Single-Cell Transcriptomic Changes in Patient-Derived Glioma and U87 Glioblastoma Cell Cultures Infected with the Oncolytic Virus VV-GMCSF-Lact
Abstract
1. Introduction
2. Results
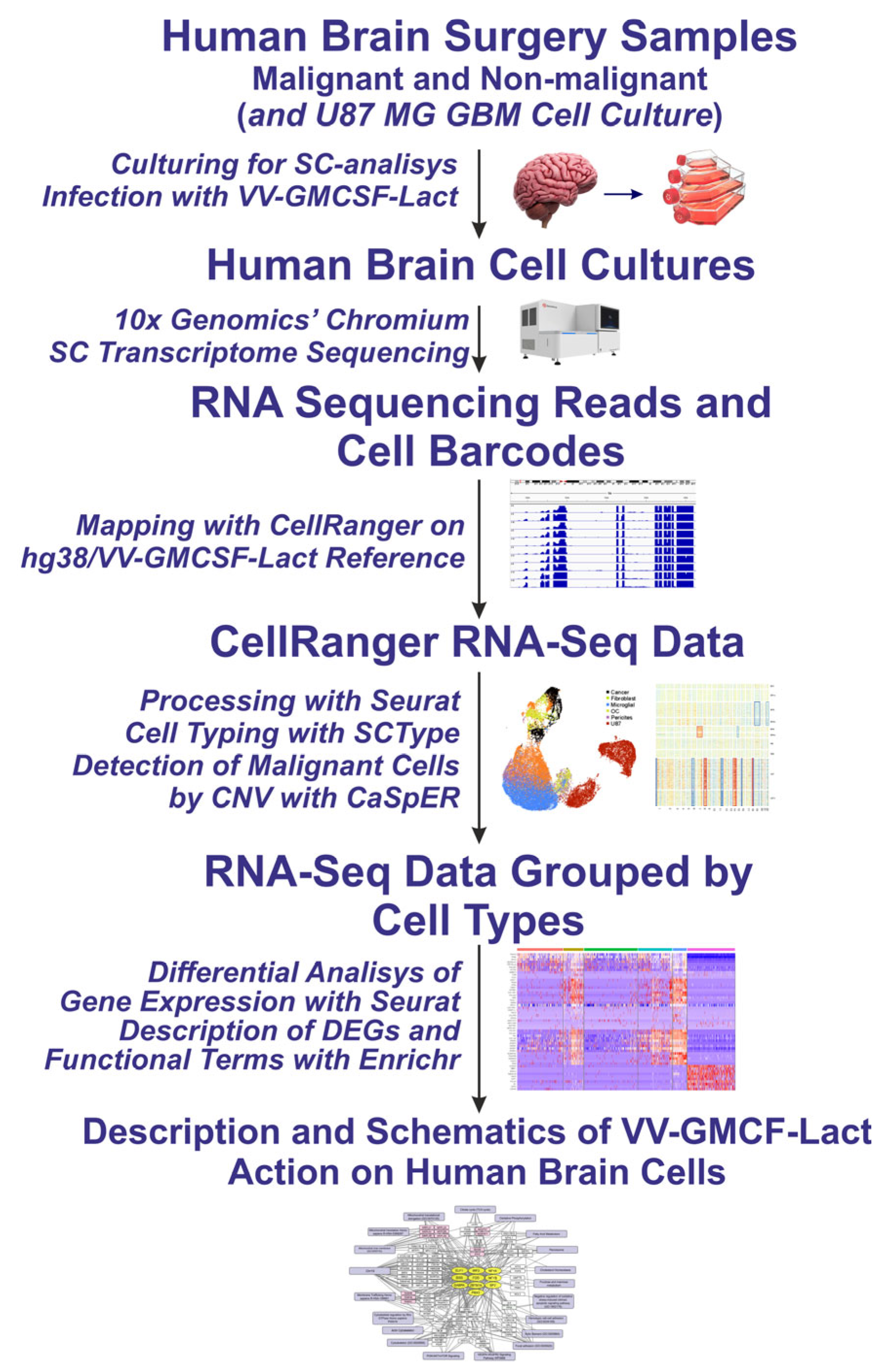
2.1. Cell Cultures and Single-Cell RNA-Seq Data
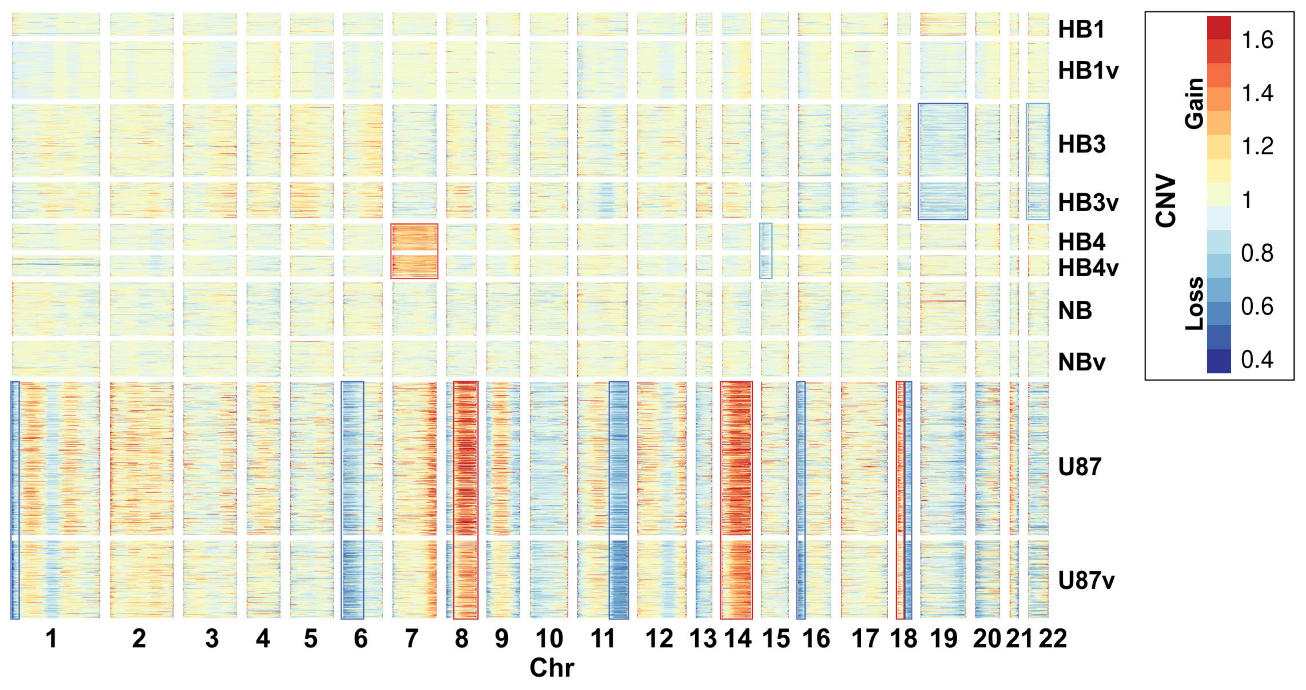
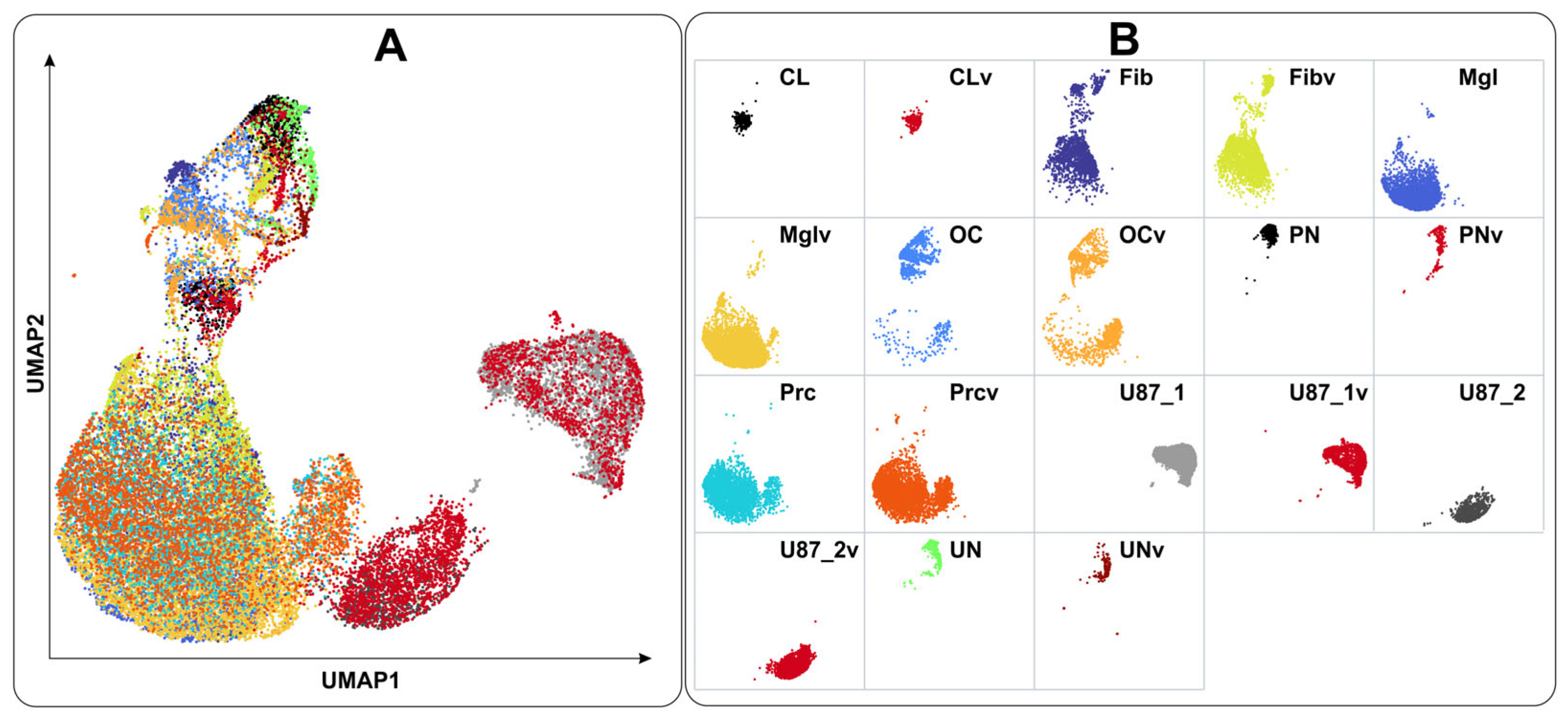
2.2. Cell Types Identified Using Single-Cell Sequencing Data
2.3. General Characteristics of the Response of Human Brain Cells to Infection with the Oncolytic Virus VV-GMCSF-Lact
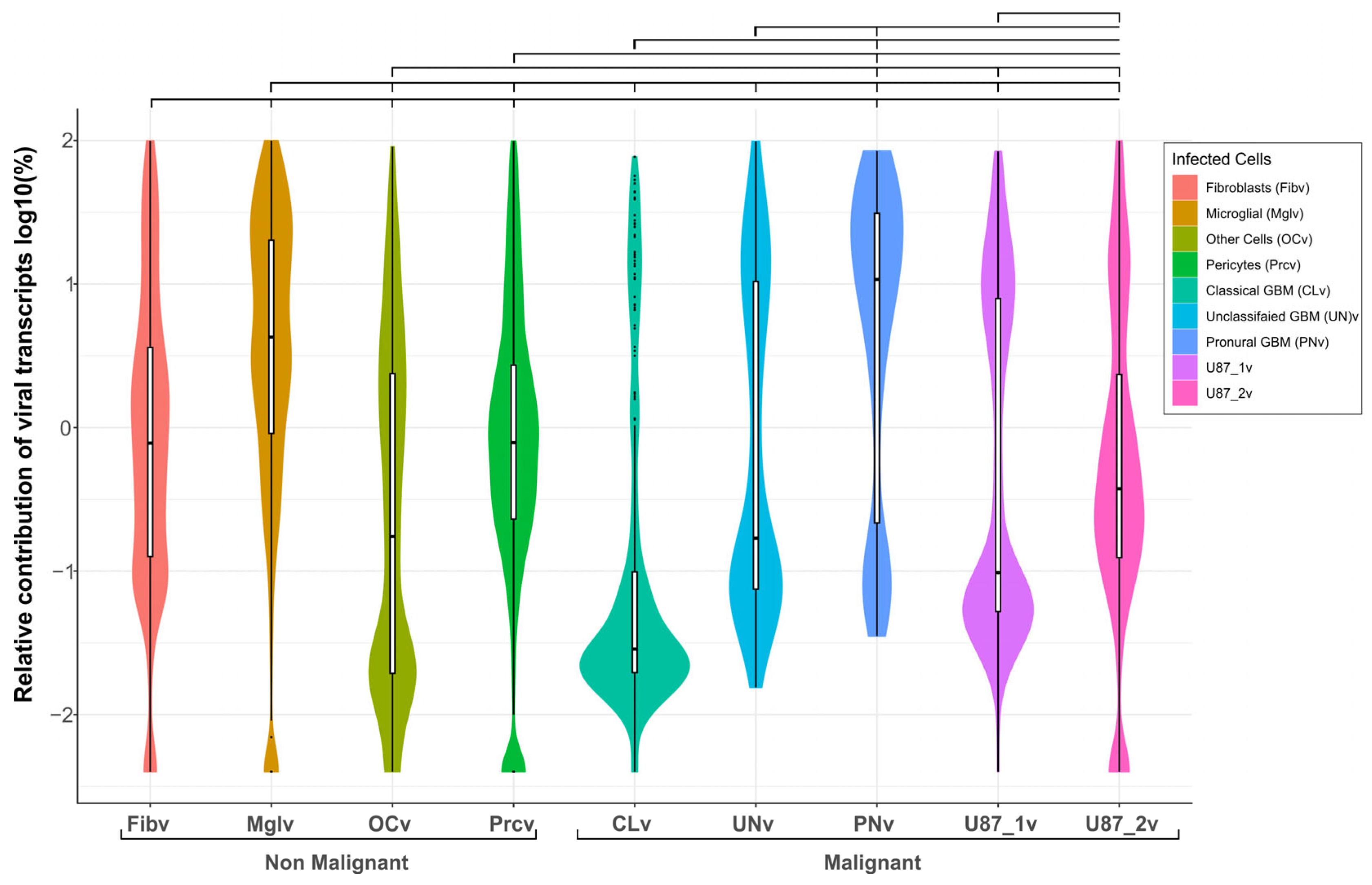
2.4. Viral Transcript Levels Indicate Cell-Type Differences in VV-GMCSF-Lact Susceptibility
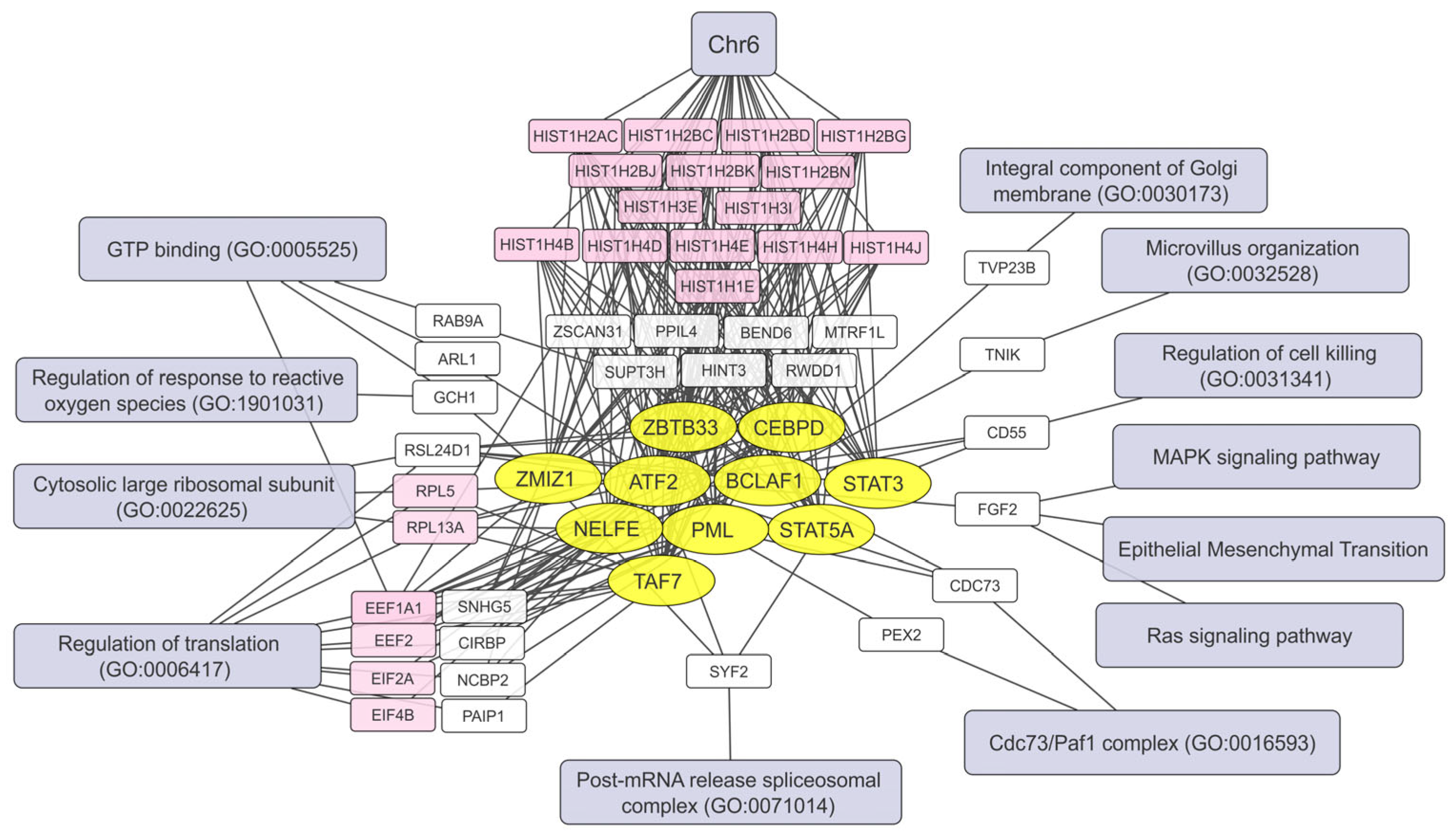
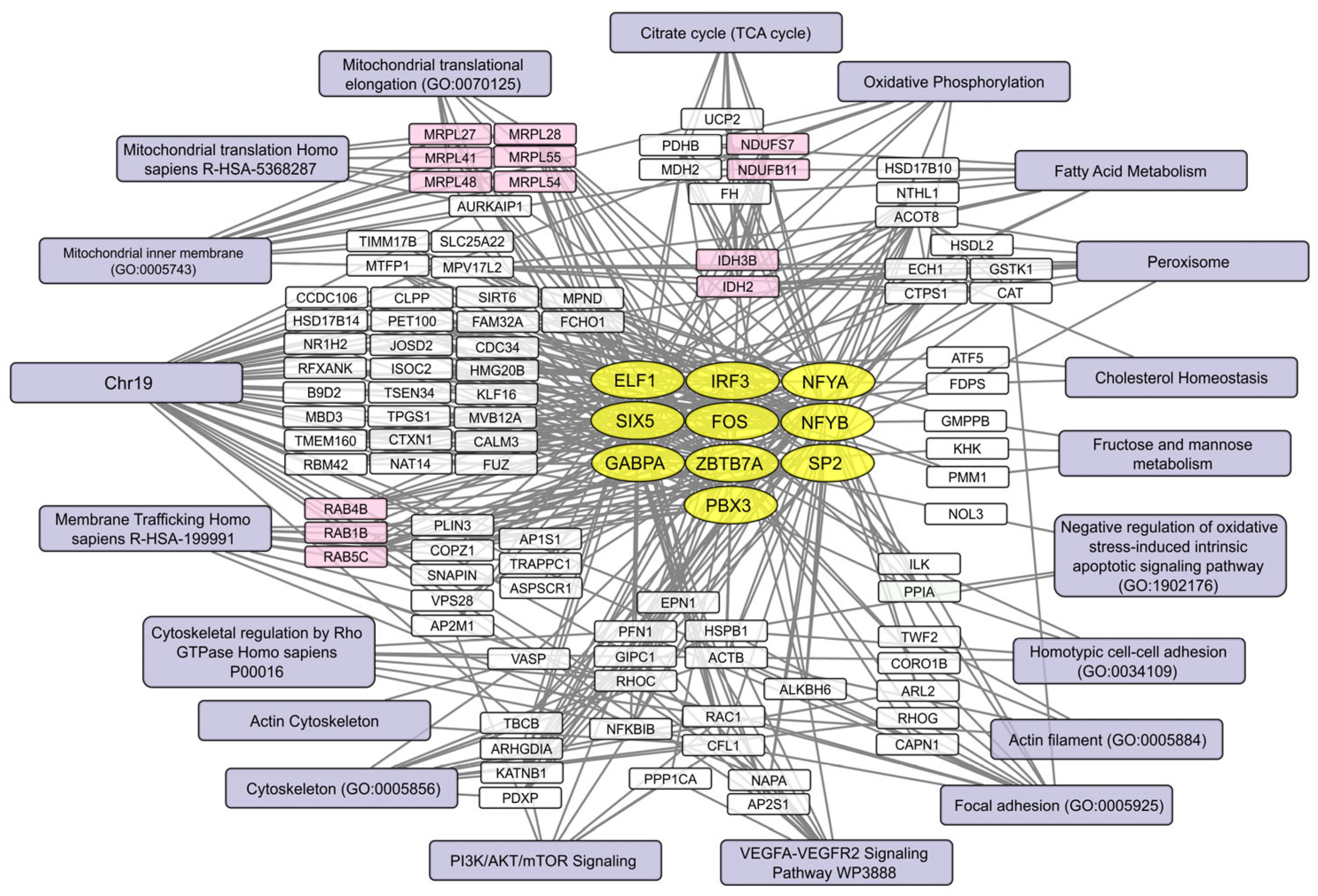
2.5. Human Brain Cell Transcripts Associated with Resistance or Susceptibility to VV-GMCSF-Lact
2.6. Pre-Infection Transcript Signatures Linked to VV-GMCSF-Lact Susceptibility in Human Brain Cells
2.7. VV Early Genes Are the Primary Viral Transcripts Linked to Infection Severity
3. Discussion
4. Materials and Methods
4.1. Immortalized Human Glioblastoma Cell Line U87 MG
4.2. Patient-Derived Cell Cultures
4.3. Oncolytic Virus VV-GMCSF-Lact
4.4. Treatment with VV-GMCSF-Lact for Single-Cell RNA Sequencing
4.5. Single-Cell RNA Sequencing
4.6. Single-Cell Transcriptome Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
Abbreviations
| CD50 | cytotoxic dose |
| CSN2 | kappa-casein gene |
| DEG | differentially expressed gene |
| GBM | glioblastoma, previously referred to as ‘glioblastoma multiforme’ |
| GM-CSF | granulocyte-macrophage colony-stimulating factor (product of the CSF2 gene) |
| HC | hierarchical clustering |
| NB | normal brain tissue cell culture |
| PCA | principal component analysis |
| PFU | plaque-forming unit |
| tk | thymidine kinase |
| U87 | U87 MG cell culture |
| VV | vaccinia virus |
| vgf | virus growth factor |
| VST | variance stabilizing transformation |
References
- Ostrom, Q.T.; Patil, N.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013–2017. Neuro. Oncol. 2020, 22, iv1–iv96, Erratum in Neuro. Oncol. 2022, 24, 1214. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro. Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Agosti, E.; Zeppieri, M.; De Maria, L.; Tedeschi, C.; Fontanella, M.M.; Panciani, P.P.; Ius, T. Glioblastoma Immunotherapy: A Systematic Review of the Present Strategies and Prospects for Advancements. Int. J. Mol. Sci. 2023, 24, 15037. [Google Scholar] [CrossRef] [PubMed]
- Hamad, A.; Yusubalieva, G.M.; Baklaushev, V.P.; Chumakov, P.M.; Lipatova, A.V. Recent Developments in Glioblastoma Therapy: Oncolytic Viruses and Emerging Future Strategies. Viruses 2023, 15, 547. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, K.; Núñez, F.J.; Haase, S.; McClellan, B.L.; Faisal, S.M.; Carney, S.V.; Yu, J.; Alghamri, M.S.; Asad, A.S.; Candia, A.J.N.; et al. Current Approaches for Glioma Gene Therapy and Virotherapy. Front. Mol. Neurosci. 2021, 14, 621831. [Google Scholar] [CrossRef] [PubMed]
- Thorne, S.; Bartlett, D.; Kirn, D. The Use of Oncolytic Vaccinia Viruses in the Treatment of Cancer: A New Role for an Old Ally? Curr. Gene Ther. 2005, 5, 429–443. [Google Scholar] [CrossRef] [PubMed]
- Beerli, C.; Yakimovich, A.; Kilcher, S.; Reynoso, G.V.; Fläschner, G.; Müller, D.J.; Hickman, H.D.; Mercer, J. Vaccinia Virus Hijacks EGFR Signalling to Enhance Virus Spread Through Rapid and Directed Infected Cell Motility. Nat. Microbiol. 2019, 4, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Kochneva, G.; Sivolobova, G.; Tkacheva, A.; Grazhdantseva, A.; Troitskaya, O.; Nushtaeva, A.; Tkachenko, A.; Kuligina, E.; Richter, V.; Koval, O. Engineering of Double Recombinant Vaccinia Virus with Enhanced Oncolytic Potential for Solid Tumor Virotherapy. Oncotarget 2016, 7, 74171–74188. [Google Scholar] [CrossRef] [PubMed]
- Koval, O.A.; Tkachenko, A.V.; Fomin, A.S.; Semenov, D.V.; Nushtaeva, A.A.; Kuligina, E.V.; Zavjalov, E.L.; Richter, V.A. Lactaptin Induces P53-Independent Cell Death Associated with Features of Apoptosis and Autophagy and Delays Growth of Breast Cancer Cells in Mouse Xenografts. PLoS ONE 2014, 9, e93921. [Google Scholar] [CrossRef] [PubMed]
- Semenov, D.V.; Vasileva, N.S.; Dymova, M.A.; Mishinov, S.V.; Savinovskaya, Y.I.; Ageenko, A.B.; Dome, A.S.; Zinchenko, N.D.; Stepanov, G.A.; Kochneva, G.V.; et al. Transcriptome Changes in Glioma Cells upon Infection with the Oncolytic Virus VV-GMCSF-Lact. Cells 2023, 12, 2616. [Google Scholar] [CrossRef] [PubMed]
- Ianevski, A.; Giri, A.K.; Aittokallio, T. Fully-Automated and Ultra-Fast Cell-Type Identification Using Specific Marker Combinations from Single-Cell Transcriptomic Data. Nat. Commun. 2022, 13, 1246. [Google Scholar] [CrossRef] [PubMed]
- Serin Harmanci, A.; Harmanci, A.O.; Zhou, X. CaSpER Identifies and Visualizes CNV Events by Integrative Analysis of Single-Cell or Bulk RNA-Sequencing Data. Nat. Commun. 2020, 11, 89. [Google Scholar] [CrossRef] [PubMed]
- Crespo, I.; Vital, A.L.; Gonzalez-Tablas, M.; Patino, M.D.C.; Otero, A.; Lopes, M.C.; de Oliveira, C.; Domingues, P.; Orfao, A.; Tabernero, M.D. Molecular and Genomic Alterations in Glioblastoma Multiforme. Am. J. Pathol. 2015, 185, 1820–1833. [Google Scholar] [CrossRef] [PubMed]
- Wortzel, I.; Seger, R. The ERK Cascade: Distinct Functions within Various Subcellular Organelles. Genes Cancer 2011, 2, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Kober, C.; Rohn, S.; Weibel, S.; Geissinger, U.; Chen, N.G.; Szalay, A.A. Microglia and Astrocytes Attenuate the Replication of the Oncolytic Vaccinia Virus LIVP 1.1.1 in Murine GL261 Gliomas by Acting as Vaccinia Virus Traps. J. Transl. Med. 2015, 13, 216. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.; Grose, R. Fibroblast Growth Factor Signalling: From Development to Cancer. Nat. Rev. Cancer 2010, 10, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Bharti, R.; Dey, G.; Lin, F.; Lathia, J.; Reizes, O. CD55 in Cancer: Complementing Functions in a Non-Canonical Manner. Cancer Lett. 2022, 551, 215935. [Google Scholar] [CrossRef] [PubMed]
- Yu, E.; Zhai, D.; Jin, C.; Gerlic, M.; Reed, J.C.; Liddington, R. Structural Determinants of Caspase-9 Inhibition by the Vaccinia Virus Protein, F1L. J. Biol. Chem. 2011, 286, 30748–30758. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, S.; Sakiyama, T.; Hasegawa, H.; Saijo, M.; Maeda, A.; Kurane, I.; Maeno, G.; Kimura, J.; Hirama, C.; Yoshida, T.; et al. An Attenuated LC16m8 Smallpox Vaccine: Analysis of Full-Genome Sequence and Induction of Immune Protection. J. Virol. 2005, 79, 11873–11891. [Google Scholar] [CrossRef] [PubMed]
- Maluquer de Motes, C.; Cooray, S.; Ren, H.; Almeida, G.M.F.; McGourty, K.; Bahar, M.W.; Stuart, D.I.; Grimes, J.M.; Graham, S.C.; Smith, G.L. Inhibition of Apoptosis and NF-ΚB Activation by Vaccinia Protein N1 Occur via Distinct Binding Surfaces and Make Different Contributions to Virulence. PLoS Pathog. 2011, 7, e1002430. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, W.P.; Tarbouriech, N.; Fender, P.; Contesto-Richefeu, C.; Peyrefitte, C.N.; Iseni, F. Crystal Structure of the Vaccinia Virus Uracil-DNA Glycosylase in Complex with DNA. J. Biol. Chem. 2015, 290, 17923–17934. [Google Scholar] [CrossRef] [PubMed]
- Senkevich, T.G.; Koonin, E.V.; Moss, B. Vaccinia Virus F16 Protein, a Predicted Catalytically Inactive Member of the Prokaryotic Serine Recombinase Superfamily, Is Targeted to Nucleoli. Virology 2011, 417, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Kotwal, G.J.; Moss, B. Vaccinia Virus Encodes Two Proteins That Are Structurally Related to Members of the Plasma Serine Protease Inhibitor Superfamily. J. Virol. 1989, 63, 600–606, Erratum in J. Virol. 1990, 64, 966. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, J.V.; Guerra, S.; Arakawa, Y.; Dodding, M.P.; Esteban, M.; Way, M. F11-Mediated Inhibition of RhoA Signalling Enhances the Spread of Vaccinia Virus in Vitro and in Vivo in an Intranasal Mouse Model of Infection. PLoS ONE 2009, 4, e8506, Erratum in PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- Qin, L.; Upton, C.; Hazes, B.; Evans, D.H. Genomic Analysis of the Vaccinia Virus Strain Variants Found in Dryvax Vaccine. J. Virol. 2011, 85, 13049–13060. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.F.; Kahn, J.S.; Esteban, M. Molecular Cloning, Encoding Sequence, and Expression of Vaccinia Virus Nucleic Acid-Dependent Nucleoside Triphosphatase Gene. Proc. Natl. Acad. Sci. USA 1986, 83, 9566–9570. [Google Scholar] [CrossRef] [PubMed]
- Reed, B.; Yakovleva, L.; Shuman, S.; Ghose, R. Characterization of DNA Binding by the Isolated N-Terminal Domain of Vaccinia Virus DNA Topoisomerase IB. Biochemistry 2017, 56, 3307–3317. [Google Scholar] [CrossRef] [PubMed]
- Plucienniczak, A.; Schroeder, E.; Zettlmeissl, G.; Streeck, R.E. Nucleotide Sequence of a Cluster of Early and Late Genes in a Conserved Segment of the Vaccinia Virus Genome. Nucleic Acids Res. 1985, 13, 985–998. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Tian, M.; Feng, Y.; Zhao, K.; Xu, J.; Liu, Y.; Shao, Y. Genomic Sequence and Virulence of Clonal Isolates of Vaccinia Virus Tiantan, the Chinese Smallpox Vaccine Strain. PLoS ONE 2013, 8, e60557. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Shen, Y.; Liang, T. Oncolytic Virotherapy: Basic Principles, Recent Advances and Future Directions. Signal Transduct. Target. Ther. 2023, 8, 156. [Google Scholar] [CrossRef] [PubMed]
- Greseth, M.D.; Traktman, P. The Life Cycle of the Vaccinia Virus Genome. Annu. Rev. Virol. 2022, 9, 239–259. [Google Scholar] [CrossRef] [PubMed]
- Condit, R.C.; Moussatche, N.; Traktman, P. In a Nutshell: Structure and Assembly of the Vaccinia Virion. Adv. Virus Res. 2006, 66, 31–124. [Google Scholar] [CrossRef] [PubMed]
- Townsley, A.C.; Weisberg, A.S.; Wagenaar, T.R.; Moss, B. Vaccinia Virus Entry into Cells via a Low-PH-Dependent Endosomal Pathway. J. Virol. 2006, 80, 8899–8908. [Google Scholar] [CrossRef] [PubMed]
- Moss, B. Poxvirus DNA Replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010199. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Bruno, D.P.; Martens, C.A.; Porcella, S.F.; Moss, B. Simultaneous High-Resolution Analysis of Vaccinia Virus and Host Cell Transcriptomes by Deep RNA Sequencing. Proc. Natl. Acad. Sci. USA 2010, 107, 11513–11518. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; DeCarvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 32, 42–56.e6, Erratum in Cancer Cell 2018, 33, 152. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.; Yan, W.; Gao, K.; Wang, Y.; Zhang, J.; You, Y. Prevalence and Clinicopathologic Characteristics of the Molecular Subtypes in Malignant Glioma: A Multi-Institutional Analysis of 941 Cases. PLoS ONE 2014, 9, e94871. [Google Scholar] [CrossRef] [PubMed]
- Delwar, Z.M.; Kuo, Y.; Wen, Y.H.; Rennie, P.S.; Jia, W. Oncolytic Virotherapy Blockade by Microglia and Macrophages Requires STAT1/3. Cancer Res. 2018, 78, 718–730. [Google Scholar] [CrossRef] [PubMed]
- Hughes, L.; Wilkins, K.; Goldsmith, C.S.; Smith, S.; Hudson, P.; Patel, N.; Karem, K.; Damon, I.; Li, Y.; Olson, V.A.; et al. A Rapid Orthopoxvirus Purification Protocol Suitable for High-Containment Laboratories. J. Virol. Methods 2017, 243, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Stuart, T.; Kowalski, M.H.; Choudhary, S.; Hoffman, P.; Hartman, A.; Srivastava, A.; Molla, G.; Madad, S.; Fernandez-Granda, C.; et al. Dictionary Learning for Integrative, Multimodal and Scalable Single-Cell Analysis. Nat. Biotechnol. 2024, 42, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lan, Y.; Xu, J.; Quan, F.; Zhao, E.; Deng, C.; Luo, T.; Xu, L.; Liao, G.; Yan, M.; et al. CellMarker: A Manually Curated Resource of Cell Markers in Human and Mouse. Nucleic Acids Res. 2019, 47, D721–D728. [Google Scholar] [CrossRef] [PubMed]
- Franzén, O.; Gan, L.-M.; Björkegren, J.L.M. PanglaoDB: A Web Server for Exploration of Mouse and Human Single-Cell RNA Sequencing Data. Database 2019, 2019, baz046. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
| Cell Culture | Gender | Age (Years) | Diagnosis | IDH1 * | Ref ** |
|---|---|---|---|---|---|
| HB1 | female | 32 | Gliosis | – *** | – |
| HB3 | male | 34 | GBM | wt | – |
| HB4 | female | 47 | GBM | wt | – |
| NB | female | 74 | – | – | [10] |
| U87 | male | 44 | GBM | wt | [10] |
| Vir Gene | Pearson Correlation Coefficient (1) | Temporal Expression (2) | Preferred Name (2). Annotation | Ref |
|---|---|---|---|---|
| F1L | 0.68 | Early | Hypothetical protein. The large subunit of mRNA capping enzyme. Transcription termination factor | [18] |
| F10L | 0.67 | Late | Ser/Thr kinase | [19] |
| C4L | 0.66 | Early/Late | Hypothetical protein | – (3) |
| N1L | 0.66 | Early/Late | Putative virulence factor. N1 inhibits both apoptosis and NF-κB activation | [20] |
| D4R | 0.64 | Early | Temporal expression. Uracil–DNA glycosylase | [21] |
| E4L | 0.63 | Early | RNA polymerase subunit | – |
| J4R | 0.63 | Early | DNA-dependent RNA polymerase subunit rpo22 | – |
| F16L | 0.61 | – | Hypothetical protein. Recombinase superfamily | [22] |
| N2L | 0.59 | Early | Putative alpha aminitin-sensitive protein | – |
| D1R | 0.59 | Early | Large subunit of mRNA capping enzyme | – |
| F14.5L | 0.58 | – | Hypothetical protein | – |
| 174 | 0.58 | – | – | – |
| C12L | 0.58 | Early | Serine protease inhibitor-like SPI-1 | [23] |
| F11L | 0.57 | – | Hypothetical protein. Inhibition of RhoA signaling | [24] |
| 201 | 0.57 | – | – | – |
| D7R | 0.57 | Early | DNA-dependent RNA polymerase subunit rpo18 | [25] |
| D11L | 0.57 | Late | ATPase, nucleoside triphosphate phosphohydrolase-I, NPH-I. Similar to VACCP-D11L; transcription elongation, termination, release factor | [26] |
| H6R | 0.56 | – | DNA topoisomerase type I | [27] |
| J3R | 0.56 | Early | Poly(A) polymerase small subunit | [28] |
| K6L | 0.56 | – | Putative monoglyceride lipase | [29] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Semenov, D.V.; Vasileva, N.S.; Menyailo, M.E.; Mishinov, S.V.; Savinovskaya, Y.I.; Ageenko, A.B.; Chesnokova, A.S.; Dymova, M.A.; Stepanov, G.A.; Kochneva, G.V.; et al. Single-Cell Transcriptomic Changes in Patient-Derived Glioma and U87 Glioblastoma Cell Cultures Infected with the Oncolytic Virus VV-GMCSF-Lact. Int. J. Mol. Sci. 2025, 26, 6983. https://doi.org/10.3390/ijms26146983
Semenov DV, Vasileva NS, Menyailo ME, Mishinov SV, Savinovskaya YI, Ageenko AB, Chesnokova AS, Dymova MA, Stepanov GA, Kochneva GV, et al. Single-Cell Transcriptomic Changes in Patient-Derived Glioma and U87 Glioblastoma Cell Cultures Infected with the Oncolytic Virus VV-GMCSF-Lact. International Journal of Molecular Sciences. 2025; 26(14):6983. https://doi.org/10.3390/ijms26146983
Chicago/Turabian StyleSemenov, Dmitriy V., Natalia S. Vasileva, Maxim E. Menyailo, Sergey V. Mishinov, Yulya I. Savinovskaya, Alisa B. Ageenko, Anna S. Chesnokova, Maya A. Dymova, Grigory A. Stepanov, Galina V. Kochneva, and et al. 2025. "Single-Cell Transcriptomic Changes in Patient-Derived Glioma and U87 Glioblastoma Cell Cultures Infected with the Oncolytic Virus VV-GMCSF-Lact" International Journal of Molecular Sciences 26, no. 14: 6983. https://doi.org/10.3390/ijms26146983
APA StyleSemenov, D. V., Vasileva, N. S., Menyailo, M. E., Mishinov, S. V., Savinovskaya, Y. I., Ageenko, A. B., Chesnokova, A. S., Dymova, M. A., Stepanov, G. A., Kochneva, G. V., Richter, V. A., & Kuligina, E. V. (2025). Single-Cell Transcriptomic Changes in Patient-Derived Glioma and U87 Glioblastoma Cell Cultures Infected with the Oncolytic Virus VV-GMCSF-Lact. International Journal of Molecular Sciences, 26(14), 6983. https://doi.org/10.3390/ijms26146983





