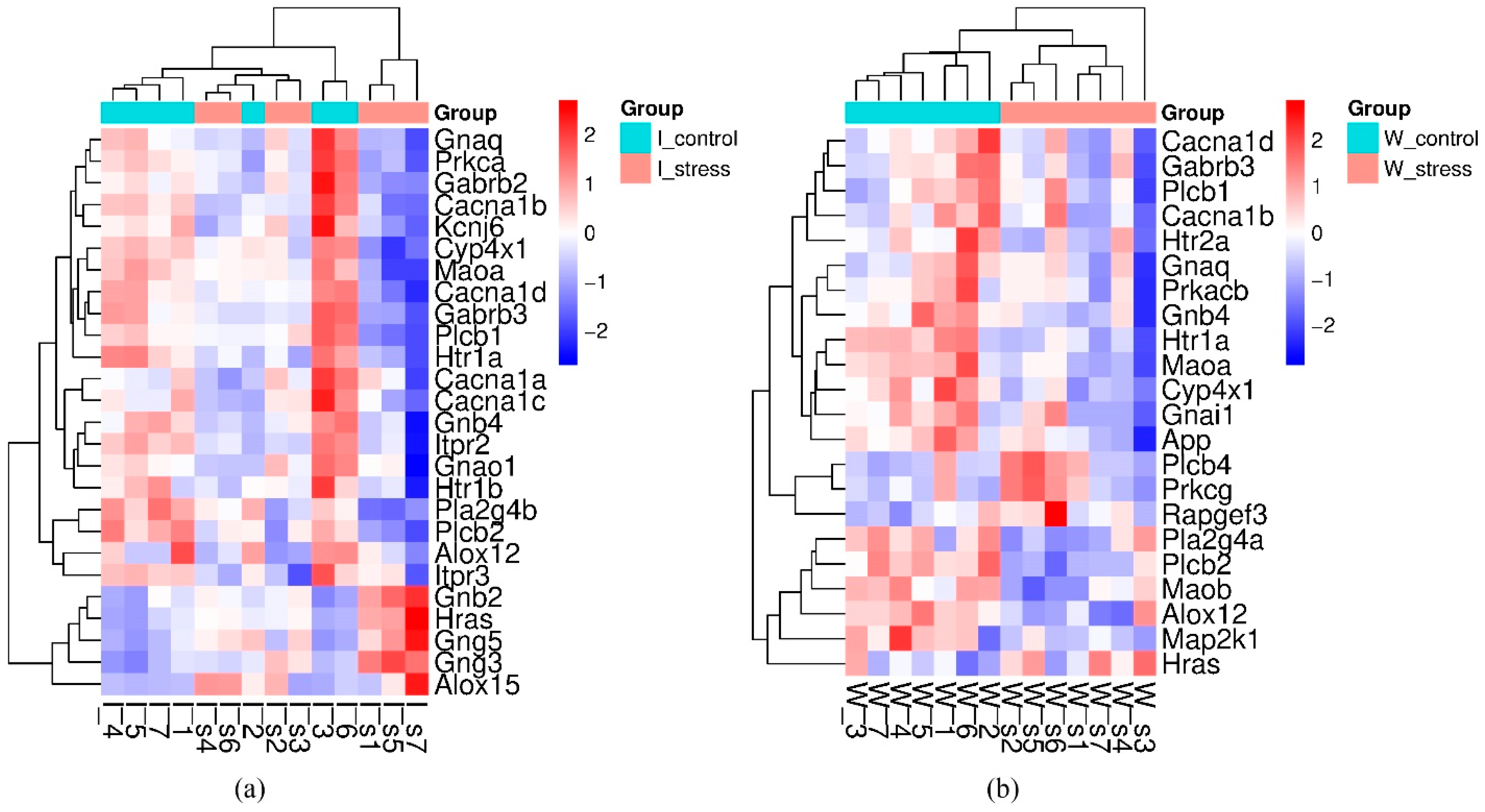




| Gene Symbol | GeneID | log2 Fold Change ISIAH_Control/ WAG_Control | padj | Description |
|---|---|---|---|---|
| Alox5 * | 25290 | −0.641 | 5.14 × 10−6 | arachidonate 5-lipoxygenase |
| App * | 54226 | −0.138 | 3.06 × 10−6 | amyloid beta precursor protein |
| Cacna1s | 682930 | −1.110 | 4.83 × 10−2 | calcium voltage-gated channel subunit alpha1 S |
| Cyp2j10 | 313373 | −1.158 | 1.30 × 10−21 | cytochrome P450, family 2, subfamily j, polypeptide 10 |
| Cyp4x1 | 246767 | −0.982 | 1.55 × 10−52 | cytochrome P450, family 4, subfamily x, polypeptide 1 |
| Gnai1 | 25686 | −0.108 | 2.07 × 10−2 | G protein subunit alpha i1 |
| Gnb3 *# | 60449 | 2.397 | 3.27 × 10−49 | G protein subunit beta 3 |
| Gnb4 | 294962 | −0.369 | 7.44 × 10−6 | G protein subunit beta 4 |
| Gng2 | 80850 | 0.111 | 1.66 × 10−2 | G protein subunit gamma 2 |
| Gng3 | 114117 | −0.191 | 7.59 × 10−4 | G protein subunit gamma 3 |
| Htr3a # | 79246 | −0.509 | 1.00 × 10−8 | 5-hydroxytryptamine receptor 3A |
| Mapk1 * | 116590 | 0.137 | 2.12 × 10−3 | mitogen-activated protein kinase 1 |
| Rapgef3 | 59326 | 0.121 | 2.61 × 10−3 | Rap guanine nucleotide exchange factor 3 |
| Slc18a1 # | 25693 | −0.638 | 1.57 × 10−2 | solute carrier family 18 member A1 |
| Gene Symbol | GeneID | ISIAH_Stress/ISIAH_Control | WAG_Stress/WAG_Control | Description | ||
|---|---|---|---|---|---|---|
| log2 Fold Change | padj | log2 Fold Change | padj | |||
| Alox12 * | 287454 | −0.716 | 3.07 × 10−2 | −0.779 | 8.06 × 10−3 | arachidonate 12-lipoxygenase, 12S type |
| Cacna1b # | 257648 | −0.290 | 8.82 × 10−9 | −0.152 | 1.28 × 10−4 | calcium voltage-gated channel subunit alpha1 B |
| Cacna1d | 29716 | −0.300 | 2.82 × 10−4 | −0.287 | 2.02 × 10−6 | calcium voltage-gated channel subunit alpha1 D |
| Cyp4x1 | 246767 | −0.363 | 1.15 × 10−3 | −0.261 | 6.10 × 10−5 | cytochrome P450, family 4, subfamily x, polypeptide 1 |
| Gabrb3 | 24922 | −0.652 | 2.88 × 10−5 | −0.405 | 2.19 × 10−3 | gamma-aminobutyric acid type A receptor subunit beta 3 |
| Gnaq # | 81666 | −0.480 | 1.41 × 10−3 | −0.307 | 1.59 × 10−2 | G protein subunit alpha q |
| Gnb4 | 294962 | −0.320 | 1.19 × 10−2 | −0.272 | 9.44 × 10−3 | G protein subunit beta 4 |
| Hras | 293621 | 0.227 | 5.62 × 10−4 | 0.106 | 4.06 × 10−2 | HRas proto-oncogene, GTPase |
| Htr1a # | 24473 | −0.575 | 3.95 × 10−3 | −0.639 | 9.84 × 10−6 | 5-hydroxytryptamine receptor 1A |
| Maoa # | 29253 | −0.162 | 9.73 × 10−3 | −0.230 | 2.83 × 10−6 | monoamine oxidase A |
| Plcb1 | 24654 | −0.286 | 7.57 × 10−3 | −0.154 | 4.39 × 10−2 | phospholipase C beta 1 |
| Plcb2 | 85240 | −0.431 | 3.41 × 10−2 | −0.596 | 2.20 × 10−4 | phospholipase C, beta 2 |
| Gene Symbol | GeneID | log2 Fold Change ISIAH_Stress/ ISIAH_Control | padj | Description |
|---|---|---|---|---|
| Alox15 * | 81639 | 1.236 | 1.98 × 10−2 | arachidonate 15-lipoxygenase |
| Cacna1a | 25398 | −0.162 | 5.36 × 10−3 | calcium voltage-gated channel subunit alpha1 A |
| Cacna1c * | 24239 | −0.211 | 3.96 × 10−2 | calcium voltage-gated channel subunit alpha1 C |
| Gabrb2 | 25451 | −0.621 | 1.19 × 10−2 | gamma-aminobutyric acid type A receptor subunit beta 2 |
| Gnao1 | 50664 | −0.232 | 1.66 × 10−3 | G protein subunit alpha o1 |
| Gnb2 | 81667 | 0.162 | 1.62 × 10−6 | G protein subunit beta 2 |
| Gng3 | 114117 | 0.185 | 4.89 × 10−3 | G protein subunit gamma 3 |
| Gng5 | 79218 | 0.150 | 3.08 × 10−2 | G protein subunit gamma 5 |
| Htr1b * | 25075 | −0.451 | 3.33 × 10−2 | 5-hydroxytryptamine receptor 1B |
| Itpr2 * | 81678 | −0.271 | 2.16 × 10−3 | inositol 1,4,5-trisphosphate receptor, type 2 |
| Itpr3 *# | 25679 | −0.389 | 9.87 × 10−4 | inositol 1,4,5-trisphosphate receptor, type 3 |
| Kcnj6 | 25743 | −0.168 | 7.57 × 10−3 | potassium inwardly-rectifying channel, subfamily J, member 6 |
| Pla2g4b | 311341 | −0.334 | 1.01 × 10−3 | phospholipase A2 group IVB |
| Prkca * | 24680 | −0.357 | 1.33 × 10−2 | protein kinase C, alpha |
| Gene Symbol | GeneID | log2 Fold Change WAG_Stress/ WAG_Control | padj | Description |
|---|---|---|---|---|
| App * | 54226 | −0.197 | 1.13 × 10−8 | amyloid beta precursor protein |
| Gnai1 | 25686 | −0.147 | 5.01 × 10−3 | G protein subunit alpha i1 |
| Htr2a * | 29595 | −0.388 | 4.97 × 10−2 | 5-hydroxytryptamine receptor 2A |
| Maob # | 25750 | −0.224 | 1.04 × 10−5 | monoamine oxidase B |
| Map2k1 | 170851 | −0.106 | 1.50 × 10−2 | mitogen-activated protein kinase kinase 1 |
| Pla2g4a | 24653 | −0.294 | 1.85 × 10−2 | phospholipase A2 group IVA |
| Plcb4 | 25031 | 0.337 | 2.73 × 10−2 | phospholipase C, beta 4 |
| Prkacb | 293508 | −0.284 | 1.04 × 10−3 | protein kinase cAMP-activated catalytic subunit beta |
| Prkcg | 24681 | 0.343 | 3.60 × 10−2 | protein kinase C, gamma |
| Rapgef3 | 59326 | 0.083 | 3.34 × 10−2 | Rap guanine nucleotide exchange factor 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Redina, O.E.; Ryazanova, M.A.; Oshchepkov, D.Y.; Makovka, Y.V.; Markel, A.L. Expression Profiles of Genes Related to Serotonergic Synaptic Function in Hypothalamus of Hypertensive and Normotensive Rats in Basal and Stressful Conditions. Int. J. Mol. Sci. 2025, 26, 7058. https://doi.org/10.3390/ijms26157058
Redina OE, Ryazanova MA, Oshchepkov DY, Makovka YV, Markel AL. Expression Profiles of Genes Related to Serotonergic Synaptic Function in Hypothalamus of Hypertensive and Normotensive Rats in Basal and Stressful Conditions. International Journal of Molecular Sciences. 2025; 26(15):7058. https://doi.org/10.3390/ijms26157058
Chicago/Turabian StyleRedina, Olga E., Marina A. Ryazanova, Dmitry Yu. Oshchepkov, Yulia V. Makovka, and Arcady L. Markel. 2025. "Expression Profiles of Genes Related to Serotonergic Synaptic Function in Hypothalamus of Hypertensive and Normotensive Rats in Basal and Stressful Conditions" International Journal of Molecular Sciences 26, no. 15: 7058. https://doi.org/10.3390/ijms26157058
APA StyleRedina, O. E., Ryazanova, M. A., Oshchepkov, D. Y., Makovka, Y. V., & Markel, A. L. (2025). Expression Profiles of Genes Related to Serotonergic Synaptic Function in Hypothalamus of Hypertensive and Normotensive Rats in Basal and Stressful Conditions. International Journal of Molecular Sciences, 26(15), 7058. https://doi.org/10.3390/ijms26157058





