Childhood Asthma Biomarkers Derived from Plasma and Saliva Exosomal miRNAs
Abstract
1. Introduction
2. Results
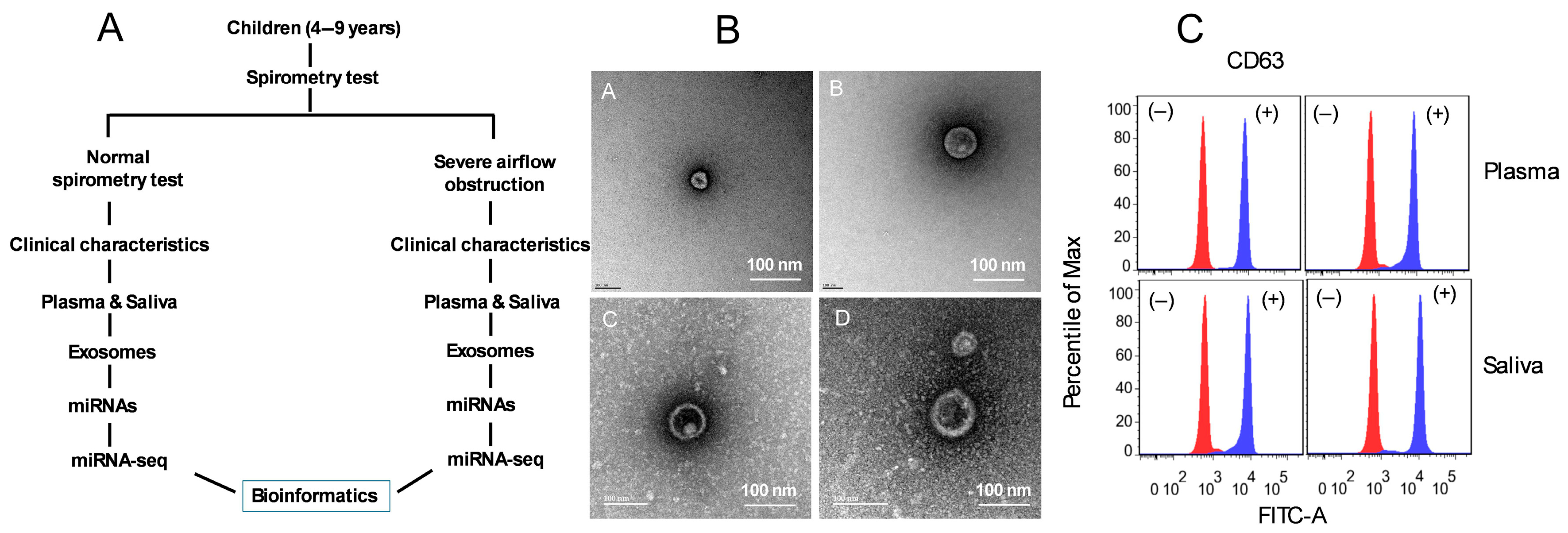
2.1. Clinical Differences in Pediatrics Severe Asthma
2.2. Characterization of Plasma- and Saliva-Derived Exosomes
2.3. Exosomal miRNA Profiling in Pediatric Plasma and Saliva
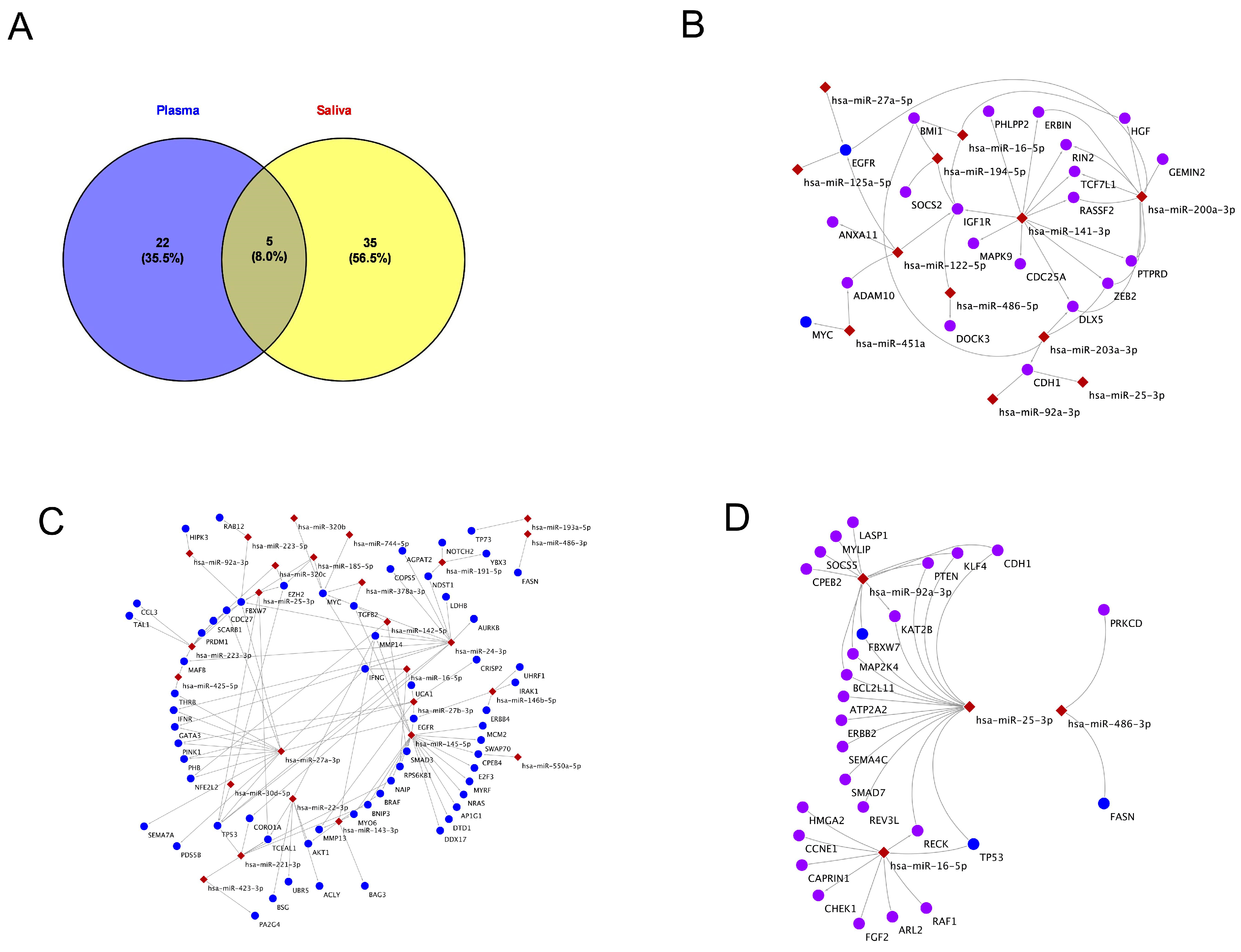
2.4. Exosomal miRNAs and Their Gene Targets in Plasma and Saliva
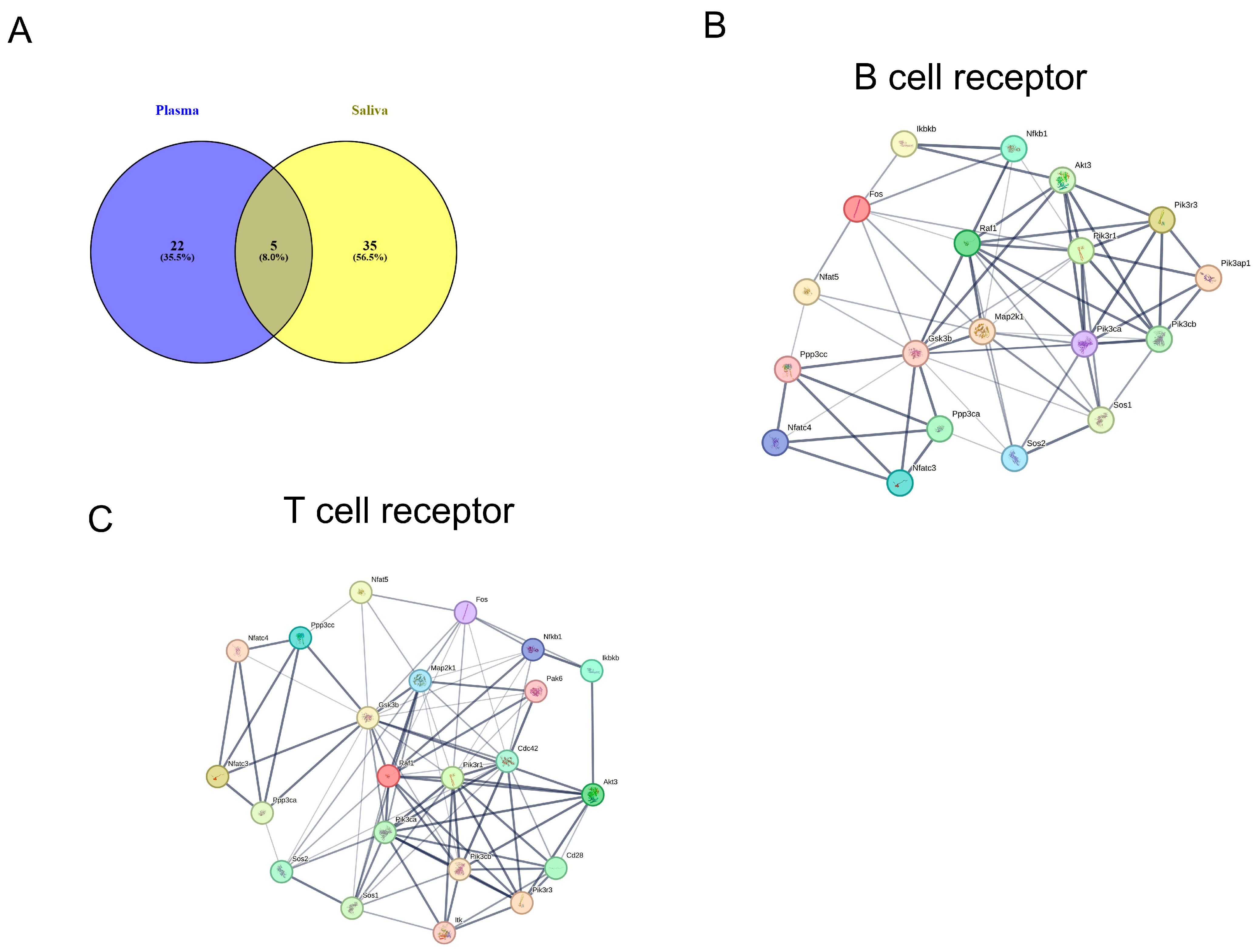
2.5. KEGG Pathway Analysis of miRNA Targets in Plasma and Saliva
2.6. KEGG Pathways of Exosomal miRNAs in Shared Plasma and Saliva
2.7. KEGG Pathway Analysis Specific to miRNAs-Plasma and Saliva
2.8. Comparative Analysis of lncRNA Expression in Plasma and Saliva
2.9. KEGG Pathways for lncRNAs in Plasma and Asthma
3. Discussion
3.1. miRNA Profiling and KEGG Pathways
3.2. KEGG Pathways for miRNAs’ Target Genes
3.3. lncRNAs Profiling and KEGG Pathways
4. Materials and Methods
4.1. Subjects
4.2. Saliva and Blood Collection
4.3. Exosome Isolations and Characterization
4.4. Small RNA-Sequencing for Plasma and Saliva
4.5. Small RNA-Seq Data Processing and Analysis
4.6. Illumina RNA Library Preparation and Sequencing
4.7. Differential Gene Expression Analysis (DGEA)
4.8. Protein–Protein Interaction (PPI) Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reddel, H.K.; Bacharier, L.B.; Bateman, E.D.; Brightling, C.E.; Brusselle, G.G.; Buhl, R.; Cruz, A.A.; Duijts, L.; Drazen, J.M.; FitzGerald, J.M.; et al. Global Initiative for Asthma Strategy 2021: Executive Summary and Rationale for Key Changes. Am. J. Respir. Crit. Care. Med. 2022, 205, 17–35. [Google Scholar] [CrossRef] [PubMed]
- Lizzo, J.M.; Goldin, J.; Cortes, S. Pediatric Asthma. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Zahran, H.S.; Bailey, C.M.; Damon, S.A.; Garbe, P.L.; Breysse, P.N. Vital Signs: Asthma in Children—United States, 2001–2016. MMWR Morb. Mortal Wkly. Rep. 2018, 67, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, T.; Castro-Rodriguez, J.A.; Brockmann, P.E. Sleep-disordered breathing in children with asthma: A systematic review on the impact of treatment. J. Asthma Allergy 2016, 9, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, N.G.; Arakawa, H.; Carlsen, K.H.; Custovic, A.; Gern, J.; Lemanske, R.; Le Souef, P.; Mäkelä, M.; Roberts, G.; Wong, G.; et al. International consensus on (ICON) pediatric asthma. Allergy 2012, 67, 976–997. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.L. Asthma in childhood: A complex, heterogeneous disease. Korean J. Pediatr. 2011, 54, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.D.; Vercelli, D. Asthma. Lancet 2013, 382, 1360–1372. [Google Scholar] [CrossRef] [PubMed]
- Calven, J.; Ax, E.; Radinger, M. The Airway Epithelium—A Central Player in Asthma Pathogenesis. Int. J. Mol. Sci. 2020, 21, 8907. [Google Scholar] [CrossRef] [PubMed]
- Hammad, H.; Lambrecht, B.N. The basic immunology of asthma. Cell 2021, 184, 1469–1485, Erratum in Cell 2021, 184, 2521–2522. [Google Scholar] [CrossRef] [PubMed]
- Quezada-Perez, E.; Guido-Bayardo, L.R.; Marin-Larrain, P.P.; Coss-Mejia, A.H. Asthma in elderly: From basic immunology to the clinic. Rev. Alerg. Mex. 2013, 60, 69–77. [Google Scholar] [PubMed]
- Ricciardolo, F.L.M.; Sprio, A.E.; Baroso, A.; Gallo, F.; Riccardi, E.; Bertolini, F.; Carriero, V.; Arrigo, E.; Ciprandi, G. Characterization of T2-Low and T2-High Asthma Phenotypes in Real-Life. Biomedicines 2021, 9, 1684. [Google Scholar] [CrossRef] [PubMed]
- Hesse, L.; Oude Elberink, J.N.G.; van Oosterhout, A.J.M.; Nawijn, M.C. Allergen immunotherapy for allergic airway diseases: Use lessons from the past to design a brighter future. Pharmacol. Ther. 2022, 237, 108115. [Google Scholar] [CrossRef] [PubMed]
- Stokes, J.R.; Casale, T.B. Characterization of asthma endotypes: Implications for therapy. Ann. Allergy Asthma Immunol. 2016, 117, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Bacharier, L.B. Biologic Therapies for Severe Asthma in School-Age Children. Respir. Care 2025, 70, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Bush, A. Update in paediatric asthma. Curr. Opin. Pulm. Med. 2025, 31, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Indolfi, C.; Klain, A.; Capuano, M.C.; Colosimo, S.; Rapillo, R.; Miraglia Del Giudice, M. Severe Asthma in School-Age Children: An Updated Appraisal on Biological Options and Challenges in This Age Group. Children 2025, 12, 167. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, S.E. Asthma phenotypes: The evolution from clinical to molecular approaches. Nat. Med. 2012, 18, 716–725. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, A.M.; Teague, W.G. National Institutes of Health/National Heart, Lung and Blood Institute’s Severe Asthma Research Program. Progressive airflow limitation is a feature of children with severe asthma. J. Allergy Clin. Immunol. 2011, 127, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder, H.; Peng, Y.Q.; Agache, I.; Diamant, Z.; Eiwegger, T.; Fokkens, W.J.; Traidl-Hoffmann, C.; Nadeau, K.; O’Hehir, R.E.; O’Mahony, L.; et al. Biomarkers for diagnosis and prediction of therapy responses in allergic diseases and asthma. Allergy 2020, 75, 3039–3068. [Google Scholar] [CrossRef] [PubMed]
- Lotvall, J.; Valadi, H. Cell to cell signalling via exosomes through esRNA. Cell Adh. Migr. 2007, 1, 156–158. [Google Scholar] [CrossRef] [PubMed]
- Schorey, J.S.; Cheng, Y.; Singh, P.P.; Smith, V.L. Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep. 2015, 16, 24–43. [Google Scholar] [CrossRef] [PubMed]
- Hough, K.P.; Chanda, D.; Duncan, S.R.; Thannickal, V.J.; Deshane, J.S. Exosomes in immunoregulation of chronic lung diseases. Allergy 2017, 72, 534–544. [Google Scholar] [CrossRef] [PubMed]
- Sastre, B.; Canas, J.A.; Rodrigo-Munoz, J.M.; Del Pozo, V. Novel Modulators of Asthma and Allergy: Exosomes and MicroRNAs. Front. Immunol. 2017, 8, 826. [Google Scholar] [CrossRef] [PubMed]
- Holgate, S.T.; Wenzel, S.; Postma, D.S.; Weiss, S.T.; Renz, H.; Sly, P.D. Asthma. Nat. Rev. Dis. Primers 2015, 1, 15025. [Google Scholar] [CrossRef] [PubMed]
- Parker, R.B.; Kohler, J.J. Regulation of intracellular signaling by extracellular glycan remodeling. ACS Chem. Biol. 2010, 5, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.J.; Lim, J.W.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev. Proteom. 2009, 6, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Alipoor, S.D.; Adcock, I.M.; Garssen, J.; Mortaz, E.; Varahram, M.; Mirsaeidi, M.; Velayati, A. The roles of miRNAs as potential biomarkers in lung diseases. Eur. J. Pharmacol. 2016, 791, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Li, K.; Hellermann, G.; Lockey, R.F.; Mohapatra, S.; Mohapatra, S. Regulating the Regulators: MicroRNA and Asthma. World Allergy Organ. J. 2011, 4, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Mera, S.; Martelo-Vidal, L.; Miguéns-Suárez, P.; Saavedra-Nieves, P.; Arias, P.; González-Fernández, C.; Mosteiro-Añón, M.; Corbacho-Abelaira, M.D.; Blanco-Aparicio, M.; Méndez-Brea, P.; et al. Serum exosome inflamma-miRs are surrogate biomarkers for asthma phenotype and severity. Allergy 2023, 78, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Joshi, T.; Chan, Y.O.; Qiao, Z.; Kheirandish-Gozal, L.; Gozal, D.; Khalyfa, A. Circulating exosomes in pediatric obstructive sleep apnea with or without neurocognitive deficits and their effects on a 3D-blood-brain barrier spheroid model. Exp. Neurol. 2025, 387, 115188. [Google Scholar] [CrossRef] [PubMed]
- Khalyfa, A.; Marin, J.M.; Sanz-Rubio, D.; Lyu, Z.; Joshi, T.; Gozal, D. Multi-Omics Analysis of Circulating Exosomes in Adherent Long-Term Treated OSA Patients. Int. J. Mol. Sci. 2023, 24, 16074. [Google Scholar] [CrossRef] [PubMed]
- Moore, W.C.; Meyers, D.A.; Wenzel, S.E.; Teague, W.G.; Li, H.; Li, X.; D’Agostino, R., Jr.; Castro, M.; Curran-Everett, D.; Fitzpatrick, A.M.; et al. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am. J. Respir. Crit. Care Med. 2010, 181, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Canas, J.A.; Sastre, B.; Rodrigo-Munoz, J.M.; Del Pozo, V. Exosomes: A new approach to asthma pathology. Clin. Chim. Acta 2019, 495, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Heffler, E.; Allegra, A.; Pioggia, G.; Picardi, G.; Musolino, C.; Gangemi, S. MicroRNA Profiling in Asthma: Potential Biomarkers and Therapeutic Targets. Am. J. Respir. Cell Mol. Biol. 2017, 57, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Dong, H.; Zhang, W.; Sun, H.; Gu, W.; Zhang, X.; Huang, L.; Yan, Y.; Hao, C.; Ji, W.; et al. MiR-493-5p inhibits Th9 cell differentiation in allergic asthma by targeting FOXO1. Respir. Res. 2022, 23, 286. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Zeng, D.; Zheng, J.; Zhao, D. MicroRNAs: Mediators and Therapeutic Targets to Airway Hyper Reactivity After Respiratory Syncytial Virus Infection. Front. Microbiol. 2018, 9, 2177. [Google Scholar] [CrossRef] [PubMed]
- Hir, S.R.; Alizadeh, Z.; Mazinani, M.; Rad, M.M.; Fazlollahi, M.R.; Kazemnejad, A.; Hosseini, A.Z.; Moin, M. Exosomal MicroRNAs as Biomarkers in Allergic Asthma. Iran J. Allergy Asthma Immunol. 2021, 20, 160–168. [Google Scholar]
- Mori, T.; Giovannelli, L.; Bilia, A.R.; Margheri, F. Exosomes: Potential Next-Generation Nanocarriers for the Therapy of Inflammatory Diseases. Pharmaceutics 2023, 15, 2276. [Google Scholar] [CrossRef] [PubMed]
- Yoo, E.J.; Ojiaku, C.A.; Sunder, K.; Panettieri, R.A., Jr. Phosphoinositide 3-Kinase in Asthma: Novel Roles and Therapeutic Approaches. Am. J. Respir. Cell Mol. Biol. 2017, 56, 700–707. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.F. p38 mitogen-activated protein kinase pathways in asthma and COPD. Chest 2011, 139, 1470–1479. [Google Scholar] [CrossRef] [PubMed]
- Kampe, M.; Lampinen, M.; Stolt, I.; Janson, C.; Stalenheim, G.; Carlson, M. PI3-kinase regulates eosinophil and neutrophil degranulation in patients with allergic rhinitis and allergic asthma irrespective of allergen challenge model. Inflammation 2012, 35, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, B.G.; Spanjer, A.I.; van der Schuyt, R.D.; Kuik, W.J.; Zaagsma, J.; Meurs, H. Focal adhesion kinase regulates collagen I-induced airway smooth muscle phenotype switching. J. Pharmacol. Exp. Ther. 2013, 346, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Ratti, M.; Lampis, A.; Ghidini, M.; Salati, M.; Mirchev, M.B.; Valeri, N.; Hahne, J.C. MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. Target Oncol. 2020, 15, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Diaz Duran, R.; Wei, H.; Kim, D.H.; Wu, J.Q. Invited Review: Long non-coding RNAs: Important regulators in the development, function and disorders of the central nervous system. Neuropathol. Appl. Neurobiol. 2019, 45, 538–556. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, S.; Shi, L.; Chen, G.; Liu, X.; Liu, M.; Guo, G. Interaction between long noncoding RNA and microRNA in lung inflammatory diseases. Immun. Inflamm. Dis 2024, 12, e1129. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, R.A.; Fraczek, M.G.; Parker, S.; Delneri, D.; O’Keefe, R.T. Non-coding RNAs and disease: The classical ncRNAs make a comeback. Biochem. Soc. Trans. 2016, 44, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.B.; Amaral, P.P.; Schlesinger, F.J.; Dinger, M.E.; Taft, R.J.; Rinn, J.L.; Ponting, C.P.; Stadler, P.F.; Morris, K.V.; Morillon, A.; et al. The reality of pervasive transcription. PLoS Biol. 2011, 9, e1000625, discussion e1001102. [Google Scholar] [CrossRef] [PubMed]
- Boateng, E.; Kovacevic, D.; Oldenburg, V.; Radinger, M.; Krauss-Etschmann, S. Role of airway epithelial cell miRNAs in asthma. Front. Allergy 2022, 3, 962693. [Google Scholar] [CrossRef] [PubMed]
- Gil-Martinez, M.; Lorente-Sorolla, C.; Naharro, S.; Rodrigo-Munoz, J.M.; Del Pozo, V. Advances and Highlights of miRNAs in Asthma: Biomarkers for Diagnosis and Treatment. Int. J. Mol. Sci. 2023, 24, 1628. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, V.; Nouri, S.; Nassaj, Z.S.; Bighash, M.; Abbasian, S.; Hagh, R.A. Long non coding RNAs reveal important pathways in childhood asthma: A future perspective. J. Mol. Histol. 2023, 54, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Liu, X.H.; Chen, X.M.; Song, X.L.; Li, W.; Huang, Y. Emerging Roles of Non-Coding RNAs in Childhood Asthma. Front. Pharmacol. 2022, 13, 856104. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, X. Long noncoding RNA antisense noncoding RNA in the INK4 locus inhibition alleviates airway remodeling in asthma through the regulation of the microRNA-7-5p/early growth response factor 3 axis. Immun. Inflamm. Dis. 2023, 11, e823. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, J. Long non-coding RNA TUG1 promotes proliferation and migration in PDGF-BB-stimulated HASMCs by regulating miR-216a-3p/SMURF2 axis. BMC Mol. Cell Biol. 2021, 22, 56. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Ding, Y.; Yin, B.; Gu, H. Long noncoding RNA LINC-PINT retards the abnormal growth of airway smooth muscle cells via regulating the microRNA-26a-5p/PTEN axis in asthma. Int. Immunopharmacol. 2021, 99, 107997. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Huang, H.; Liang, L.; Lu, H.; Liang, D. Long non-coding RNA (LncRNA) non-coding RNA activated by DNA damage (NORAD) knockdown alleviates airway remodeling in asthma via regulating miR-410-3p/RCC2 and inhibiting Wnt/beta-catenin pathway. Heliyon 2024, 10, e23860. [Google Scholar] [CrossRef] [PubMed]
- Dai, B.; Sun, F.; Cai, X.; Li, C.; Liu, F.; Shang, Y. Long noncoding RNA PTTG3P/miR-192-3p/CCNB1 axis is a potential biomarker of childhood asthma. Int. Immunopharmacol. 2021, 101, 108229. [Google Scholar] [CrossRef]
- Li, W.; Wang, X.; Sun, S.; An, H. Long non-coding RNA colorectal neoplasia differentially expressed correlates negatively with miR-33a and miR-495 and positively with inflammatory cytokines in asthmatic children. Clin. Respir. J. 2021, 15, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ye, S.; Lu, Y. Long non-coding RNA NEAT1 overexpression associates with increased exacerbation risk, severity, and inflammation, as well as decreased lung function through the interaction with microRNA-124 in asthma. J. Clin. Lab. Anal. 2020, 34, e23023. [Google Scholar] [CrossRef] [PubMed]
- Francisco, B.; Ner, Z.; Ge, B.; Hewett, J.; Konig, P. Sensitivity of different spirometric tests for detecting airway obstruction in childhood asthma. J. Asthma 2015, 52, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Khalyfa, A.; Masa, J.F.; Qiao, Z.; Gonzalez, M.; Marti, S.; Khalyfa, A.A.; Kheirandish-Gozal, L.; Gozal, D. Plasma exosomes in obesity hypoventilation syndrome patients drive lung cancer cell malignant properties: Effect of long-term adherent CPAP treatment. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166479. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Vermilya, A.; Clogston, J.D. Particle Size and Concentration Measurement Using the Spectradyne nCS1 Instrument. Methods Mol. Biol. 2024, 2789, 45–52. [Google Scholar] [PubMed]
- Patil, A.H.; Halushka, M.K. miRge3.0: A comprehensive microRNA and tRF sequencing analysis pipeline. NAR Genom. Bioinform. 2021, 3, lqab068. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Frankish, A.; Carbonell-Sala, S.; Diekhans, M.; Jungreis, I.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Arnan, C.; Barnes, I.; et al. GENCODE: Reference annotation for the human and mouse genomes in 2023. Nucleic Acids Res. 2023, 51, D942–D949. [Google Scholar] [CrossRef] [PubMed]
- The RNAcentral Consortium. RNAcentral: A hub of information for non-coding RNA sequences. Nucleic Acids Res. 2019, 47, D1250–D1251. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Volders, P.J.; Helsens, K.; Wang, X.; Menten, B.; Martens, L.; Gevaert, K.; Vandesompele, J.; Mestdagh, P. LNCipedia: A database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013, 41, D246–D251. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, L.; Feng, C.; Qin, Y.; Xiao, J.; Zhang, Z.; Ma, L. LncBook 2.0: Integrating human long non-coding RNAs with multi-omics annotations. Nucleic Acids Res. 2023, 51, D186–D191. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Y.; Fang, J.; Xu, Z.; Zhang, H.; Mao, M.; Chen, Y.; Zhang, L.; Pian, C. NcPath: A novel platform for visualization and enrichment analysis of human non-coding RNA and KEGG signaling pathways. Bioinformatics 2023, 39, btac812. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Gustavsen, J.A.; Pai, S.; Isserlin, R.; Demchak, B.; Pico, A.R. RCy3: Network biology using Cytoscape from within R. F1000Research 2019, 8, 1774. [Google Scholar] [CrossRef] [PubMed]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]

| Items | Normal Lung Function (NLF) | Severe Airflow Obstruction (SAO) | p-Value |
|---|---|---|---|
| Age | 10.71 ± 1.33 | 13.81 ± 2.67 | 0.006 |
| FVC | 2.58 ± 0.43 | 3.93 ± 1.51 | 0.01973155 |
| FVC% Pred | 112.51 ± 10.48 | 108.91 ± 16.01 | 0.579436397 |
| FEV1 | 2.23 ± 0.37 | 2.53 ± 1.05 | 0.427157081 |
| FEV1% Pred | 112.51 ± 9.81 | 82.11 ± 17.81 | 0.000287177 |
| FEV1/FVC Ratio | 86.52 ± 1.63 | 64.52 ± 7.77 | 1.42353 × 10−7 |
| FEF75% Pred | 124.74 ± 19.09 | 39.72 ± 10.61 | 7.83717 × 10−10 |
| IgE | 72.11 ± 62.54 | 288.12 ± 223.19 | 0.01 |
| Name | Log2 Fold Change | p-Value | Adjusted p-Value |
|---|---|---|---|
| hsa-miR-451b-5p | 6.873 | <0.0001 | <0.0001 |
| hsa-miR-7706 | 5.907 | 0.003 | 0.02 |
| hsa-miR-195-3p | 5.825 | <0.0001 | 0.0004 |
| hsa-miR-141-3p | 5.218 | 0.004 | 0.03 |
| hsa-miR-3158-5p | 4.886 | <0.0001 | <0.0001 |
| hsa-miR-3158-3p | 4.584 | <0.0001 | <0.0001 |
| hsa-miR-375-3p | 4.126 | <0.0001 | <0.0001 |
| hsa-miR-501-3p | 3.835 | <0.0001 | <0.0001 |
| hsa-miR-200a-3p | 3.760 | <0.0001 | <0.0001 |
| hsa-miR-451a | 2.626 | <0.0001 | <0.0001 |
| hsa-miR-122-5p | 1.985 | <0.0001 | <0.0001 |
| hsa-miR-192-5p | 1.961 | <0.0001 | <0.0001 |
| hsa-miR-122b-3p | 1.925 | <0.0001 | <0.0001 |
| hsa-miR-16-2-3p | 1.851 | <0.0001 | <0.0001 |
| hsa-miR-194-5p | 1.653 | 0.0001 | 0.001 |
| hsa-miR-486-3p | 1.627 | <0.0001 | 0.0001 |
| hsa-miR-16-5p | 1.601 | <0.0001 | <0.0001 |
| hsa-miR-25-3p | 1.594 | <0.0001 | <0.0001 |
| hsa-miR-486-5p | 1.525 | <0.0001 | 0.0002 |
| hsa-miR-92a-3p | 1.068 | <0.0001 | <0.0001 |
| hsa-miR-27a-5p | −1.131 | 0.001 | 0.01 |
| hsa-miR-125a-5p | −1.168 | <0.0001 | 0.0006 |
| hsa-miR-203a-3p | −1.384 | 0.0006 | 0.006 |
| hsa-miR-4433a-3p | −1.662 | 0.003 | 0.02 |
| hsa-miR-203b-5p | −1.705 | <0.0001 | 0.0006 |
| hsa-miR-203b-5p | −1.705 | <0.0001 | 0.0006 |
| hsa-miR-1246 | −1.746 | 0.0001 | 0.001 |
| hsa-miR-4665-5p | −7.627 | 0.003 | 0.02 |
| MiRNA | Log2 Fold Change | p-Value | Adjusted p-Value |
|---|---|---|---|
| hsa-miR-550a-3-5p | 3.063 | 0.009 | 0.04 |
| hsa-miR-486-3p | 2.363 | 0.0009 | 0.007 |
| hsa-miR-3615-3p | 2.261 | <0.0001 | 0.0003 |
| hsa-miR-106b-3p | 2.234 | 0.0007 | 0.0055 |
| hsa-miR-25-3p | 2.084 | <0.0001 | 0.0001 |
| hsa-miR-140-3p | 2.004 | <0.0001 | 0.0001 |
| hsa-miR-423-3p | 1.977 | <0.0001 | 0.0003 |
| hsa-miR-3184-5p | 1.963 | <0.0001 | 0.0003 |
| hsa-miR-629-5p | 1.909 | <0.0001 | 0.0001 |
| hsa-miR-223-3p | 1.880 | 0.0001 | 0.001 |
| hsa-miR-92a-3p | 1.837 | <0.0001 | 0.0001 |
| hsa-miR-142-5p | 1.818 | <0.0001 | 0.0001 |
| hsa-miR-345-5p | 1.812 | <0.0001 | 0.0001 |
| hsa-miR-501-3p | 1.794 | 0.006 | 0.03 |
| hsa-miR-425-5p | 1.751 | <0.0001 | 0.0001 |
| hsa-miR-223-5p | 1.629 | <0.0001 | 0.0002 |
| hsa-miR-146b-5p | 1.624 | <0.0001 | 0.0007 |
| hsa-let-7d-3p | 1.618 | <0.0001 | 0.0004 |
| hsa-miR-24-3p | 1.609 | <0.0001 | 0.0001 |
| hsa-miR-140-5p | 1.578 | 0.0008 | 0.006 |
| hsa-miR-199a-3p | 1.559 | 0.0003 | 0.003 |
| hsa-miR-744-5p | 1.552 | 0.002 | 0.01 |
| hsa-miR-221-3p | 1.541 | <0.0001 | 0.0002 |
| hsa-miR-145-5p | 1.528 | 0.004 | 0.02 |
| hsa-miR-3074-5p | 1.512 | <0.0001 | 0.0001 |
| hsa-miR-23a-3p | 1.483 | 0.0002 | 0.002 |
| hsa-miR-143-3p | 1.478 | 0.0004 | 0.003 |
| hsa-miR-193a-5p | 1.447 | 0.0001 | 0.001 |
| hsa-miR-941 | 1.373 | 0.001 | 0.01 |
| hsa-miR-378a-3p | 1.340 | <0.0001 | 0.0003 |
| hsa-miR-1307-3p | 1.298 | 0.0007 | 0.005 |
| hsa-miR-320a-3p | 1.279 | 0.004 | 0.02 |
| hsa-miR-652-3p | 1.273 | 0.002 | 0.01 |
| hsa-miR-191-5p | 1.248 | 0.003 | 0.02 |
| hsa-miR-185-5p | 1.235 | 0.002 | 0.01 |
| hsa-miR-16-5p | 1.197 | 0.004 | 0.02 |
| hsa-miR-27a-3p | 1.150 | 0.001 | 0.007 |
| hsa-miR-22-3p | 1.084 | 0.0001 | 0.001 |
| hsa-miR-30d-5p | 1.025 | 0.003 | 0.01 |
| hsa-miR-103b | 1.015 | 0.01 | 0.04 |
| Plasma | Saliva | ||
|---|---|---|---|
| Name | Log2 Fold Change | Adjusted p-Value | Log2 Fold Change |
| hsa-miR-501-3p | 3.83 | 1.57 × 10−5 | 1.79 |
| hsa-miR-486-3p | 1.62 | 0.0001 | 2.36 |
| hsa-miR-16-5p | 1.60 | 1.83 × 10−15 | 1.19 |
| hsa-miR-25-3p | 1.59 | 9.64 × 10−18 | 2.08 |
| hsa-miR-92a-3p | 1.067 | 3.15 × 10−7 | 1.83 |
| KEGG Pathway | KEGG IDs | p-Value | #Genes | #miRNAs |
|---|---|---|---|---|
| PI3K-Akt signaling pathway | hsa04151 | 2.11 × 10−12 | 61 | 5 |
| Focal adhesion | hsa04510 | 4.44 × 10−12 | 43 | 5 |
| Wnt signaling pathway | hsa04310 | 2.27 × 10−9 | 35 | 5 |
| Non-small cell lung cancer | hsa05223 | 7.41 × 10−8 | 15 | 5 |
| Regulation of actin cytoskeleton | hsa04810 | 1.36 × 10−7 | 39 | 5 |
| B cell receptor signaling pathway | hsa04662 | 1.48 × 10−7 | 19 | 5 |
| Small cell lung cancer | hsa05222 | 6.48 × 10−7 | 19 | 5 |
| Insulin signaling pathway | hsa04910 | 8.42 × 10−7 | 27 | 5 |
| Neurotrophin signaling pathway | hsa04722 | 9.18 × 10−7 | 25 | 5 |
| T cell receptor signaling pathway | hsa04660 | 9.23 × 10−7 | 23 | 5 |
| MAPK signaling pathway | hsa04010 | 2.31 × 10−6 | 44 | 5 |
| Acute myeloid leukemia | hsa05221 | 3.98 × 10−6 | 15 | 5 |
| Long-term potentiation | hsa04720 | 5.06 × 10−6 | 16 | 5 |
| Dopaminergic synapse | hsa04728 | 9.10 × 10−6 | 26 | 5 |
| Ubiquitin mediated proteolysis | hsa04120 | 9.52 × 10−6 | 26 | 5 |
| Glioma | hsa05214 | 1.19 × 10−5 | 16 | 5 |
| VEGF signaling pathway | hsa04370 | 1.59 × 10−5 | 15 | 5 |
| Chronic myeloid leukemia | hsa05220 | 0.00017042 | 16 | 5 |
| Type II diabetes mellitus | hsa04930 | 0.000395913 | 11 | 5 |
| Renal cell carcinoma | hsa05211 | 0.000395913 | 16 | 5 |
| Endocytosis | hsa04144 | 0.000453408 | 32 | 5 |
| ErbB signaling pathway | hsa04012 | 0.000758374 | 16 | 5 |
| Fc gamma R-mediated phagocytosis | hsa04666 | 0.001040455 | 17 | 5 |
| Axon guidance | hsa04360 | 0.001191295 | 24 | 5 |
| HIF-1 signaling pathway | hsa04066 | 0.002165998 | 19 | 5 |
| Cholinergic synapse | hsa04725 | 0.002246121 | 21 | 5 |
| Bacterial invasion of epithelial cells | hsa05100 | 0.004172215 | 13 | 5 |
| Calcium signaling pathway | hsa04020 | 0.00556725 | 27 | 5 |
| Gap junction | hsa04540 | 0.00633416 | 16 | 5 |
| Glutamatergic synapse | hsa04724 | 0.007451913 | 19 | 5 |
| Phosphatidylinositol signaling system | hsa04070 | 0.008247675 | 14 | 5 |
| GnRH signaling pathway | hsa04912 | 0.009589913 | 15 | 5 |
| Viral carcinogenesis | hsa05203 | 0.01249035 | 25 | 5 |
| Gastric acid secretion | hsa04971 | 0.02177694 | 12 | 5 |
| Adherens junction | hsa04520 | 0.02235759 | 14 | 5 |
| Melanogenesis | hsa04916 | 0.02235759 | 16 | 5 |
| Osteoclast differentiation | hsa04380 | 0.02479279 | 19 | 5 |
| Cell cycle | hsa04110 | 0.03556233 | 21 | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalyfa, A.; Verma, M.; Alexander, M.M.; Qiao, Z.; Rood, T.; Kapoor, R.; Joshi, T.; Gozal, D.; Francisco, B.D. Childhood Asthma Biomarkers Derived from Plasma and Saliva Exosomal miRNAs. Int. J. Mol. Sci. 2025, 26, 7043. https://doi.org/10.3390/ijms26157043
Khalyfa A, Verma M, Alexander MM, Qiao Z, Rood T, Kapoor R, Joshi T, Gozal D, Francisco BD. Childhood Asthma Biomarkers Derived from Plasma and Saliva Exosomal miRNAs. International Journal of Molecular Sciences. 2025; 26(15):7043. https://doi.org/10.3390/ijms26157043
Chicago/Turabian StyleKhalyfa, Abdelnaby, Mohit Verma, Meghan M. Alexander, Zhuanhong Qiao, Tammy Rood, Ragini Kapoor, Trupti Joshi, David Gozal, and Benjamin D. Francisco. 2025. "Childhood Asthma Biomarkers Derived from Plasma and Saliva Exosomal miRNAs" International Journal of Molecular Sciences 26, no. 15: 7043. https://doi.org/10.3390/ijms26157043
APA StyleKhalyfa, A., Verma, M., Alexander, M. M., Qiao, Z., Rood, T., Kapoor, R., Joshi, T., Gozal, D., & Francisco, B. D. (2025). Childhood Asthma Biomarkers Derived from Plasma and Saliva Exosomal miRNAs. International Journal of Molecular Sciences, 26(15), 7043. https://doi.org/10.3390/ijms26157043








