Toward Safer Biotherapeutics: Expression and Characterization of a Humanized Chimeric L-Asparaginase in E. coli
Abstract
1. Introduction
1.1. L-Asparaginase (ASNase): Sources of Obtention, Structural Characteristics, and Mechanism of Action
1.2. Main Adverse Responses to L-Asparaginase (ASNase) Treatment
1.3. Approaches to Reduce the Immunogenicity of L-Asparaginase
2. Results and Discussion
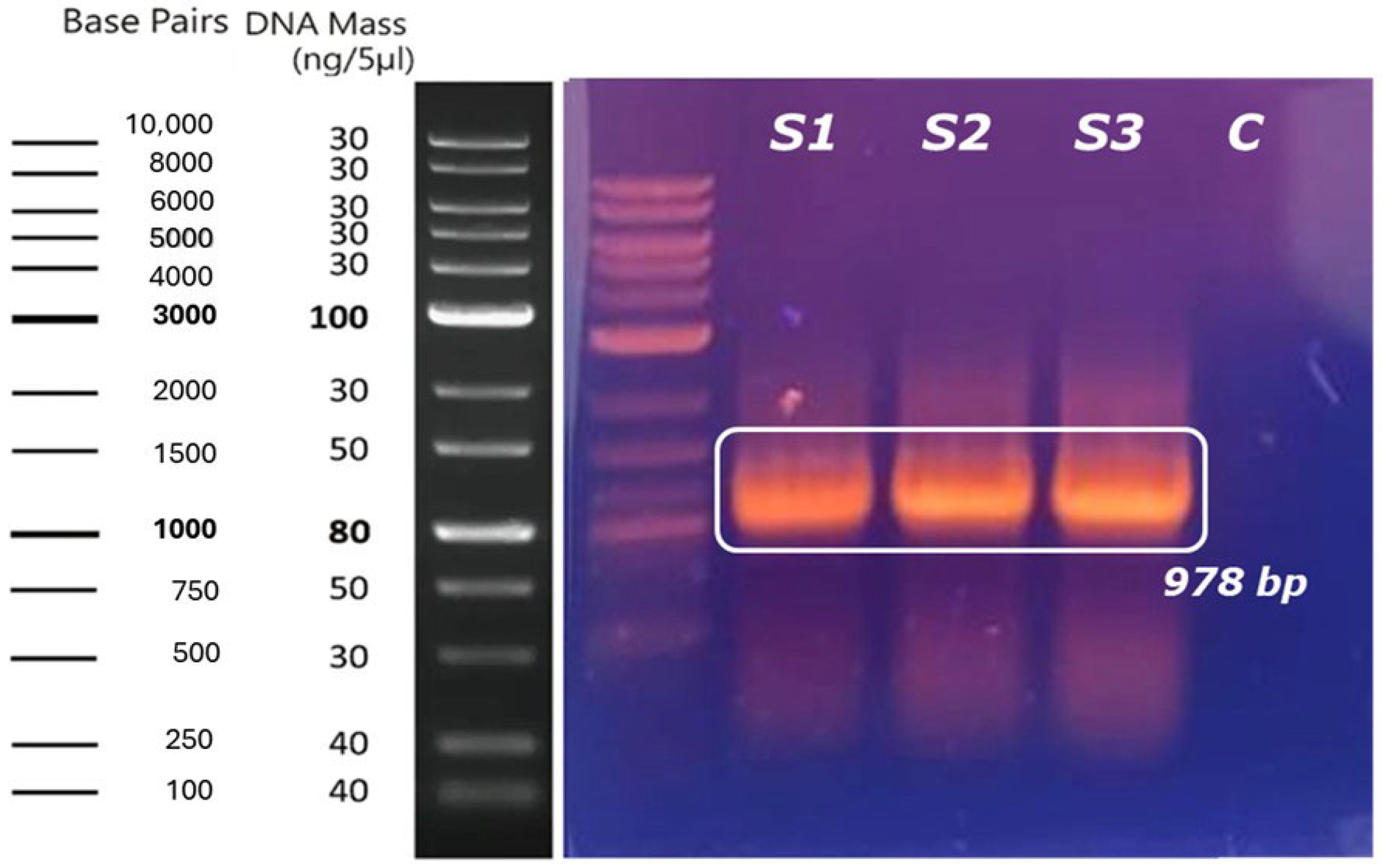
2.1. Obtaining the Recombinant Plasmid with the Inserted Gene of Interest
2.2. Digestion and Ligation of the Gene of Interest in the Expression Vector
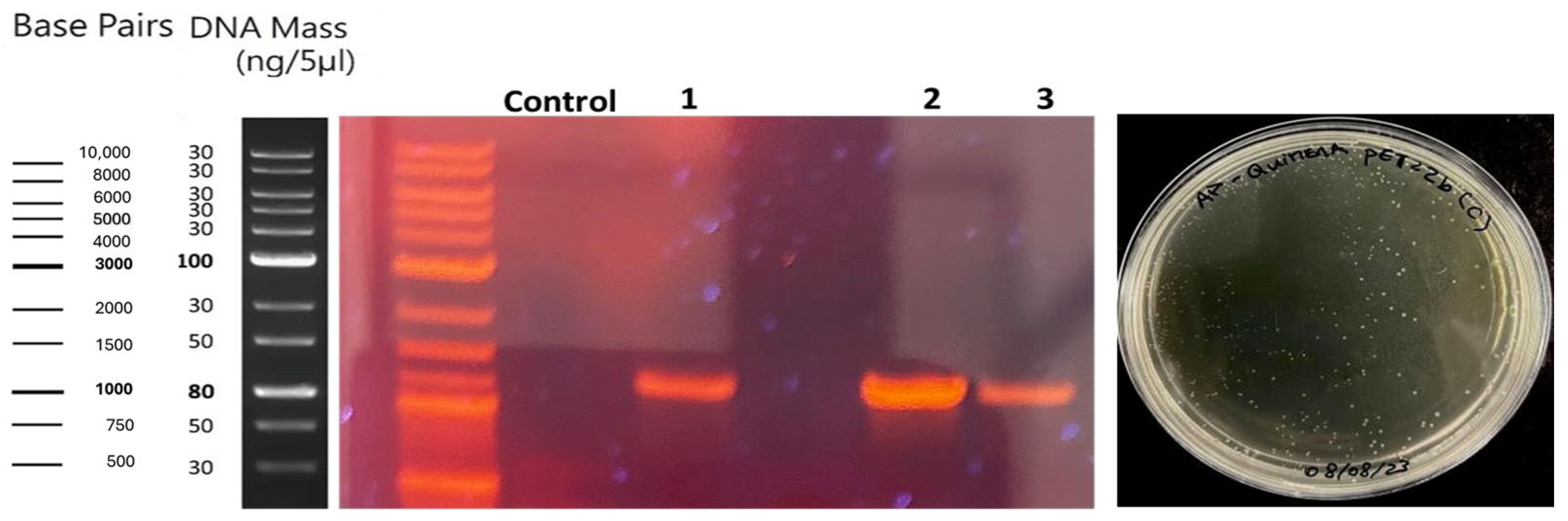
2.3. Transformation
Transformation of BL21 and Rossetta Strains and Chimeric Protein Expression Assays of Chimeric ASNase Chimeric Protein in E. coli BL21 and Rosetta
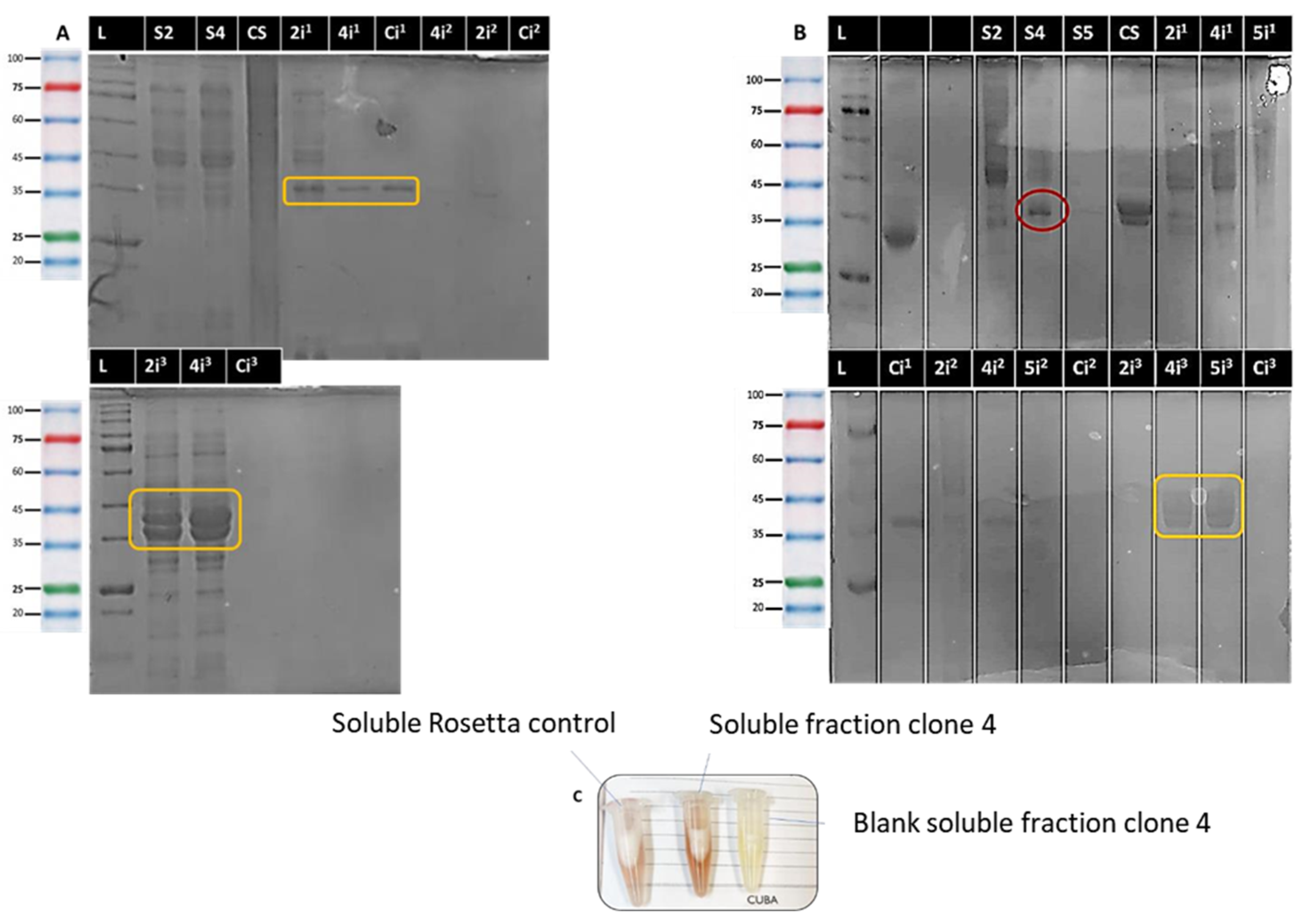
2.4. Optimization
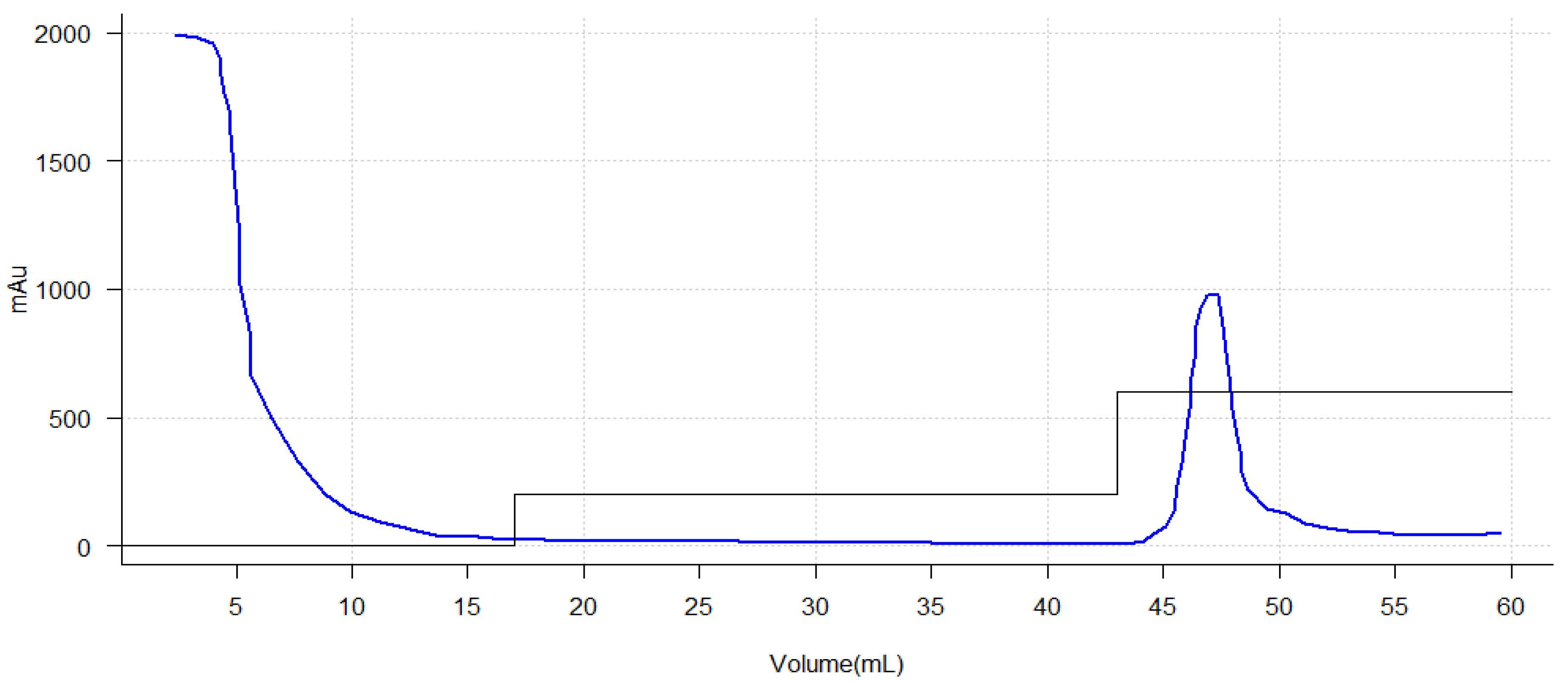
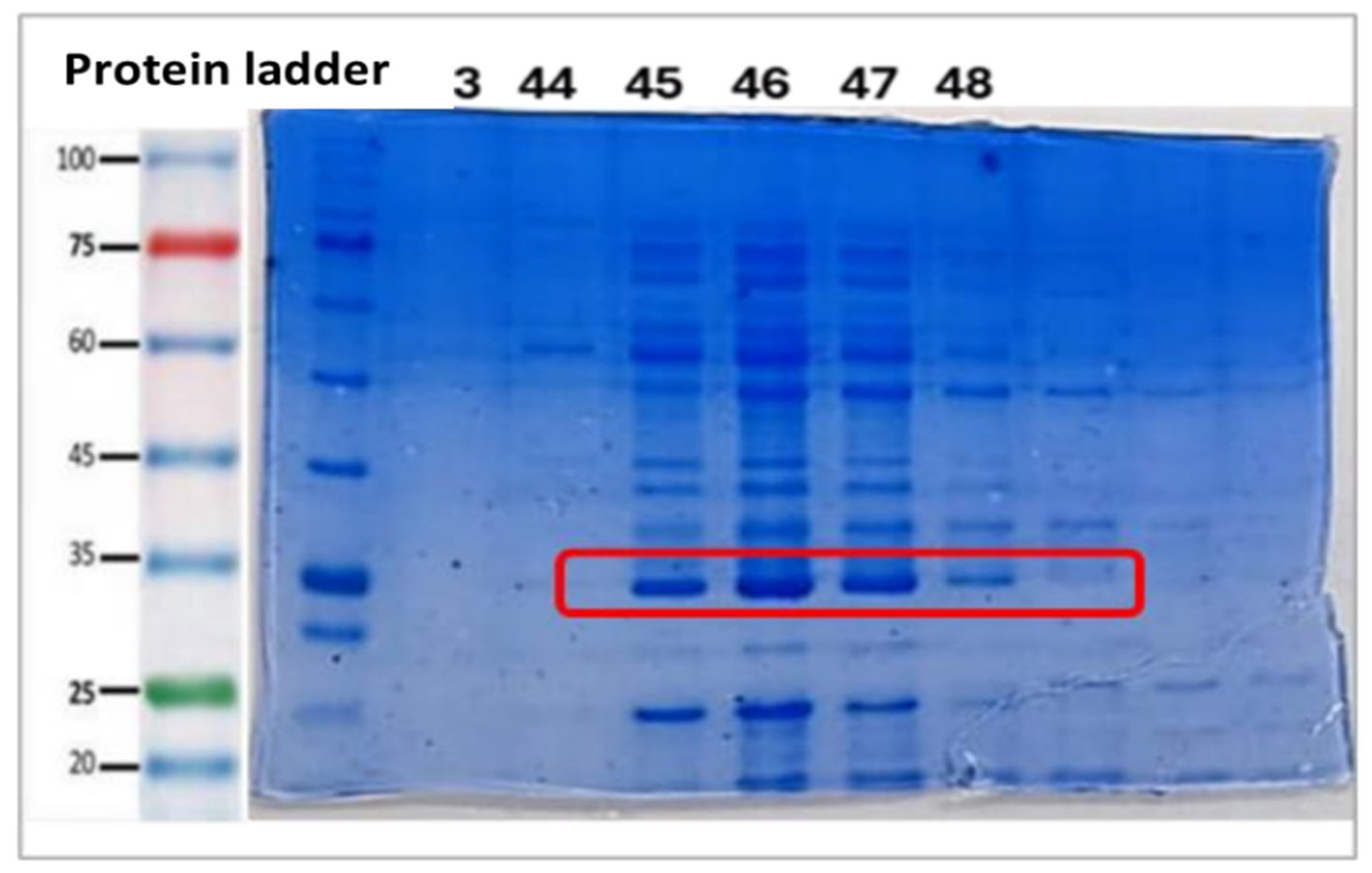
2.5. Culture Scale-Up and Purification
3. Materials and Methods
3.1. Preparation of Electrocompetent E. coli Cells (DH5α, BL21, Rosetta)
3.2. Obtaining the Gene of Interest
3.3. Amplification of the Gene of Interest
3.4. Plasmid and Insert Digestion
3.5. Plasmid and Insert Ligation
3.6. Transformation of E. coli (DH5α, BL21, Rosetta) Electrocompetent
3.7. Cloning of Plasmid pET-22b with the Inserted Gene
3.8. Amplification and Selection of the Transformed Plasmid (Verification of Transformation)
3.9. DNA Quantification by UV Spectrophotometry
3.10. Transformation of BL21 and Rosetta Strains and Amplification of Selected Positive Clones
3.11. Induction of Protein Expression in the Periplasm by IPTG
3.12. Quantification of Proteins by UV Spectrophotometry
3.13. Detection of Asparaginase Activity by Qualitative Hydroxylamine Assay
3.14. Measurement of Activity by Nessler’s Assay
3.15. Optimization of the Process of Induction of Chimeric Protein Expression by IPTG
3.16. Scale-Up of the Culture to 1L and Induction of Protein Expression in the Periplasm by IPTG
3.17. Protein Extraction
Sonication
3.18. Purification of the Engineered Chimeric Protein
AKTA Star Affinity Chromatography
3.19. 12% Polyacrylamide Gel Electrophoresis for Identification of the Chimeric Enzyme
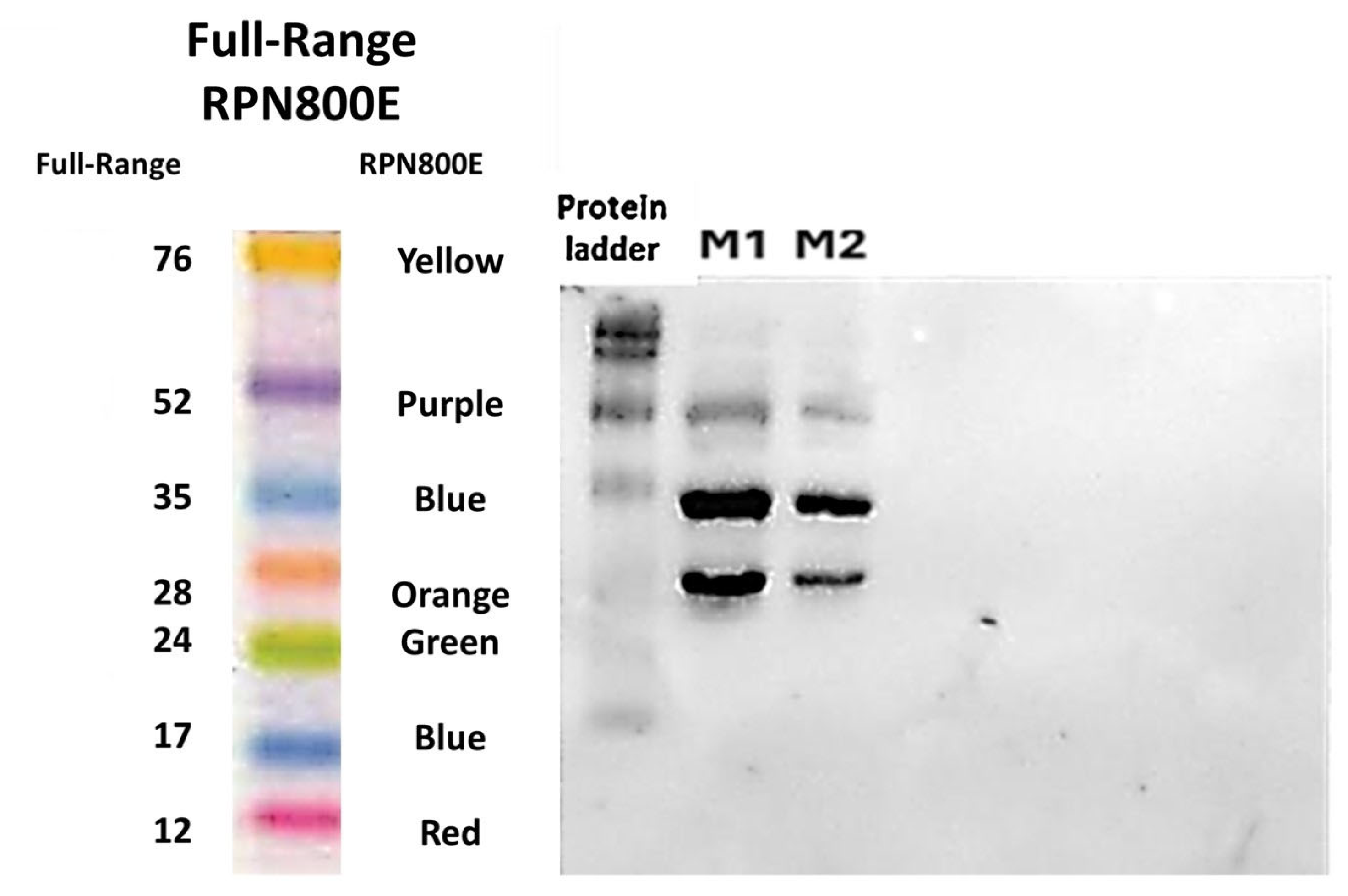
3.20. Protein Identification by Western Blot
3.21. Statistical Analysis
Full Factorial Design
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- SEPEAP. Consejo Editorial Subdirectores Ejecutivos Pediatría Integral. 2016. Available online: www.sepeap.org (accessed on 3 June 2025).
- Ceppi, F.; Cazzaniga, G.; Colombini, A.; Biondi, A.; Conter, V. Risk factors for relapse in childhood acute lymphoblastic leukemia: Prediction and prevention. Expert Rev. Hematol. 2015, 8, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, A.; Miyake, K.; Nordlund, J.; Syvänen, A.-C.; van der Weyden, L.; Honda, H.; Yamasaki, N.; Nagamachi, A.; Inaba, T.; Ikawa, T.; et al. Association of aberrant ASNS imprinting with asparaginase sensitivity and chromosomal abnormality in childhood BCP-ALL. Blood 2020, 136, 2319–2333. [Google Scholar] [CrossRef] [PubMed]
- Pui, C.-H.; Yang, J.J.; Hunger, S.P.; Pieters, R.; Schrappe, M.; Biondi, A.; Vora, A.; Baruchel, A.; Silverman, L.B.; Schmiegelow, K.; et al. Childhood Acute Lymphoblastic Leukemia: Progress Through Collaboration. J. Clin. Oncol. 2015, 33, 2938–2948. [Google Scholar] [CrossRef] [PubMed]
- Baruchel, A.; Brown, P.; Rizzari, C.; Silverman, L.; van der Sluis, I.; Wolthers, B.O.; Schmiegelow, K. Increasing completion of asparaginase treatment in childhood acute lymphoblastic leukaemia (ALL): Summary of an expert panel discussion. ESMO Open 2020, 5, e000977. [Google Scholar] [CrossRef] [PubMed]
- Pieters, R.; Hunger, S.P.; Boos, J.; Rizzari, C.; Silverman, L.; Baruchel, A.; Goekbuget, N.; Schrappe, M.; Pui, C. L-asparaginase treatment in acute lymphoblastic leukemia. Cancer 2011, 117, 238–249. [Google Scholar] [CrossRef] [PubMed]
- Völler, S.; Pichlmeier, U.; Zens, A.; Hempel, G. Pharmacokinetics of recombinant asparaginase in children with acute lymphoblastic leukemia. Cancer Chemother. Pharmacol. 2018, 81, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Ettinger, A.R. Pegaspargase (Oncaspar). J. Pediatr. Oncol. Nurs. 1995, 12, 46–48. [Google Scholar] [CrossRef] [PubMed]
- Kloos, R.; Van Der Sluis, I.M.; Mastrobattista, E.; Hennink, W.; Pieters, R.; Verhoef, J.J. Acute lymphoblastic leukaemia patients treated with PEGasparaginase develop antibodies to PEG and the succinate linker. Br. J. Haematol. 2020, 189, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Avramis, V.I.; Tiwari, P.N. Asparaginase (native ASNase or pegylated ASNase) in the treatment of acute lymphoblastic leukemia. Int. J. Nanomed. 2006, 1, 241–254. [Google Scholar]
- Lin, T.; Dumas, T.; Kaullen, J.; Berry, N.S.; Choi, M.R.; Zomorodi, K.; Silverman, J.A. Population Pharmacokinetic Model Development and Simulation for Recombinant Erwinia Asparaginase Produced in Pseudomonas fluorescens (JZP-458). Clin. Pharmacol. Drug Dev. 2021, 2021, 1503–1513. [Google Scholar] [CrossRef] [PubMed]
- Batool, T.; Makky, E.A.; Jalal, M.; Yusoff, M.M. A Comprehensive Review on L-Asparaginase and Its Applications. Appl. Biochem. Biotechnol. 2016, 178, 900–923. [Google Scholar] [CrossRef] [PubMed]
- Kidd, J.G. Regression of transplanted lymphomas induced in vivo by means of normal guinea pig serum: I. Course of transplanted cancers of various kinds in mice and rats given guinea pig serum, horse serum, or rabbit serum. J. Exp. Med. 1953, 98, 565–582. [Google Scholar] [CrossRef] [PubMed]
- Broome, J.D. Evidence that the L-Asparaginase Activity of Guinea Pig Serum is responsible for its Antilymphoma Effects. Nature 1961, 191, 1114–1115. [Google Scholar] [CrossRef]
- Asthana, N.; Azrni, W. Microbial L-Asparaginase: A Potent Antitumour Enzyme. Indian J. Biotechnol. 2003, 2, 184–194. [Google Scholar]
- Krishna, K.; Patro, R.; Satpathy, S.; Gupta, N. Evaluation of some fungi for L-asparaginase production. Indian J. Fundam. Appl. Life Sci. 2011, 1, 219–221. [Google Scholar]
- Sanches, M.; Krauchenco, S.; Polikarpov, I. Structure, Substrate Complexation and Reaction Mechanism of Bacterial Asparaginases. Curr. Chem. Biol. 2008, 1, 75–86. [Google Scholar]
- Borek, D.; Jaskólski, M. Sequence analysis of enzymes with asparaginase activity. Acta Biochim. Pol. 2001, 48, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Michalska, K.; Bujacz, G.; Jaskolski, M. Crystal Structure of Plant Asparaginase. J. Mol. Biol. 2006, 360, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Ortuño-Olea, L.; Durán-Vargas, S. The L-asparagine operon of Rhizobium etli contains a gene encoding an atypical asparaginase. FEMS. Microbiol. Lett. 2000, 189, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Belviso, S.; Iuliano, R.; Amato, R.; Perrotti, N.; Menniti, M. The human asparaginase enzyme (ASPG) inhibits growth in leukemic cells. PLoS ONE 2017, 12, e0178174. [Google Scholar] [CrossRef] [PubMed]
- Lubkowski, J.; Wlodawer, A. Structural and biochemical properties of L-asparaginase. FEBS J. 2021, 288, 4183–4209. [Google Scholar] [CrossRef] [PubMed]
- Swain, A.L.; Jask6lski, M.; Houssett, D.; Rao, J.K.M.; Wlodawert, A. Crystal structure of Escherichia coli L-asparaginase, an enzyme used in cancer therapy (amidohydrolase/leukemia/active site/aspartate). Proc. Natl. Acad. Sci. USA 1993, 90, 1474–1478. [Google Scholar] [CrossRef] [PubMed]
- Labrou, N.E.; Papageorgiou, A.C.; Avramis, V.I. Structure-Function Relationships and Clinical Applications of L-Asparaginases. Curr. Med. Chem. 2010, 17, 2183–2195. [Google Scholar] [CrossRef] [PubMed]
- Chand, S.; Mahajan, R.V.; Prasad, J.P.; Sahoo, D.K.; Mihooliya, K.N.; Dhar, M.S.; Sharma, G. A comprehensive review on microbial L-asparaginase: Bioprocessing, characterization, and industrial applications. Biotechnol. Appl. Biochem. 2020, 67, 619–647. [Google Scholar] [CrossRef] [PubMed]
- Beckett, A.; Gervais, D. What Makes A Good New Ther. L-Asparaginase? World J. Microbiol. Biotechnol. 2019, 35, 152. [Google Scholar] [CrossRef] [PubMed]
- MOrabi, H.; El-Fakharany, E.M.; Abdelkhalek, E.S.; Sidkey, N.M. L-asparaginase and L-glutaminase: Sources, production, and applications in medicine and industry. J. Microbiol. Biotechnol. Food Sci. 2021, 2021, 179–190. [Google Scholar] [CrossRef]
- Müller, H.J.; Boos, J. Use of L-asparaginase in childhood ALL. Crit. Rev. Oncol. Hematol. 1998, 28, 97–113. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Batra, S.; Zhang, J. Asparagine: A Metabolite to Be Targeted in Cancers. Metabolites 2021, 11, 402. [Google Scholar] [CrossRef] [PubMed]
- Ettinger, L.J.; Ettinger, A.G.; Avramis, W.I.; Gaynon, P.S. Acute Lymphoblastic Leukaemia. BioDrugs 1997, 7, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Mehta, R.K.; Verma, S.; Pati, R.; Sengupta, M.; Khatua, B.; Jena, R.K.; Sethy, S.; Kar, S.K.; Mandal, C.; Roehm, K.H.; et al. Mutations in Subunit Interface and B-cell Epitopes Improve Antileukemic Activities of Escherichia coli Asparaginase-II: Evaluation of immunogenicity in mice. J. Biol. Chem. 2014, 289, 3555–3570. [Google Scholar] [CrossRef] [PubMed]
- Krishna, M.; Nadler, S.G. Immunogenicity to Biotherapeutics–The Role of Anti-drug Immune Complexes. Front. Immunol. 2016, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, M.H.G.; da Silva Fiúza, T.; de Morais, S.B.; de Souza, T.d.A.C.B.; Trevizani, R. Circumventing the side effects of L-asparaginase. In Biomedicine and Pharmacotherapy; Elsevier Masson s.r.l.: Amsterdam, The Netherlands, 2021; Volume 139. [Google Scholar]
- Fung, M.K.L.; Chan, G.C.F. Drug-induced amino acid deprivation as strategy for cancer therapy. J. Hematol. Oncol. 2017, 10, 144. [Google Scholar] [CrossRef] [PubMed]
- Offman, M.N.; Krol, M.; Patel, N.; Krishnan, S.; Liu, J.; Saha, V.; Bates, P.A. Rational engineering of L-asparaginase reveals importance of dual activity for cancer cell toxicity. Blood 2011, 117, 1614–1621. [Google Scholar] [CrossRef] [PubMed]
- Peterson, R.G.; Handschumacher, R.E.; Mitchell, M.S. Immunological responses to L-asparaginase. J. Clin. Investig. 1971, 50, 1080–1090. [Google Scholar] [CrossRef] [PubMed]
- Battistel, A.P.; Rocha BSda Santos MTdos Daudt, L.E.; Michalowski, M.B. Allergic reactions to asparaginase: Retrospective cohort study in pediatric patients with acute lymphoid leukemia. Hematol. Transfus. Cell Ther. 2021, 43, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Zalewska-Szewczyk, B.; Andrzejewski, W.; Młynarski, W.; Jędrychowska-Dańska, K.; Witas, H.; Bodalski, J. The anti-asparagines antibodies correlate with L-asparagines activity and may affect clinical outcome of childhood acute lymphoblastic leukemia. Leuk. Lymphoma 2007, 48, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Rodríguez, G.; Jaime-Pérez, J.C.; Salinas-Carmona, M.C.; González-Díaz, S.N.; Castro-Corona, Á.; Cavazos-González, R.; Treviño-Villarreal, H.; Heredia-Salazar, A.C.; Gómez-Almaguer, D. Do immunoglobulin G and immunoglobulin E anti-L-asparaginase antibodies have distinct implications in children with acute lymphoblastic leukemia? A cross-sectional study. Rev. Bras. Hematol. Hemoter. 2017, 39, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Burke, M.J.; Zalewska-Szewczyk, B. Hypersensitivity reactions to asparaginase therapy in acute lymphoblastic leukemia: Immunology and clinical consequences. In Future Oncology; Future Medicine Ltd.: London, UK, 2022; Volume 18, pp. 1285–1299. [Google Scholar]
- Belén, L.H.; Lissabet, J.B.; Rangel-Yagui, C.d.O.; Effer, B.; Monteiro, G.; Pessoa, A.; Avendaño, J.G.F. A structural in silico analysis of the immunogenicity of L-asparaginase from Escherichia coli and Erwinia carotovora. Biologicals 2019, 59, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Avramis, V.I.; Panosyan, E.H. Pharmacokinetic/Pharmacodynamic Relationships of Asparaginase Formulations. Clin. Pharmacokinet. 2005, 44, 367–393. [Google Scholar] [CrossRef] [PubMed]
- Asselin, B.L.; Whitin, J.C.; Coppola, D.J.; Rupp, I.P.; Sallan, S.E.; Cohen, H.J. Comparative pharmacokinetic studies of three asparaginase preparations. J. Clin. Oncol. 1993, 11, 1780–1786. [Google Scholar] [CrossRef] [PubMed]
- Keating, M.J.; Holmes, R.; Lerner, S.; Ho, D.H. L-Asparaginase and PEG Asparaginase—Past, Present, and Future. Leuk Lymphoma 1993, 10 (Suppl. S1), 153–157. [Google Scholar] [CrossRef] [PubMed]
- Rogers, T.H.; Babensee, J.E. Altered adherent leukocyte profile on biomaterials in Toll-like receptor 4 deficient mice. Biomaterials 2010, 31, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Pokrovsky, V.S.; Kazanov, M.D.; Dyakov, I.N.; Pokrovskaya, M.V.; Aleksandrova, S.S. Comparative immunogenicity and structural analysis of epitopes of different bacterial L-asparaginases. BMC Cancer 2016, 16, 89. [Google Scholar] [CrossRef] [PubMed]
- Zalewska-Szewczyk, B.; Gach, A.; Wyka, K.; Bodalski, J.; Młynarski, W. The cross-reactivity of anti-asparaginase antibodies against different L-asparaginase preparations. Clin. Exp. Med. 2009, 9, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Marini, B.L.; Perissinotti, A.J.; Bixby, D.L.; Brown, J.; Burke, P.W. Catalyzing improvements in ALL therapy with asparaginase. Blood Rev. 2017, 31, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Vrooman, L.M.; Supko, J.G.; Neuberg, D.S.; Asselin, B.L.; Athale, U.H.; Clavell, L.; Kelly, K.M.; Laverdière, C.; Michon, B.; Schorin, M.; et al. Erwinia asparaginase after allergy to E. coli asparaginase in children with acute lymphoblastic leukemia. Pediatr. Blood Cancer 2010, 54, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Rigouin, C.; Nguyen, H.A.; Schalk, A.M.; Lavie, A. Discovery of human-like L-asparaginases with potential clinical use by directed evolution. Sci. Rep. 2017, 7, 10224. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.L.; Guimarães, G.M.; Polakiewicz, B.; de Moraes Pitombo, R.N.; Abrahão-Neto, J. Effects of polyethylene glycol attachment on physicochemical and biological stability of E. coli L-asparaginase. Int. J. Pharm. 2002, 237, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Wenner, K.A.; Vieira Pinheiro, J.P.; Escherich, G.; Wessalowski, R.; Jorch, N.; Wolff, J.; Stehn, M.; Kohlschutter, A.; Boos, J.; Jenka-Schaub, G.E. Asparagine Concentration in Plasma After 2500 IU/m2 PEG-Asparaginase i.v. in Child. Acute Lymphoblastic Leukemia. Klin. Padiatr. 2005, 217, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.; Kirkwood, A.A.; Dey, A.; Marks, D.I.; McMillan, A.K.; Menne, T.F.; Micklewright, L.; Patrick, P.; Purnell, S.; Rowntree, C.J.; et al. Pegylated-asparaginase during induction therapy for adult acute lymphoblastic leukaemia: Toxicity data from the UKALL14 trial. Leukemia 2017, 31, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Asselin, B.; Rizzari, C. Asparaginase pharmacokinetics and implications of therapeutic drug monitoring. Leuk. Lymphoma 2015, 56, 2273–2280. [Google Scholar] [CrossRef] [PubMed]
- Alshiekh-Nasany, R.; Douer, D. L-Carnitine for Treatment of Pegasparaginase-Induced Hepatotoxicity. Acta Haematol. 2016, 135, 208–210. [Google Scholar] [CrossRef] [PubMed]
- Zha, D. Glycoengineered Pichia-based expression of monoclonal antibodies. In Glycosylation Engineering of Biopharmaceuticals; Springer: Berlin/Heidelberg, Germany, 2013; pp. 31–43. [Google Scholar]
- Effer, B.; Lima, G.M.; Cabarca, S.; Pessoa, A.; Farías, J.G.; Monteiro, G. L-Asparaginase from E. Chrysanthemi expressed in glycoswitch®: Effect of His-Tag fusion on the extracellular expression. Prep. Biochem. Biotechnol. 2019, 49, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Laukens, B.; Wachter CDe Callewaert, N. Engineering the Pichia pastoris N-glycosylation pathway using the GlycoSwitch technology. In Glyco-Engineering; Springer: Berlin/Heidelberg, Germany, 2015; pp. 103–122. [Google Scholar]
- Effenberger, I.; Harport, M.; Pfannstiel, J.; Klaiber, I.; Schaller, A. Expression in Pichia Pastoris and Characterization of Two Novel Dirigent Proteins for Atropselective Formation of Gossypol. Appl. Microbiol. Biotechnol. 2017, 101, 2021–2032. [Google Scholar] [CrossRef] [PubMed]
- Moola, Z.B.; Scawen, M.D.; Atkinson, T.; Nicholls, D.J. Erwinia chrysanthemil-asparaginase: Epitope mapping and production of antigenically modified enzymes. Biochem. J. 1994, 302, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Belén, L.H.; Beltrán Lissabet, J.F.; de Oliveira Rangel-Yagui, C.; Monteiro, G.; Pessoa, A.; Farías, J.G. Immunogenicity assessment of fungal L-asparaginases: An in silico approach. SN Appl. Sci. 2020, 2, 222. [Google Scholar] [CrossRef]
- Ramya, L.N.; Pulicherla, K.K. Studies on Deimmunization of Antileukaemic L-Asparaginase to have Reduced Clinical Immunogenicity- An in silico Approach. Pathol. Oncol. Res. 2015, 21, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Maggi, M.; Mittelman, S.D.; Parmentier, J.H.; Colombo, G.; Meli, M.; Whitmire, J.M.; Merrell, D.S.; Whitelegge, J.; Scotti, C. A protease-resistant Escherichia coli asparaginase with outstanding stability and enhanced anti-leukaemic activity in vitro. Sci. Rep. 2017, 7, 14479. [Google Scholar] [CrossRef] [PubMed]
- Mahboobi, M.; Sedighian, H.; Hedayati, C.H.M.; Bambai, B.; Soofian, S.E.; Amani, J. Applying bioinformatic tools for modeling and modifying type II E. coli L-Asparginase to present a better therapeutic agent/drug for acute lymphoblastic leukemia. Int. J. Cancer Manag. 2017, 10, e5785. [Google Scholar] [CrossRef]
- Jianhua, C.; Yujun, W.; Ruibo, J.; Min, W.; Wutong, W. Probing the antigenicity of E. colil-Asparaginase by Mutational analysis. Mol. Biotechnol. 2006, 33, 57–65. [Google Scholar] [CrossRef]
- Thomas, X.; Le Jeune, C. Erythrocyte encapsulated L-asparaginase (GRASPA) in acute leukemia. Int. J. Hematol. Oncol. 2016, 5, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Ben-Yehudah, A.; Lorberboum-Galski, H. Targeted cancer therapy with gonadotropin-releasing hormone chimeric proteins. Expert Rev. Anticancer Ther. 2004, 4, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Lorberboum-Galski, H. Human toxin-based recombinant immunotoxins/chimeric proteins as a drug delivery system for targeted treatment of human diseases. Expert Opin. Drug Deliv. 2011, 8, 605–621. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Park, T.; Hong, E.; Amatya, R.; Park, K.-A.; Park, Y.-H.; Min, K.A.; Jin, M.; Lee, S.; Hwang, S.; et al. Genetic engineering of novel super long-acting Exendin-4 chimeric protein for effective treatment of metabolic and cognitive complications of obesity. Biomaterials 2020, 257, 120250. [Google Scholar] [CrossRef] [PubMed]
- Nie, T.; Meng, F.; Zhou, L.; Lu, F.; Bie, X.; Lu, Z.; Lu, Y. In Silico Development of Novel Chimeric Lysins with Highly Specific Inhibition against Salmonella by Computer-Aided Design. J. Agric. Food Chem. 2021, 69, 3751–3760. [Google Scholar] [CrossRef] [PubMed]
- Newsted, W.J.; Ramjeesingh, M.; Zywulko, M.; Rothstein, S.J.; Shami, E.Y. Engineering resistance to trypsin inactivation into L-asparaginase through the production of a chimeric protein between the enzyme and a protective single-chain antibody. Enzym. Microb. Technol. 1995, 17, 757–764. [Google Scholar] [CrossRef] [PubMed]
- Gaofu, Q.; Rongyue, C.; Dan, M.; Xiuyun, Z.; Xuejun, W.; Jie, W.; Jingjing, L. Asparaginase Display of Human Cholesteryl Ester Transfer Protein (CETP) B Cell Epitopes for Inducing High Titers of Anti-CETP Antibodies In Vivo. Protein Pept. Lett. 2006, 13, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Pedroso, A.; Belén, L.H.; Beltrán, J.F.; Castillo, R.L.; Pessoa, A.; Pedroso, E.; Farías, J.G. In Silico Design of a Chimeric Humanized L-asparaginase. Int. J. Mol. Sci. 2023, 24, 7550. [Google Scholar] [CrossRef] [PubMed]
- Gilje, B.; Heikkilä, R.; Oltedal, S.; Tjensvoll, K.; Nordgård, O. High-fidelity DNA polymerase enhances the sensitivity of a peptide nucleic acid clamp PCR assay for K-ras mutations. J. Mol. Diagnostics 2008, 10, 325–331. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jang, M.; Kim, S. Inhibition of Non-specific Amplification in Loop-Mediated Isothermal Amplification via Tetramethylammonium Chloride. Biochip J. 2022, 16, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Alicia Farma, S.; Handayani, D.; Hilda Putri, D. Optimization of Annealing Temperature of HIF-1 A and 18s rRNA in Blood of Swimming Athletes Using RT-PCR 2020. Available online: http://tmcalculator.neb.com/ (accessed on 3 June 2025).
- So, K.Y.K.; Fong, J.J.; Lam, I.P.Y.; Dudgeon, D. Pitfalls during in silico prediction of primer specificity for eDNA surveillance. Ecosphere 2020, 11, e03193. [Google Scholar] [CrossRef]
- Zhou, Y.; Bo, F.; Tian, T.; Wu, B.; Zhu, B. Excessive addition split peak formed by the non-templated nucleotide addition property of Taq DNA polymerase after PCR amplification. Front. Bioeng. Biotechnol. 2023, 11, 1180542. [Google Scholar] [CrossRef] [PubMed]
- Barboza-Fallas, L.; Guillen-Watson, R.; Rivas-Solano, O.; Peraza-Moraga, J. Optimización de temperatura de anillamiento de PCR multiplex para la detección de Listeria monocytogenes. Tecnol. Marcha 2019, 32, 37–42. [Google Scholar]
- Farrell, R.E. Chapter 8—RT-PCR: A Science and an Art Form. In RNA Methodologies, 5th ed.; Farrell, R.E., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 209–281. Available online: https://www.sciencedirect.com/science/article/pii/B9780128046784000087 (accessed on 3 June 2025).
- Herzer, S. DNA Purification. In Molecular Biology Problem Solver; Wiley: Hoboken, NJ, USA, 2001; pp. 167–195. [Google Scholar] [CrossRef]
- Gingeras, T.R.; Myers, P.A.; Olson, J.A.; Hanberg, F.A.; Roberts, R.J. A new specific endonuclease present in Xanthomonas holcicola, Xanthomonas papavericola and Brevibacterium luteum. J. Mol. Biol. 1978, 118, 113–122. Available online: https://www.sciencedirect.com/science/article/pii/0022283678902474 (accessed on 3 June 2025). [CrossRef] [PubMed]
- Arber, W. Restriction endonucleases. Angew. Chem. Int. Ed. Engl. 1978, 17, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Jung, V.; Pestka, S.B.; Pestka, S. Efficient cloning of PCR generated DNA containing terminal restriction endonuclease recognition sites. Nucleic Acids Res. 1990, 18, 6156. [Google Scholar] [CrossRef] [PubMed]
- Pabón, L.C.V.; Castaño, D.M.; Gutiérrez, F.A.A. Clonación y expresión en Escherichia coli de genes de celulasas de Clostridium IBUN 22A. Rev. Colomb. Biotecnol. 2002, 4, 29–35. [Google Scholar]
- El-Mogy, M.A.; Haj-Ahmad, Y. Effect of DNA Contaminants on Calcium Phosphate-Based DNA Delivery and Gene Expression. J. Biotech Res. 2012, 4, 619–647. [Google Scholar]
- Sambrook, J.; Russell, D.W. Detection of DNA in agarose gels. In Molecular Cloning, a Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; pp. 5–14. [Google Scholar]
- Sambrook, J.; Russell, D.W. Chapter 8—Molecular Cloning. In In Vitro Amplification of DNA by the Polymerase Chain Reaction; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; Volume 2. [Google Scholar]
- Iglesias, J.O. Comprendiendo la resistencia a antibióticos. Rev. De Investig. Y Educ. En Cienc. De La Salud (RIECS) 2019, 4, 84–89. [Google Scholar] [CrossRef]
- Liu, X.; Liu, L.; Wang, Y.; Wang, X.; Ma, Y.; Li, Y. The Study on the factors affecting transformation efficiency of E. coli. Competent cells. Cell 2014, 5, x106. [Google Scholar]
- Jonasson, P.; Liljeqvist, S.; Nygren Pke Ståhl, S. Genetic design for facilitated production and recovery of recombinant proteins in Escherichia coli. Biotechnol. Appl. Biochem. 2002, 35, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Keum, K.C.; Lee, S.Y. Production of recombinant proteins by high cell density culture of Escherichia coli. Chem. Eng. Sci. 2006, 61, 876–885. [Google Scholar] [CrossRef]
- de Moura, W.A.F.; Schultz, L.; Breyer, C.A.; de Oliveira, A.L.P.; Tairum, C.A.; Fernandes, G.C.; Toyama, M.H.; Pessoa, A., Jr.; Monteiro, G.; de Oliveira, M.A. Functional and structural evaluation of the antileukaemic enzyme L-asparaginase II expressed at low temperature by different Escherichia coli strains. Biotechnol. Lett. 2020, 42, 2333–2344. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, W.; Wu, H.; Zhang, W.; Guang, C.; Mu, W. Microbial production, molecular modification, and practical application of L-Asparaginase: A review. Int. J. Biol. Macromol. 2021, 186, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Falero-Morejón, A.; Brito-Molina, S.; González-Acevedo-Forte, D.; Serrano-Rivero, Y.; Aldama-Paz, G.; Pimienta-Rodriguez, E.; Domínguez, K.M. Purificación de los cuerpos de inclusión que contienen a la proteína L1 del Virus del Papiloma Humano 16 producidos en la cepa Rosetta de Escherichia coli. Rev. CENIC Cienc. Biológicas 2015, 46, 344–348. [Google Scholar]
- Moreno-González, P.A.; Diaz, G.J.; Ramírez-Hernández, M.H. Production and Purification of Avian Antibodies (IgYs) from Inclusion Bodies of a Recombinant Protein Central in NAD+ Metabolism; Revista Colombiana de Química: Havana, Cuba, 2013. [Google Scholar]
- Xia, X.X.; Qian, Z.G.; Lee, S.Y. Comparative proteomic and genetic analyses reveal unidentified mutations in Escherichia coli XL1-Blue and DH5α. FEMS Microbiol. Lett. 2011, 314, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.T.; Verma, C.S.; Lane, D.P.; Gan, S.K.E. A comparison and optimization of methods and factors affecting the transformation of Escherichia coli. Biosci. Rep. 2013, 33, e00086. [Google Scholar] [CrossRef] [PubMed]
- Liu, I.; Liu, M.; Shergill, K. The effect of spheroplast formation on the transformation efficiency in Escherichia coli DH5α. J. Exp. Microbiol. Immunol. 2006, 9, 81–85. [Google Scholar]
- Baneyx, F.; Mujacic, M. Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 2004, 22, 1399–1408. [Google Scholar] [CrossRef] [PubMed]
- Dastmalchi, M.; Alizadeh, M.; Jamshidi-Kandjan, O.; Rezazadeh, H.; Hamzeh-Mivehroud, M.; Farajollahi, M.M.; Dastmalchi, S. Expression and Biological Evaluation of an Engineered Recombinant L-asparaginase Designed by In Silico Method Based on Sequence of the Enzyme from Escherichia coli. Adv. Pharm. Bull. 2023, 13, 827–836. Available online: https://apb.tbzmed.ac.ir/Article/apb-39578 (accessed on 3 June 2025). [CrossRef] [PubMed]
- Gao, X.; Peng, S.; Mei, S.; Liang, K.; Khan, M.S.I.; Vong, E.G.; Zhan, J. Expression and functional identification of recombinant SARS-CoV-2 receptor binding domain (RBD) from E.coli. system. Prep. Biochem. Biotechnol. 2022, 52, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Afroze, B.; Abbas, R.; Khan, M.A.; Akram, M.; Tahir, S.; Bakht, S.; Munir, A.; Shahid, A.A. Method for efficient soluble expression and purification of recombinant human interleukin-15. Protein Expr. Purif. 2021, 177, 105746. [Google Scholar] [CrossRef] [PubMed]
- Abdollahi, S.; Morowvat, M.H.; Savardashtaki, A.; Irajie, C.; Najafipour, S.; Ghasemi, Y. Evaluating five Escherichia coli derivative strains as a platform for arginine deiminase overproduction. Recent Pat. Biotechnol. 2022, 16, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.D.; Hatahet, F.; Salo, K.E.H.; Enlund, E.; Zhang, C.; Ruddock, L.W. Pre-expression of a sulfhydryl oxidase significantly increases the yields of eukaryotic disulfide bond containing proteins expressed in the cytoplasm of E. coli. Microb. Cell Factories 2011, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Rosano, G.L.; Morales, E.S.; Ceccarelli, E.A. New tools for recombinant protein production in Escherichia coli: A 5-year update. Protein Sci. 2019, 28, 1412–1422. [Google Scholar] [CrossRef] [PubMed]
- Ohnuma, T.; Holland, J.F.; Freeman, A.; Sinks, L.F. Biochemical and Pharmacological Studies with Asparaginase in Man1. Cancer Res. 1970, 30, 2297–2305. [Google Scholar] [PubMed]
- Tegel, H.; Tourle, S.; Ottosson, J.; Persson, A. Increased levels of recombinant human proteins with the. Escherichia coli. Protein Expr. Purif. 2010, 69, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Caetano, L.F. Produção e Caracterização Preliminar de Variantes de L-Asparaginase II de Escherichia coli de Menor Potencial Imunogênico: Combinação de Estudos In Silico e In Vitro; Universidade Federal do Ceará: Fortaleza, Brasil, 2020. [Google Scholar]
- Bhatia, S.K.; Vivek, N.; Kumar, V.; Chandel, N.; Thakur, M.; Kumar, D.; Yang, Y.-H.; Pugazendhi, A.; Kumar, G. Molecular biology interventions for activity improvement and production of industrial enzymes. Bioresour. Technol. 2021, 324, 124596. [Google Scholar] [CrossRef] [PubMed]
- Oviedo-Trespalacios, O.; Peñabaena, R.P. Optimización de sistemas simulados a través de técnicas de superficie de respuesta. Ingeniare Rev. Chil. Ing. 2015, 23, 421–428. [Google Scholar] [CrossRef]
- Lopes, W.; dos Santos, B.A.F.; Sampaio, A.L.F.; Fontão, A.P.G.A.; Nascimento, H.J.; Jurgilas, P.B.; Torres, F.A.G.; Bon, E.P.d.S.; Almeida, R.V.; Ferrara, M.A. Expression, purification, and characterization of asparaginase II from Saccharomyces cerevisiae in Escherichia coli. Protein Expr. Purif. 2019, 159, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Soleymani, B.; Mostafaie, A. Analysis of methods to improve the solubility of recombinant bovine sex determining region Y protein. Rep. Biochem. Mol. Biol. 2019, 8, 227. [Google Scholar]
- Sandén, A.M.; Boström, M.; Markland, K.; Larsson, G. Solubility and proteolysis of the Zb-MalE and Zb-MalE31 proteins during overproduction in Escherichia coli. Biotechnol. Bioeng. 2005, 90, 239–247. [Google Scholar] [CrossRef] [PubMed]
- San-Miguel, T.; Pérez-Bermúdez, P.; Gavidia, I. Production of soluble eukaryotic recombinant proteins in E. coli is favoured in early log-phase cultures induced at low temperature. SpringerPlus 2013, 2, 89. [Google Scholar] [CrossRef] [PubMed]
- Costa, I.M.; Moura, D.C.; Lima, G.M.; Pessoa, A.; dos Santos, C.O.; de Oliveira, M.A.; Monteiro, G. Engineered asparaginase from enhances asparagine hydrolase activity and diminishes enzyme immunoreactivity—A new promise to treat acute lymphoblastic leukemia. J. Chem. Technol. Biotechnol. 2022, 97, 228–239. [Google Scholar] [CrossRef]
- Sinha, J.; Plantz, B.A.; Inan, M.; Meagher, M.M. Causes of proteolytic degradation of secreted recombinant proteins produced in methylotrophic yeast Pichia pastoris: Case study with recombinant ovine interferon-τ. Biotechnol. Bioeng. 2005, 89, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Meena, B.; Anburajan, L.; Vinithkumar, N.V.; Shridhar, D.; Raghavan, R.V.; Dharani, G.; Kirubagaran, R. Molecular expression of L-asparaginase gene from Nocardiopsis alba NIOT-VKMA08 in Escherichia coli: A prospective recombinant enzyme for leukaemia chemotherapy. Gene 2016, 590, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Muneer, F.; Siddique, M.H.; Azeem, F.; Rasul, I.; Muzammil, S.; Zubair, M.; Afzal, M.; Nadeem, H. Microbial L-asparaginase: Purification, characterization and applications. Arch. Microbiol. 2020, 202, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Arias, A. Las bacterias como sistema de expresión de proteínas heterólogas terapéuticas: Una revisión bibliográfica. Hechos Microbiológicos 2013, 4, 106–116. [Google Scholar]
- Pourhossein, M.; Korbekandi, H. Cloning, expression, purification and characterisation of Erwinia carotovora L-asparaginase in Escherichia coli. Adv. Biomed. Res. 2014, 3, 82. [Google Scholar] [CrossRef] [PubMed]
- García, J.; Santana, Z.; Zumalacárregui, L.; Quintana, M.; González, D.; Furrazola, G.; Cruz, O. Estrategias de obtención de proteínas recombinantes en Escherichia coli. Vaccimonitor 2013, 22, 30–39. [Google Scholar]
- Rozkov, A.; Högskolan, K.T. Control of Proteolysis of Recombinant Proteins in Escherichia coli; Royal Institute of Technology: Stockholm, Sweden, 2001. [Google Scholar]
- Gottesman, S. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 1996, 30, 465–506. [Google Scholar] [CrossRef] [PubMed]
- Farfán-García, A.E.; Ariza-Rojas, S.C.; Vargas-Cárdenas, F.A.; Vargas-Remolina, L.V. Mecanismos de virulencia de Escherichia coli enteropatógena. Rev. Chil. Infectol. 2016, 33, 438–450. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Sluis-Cremer, N.; Tachedjian, G. Dimerization of human immunodeficiency virus type 1 reverse transcriptase as an antiviral target. Curr. Pharm. Des. 2006, 12, 1879–1894. [Google Scholar] [CrossRef] [PubMed]
- Strzelczyk, P.; Zhang, D.; Alexandratos, J.; Piszczek, G.; Wlodawer, A.; Lubkowski, J. The dimeric form of bacterial l-asparaginase YpAI is fully active. FEBS J. 2023, 290, 780–795. [Google Scholar] [CrossRef] [PubMed]
- Pérez de Castro, A.M. Reacción en Cadena de la Polimerasa (Polymerase Chain Reaction, PCR); Universitat Politècnica de València: Valencia, Italy, 2011. [Google Scholar]
- Flores-Santos, J.C.; Moguel, I.S.; Monteiro, G.; Pessoa, A.; Vitolo, M. Improvement in extracellular secretion of recombinant L-asparaginase II by Escherichia coli BL21 (DE3) using glycine and n-dodecane. Braz. J. Microbiol. 2021, 52, 1247–1255. [Google Scholar] [CrossRef] [PubMed]
- Bahreini, E.; Aghaiypour, K.; Abbasalipourkabir, R.; Goodarzi, M.T.; Saidijam, M.; Safavieh, S.S. An optimized protocol for overproduction of recombinant protein expression in Escherichia coli. Prep. Biochem. Biotechnol. 2014, 44, 510–528. [Google Scholar] [CrossRef] [PubMed]
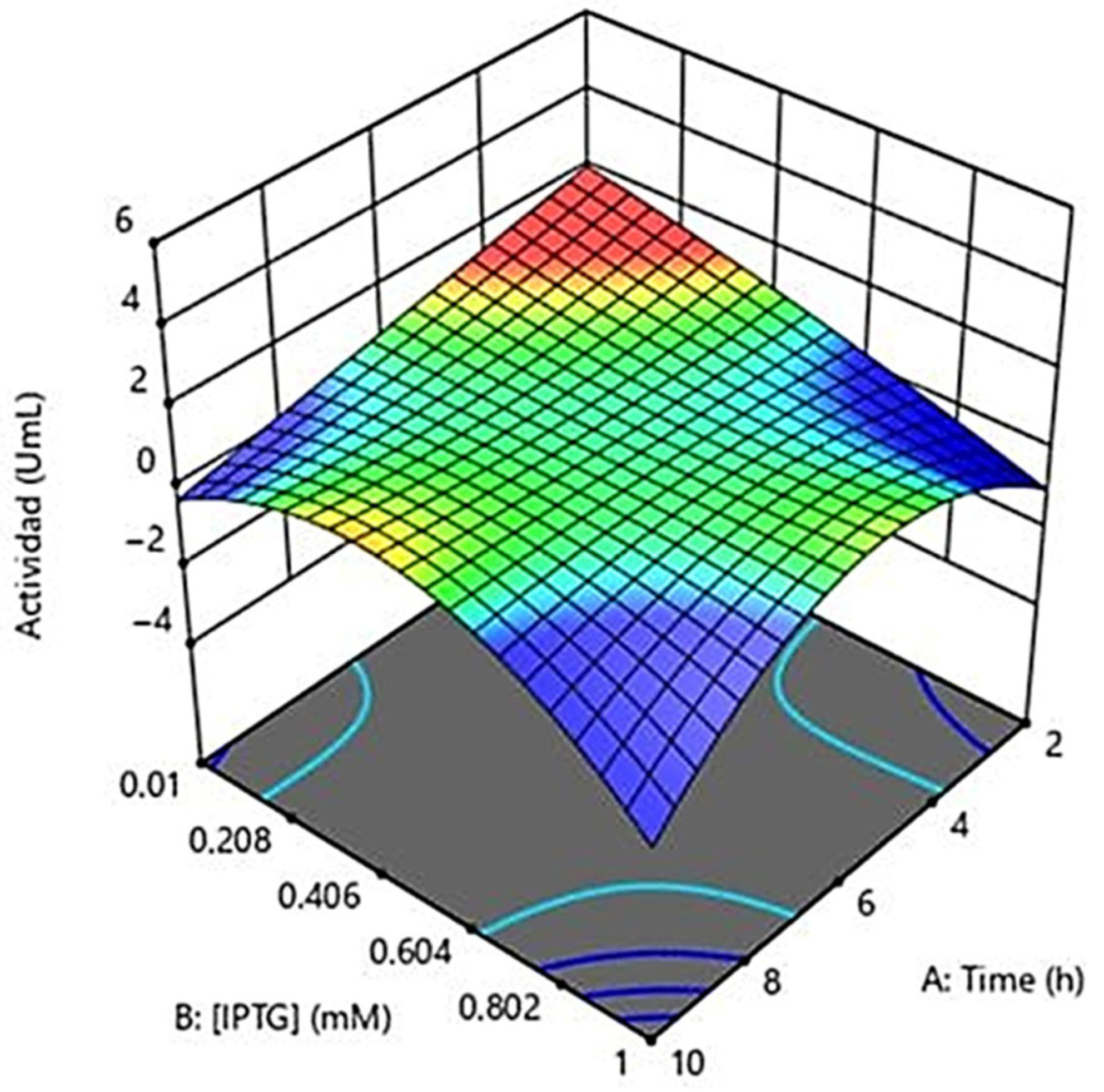
| STD | Run | Time | IPTG [mM] | Temperature °C |
|---|---|---|---|---|
| 13 | 3 | 6 | 0.505 | 20 |
| 1 | 4 | 4 | 0.2575 | 24.25 |
| 2 | 6 | 8 | 0.2575 | 24.25 |
| 3 | 16 | 4 | 0.7525 | 24.25 |
| 4 | 2 | 8 | 0.7525 | 24.25 |
| 9 | 13 | 2 | 0.505 | 28.5 |
| 10 | 19 | 10 | 0.505 | 28.5 |
| 11 | 5 | 6 | 0.01 | 28.5 |
| 12 | 17 | 6 | 1 | 28.5 |
| 15 | 15 | 6 | 0.505 | 28.5 |
| 16 | 20 | 6 | 0.505 | 28.5 |
| 17 | 10 | 6 | 0.505 | 28.5 |
| 18 | 12 | 6 | 0.505 | 28.5 |
| 19 | 1 | 6 | 0.505 | 28.5 |
| 20 | 14 | 6 | 0.505 | 28.5 |
| 5 | 8 | 4 | 0.2575 | 32.75 |
| 6 | 18 | 8 | 0.2575 | 32.75 |
| 7 | 7 | 4 | 0.7525 | 32.75 |
| 8 | 11 | 8 | 0.7525 | 32.75 |
| 14 | 9 | 6 | 0.505 | 37 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pedroso, A.; Miranda, J.; Lefin, N.; Effer, B.; Reyanldo, E.P.; Calle, Y.; Monteiro, G.; Pessoa, A., Jr.; Farias, J.G. Toward Safer Biotherapeutics: Expression and Characterization of a Humanized Chimeric L-Asparaginase in E. coli. Int. J. Mol. Sci. 2025, 26, 6919. https://doi.org/10.3390/ijms26146919
Pedroso A, Miranda J, Lefin N, Effer B, Reyanldo EP, Calle Y, Monteiro G, Pessoa A Jr., Farias JG. Toward Safer Biotherapeutics: Expression and Characterization of a Humanized Chimeric L-Asparaginase in E. coli. International Journal of Molecular Sciences. 2025; 26(14):6919. https://doi.org/10.3390/ijms26146919
Chicago/Turabian StylePedroso, Alejandro, Javiera Miranda, Nicolás Lefin, Brian Effer, Enrique Pedroso Reyanldo, Yolanda Calle, Gisele Monteiro, Adalberto Pessoa, Jr., and Jorge G. Farias. 2025. "Toward Safer Biotherapeutics: Expression and Characterization of a Humanized Chimeric L-Asparaginase in E. coli" International Journal of Molecular Sciences 26, no. 14: 6919. https://doi.org/10.3390/ijms26146919
APA StylePedroso, A., Miranda, J., Lefin, N., Effer, B., Reyanldo, E. P., Calle, Y., Monteiro, G., Pessoa, A., Jr., & Farias, J. G. (2025). Toward Safer Biotherapeutics: Expression and Characterization of a Humanized Chimeric L-Asparaginase in E. coli. International Journal of Molecular Sciences, 26(14), 6919. https://doi.org/10.3390/ijms26146919








