Disruption Events in the HPV18 E1 and E2 Genes in Precancerous Cervical Lesions
Abstract
1. Introduction
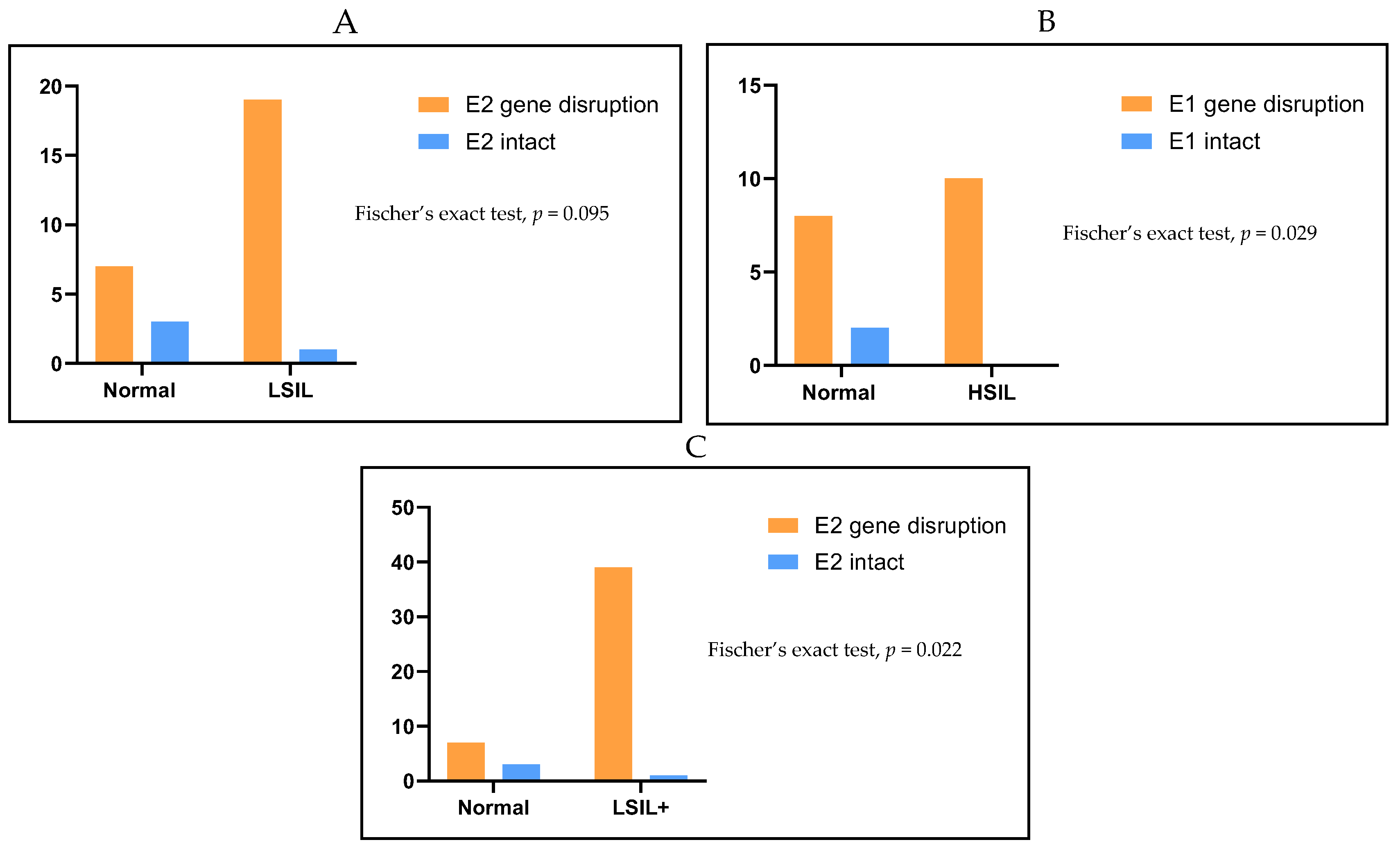
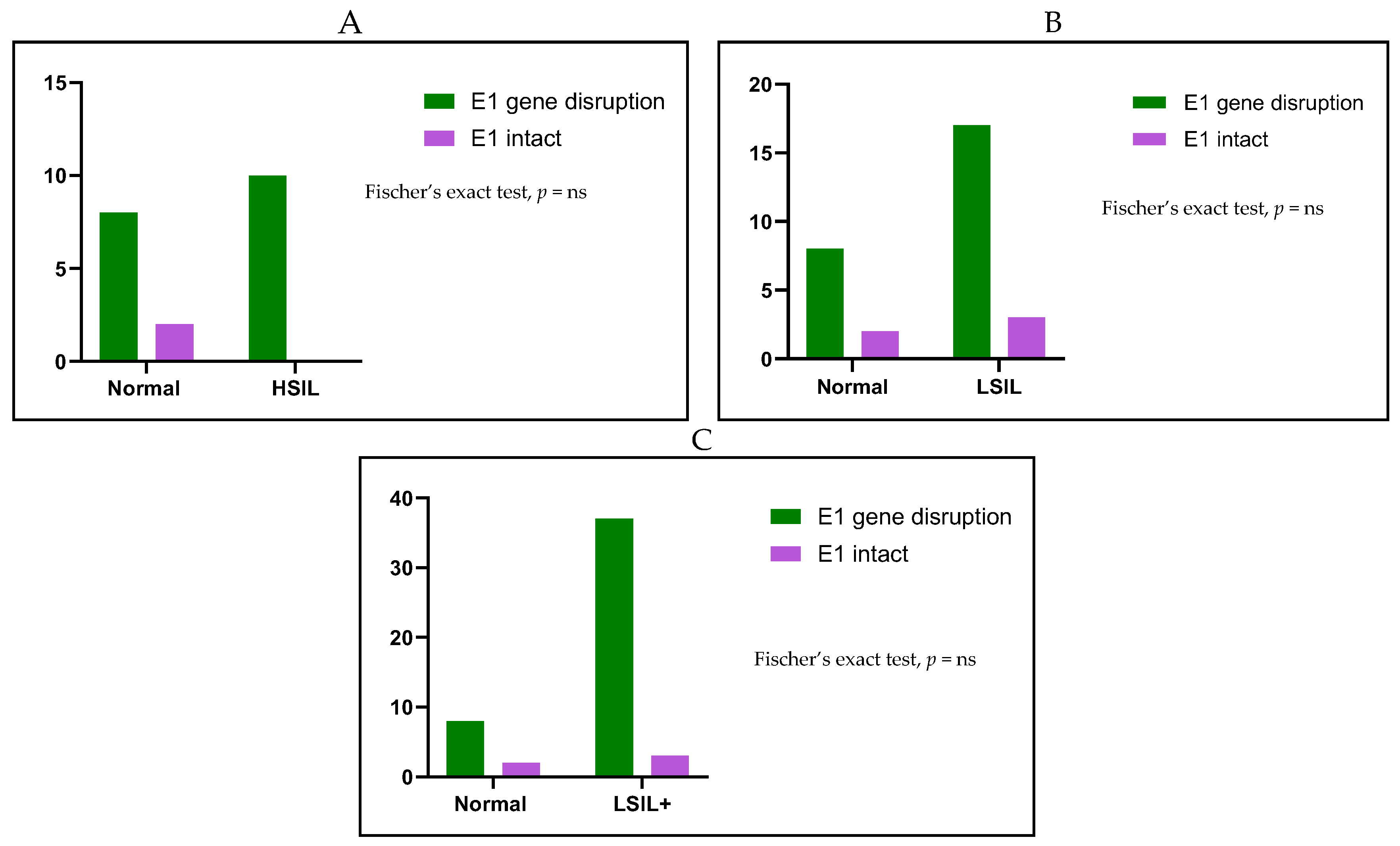
2. Results
3. Discussion
4. Materials and Methods
4.1. Cervical Samples
4.2. DNA Isolation and HPV Genotyping
4.3. Amplification of HPV18 E6 and E7 Genes
4.4. Primer Validation
4.5. PCR Amplification of HPV18 Ε1 Gene and HPV18 Ε2 Gene
4.6. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- zur Hausen, H. Papillomavirus infections—A major cause of human cancers. Biochim. Biophys. Acta 1996, 1288, F55–F78. [Google Scholar] [CrossRef] [PubMed]
- Van Doorslaer, K.; Chen, Z.; Bernard, H.U.; Chan, P.K.S.; DeSalle, R.; Dillner, J.; Forslund, O.; Haga, T.; McBride, A.A.; Villa, L.L.; et al. ICTV Virus Taxonomy Profile: Papillomaviridae. J. Gen. Virol. 2018, 99, 989–990. [Google Scholar] [CrossRef] [PubMed]
- Bernard, H.U.; Burk, R.D.; Chen, Z.; van Doorslaer, K.; zur Hausen, H.; de Villiers, E.M. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 2010, 401, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Bernard, H.U.; Calleja-Macias, I.E.; Dunn, S.T. Genome variation of human papillomavirus types: Phylogenetic and medical implications. Int. J. Cancer 2006, 118, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Bletsa, G.; Zagouri, F.; Amoutzias, G.D.; Nikolaidis, M.; Zografos, E.; Markoulatos, P.; Tsakogiannis, D. Genetic variability of the HPV16 early genes and LCR. Present and future perspectives. Expert Rev. Mol. Med. 2021, 23, e19. [Google Scholar] [CrossRef] [PubMed]
- Tsakogiannis, D.; Darmis, F.; Gortsilas, P.; Ruether, I.G.; Kyriakopoulou, Z.; Dimitriou, T.G.; Amoutzias, G.; Markoulatos, P. Nucleotide polymorphisms of the human papillomavirus 16 E1 gene. Arch. Virol. 2014, 159, 51–63. [Google Scholar] [CrossRef] [PubMed]
- de Villiers, E.M.; Fauquet, C.; Broker, T.R.; Bernard, H.U.; zur Hausen, H. Classification of papillomaviruses. Virology 2004, 324, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Mirabello, L.; Clarke, M.A.; Nelson, C.W.; Dean, M.; Wentzensen, N.; Yeager, M.; Cullen, M.; Boland, J.F.; Workshop, N.H.; Schiffman, M.; et al. The Intersection of HPV Epidemiology, Genomics and Mechanistic Studies of HPV-Mediated Carcinogenesis. Viruses 2018, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, C. Human Papillomavirus-Related Cancers. Adv. Exp. Med. Biol. 2017, 1018, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Bruni, L.A.G.; Serrano, B.; Mena, M.; Collado, J.J.; Gómez, D.; Muñoz, J.; Bosch, F.X.; de Sanjosé, S.; ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre). Human Papillomavirus and Related Diseases in the World. 2023. Available online: https://hpvcentre.net/statistics/reports/XWX.pdf (accessed on 15 September 2023).
- Tsakogiannis, D.; Zografos, E.; Tzioga, L.; Zografos, C.G.; Zagouri, F.; Bletsa, G. Prevalence and Genotype Distribution of High-Risk HPV Genotypes Among Women in Greece: A Retrospective Analysis of 3500 Women. Cancers 2025, 17, 1267. [Google Scholar] [CrossRef] [PubMed]
- Zygouras, I.; Leventakou, D.; Pouliakis, A.; Panagiotou, S.; Tsakogiannis, D.; Konstantopoulos, G.; Logotheti, E.; Samaras, M.; Kyriakopoulou, Z.; Beloukas, A.; et al. Human Papillomavirus 16 DNA Methylation Patterns and Investigation of Integration Status in Head and Neck Cancer Cases. Int. J. Mol. Sci. 2023, 24, 14593. [Google Scholar] [CrossRef] [PubMed]
- Prati, B.; Marangoni, B.; Boccardo, E. Human papillomavirus and genome instability: From productive infection to cancer. Clinics 2018, 73, e539s. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Sun, Q.; Tian, Q.; Shi, H.; Yang, H.; Ren, J. Human papillomavirus 16 E6/E7 contributes to immune escape and progression of cervical cancer by regulating miR-142-5p/PD-L1 axis. Arch. Biochem. Biophys. 2022, 731, 109449. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Zapien, D.; Ruiz, F.X.; Poirson, J.; Mitschler, A.; Ramirez, J.; Forster, A.; Cousido-Siah, A.; Masson, M.; Vande Pol, S.; Podjarny, A.; et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature 2016, 529, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Olmedo-Nieva, L.; Munoz-Bello, J.O.; Contreras-Paredes, A.; Lizano, M. The Role of E6 Spliced Isoforms (E6*) in Human Papillomavirus-Induced Carcinogenesis. Viruses 2018, 10, 45. [Google Scholar] [CrossRef] [PubMed]
- Dick, F.A.; Goodrich, D.W.; Sage, J.; Dyson, N.J. Non-canonical functions of the RB protein in cancer. Nat. Rev. Cancer 2018, 18, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Kundu, R. Human Papillomavirus E6 and E7: The Cervical Cancer Hallmarks and Targets for Therapy. Front. Microbiol. 2019, 10, 3116. [Google Scholar] [CrossRef] [PubMed]
- Akagi, K.; Li, J.; Broutian, T.R.; Padilla-Nash, H.; Xiao, W.; Jiang, B.; Rocco, J.W.; Teknos, T.N.; Kumar, B.; Wangsa, D.; et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2014, 24, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Albert Einstein College of Medicine; Analytical Biological Services; Barretos Cancer Hospital; Baylor College of Medicine; Beckman Research Institute of City of Hope; Buck Institute for Research on Aging; Canada’s Michael Smith Genome Sciences Centre; Harvard Medical School; Helen F. Graham Cancer Center & Research Institute at Christiana Care Health Services; et al. Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Tsakogiannis, D.; Gartzonika, C.; Levidiotou-Stefanou, S.; Markoulatos, P. Molecular approaches for HPV genotyping and HPV-DNA physical status. Expert Rev. Mol. Med. 2017, 19, e1. [Google Scholar] [CrossRef] [PubMed]
- Tsakogiannis, D.; Gortsilas, P.; Kyriakopoulou, Z.; Ruether, I.G.; Dimitriou, T.G.; Orfanoudakis, G.; Markoulatos, P. Sites of disruption within E1 and E2 genes of HPV16 and association with cervical dysplasia. J. Med. Virol. 2015, 87, 1973–1980. [Google Scholar] [CrossRef] [PubMed]
- Cricca, M.; Venturoli, S.; Leo, E.; Costa, S.; Musiani, M.; Zerbini, M. Disruption of HPV 16 E1 and E2 genes in precancerous cervical lesions. J. Virol. Methods 2009, 158, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Ehrig, F.; Hafner, N.; Driesch, C.; Kraus Christiansen, I.; Beer, K.; Schmitz, M.; Runnebaum, I.B.; Durst, M. Differences in Stability of Viral and Viral-Cellular Fusion Transcripts in HPV-Induced Cervical Cancers. Int. J. Mol. Sci. 2019, 21, 112. [Google Scholar] [CrossRef] [PubMed]
- Oyervides-Munoz, M.A.; Perez-Maya, A.A.; Rodriguez-Gutierrez, H.F.; Gomez-Macias, G.S.; Fajardo-Ramirez, O.R.; Trevino, V.; Barrera-Saldana, H.A.; Garza-Rodriguez, M.L. Understanding the HPV integration and its progression to cervical cancer. Infect. Genet. Evol. 2018, 61, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Tsakogiannis, D.; Kyriakopoulou, Z.; Ruether, I.G.A.; Amoutzias, G.D.; Dimitriou, T.G.; Diamantidou, V.; Kotsovassilis, C.; Markoulatos, P. Determination of human papillomavirus 16 physical status through E1/E6 and E2/E6 ratio analysis. J. Med. Microbiol. 2014, 63, 1716–1723. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Weng, D.; Chen, Y.; Wang, Y.; Li, X.; Wang, X.; Cao, C.; Gong, D.; Zeng, Z.; Wu, Q.; et al. Risk Assessment and Triage Strategy of Cervical Cancer Primary Screening On HPV Integration Status. J. Natl. Cancer Cent. 2024, 4, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Vinokurova, S.; Wentzensen, N.; Kraus, I.; Klaes, R.; Driesch, C.; Melsheimer, P.; Kisseljov, F.; Durst, M.; Schneider, A.; von Knebel Doeberitz, M. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Res. 2008, 68, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Lagstrom, S.; Lovestad, A.H.; Umu, S.U.; Ambur, O.H.; Nygard, M.; Rounge, T.B.; Christiansen, I.K. HPV16 and HPV18 type-specific APOBEC3 and integration profiles in different diagnostic categories of cervical samples. Tumour Virus Res. 2021, 12, 200221. [Google Scholar] [CrossRef] [PubMed]
- Kamal, M.; Lameiras, S.; Deloger, M.; Morel, A.; Vacher, S.; Lecerf, C.; Dupain, C.; Jeannot, E.; Girard, E.; Baulande, S.; et al. Human papilloma virus (HPV) integration signature in Cervical Cancer: Identification of MACROD2 gene as HPV hot spot integration site. Br. J. Cancer 2021, 124, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Cheung, J.L.; Cheung, T.H.; Ng, C.W.; Yu, M.Y.; Wong, M.C.; Siu, S.S.; Yim, S.F.; Chan, P.K. Analysis of human papillomavirus type 18 load and integration status from low-grade cervical lesion to invasive cervical cancer. J. Clin. Microbiol. 2009, 47, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qiu, K.; Ren, J.; Zhao, Y.; Cheng, P. Roles of human papillomavirus in cancers: Oncogenic mechanisms and clinical use. Signal Transduct. Target. Ther. 2025, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- Baba, S.; Taguchi, A.; Kawata, A.; Hara, K.; Eguchi, S.; Mori, M.; Adachi, K.; Mori, S.; Iwata, T.; Mitsuhashi, A.; et al. Differential expression of human papillomavirus 16-, 18-, 52-, and 58-derived transcripts in cervical intraepithelial neoplasia. Virol. J. 2020, 17, 32. [Google Scholar] [CrossRef] [PubMed]
- Reidy, P.M.; Dedo, H.H.; Rabah, R.; Field, J.B.; Mathog, R.H.; Gregoire, L.; Lancaster, W.D. Integration of human papillomavirus type 11 in recurrent respiratory papilloma-associated cancer. Laryngoscope 2004, 114, 1906–1909. [Google Scholar] [CrossRef] [PubMed]
- Egawa, N.; Doorbar, J. The low-risk papillomaviruses. Virus Res. 2017, 231, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Donne, A.J.; Hampson, L.; Homer, J.J.; Hampson, I.N. The role of HPV type in Recurrent Respiratory Papillomatosis. Int. J. Pediatr. Otorhinolaryngol. 2010, 74, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Porter, V.L.; Marra, M.A. The Drivers, Mechanisms, and Consequences of Genome Instability in HPV-Driven Cancers. Cancers 2022, 14, 4623. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.I.; Constandinou-Williams, C.; Wen, K.; Young, L.S.; Roberts, S.; Murray, P.G.; Woodman, C.B. Disruption of the E2 gene is a common and early event in the natural history of cervical human papillomavirus infection: A longitudinal cohort study. Cancer Res. 2009, 69, 3828–3832. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.W.; Chao, S.L.; Lee, B.H. Integration of human papillomavirus type-16 and type-18 is a very early event in cervical carcinogenesis. J. Clin. Pathol. 2008, 61, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, Y.; Zhang, R.; Cai, Y.; Yang, X.; Wang, Z.; Li, Y.; Cheng, X.; Ye, X.; Xiang, Y.; et al. Preferential sites for the integration and disruption of human papillomavirus 16 in cervical lesions. J. Clin. Virol. 2013, 56, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Amaro-Filho, S.M.; Pereira Chaves, C.B.; Felix, S.P.; Basto, D.L.; de Almeida, L.M.; Moreira, M.A.M. HPV DNA methylation at the early promoter and E1/E2 integrity: A comparison between HPV16, HPV18 and HPV45 in cervical cancer. Papillomavirus Res. 2018, 5, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Casas, I.; Powell, L.; Klapper, P.E.; Cleator, G.M. New method for the extraction of viral RNA and DNA from cerebrospinal fluid for use in the polymerase chain reaction assay. J. Virol. Methods 1995, 53, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Daskou, M.; Tsakogiannis, D.; Dimitriou, T.G.; Manali, M.; Apti, C.; Amoutzias, G.D.; Mossialos, D.; Kottaridi, C.; Markoulatos, P. Alpha 2-stage, nested-like nucleic acid amplification method (IsoPCR) for the highly sensitive detection of HPV16 and HPV18 DNA. Mol. Cell. Probes 2019, 45, 1–7. [Google Scholar] [CrossRef] [PubMed]
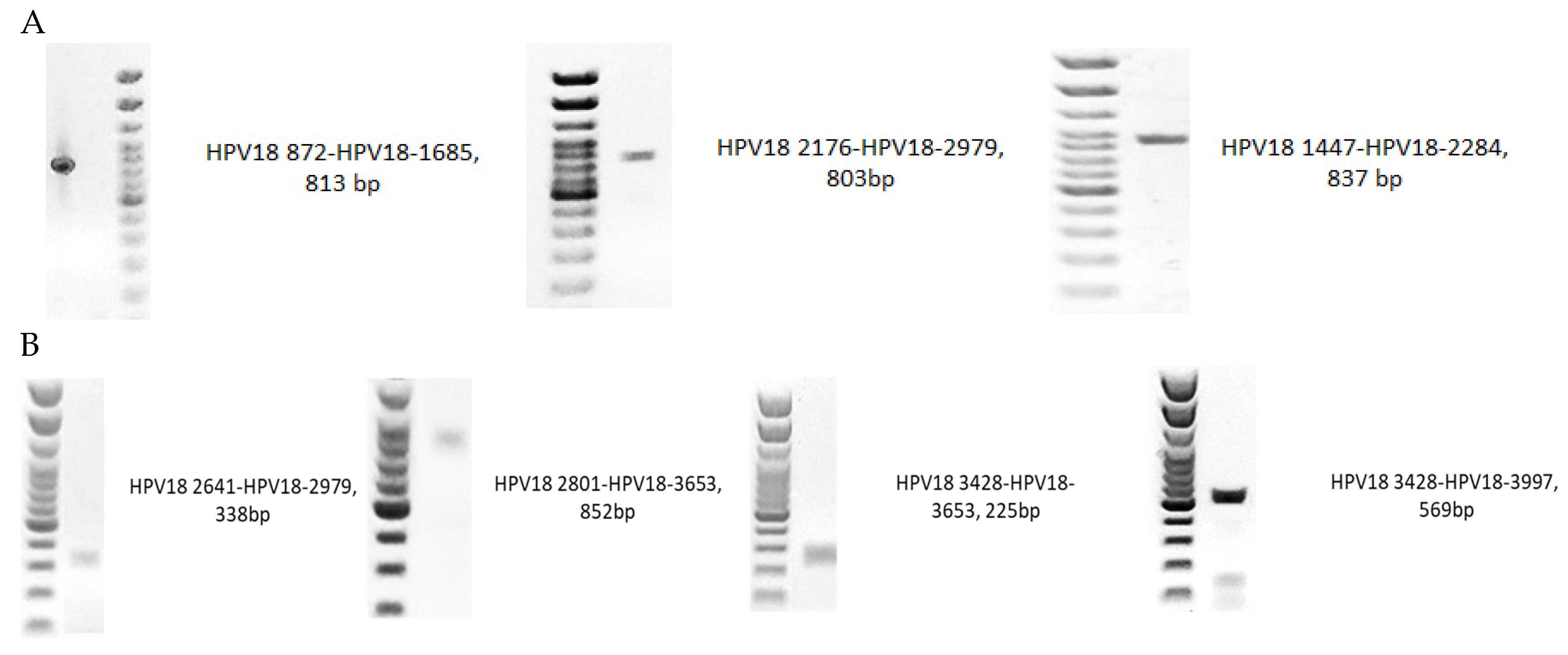


| HPV18 | Primers | Tm (°C) | PCR Product Length (bp) | Position (nt) | Sequence (5′–3′) |
|---|---|---|---|---|---|
| E6 and E7 | HPV18-31 | 57 | 857 | 31 | AAAAGGGAGTAACCGAAAACG |
| HPV18-888 | 60 | 888 | CACGGACACACAAAGGACAG | ||
| E1 | HPV18-872 | 60 | 813 | 872 | GTCCTTTGTGTGTCCGTGGT |
| HPV18-1685 | 58 | 1685 | TCCTTCTGCTATTGTTGGGTTT | ||
| HPV18-1447 | 62 | 837 | 1447 | GCAGTGTAGACGGTACAAGTGA | |
| HPV18-2284 | 56 | 2284 | TGTTGGTATCGCAGGAATTG | ||
| HPV18-2176 | 54 | 803 | 2176 | AGCCCAAAAACGACAAATGA | |
| HPV18-2979 | 56 | 2979 | TGCCATGTTCCCTTGCTG | ||
| HPV18-2641 | 58 | 338 | 2641 | TCCAGCAAAGGATAATAGATGG | |
| HPV18-2979 | 56 | 2979 | TGCCATGTTCCCTTGCTG | ||
| E2 | HPV18-2801 | 57 | 852 | 2801 | GCACGAGGAAGAGAAGATG |
| HPV18-3653 | 56 | 3653 | CGTCTTTTGTTGTTGCCTGT | ||
| HPV18-3428 | 60 | 225 | 3428 | TGTGCAGTACCAGTGACGAC | |
| HPV18-3653 | 56 | 3653 | CGTCTTTTGTTGTTGCCTGT | ||
| HPV18-3428 | 60 | 569 | 3428 | TGTGCAGTACCAGTGACGAC | |
| HPV18-3997 | 60 | 3997 | GGACATGGCAGCACACATAC |
| E1 Gene Disruption | E1 Intact | Total | |
| n (%) | n (%) | n (%) | |
| Normal | 8 (80) | 2 (20) | 10 (100) |
| LSIL | 17 (85) | 3 (15) | 20 (100) |
| HSIL | 20 (100) | 0 (0) | 10 (100) |
| Total | 45 (90) | 5 (10) | 50 (100) |
| E2 Gene Disruption | E2 Intact | Total | |
| n (%) | n (%) | n (%) | |
| Normal | 7 (70) | 3 (30) | 10 (100) |
| LSIL | 19 (95) | 1 (5) | 20 (100) |
| HSIL | 20 (100) | 0 (0) | 20 (100) |
| Total | 46 (92) | 4 (8) | 50 (100) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agnanti, E.; Tsakogiannis, D.; Papadopoulos, T.; Arvanitidis, K.I.; Kyriakopoulou, Z.; Karakasiliotis, I.; Kottaridi, C. Disruption Events in the HPV18 E1 and E2 Genes in Precancerous Cervical Lesions. Int. J. Mol. Sci. 2025, 26, 6974. https://doi.org/10.3390/ijms26146974
Agnanti E, Tsakogiannis D, Papadopoulos T, Arvanitidis KI, Kyriakopoulou Z, Karakasiliotis I, Kottaridi C. Disruption Events in the HPV18 E1 and E2 Genes in Precancerous Cervical Lesions. International Journal of Molecular Sciences. 2025; 26(14):6974. https://doi.org/10.3390/ijms26146974
Chicago/Turabian StyleAgnanti, Eirini, Dimitris Tsakogiannis, Theologos Papadopoulos, Konstantinos I. Arvanitidis, Zaharoula Kyriakopoulou, Ioannis Karakasiliotis, and Christine Kottaridi. 2025. "Disruption Events in the HPV18 E1 and E2 Genes in Precancerous Cervical Lesions" International Journal of Molecular Sciences 26, no. 14: 6974. https://doi.org/10.3390/ijms26146974
APA StyleAgnanti, E., Tsakogiannis, D., Papadopoulos, T., Arvanitidis, K. I., Kyriakopoulou, Z., Karakasiliotis, I., & Kottaridi, C. (2025). Disruption Events in the HPV18 E1 and E2 Genes in Precancerous Cervical Lesions. International Journal of Molecular Sciences, 26(14), 6974. https://doi.org/10.3390/ijms26146974







