APOE Genotype-Stratified Meta-Analysis of Cognitive Decline Reveals Novel Loci for Language and Global Cognitive Function in Older Adults
Abstract
1. Introduction
2. Results
2.1. Gene-Based Analysis
2.2. Gene Mapping
2.3. Gene Set Enrichment Analysis
2.4. Cognitive Ability and AD
2.5. Comparison with AD-Associated SNPs
3. Discussion
4. Materials and Methods
4.1. Study Cohorts
4.2. Cognitive Assessment
4.3. Cognitive Decline Slope
4.4. Genotyping, Imputation, and Quality Control
4.5. Genome-Wide Association and Meta-Analysis
4.6. Genomic Risk Locus Characterization
4.7. Gene Mapping
4.8. Gene-Based Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s Disease |
| APOE | Apolipoprotein E |
| CDR | Clinical Dementia Rating |
| MYHAT | The Monongahela-Youghiogheny Healthy Aging Team |
| MoVIES | Monongahela Valley Independent Elders Survey |
| GEM | Gingko Evaluation of Memory |
| LD | Linear Disequilibrium |
| MAF | Minor Allele Frequency |
References
- Lindenberger, U. Human cognitive aging: Corriger la fortune? Science 2014, 346, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Mcclearn, G.E. Substantial Genetic Influence on Cognitive Abilities in Twins 80 or More Years Old. Science 1997, 276, 1560–1563. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.A.; Finkel, D. A Meta-analysis of Heritability of Cognitive Aging: Minding the “Missing Heritability” Gap. Neuropsychol. Rev. 2015, 25, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Swan, G.E.; Carmelli, D.; Reed, T.; Harshfield, G.A.; Fabsitz, R.R.; Eslinger, P.J. Heritability of Cognitive Performance in Aging Twins The National Heart, Lung, and Blood Institute Twin Study. Arch. Neurol. 1990, 47, 259–262. [Google Scholar] [CrossRef] [PubMed]
- Malanchini, M.; Rimfeld, K.; Allegrini, A.G.; Ritchie, S.J.; Plomin, R. Cognitive ability and education: How behavioural genetic research has advanced our knowledge and understanding of their association. Neurosci. Biobehav. Rev. 2020, 111, 229–245. [Google Scholar] [CrossRef] [PubMed]
- Kamboh, M.I.; Fan, K.-H.; Yan, Q.; Beer, J.C.; Snitz, B.E.; Wang, X.; Chang, C.-C.H.; Demirci, F.Y.; Feingold, E.; Ganguli, M. Population-based genome-wide association study of cognitive decline in older adults free of dementia: Identification of a novel locus for the attention domain. Neurobiol. Aging 2019, 84, 239.e15–239.e24. [Google Scholar] [CrossRef] [PubMed]
- Warrier, V.; Grasby, K.L.; Uzefovsky, F.; Toro, R.; Smith, P.; Chakrabarti, B.; Khadake, J.; Mawbey-Adamson, E.; Litterman, N.; Hottenga, J.-J.; et al. Genome-wide meta-analysis of cognitive empathy: Heritability, and correlates with sex, neuropsychiatric conditions and cognition. Mol. Psychiatry 2018, 23, 1402–1409. [Google Scholar] [CrossRef] [PubMed]
- Acharya, V.; Fan, K.H.; Snitz, B.E.; Ganguli, M.; Dekosky, S.T.; Lopez, O.L.; Feingold, E.; Kamboh, M.I. Meta-analysis of age-related cognitive decline reveals a novel locus for the attention domain and implicates a COVID-19-related gene for global cognitive function. Alzheimer’s Dement. 2023, 19, 5010–5022. [Google Scholar] [CrossRef] [PubMed]
- Kamboh, M.I. Genomics and Functional Genomics of Alzheimer’s Disease. Neurotherapeutics 2022, 19, 152–172. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, K.L.; Tybjærg-Hansen, A.; Nordestgaard, B.G.; Frikke-Schmidt, R. Absolute 10-year risk of dementia by age, sex and APOE genotype: A population-based cohort study. Cmaj 2018, 190, E1033–E1041. [Google Scholar] [CrossRef] [PubMed]
- Reiman, E.M.; Arboleda-Velasquez, J.F.; Quiroz, Y.T.; Huentelman, M.J.; Beach, T.G.; Caselli, R.J.; Chen, Y.; Su, Y.; Myers, A.J.; Hardy, J.; et al. Exceptionally low likelihood of Alzheimer’s dementia in APOE2 homozygotes from a 5,000-person neuropathological study. Nat. Commun. 2020, 11, 667. [Google Scholar] [CrossRef] [PubMed]
- Rawle, M.J.; Davis, D.; Bendayan, R.; Wong, A.; Kuh, D.; Richards, M. Apolipoprotein-E (Apoe) ε4 and cognitive decline over the adult life course. Transl. Psychiatry 2018, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Reas, E.T.; Laughlin, G.A.; Bergstrom, J.; Kritz-Silverstein, D.; Barrett-Connor, E.; McEvoy, L.K. Effects of APOE on cognitive aging in community-dwelling older adults. Neuropsychology 2019, 33, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Riaz, M.; Huq, A.; Ryan, J.; Orchard, S.G.; Tiller, J.; Lockery, J.; Woods, R.L.; Wolfe, R.; Renton, A.E.; Goate, A.M.; et al. Effect of APOE and a polygenic risk score on incident dementia and cognitive decline in a healthy older population. Aging Cell 2021, 20, e13384. [Google Scholar] [CrossRef] [PubMed]
- Makkar, S.R.; Lipnicki, D.M.; Crawford, J.D.; Kochan, N.A.; Castro-Costa, E.; Lima-Costa, M.F.; Diniz, B.S.; Brayne, C.; Stephan, B.; Matthews, F.; et al. APOE ε4 and the Influence of Sex, Age, Vascular Risk Factors, and Ethnicity on Cognitive Decline. J. Gerontol. A Biol. Sci. Med. Sci. 2020, 75, 1863–1873. [Google Scholar] [CrossRef] [PubMed]
- Montagne, A.; Nation, D.A.; Sagare, A.P.; Barisano, G.; Sweeney, M.D.; Chakhoyan, A.; Pachicano, M.; Joe, E.; Nelson, A.R.; D’Orazio, L.M.; et al. APOE4 leads to blood–brain barrier dysfunction predicting cognitive decline. Nature 2020, 581, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Sweigart, B.; Andersen, S.L.; Gurinovich, A.; Cosentino, S.; Schupf, N.; Perls, T.T.; Sebastiani, P. APOE E2/E2 Is Associated with Slower Rate of Cognitive Decline with Age. J. Alzheimer’s Dis. 2021, 83, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.-H.; Feingold, E.; Rosenthal, S.L.; Demirci, F.Y.; Ganguli, M.; Lopez, O.L.; Kamboh, M.I. Whole-Exome Sequencing Analysis of Alzheimer’s Disease in Non-APOE*4 Carriers. J. Alzheimer’s Dis. 2020, 76, 1553–1565. [Google Scholar] [CrossRef] [PubMed]
- Jun, G.; Ibrahim-Verbaas, C.A.; Vronskaya, M.; Lambert, J.C.; Chung, J.; Naj, A.C.; Kunkle, B.W.; Wang, L.-S.; Bis, J.C.; Bellenguez, C.; et al. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol. Psychiatry 2016, 21, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Jun, G.R.; Zhang, X.; Chung, J.; Naj, A.C.; Chen, Y.; Bellenguez, C.; Hamilton-Nelson, K.; Martin, E.R.; Kunkle, B.W.; et al. Analysis of Whole-Exome Sequencing Data for Alzheimer Disease Stratified by APOE Genotype. JAMA Neurol. 2019, 76, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Võsa, U.; Claringbould, A.; Westra, H.-J.; Bonder, M.J.; Deelen, P.; Zeng, B.; Kirsten, H.; Saha, A.; Kreuzhuber, R.; Yazar, S.; et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat. Genet. 2021, 53, 1300–1310. [Google Scholar] [CrossRef] [PubMed]
- Bellenguez, C.; Küçükali, F.; Jansen, I.E.; Kleineidam, L.; Moreno-Grau, S.; Amin, N.; Naj, A.C.; Martin-Campos, R.; Grenier-Boley, B.; Andrade, V.; et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 2022, 54, 412–436. [Google Scholar] [CrossRef] [PubMed]
- Sherva, R.; Gross, A.; Mukherjee, S.; Koesterer, R.; Amouyel, P.; Bellenguez, C.; Dufouil, C.; Bennett, D.A.; Chibnik, L.; Cruchaga, C.; et al. Genome-wide association study of rate of cognitive decline in Alzheimer’s disease patients identifies novel genes and pathways. Alzheimer’s Dement. 2020, 16, 1134–1145. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.J.; Wedow, R.; Okbay, A.; Kong, E.; Maghzian, O.; Zacher, M.; Nguyen-Viet, T.A.; Bowers, P.; Sidorenko, J.; Linnér, R.K.; et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 2018, 50, 1112–1121. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, A.; Bui, T.T.; Helmy, M.; Selvarajoo, K. Identifying toggle genes from transcriptome-wide scatter: A new perspective for biological regulation. Genomics 2022, 114, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Poliseno, L. Pseudogenes: Newly discovered players in human cancer. Sci. Signal. 2012, 5, re5. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, X.; Zhao, D.; Ruan, X.; Su, R.; Shang, X.; Wang, D.; Yang, C.; Xue, Y. Pseudogene ACTBP2 increases blood–brain barrier permeability by promoting KHDRBS2 transcription through recruitment of KMT2D/WDR5 in Aβ1–42 microenvironment. Cell Death Discov. 2021, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y.-K.; Duman, J.G.; Tolias, K.F. The Adhesion-GPCR BAI1 Promotes Excitatory Synaptogenesis by Coordinating Bidirectional Trans-synaptic Signaling. J. Neurosci. 2018, 38, 8388–8406. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.-S.; Li, G.; Li, S.; Gao, W.; Chen, G.; Gan, S.; Zhang, M.; Li, H.; Wu, S.; Du, Y. G protein-coupled receptors in neurodegenerative diseases and psychiatric disorders. Signal Transduct. Target. Ther. 2023, 8, 177. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Li, C.; Swanson, A.M.; Villalba, R.M.; Guo, J.; Zhang, Z.; Matheny, S.; Murakami, T.; Stephenson, J.R.; Daniel, S.; et al. BAI1 regulates spatial learning and synaptic plasticity in the hippocampus. J. Clin. Investig. 2015, 125, 1497–1508. [Google Scholar] [CrossRef] [PubMed]
- Amor, D.J.; Marsh, A.P.L.; Storey, E.; Tankard, R.; Gillies, G.; Delatycki, M.B.; Pope, K.; Bromhead, C.; Leventer, R.J.; Bahlo, M.; et al. Heterozygous mutations in HSD17B4 cause juvenile peroxisomal D-bifunctional protein deficiency. Neurol. Genet. 2016, 2, e114. [Google Scholar] [CrossRef] [PubMed]
- Pierce, S.B.; Walsh, T.; Chisholm, K.M.; Lee, M.K.; Thornton, A.M.; Fiumara, A.; Opitz, J.M.; Levy-Lahad, E.; Klevit, R.E.; King, M.-C. Mutations in the DBP-Deficiency Protein HSD17B4 Cause Ovarian Dysgenesis, Hearing Loss, and Ataxia of Perrault Syndrome. Am. J. Hum. Genet. 2010, 87, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, Y.; Morino, H.; Miyamoto, R.; Kurashige, T.; Kume, K.; Mizuno, N.; Kanaya, Y.; Tada, Y.; Ohsawa, R.; Yokota, K.; et al. Biallelic mutation of HSD17B4 induces middle age-onset spinocerebellar ataxia. Neurol. Genet. 2020, 6, e396. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, S.E.; Fakiola, M.; Tang, D.; Scaman, E.; Syn, G.; Francis, R.W.; Coates, H.L.; Anderson, D.; Lassmann, T.; Cordell, H.J.; et al. Common and Rare Genetic Variants That Could Contribute to Severe Otitis Media in an Australian Aboriginal Population. Clin. Infect. Dis. 2021, 73, 1860–1870. [Google Scholar] [CrossRef] [PubMed]
- Shojaei, E.; Jafari, Z.; Gholami, M. Effect of Early Intervention on Language Development in Hearing-Impaired Children. Iran. J. Otorhinolaryngol. 2016, 28, 13–21. [Google Scholar] [PubMed]
- Jiang, K.; Armstrong, N.M.; Agrawal, Y.; Gross, A.L.; Schrack, J.A.; Lin, F.R.; Ferrucci, L.; Resnick, S.M.; Deal, J.A.; Powell, D.S. Associations of audiometric hearing and speech-in-noise performance with cognitive decline among older adults: The Baltimore Longitudinal Study of Aging (BLSA). Front. Neurol. 2022, 13, 1029851. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.R.; Jiang, K.; Lin, F.R.; Deal, J.A.; Reed, N.S. Hearing loss and dementia prevalence in older adults in the US. JAMA 2023, 329, 171–173. [Google Scholar] [CrossRef] [PubMed]
- Living, G.; Huntley, J.; Liu, K.Y.; Costaferda, S.G.; Selbæk, G.; Suvarna, A. Dementia prevention, intervention, and care: 2024 report of the Lancet standing Commission. Lancet 2024, 404, 572–628. [Google Scholar] [CrossRef] [PubMed]
- Ermis Akyuz, E.; Bell, S.M. The Diverse Role of CUB and Sushi Multiple Domains 1 (CSMD1) in Human Diseases. Genes 2022, 13, 2332. [Google Scholar] [CrossRef] [PubMed]
- Koiliari, E.; Roussos, P.; Pasparakis, E.; Lencz, T.; Malhotra, A.; Siever, L.J.; Giakoumaki, S.G.; Bitsios, P. The CSMD1 genome-wide associated schizophrenia risk variant rs10503253 affects general cognitive ability and executive function in healthy males. Schizophr. Res. 2014, 154, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Fu, A.K.Y.; Hung, K.-W.; Yuen, M.Y.F.; Zhou, X.; Mak, D.S.Y.; Chan, I.C.W.; Cheung, T.H.; Zhang, B.; Fu, W.-Y.; Liew, F.; et al. IL-33 ameliorates Alzheimer’s disease-like pathology and cognitive decline. Proc. Natl. Acad. Sci. USA 2016, 113, E2705–E2713. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhou, X.; Wong, H.Y.; Ouyang, L.; Ip, F.C.F.; Chau, V.M.N.; Lau, S.-F.; Wu, W.; Wong, D.Y.K.; Seo, H.; et al. An IL1RL1 genetic variant lowers soluble ST2 levels and the risk effects of APOE-ε4 in female patients with Alzheimer’s disease. Nat. Aging 2022, 2, 616–634. [Google Scholar] [CrossRef] [PubMed]
- Zong, X.; Fan, Q.; Zhang, H.; Yang, Q.; Xie, H.; Chen, Q.; Zhang, R.; Tao, R. Soluble ST2 levels for predicting the presence and severity of metabolic syndrome. Endocr. Connect. 2021, 10, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Okbay, A.; Wu, Y.; Wang, N.; Jayashankar, H.; Bennett, M.; Nehzati, S.M.; Sidorenko, J.; Kweon, H.; Goldman, G.; Gjorgjieva, T.; et al. Polygenic prediction of educational attainment within and between families from genome-wide association analyses in 3 million individuals. Nat. Genet. 2022, 54, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Homann, J.; Osburg, T.; Ohlei, O.; Dobricic, V.; Deecke, L.; Bos, I.; Vandenberghe, R.; Gabel, S.; Scheltens, P.; Teunissen, C.E.; et al. Genome-Wide Association Study of Alzheimer’s Disease Brain Imaging Biomarkers and Neuropsychological Phenotypes in the European Medical Information Framework for Alzheimer’s Disease Multimodal Biomarker Discovery Dataset. Front. Aging Neurosci. 2022, 14, 840651. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, S.; Sleyp, Y.; Hoskens, H.; Indencleef, K.; Spence, J.P.; Bruffaerts, R.; Radwan, A.; Eller, R.J.; Richmond, S.; Shriver, M.D.; et al. Shared heritability of human face and brain shape. Nat. Genet. 2021, 53, 830–839. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liang, Z.; Li, G. Rab22 controls NGF signaling and neurite outgrowth in PC12 cells. Mol. Biol. Cell 2011, 22, 3853–3860. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Zhao, X.; He, L.; Ding, Y.; Xu, W.; Lin, S.; Fang, S.; Yang, W.; Sung, K.; Spencer, B.; et al. Upregulation of RIN3 induces endosomal dysfunction in Alzheimer’s disease. Transl. Neurodegener. 2020, 9, 26. [Google Scholar] [CrossRef] [PubMed]
- Schott, J.M.; Crutch, S.J.; Carrasquillo, M.M.; Uphill, J.; Shakespeare, T.J.; Ryan, N.S.; Yong, K.X.; Lehmann, M.; Ertekin-Taner, N.; Graff-Radford, N.R.; et al. Genetic risk factors for the posterior cortical atrophy variant of Alzheimer’s disease. Alzheimer’s Dementia. 2016, 12, 862–871. [Google Scholar] [CrossRef] [PubMed]
- Schott, J.M.; Crutch, S.J. Posterior Cortical Atrophy. Contin. Lifelong Learn. Neurol. 2019, 25, 52–75. [Google Scholar] [CrossRef] [PubMed]
- Golanska, E.; Sieruta, M.; Gresner, S.M.; Pfeffer, A.; Chodakowska-Zebrowska, M.; Sobow, T.M.; Klich, I.; Mossakowska, M.; Szybinska, A.; Barcikowska, M.; et al. APBB2 genetic polymorphisms are associated with severe cognitive impairment in centenarians. Exp. Gerontol. 2013, 48, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Dubé, J.B.; Johansen, C.T.; Robinson, J.F.; Lindsay, J.; Hachinski, V.; Hegele, R.A. Genetic determinants of “cognitive impairment, no dementia”. J. Alzheimer’s Dis. 2013, 33, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Wei, X.; Lin, L.; Yuan, F.; Bi, Y.; Guo, Z.; Liu, L.; Ji, L.; Yang, X.; Han, K.; et al. A novel heterozygous missense variant of the ARID4A gene identified in Han Chinese families with schizophrenia-diagnosed siblings that interferes with DNA-binding activity. Mol. Psychiatry 2022, 27, 2777–2786. [Google Scholar] [CrossRef] [PubMed]
- Sim, K.; Chan, W.Y.; Woon, P.S.; Low, H.Q.; Lim, L.; Yang, G.L.; Lee, J.; Chong, S.A.; Sithoh, Y.-Y.; Chan, Y.H.; et al. ARVCF genetic influences on neurocognitive and neuroanatomical intermediate phenotypes in Chinese patients with schizophrenia. J. Clin. Psychiatry 2012, 73, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Yin, H.; Xu, Y.; Zhu, Q.; Luo, J.; Wang, X.; Chen, G. Increased expression of calponin-3 in epileptic patients and experimental rats. Exp. Neurol. 2012, 233, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Furuta, N.; Makioka, K.; Fujita, Y.; Ikeda, M.; Takatama, M.; Matsuoka, M.; Okamoto, K. Reduced expression of BTBD10 in anterior horn cells with Golgi fragmentation and pTDP-43-positive inclusions in patients with sporadic amyotrophic lateral sclerosis. Neuropathology 2013, 33, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Bello, A.I.; Goswami, R.; Brown, S.L.; Costanzo, K.; Shores, T.; Allan, S.; Odah, R.; Mohan, R.D. Deubiquitinases in Neurodegeneration. Cells 2022, 11, 556. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Liu, G.A.; Perez, E.; Rainer, R.D.; Febo, M.; Cruz-Almeida, Y.; Ebner, N.C. Systemic Inflammation Mediates Age-Related Cognitive Deficits. Front. Aging Neurosci. 2018, 10, 236. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xiao, D.; Mao, Q.; Xia, H. Role of neuroinflammation in neurodegeneration development. Signal Transduct. Target. Therapy 2023, 8, 267. [Google Scholar] [CrossRef] [PubMed]
- Dunn, G.A.; Nigg, J.T.; Sullivan, E.L. Neuroinflammation as a risk factor for attention deficit hyperactivity disorder. Pharmacol. Biochem. Behav. 2019, 182, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Fiani, B.; Zhu, L.; Musch, B.L.; Briceno, S.; Andel, R.; Sadeq, N.; Ansari, A.Z. The Neurophysiology of Caffeine as a Central Nervous System Stimulant and the Resultant Effects on Cognitive Function. Cureus 2021, 13, e15032. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Vrontakis, M.; Parkinson, F.; Chelikani, P. Functional bitter taste receptors are expressed in brain cells. Biochem. Biophys. Res. Commun. 2011, 406, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.C.; Costa, A.R.; Gonçalves, I.; Quintela, T.; Preissner, R.; Santos, C.R.A. The druggability of bitter taste receptors for the treatment of neurodegenerative disorders. Biochem. Pharmacol. 2022, 197, 114915. [Google Scholar] [CrossRef] [PubMed]
- Tiroch, J.; Sterneder, S.; Di Pizio, A.; Lieder, B.; Hoelz, K.; Holik, A.-K.; Pignitter, M.; Behrens, M.; Somoza, M.; Ley, J.P.; et al. Bitter Sensing TAS2R50 Mediates the trans-Resveratrol-Induced Anti-inflammatory Effect on Interleukin 6 Release in HGF-1 Cells in Culture. J. Agric. Food Chem. 2021, 69, 13339–13349. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, M.; Simstich, S.; Fauler, G.; Hofer, E.; Fritz-Petrin, E.; Herrmann, W.; Schmidt, R. The relationship between plasma free fatty acids, cognitive function and structural integrity of the brain in middle-aged healthy humans. Aging 2021, 13, 22078–22091. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Shue, F.; Zhao, N.; Shinohara, M.; Bu, G. APOE2: Protective mechanism and therapeutic implications for Alzheimer’s disease. Mol. Neurodegener. 2020, 15, 63. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Boyle, P.A.; Nag, S.; Leurgans, S.; Buchman, A.S.; Wilson, R.S.; Arvanitakis, Z.; Farfel, J.M.; Jager, P.L.D.; Bennett, D.A.; et al. APOE and cerebral amyloid angiopathy in community-dwelling older persons. Neurobiol. Aging 2015, 36, 2946–2953. [Google Scholar] [CrossRef] [PubMed]
- Dekosky, S.T.; Fitzpatrick, A.; Ives, D.G.; Saxton, J.; Williamson, J.; Lopez, O.L.; Burke, G.; Fried, L.; Kuller, L.H.; Robbins, J.; et al. The Ginkgo Evaluation of Memory (GEM) study: Design and baseline data of a randomized trial of Ginkgo biloba extract in prevention of dementia. Contemp. Clin. Trials. 2006, 27, 238–253. [Google Scholar] [CrossRef] [PubMed]
- Dekosky, S.T.; Williamson, J.D.; Fitzpatrick, A.L.; Kronmal, R.A.; Ives, D.G.; Saxton, J.A.; Lopez, O.L.; Burke, G.; Carlson, M.C.; Fried, L.P.; et al. Ginkgo biloba for prevention of dementia: A randomized controlled trial. JAMA 2008, 300, 2253–2262. [Google Scholar] [CrossRef] [PubMed]
- Ganguli, M.; Snitz, B.; Bilt, J.V.; Chang, C.-C.H. How much do depressive symptoms affect cognition at the population level? The monongahela-youghiogheny healthy aging team (MYHAT) study. Int. J. Geriatr. Psychiatry 2009, 24, 1277–1284. [Google Scholar] [CrossRef] [PubMed]
- Ganguli, M.; Snitz, B.E.; Lee, C.-W.; Vanderbilt, J.; Saxton, J.A.; Chang, C.-C.H. Age and education effects and norms on a cognitive test battery from a population-based cohort: The Monongahela–Youghiogheny Healthy Aging Team. Aging Ment. Health 2010, 14, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Ganguli, M.; Seaberg, E.C.; Ratcliff, G.G.; Belle, S.H.; DeKosky, S.T. Cognitive stability over 2 years in a rural elderly population: The MoVIES project. Neuroepidemiology 1996, 15, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; Bakker, P.I.W.B.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.D.; Fan, K.H.; Aslam, M.M.; Snitz, B.E.; DeKosky, S.T.; Lopez, O.L.; Feingold, E.; Kamboh, M.I. Genome-Wide Association Study of Incident Dementia in a Community-Based Sample of Older Subjects. J. Alzheimer’s Dis. 2022, 88, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Willer, C.J.; Li, Y.; Abecasis, G.R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010, 26, 2190–2191. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Taskesen, E.; Van Bochoven, A.; Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef] [PubMed]
- Giusti-Rodriguez, P.M.; Sullivan, P.F. Using three-dimensional regulatory chromatin interactions from adult and fetal cortex to interpret genetic results for psychiatric disorders and cognitive traits. bioRxiv 2019. [Google Scholar] [CrossRef]

| Gingko Evaluation of Memory (GEM) | Monongahela-Youghiogheny Healthy Aging Team (MYHAT) | Monongahela Valley Independent Elders Survey (MoVIES) | |
|---|---|---|---|
| Total Participants | 1898 | 745 | 378 |
| Age (Mean (SD)) | 78.13 ± 3.04 | 77.15 ± 7.32 | 77.46 ± 4.23 |
| Female (N, %) | 840 (44.25) | 451 (60.53) | 254 (67.19) |
| Education (Years) | 14.4 (2.9) | 13.04 (2.51) | 11.81 (2.27) |
| Baseline CDR, (N, %) | |||
| 0 | 1263 (66.54) | 570 (76.51) | 378 (100) |
| 0.5 | 635 (33.45) | 175 (23.48) | 0 (0) |
| Last CDR, (N, %) | |||
| 0 | 939 (49.47) | 501 (67.24) | 378 (100) |
| 0.5 | 959 (50.52) | 220 (29.53) | 0 (0) |
| ≥1 | 0 (0) | 24 (3.22) | 0 (0) |
| APOE2 (N) | 258 | 100 | 55 |
| Age (Mean (SD)) | 78.45 ± 3.25 | 79.02 ± 7.59 | 77.70 ± 5.16 |
| Female (N, %) | 118 (45.73) | 52 (52) | 46 (95) |
| Education (Years) | 14.19 | 13 | 11.89 |
| Baseline CDR (N, %) | |||
| 0 | 162 (62.79) | 80 (80) | 55 (100) |
| 0.5 | 96 (37.2) | 20 (20) | 0 (0) |
| ≥1 | 0 (0) | ||
| Last CDR (N, %) | |||
| 0 | 128 (49.61) | 70 (70) | 55 (100) |
| 0.5 | 130 (50.38) | 28 (28) | 0 (0) |
| ≥1 | 0 (0) | 2 (2) | 0 (0) |
| APOE 3/3 (N) | 1237 | 498 | 260 |
| Age (Mean (SD)) | 78.18 ± 3.06 | 77.22 ± 7.31 | 77.49 ± 4.18 |
| Female (%) | 531 (42.92) | 305 (61.24) | 169 (65) |
| Education (years) | 14.40 | 13.02 | 11.73 |
| Baseline CDR (N, %) | |||
| 0 | 840 (67.90) | 382 (76.7) | 260 (100) |
| 0.5 | 397 (32.09) | 116 (23.2) | 0 (0) |
| ≥1 | 0 (0) | 0 (0) | 0 (0) |
| Last CDR (N, %) | |||
| 0 | 615 (49.71) | 340 (68.3) | 260 (100) |
| 0.5 | 622 (50.27) | 145 (29.1) | 0 (0) |
| ≥1 | 0 (0) | 13 (2.6) | 0 (0) |
| APOE4 (N) | 367 | 138 | 56 |
| Age (Mean (SD)) | 77.74 ± 2.65 | 75.71 ± 7.02 | 76.96 ± 3.37 |
| Female (N, %) | 174 (47.70) | 88 (63.76) | 34 (60.71) |
| Education (years) | 14.48 | 13.21 | 12.17 |
| Baseline CDR (N, %) | |||
| 0 | 238 (64.9) | 102 (73.9) | 56 (100) |
| 0.5 | 129 (35.1) | 36 (26.1) | 0 (0) |
| ≥1 | 0 (0) | 0 (0) | 0 (0) |
| Last CDR (N, %) | |||
| 0 | 181 (49.3) | 84 (60.9) | 56 (100) |
| 0.5 | 186 (50.7) | 46 (33.3) | 0 (0) |
| ≥1 | 0 (0) | 8 (5.8) | 0 (0) |
| APOE Subgroups | Domain | SNP | Chr:Position | A1/A2 | Meta-Analysis | Loc | Gene | ||
|---|---|---|---|---|---|---|---|---|---|
| MAF | Beta | p | |||||||
| APOE4 Group | Attention | rs62371993 | 5:42622849 | C/A | 0.04 | −0.77 | 1.87 × 10−7 | intronic | GHR |
| rs7320036 | 13:85848704 | C/T | 0.14 | 0.42 | 4.44 × 10−7 | intergenic | LINC00375, LINC00351 | ||
| rs62253001 | 3:69767057 | G/A | 0.07 | 0.57 | 6.60 × 10−7 | intergenic | FRMD4B, MITF | ||
| rs4732363 | 7:138990811 | T/C | 0.49 | 0.28 | 8.27 × 10−7 | UTR3 | UBN2 | ||
| Executive Function | rs293879 | 8:4586377 | T/G | 0.32 | 0.32 | 8.49 × 10−8 | intronic | CSMD1 | |
| rs62499971 | 8:20125115 | T/C | 0.19 | −0.36 | 2.01 × 10−7 | intronic | LZTS1 | ||
| rs2943674 | 1:85105006 | C/T | 0.10 | −0.45 | 7.31 × 10−7 | intergenic | LINC01555, SSX2IP | ||
| Memory | rs72845557 | 17:60458122 | C/A | 0.12 | 0.46 | 6.60 × 10−7 | intronic | EFCAB3 | |
| Language | rs13187183 | 5:117976098 | C/T | 0.06 | 0.69 | 3.79 × 10−8 | intergenic | LINC02215, DTWD2 | |
| rs4888647 | 16:73748328 | C/T | 0.09 | −0.48 | 9.52 × 10−7 | intergenic | LINC01568, LOC101928035 | ||
| Global | rs62297316 | 4:5376604 | G/C | 0.02 | 1.21 | 1.58 × 10−7 | intronic | STK32B | |
| rs12746598 | 1:240305899 | T/C | 0.34 | −0.30 | 9.81 × 10−7 | intronic | FMN2 | ||
| APOE 3/3
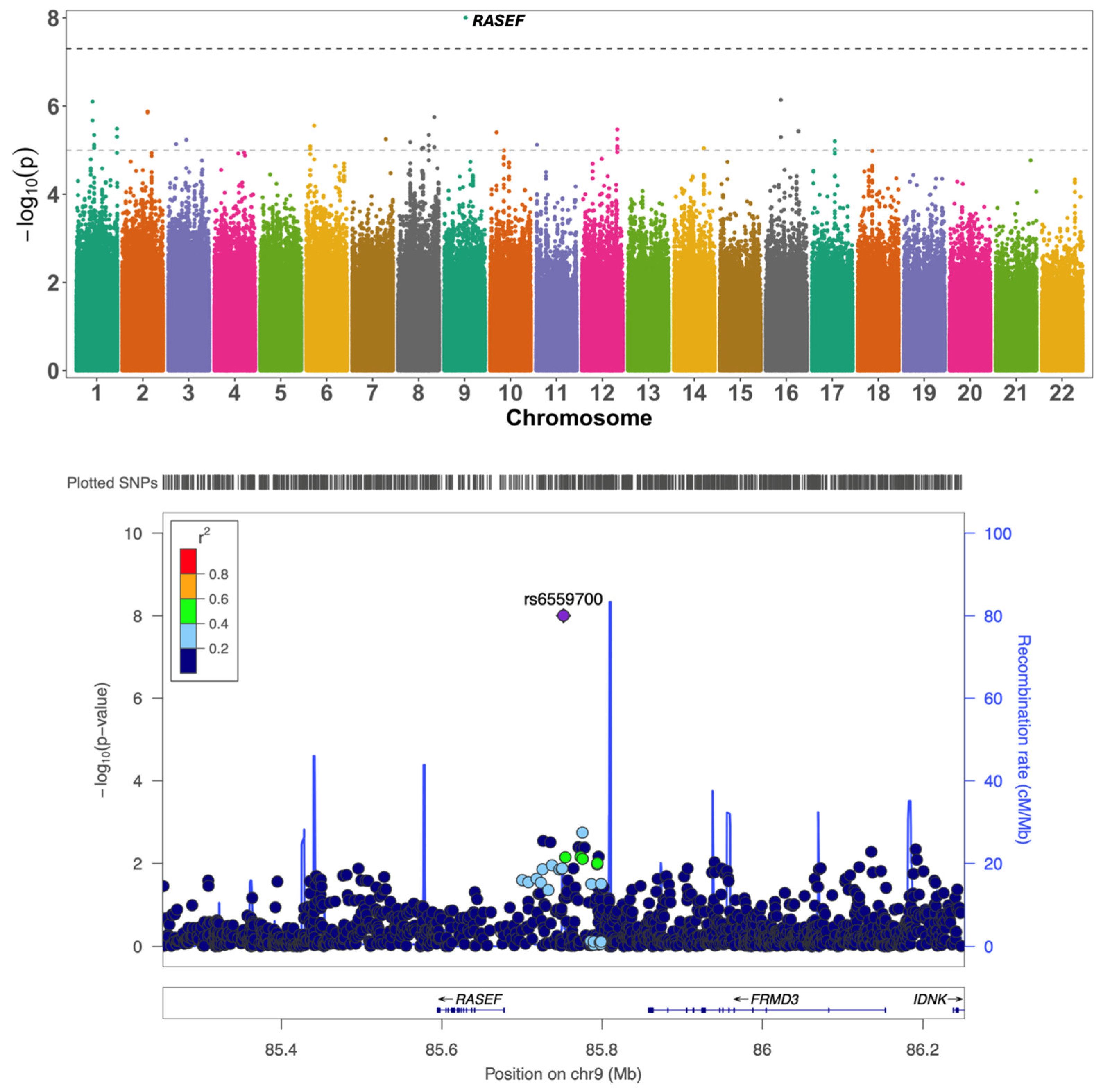
Group | Attention | rs6559700 | 9:85751913 | A/G | 0.09 | −0.29 | 9.95 × 10−9 | intergenic | RASEF, FRMD3 |
| rs77848581 | 16:26919840 | C/T | 0.01 | 0.71 | 7.23 × 10−7 | intergenic | HS3ST4, C16orf82 | ||
| Executive Function | rs62320280 | 4:131795996 | T/C | 0.13 | −0.23 | 5.19 × 10−7 | intergenic | LINC02479, SNHG27 | |
| rs112356643 | 13:55089341 | T/C | 0.02 | −0.50 | 6.99 × 10−7 | intergenic | MIR1297, MIR5007 | ||
| Memory | rs3865460 | 19:41447928 | G/A | 0.21 | 0.18 | 5.35 × 10−7 | lncRNA intronic | CYP2B7P | |
| rs151090836 | 7:38728382 | C/T | 0.03 | −0.46 | 5.56 × 10−7 | intergenic | FAM183BP, VPS41 | ||
| rs426171 | 13:110557648 | C/A | 0.35 | −0.15 | 6.04 × 10−7 | intergenic | IRS2, LINC00396 | ||
| rs116725560 | 4:8374858 | T/C | 0.05 | −0.35 | 7.73 × 10−7 | intronic | ACOX3 | ||
| Visuospatial Function | rs58302405 | 2:218000504 | T/C | 0.03 | −0.46 | 2.69 × 10−7 | intergenic | LINC01921, DIRC3-AS1 | |
| rs62521843 | 8:124198306 | A/G | 0.03 | −0.44 | 5.17 × 10−7 | intronic | FAM83A | ||
| rs139851690 | 10:2600111 | C/T | 0.02 | 0.56 | 9.48 × 10−7 | intergenic | LINC02645, LOC101927824 | ||
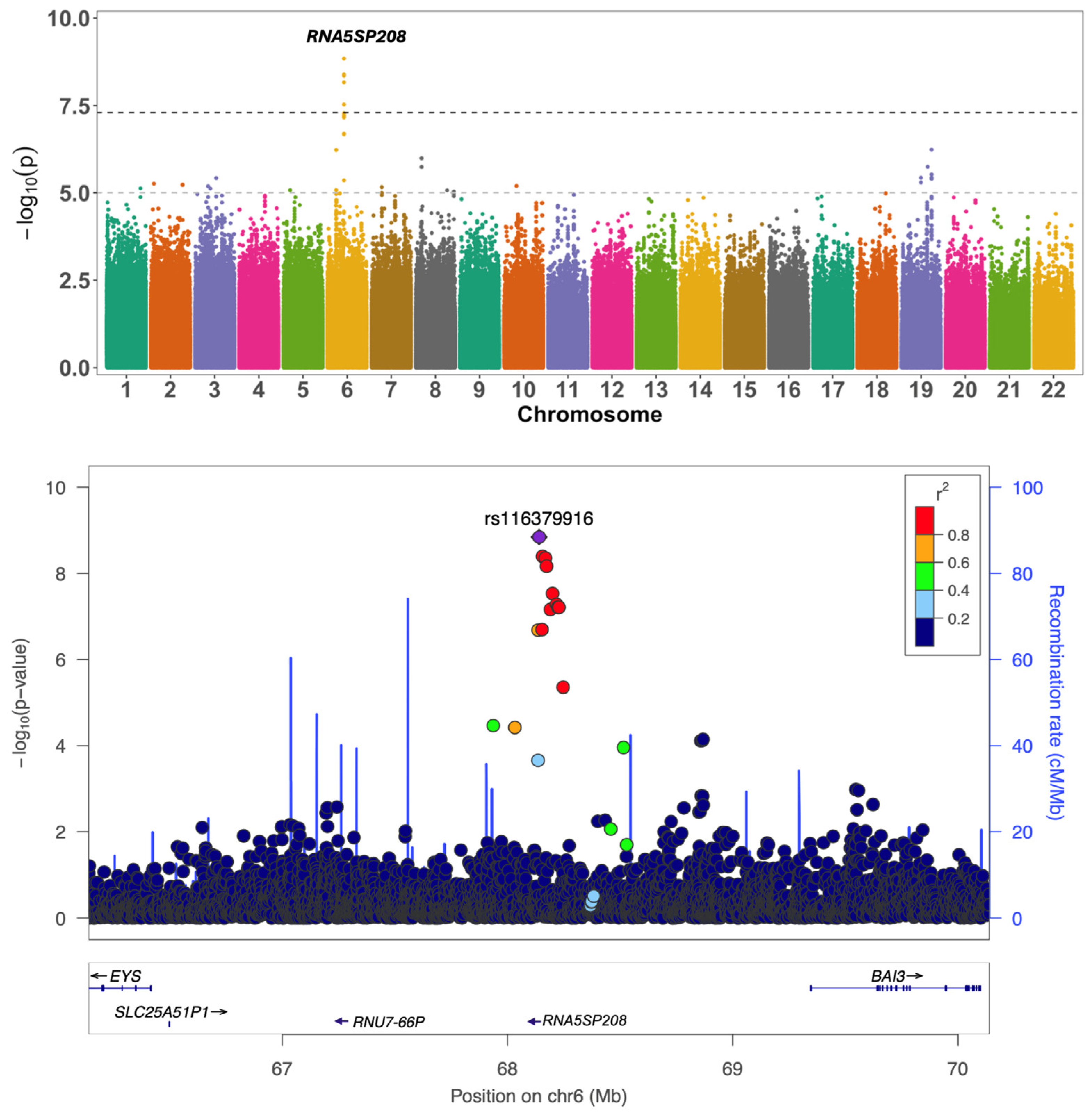
| Global Function | rs116379916 | 6:68140492 | T/G | 0.03 | −0.51 | 1.44 × 10−9 | intergenic | RNU7-66P, RNA5SP208 | |
| APOE2 Group | Attention | rs77127114 | 2:103000000 | A/T | 0.04 | 0.86 | 8.64 × 10−8 | intronic | IL1RL1 |
| rs72706424 | 14:93522585 | A/G | 0.05 | 0.71 | 2.17 × 10−7 | intronic | ITPK1 | ||
| rs11465596 | 2:103000000 | A/C | 0.10 | 0.53 | 4.67 × 10−7 | intronic | IL18R1 | ||
| rs11787153 | 8:139697286 | C/T | 0.25 | 0.36 | 8.22 × 10−7 | intronic | COL22A1 | ||
| rs4541656 | 5:123971932 | A/C | 0.34 | 0.33 | 9.84 × 10−7 | downstream | ZNF608 | ||
| Executive Function | rs35871159 | 7:30781232 | T/C | 0.36 | 0.33 | 2.97 × 10−7 | intergenic | CRHR2, INMT | |
| rs13339093 | 16:78619484 | C/T | 0.06 | 0.69 | 3.65 × 10−7 | intronic | WWOX | ||
| rs16921936 | 12:19924660 | G/T | 0.04 | −0.81 | 3.94 × 10−7 | intergenic | AEBP2, LINC02398 | ||
| Language | rs116191836 | 5:113000000 | T/C | 0.02 | 1.41 | 5.66 × 10−8 | intergenic | YTHDC2, KCNN2 | |
| Visuospatial Function | rs10849223 | 12:5384521 | C/T | 0.14 | 0.51 | 2.98 × 10−7 | intergenic | LINC02443, NTF3 | |
| rs12319445 | 12:92357858 | G/A | 0.02 | −1.14 | 3.30 × 10−7 | intergenic | DCN, LINC01619 | ||
| Global Function | rs75168743 | 14:44488668 | C/T | 0.07 | 0.63 | 1.65 × 10−7 | intergenic | NONE, FSCB | |
| rs7998149 | 13:79077882 | G/A | 0.48 | −0.34 | 1.85 × 10−7 | ncRNA intronic | OBI1-AS1 | ||
| rs12641677 | 4:182290596 | G/A | 0.10 | −0.52 | 4.25 × 10−7 | intergenic | LINC02500, TEMN3-AS1 | ||
| Domain | APOE2 Carriers | APOE 3/3 | APOE4 Carriers |
|---|---|---|---|
| Attention | 65 ZNF608, ARID4A | 118 CNN3, RP4-639F20.1 | 120 USP36, IRF1 |
| Executive Function | 82 - * | 81 CTB-129O4.1, PDP1 | 150 LZTS1, KAT2A, RAB5C, ARVCF, DGCR8 |
| Memory | 74 VANGL1, PTPN14 | 38 APBB2 | 153 PPP2R5A, TMEM206, BTBD10 |
| Language | 78 MSRA, CTB113P19.3 | 64 ASNSD1, OSGEPL1, OSGEPL1-AS1, ORMDL1, PRR5L | 89 PDE6B |
| Visuospatial Function | 147 ZNF69, ZNF788, ZNF20, RSL24D1P8 | 41 - | 122 PIGCP1, CSTF3, HIPK3, SAMD4A |
| Global Function | 88 - | 95 - | 79 - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Acharya, V.; Fan, K.-H.; Snitz, B.E.; Ganguli, M.; DeKosky, S.T.; Lopez, O.L.; Feingold, E.; Kamboh, M.I. APOE Genotype-Stratified Meta-Analysis of Cognitive Decline Reveals Novel Loci for Language and Global Cognitive Function in Older Adults. Int. J. Mol. Sci. 2025, 26, 6940. https://doi.org/10.3390/ijms26146940
Acharya V, Fan K-H, Snitz BE, Ganguli M, DeKosky ST, Lopez OL, Feingold E, Kamboh MI. APOE Genotype-Stratified Meta-Analysis of Cognitive Decline Reveals Novel Loci for Language and Global Cognitive Function in Older Adults. International Journal of Molecular Sciences. 2025; 26(14):6940. https://doi.org/10.3390/ijms26146940
Chicago/Turabian StyleAcharya, Vibha, Kang-Hsien Fan, Beth E. Snitz, Mary Ganguli, Steven T. DeKosky, Oscar L. Lopez, Eleanor Feingold, and M. Ilyas Kamboh. 2025. "APOE Genotype-Stratified Meta-Analysis of Cognitive Decline Reveals Novel Loci for Language and Global Cognitive Function in Older Adults" International Journal of Molecular Sciences 26, no. 14: 6940. https://doi.org/10.3390/ijms26146940
APA StyleAcharya, V., Fan, K.-H., Snitz, B. E., Ganguli, M., DeKosky, S. T., Lopez, O. L., Feingold, E., & Kamboh, M. I. (2025). APOE Genotype-Stratified Meta-Analysis of Cognitive Decline Reveals Novel Loci for Language and Global Cognitive Function in Older Adults. International Journal of Molecular Sciences, 26(14), 6940. https://doi.org/10.3390/ijms26146940





