Identifying Cortical Molecular Biomarkers Potentially Associated with Learning in Mice Using Artificial Intelligence
Abstract
1. Introduction
1.1. Closely Related Work
1.2. Hypothesis
2. Results
2.1. Predicting Whether a Mouse Was Shocked to Learn
2.2. Results of the Alternative Approach to Detecting Potentially Learning-Linked Proteins
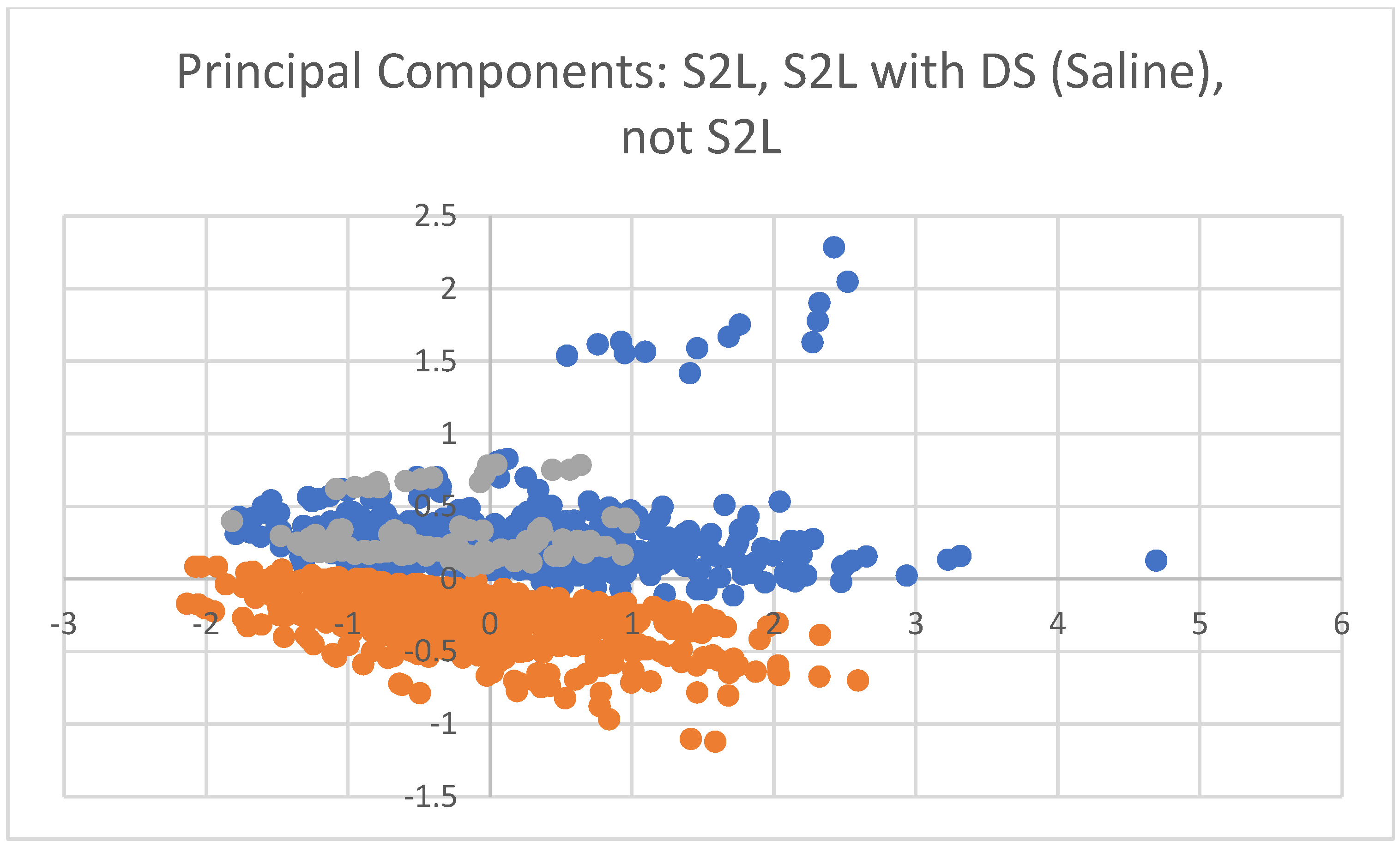
2.3. Visualization of Findings
3. Discussion
3.1. Protein Expression Potential Significance
3.2. Potential Implications: Causality and Correlation
3.3. Discussion of Alternative Analysis
3.4. Literature Comparison
3.5. Machine Learning and Feature Selection
3.6. Strengths, Limitations, and Future Work
4. Materials and Methods
4.1. Dataset Description
4.2. Machine Learning
4.3. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| acc | accuracy |
| AI | artificial intelligence |
| assoc | filter-based association FS |
| auroc | area under the receiver operating characteristic curve |
| bal-acc | balanced accuracy |
| BCL2 | B-cell lymphoma 2 |
| BDNF | brain-derived neurotrophic factor |
| DS | Down syndrome |
| embed_lgbm | embedded lgbm FS |
| embed_linear | embedded linear FS |
| f1 | F1 score |
| FS | feature selection |
| H3AcK18 | histone H3 acetylation at lysine 18 |
| KNN | k-nearest neighbor |
| lgbm | Light Gradient-Boosting Machine |
| lr | logistic regression |
| ML | Machine Learning |
| NDMA | N-methyl-D-aspartate receptor |
| npv | Negative Predictive Value |
| NR2A | the subunit of the NDMA receptor |
| pERK | protein kinase R-like endoplasmic reticulum kinase |
| ppv | positive predictive value |
| pred | filter-based prediction FS |
| rf | random forest |
| S2L | shocked to learn |
| sens | sensitivity |
| sgd | stochastic gradient descent |
| SOD1 | superoxide dismutase 1 |
| spec | specificity |
| SOM | self-organizing map |
| wrap | wrapper-based redundancy aware FS |
References
- Adolphs, R. The unsolved problems of neuroscience. Trends Cogn. Sci. 2015, 19, 173–175. [Google Scholar] [CrossRef]
- Mice Protein. OpenML. Available online: https://www.openml.org/search?type=data&sort=runs&status=active&qualities.NumberOfInstances=between_1000_10000&order=desc&id=40966 (accessed on 30 September 2024).
- Abukhaled, Y.; Hatab, K.; Awadhalla, M.; Hamdan, H. Understanding the genetic mechanisms and cognitive impairments in Down syndrome: Towards a holistic approach. J. Neurol. 2023, 271, 87–104. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.M.; Dhanasekaran, A.R.; Block, A.; Tong, S.; Costa, A.C.S.; Stasko, M.; Gardiner, K.J. Protein dynamics associated with failed and rescued learning in the Ts65Dn mouse model of Down syndrome. PLoS ONE 2015, 10, e0119491. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, J.; Wang, X.; Zhu, J.; Liu, Q.; Shi, Z.; Chambers, M.C.; Zimmerman, L.J.; Shaddox, K.F.; Kim, S.; et al. Proteogenomic characterization of human colon and rectal cancer. Nature 2014, 513, 382–387. [Google Scholar] [CrossRef] [PubMed]
- AlQuraishi, M. End-to-End Differentiable Learning of Protein Structure. Cell Syst. 2019, 8, 292–301.e3. [Google Scholar] [CrossRef]
- Jin, B.; Fei, G.; Sang, S.; Zhong, C. Identification of biomarkers differentiating Alzheimer’s disease from other neurodegenerative diseases by integrated bioinformatic analysis and machine-learning strategies. Front. Mol. Neurosci. 2023, 16, 1152279. [Google Scholar] [CrossRef]
- Ching, T.; Himmelstein, D.S.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Do, B.T.; Way, G.P.; Ferrero, E.; Agapow, P.-M.; Zietz, M.; Hoffman, M.M.; et al. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface 2018, 15, 20170387. [Google Scholar] [CrossRef]
- Higuera, C.; Gardiner, K.J.; Cios, K.J. Self-Organizing Feature Maps Identify Proteins Critical to Learning in a Mouse Model of Down Syndrome. PLoS ONE 2015, 10, e0129126. [Google Scholar] [CrossRef]
- Bati, C.T.; Ser, G. Evaluation of Machine Learning Hyperparameters Performance for Mice Protein Expression Data in Different Situations. Eur. J. Tech. 2021, 11, 255–263. [Google Scholar] [CrossRef]
- Gemci, F.; Ibrikci, T. Classification of Down Syndrome of Mice Protein Dataset on MongoDB Database. Balk. J. Electr. Comput. Eng. 2018, 6, 44–49. [Google Scholar] [CrossRef]
- Saringat, M.; Mustapha, A.; Andeswari, R. Comparative Analysis of Mice Protein Expression: Clustering and Classification Approach. Int. J. Integr. Eng. 2018, 10, 26–30. [Google Scholar] [CrossRef]
- Ahmed, M.M.; Dhanasekaran, A.R.; Block, A.; Tong, S.; Costa, A.C.S.; Gardiner, K.J. Protein Profiles Associated With Context Fear Conditioning and Their Modulation By Memantine. Mol. Cell. Proteom. 2014, 13, 919–937. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Merschbaecher, K.; Haettig, J.; Mueller, U. Acetylation-mediated suppression of transcription-independent memory: Bidirectional modulation of memory by acetylation. PLoS ONE 2012, 7, e45131. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Ounallah-Saad, H.; Chakraborty, D.; Hleihil, M.; Sood, R.; Barrera, I.; Edry, E.; Chandran, S.K.; de Leon, S.B.T.; Kaphzan, H.; et al. Local Inhibition of PERK Enhances Memory and Reverses Age-Related Deterioration of Cognitive and Neuronal Properties. J. Neurosci. 2018, 38, 648–658. [Google Scholar] [CrossRef] [PubMed]
- Quarta, E.; Bravi, R.; Scambi, I.; Mariotti, R.; Minciacchi, D. Increased anxiety-like behavior and selective learning impairments are concomitant to loss of hippocampal interneurons in the presymptomatic SOD1(G93A) ALS mouse model. J. Comp. Neurol. 2015, 523, 1622–1638. [Google Scholar] [CrossRef]
- Bathina, S.; Das, U.N. Brain-derived neurotrophic factor and its clinical implications. Arch. Med. Sci. 2015, 11, 1164–1178. [Google Scholar] [CrossRef]
- Petralia, R.S.; Wang, Y.X.; Wenthold, R.J. The NMDA receptor subunits NR2A and NR2B show histological and ultrastructural localization patterns similar to those of NR1. J. Neurosci. 1994, 14, 6102–6120. [Google Scholar] [CrossRef]
- BCL2. National Cancer Institute. Available online: https://www.cancer.gov/publications/dictionaries/cancer-terms/def/bcl2 (accessed on 17 April 2025).
- Hardwick, J.M.; Soane, L. Multiple Functions of BCL-2 Family Proteins. Cold Spring Harb. Perspect. Biol. 2013, 5, a008722. [Google Scholar] [CrossRef]
- Schroer, J.; Warm, D.; De Rosa, F.; Luhmann, H.J.; Sinning, A. Activity-dependent regulation of the BAX/BCL-2 pathway protects cortical neurons from apoptotic death during early development. Cell. Mol. Life Sci. 2023, 80, 175. [Google Scholar] [CrossRef]
- Ertürk, A.; Wang, Y.; Sheng, M. Local pruning of dendrites and spines by caspase-3-dependent and proteasome-limited mechanisms. J. Neurosci. 2014, 34, 1672–1688. [Google Scholar] [CrossRef]
- Singh, K.K.; Park, K.J.; Hong, E.J.; Kramer, B.M.; Greenberg, M.E.; Kaplan, D.R.; Miller, F.D. Developmental axon pruning mediated by BDNF-p75NTR-dependent axon degeneration. Nat. Neurosci. 2008, 11, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Personius, K.E.; Slusher, B.S.; Udin, S.B. Neuromuscular NDMA Receptors Modulate Developmental Synapse Elimination. J. Neurosci. 2016, 36, 8783–8789. [Google Scholar] [CrossRef] [PubMed]
- Chambers, R.A.; Potenza, M.N.; Hoffman, R.E.; Miranker, W. Simulated Apoptosis/Neurogenesis Regulates Learning and Memory Capabilities of Adaptive Neural Networks. Neuropsychopharmacology 2004, 29, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Berntson, G.G.; Khalsa, S.S. Neural Circuits of Interoception. Trends Neurosci. 2021, 44, 17–28. [Google Scholar] [CrossRef]
- Ackerman, S. Chapter 6 The Development and Shaping of the Brain. In Discovering the Brain; National Academies Press: Washington, DC, USA, 1992. [Google Scholar]
- Azevedo, F.A.C.; Carvalho, L.R.B.; Grinberg, L.T.; Farfel, J.M.; Ferretti, R.E.L.; Leite, R.E.P.; Filho, W.J.; Lent, R.; Herculano-Houzel, S. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurol. 2009, 513, 532–541. [Google Scholar] [CrossRef]
- Andrade-Moraes, C.H.; Oliveira-Pinto, A.V.; Castro-Fonseca, E.; da Silva, C.G.; Guimarães, D.M.; Szczupak, D.; Parente-Bruno, D.R.; Carvalho, L.R.B.; Polichiso, L.; Gomes, B.V.; et al. Cell number changes in Alzheimer’s disease relate to dementia, not to plaques and tangles. Brain 2013, 136, 3738–3752. [Google Scholar] [CrossRef]
- Goriely, A. Eighty-six billion and counting: Do we know the number of neurons in the human brain? Brain 2025, 148, 689–691. [Google Scholar] [CrossRef]
- Huang, X. DataCleaningForMiceProtein. Available online: https://github.com/raymondstfx/DataCleaningForMiceProtein (accessed on 1 January 2025).
- stfxecutables, “df-analyze,” GitHub. 2021. Available online: https://github.com/stfxecutables/df-analyze (accessed on 1 November 2024).
- Levman, J.; Jennings, M.; Rouse, E.; Berger, D.; Kabaria, P.; Nangaku, M.; Gondra, I.; Takahashi, E. A Morphological Study of Schizophrenia with Magnetic Resonance Imaging, Advanced Analytics, and Machine Learning. Front. Neurosci. 2022, 16, 926426. [Google Scholar] [CrossRef]
- Figueroa, J.; Etim, P.; Shibu, A.; Berger, D.; Levman, J. Diagnosing and Characterizing Chronic Kidney Disease with Machine Learning: The Value of Clinical Patient Characteristics as Evidenced from an Open Dataset. Electronics 2024, 13, 4326. [Google Scholar] [CrossRef]
- Saville, K.; Berger, D.; Levman, J. Mitigating Bias Due to Race and Gender in Machine Learning Predictions of Traffic Stop Outcomes. Information 2024, 15, 687. [Google Scholar] [CrossRef]
- Joseph, M.; Raj, H. GANDALF: Gated Adaptive Network for Deep Automated Learning of Features. arXiv 2022, arXiv:2207.08548. [Google Scholar] [CrossRef]
- Berger, D. Redundancy-Aware Feature Selection. Available online: https://github.com/stfxecutables/df-analyze/tree/experimental?tab=readme-ov-file#redundancy-aware-feature-selection-new (accessed on 23 December 2024).
| Model | Selection | Embed Selector | Acc | AUROC | Bal-Acc | F1 | NPV | PPV | Sens | Spec |
|---|---|---|---|---|---|---|---|---|---|---|
| lr | wrap | none | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| sgd | wrap | none | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| lr | embed_lgbm | lgbm | 0.993 | 1.000 | 0.993 | 0.993 | 1.000 | 0.984 | 0.993 | 0.987 |
| sgd | embed_lgbm | lgbm | 0.992 | 0.996 | 0.993 | 0.992 | 1.000 | 0.984 | 0.993 | 0.987 |
| lgbm | embed_linear | linear | 0.983 | 1.000 | 0.983 | 0.983 | 0.980 | 0.988 | 0.983 | 0.982 |
| lgbm | assoc | none | 0.981 | 1.000 | 0.981 | 0.981 | 0.980 | 0.984 | 0.981 | 0.979 |
| sgd | pred | none | 0.981 | 1.000 | 0.983 | 0.981 | 0.964 | 1.000 | 0.983 | 1.000 |
| lr | embed_linear | linear | 0.981 | 1.000 | 0.983 | 0.981 | 0.972 | 0.988 | 0.983 | 0.990 |
| lgbm | wrap | none | 0.980 | 1.000 | 0.980 | 0.979 | 0.975 | 0.984 | 0.980 | 0.980 |
| lgbm | pred | none | 0.980 | 1.000 | 0.979 | 0.979 | 0.980 | 0.981 | 0.979 | 0.976 |
| lgbm | none | none | 0.980 | 1.000 | 0.979 | 0.979 | 0.980 | 0.981 | 0.979 | 0.976 |
| lgbm | embed_lgbm | lgbm | 0.980 | 1.000 | 0.979 | 0.979 | 0.980 | 0.981 | 0.979 | 0.976 |
| sgd | embed_linear | linear | 0.975 | 1.000 | 0.978 | 0.975 | 0.956 | 0.991 | 0.978 | 0.993 |
| lr | pred | none | 0.975 | 1.000 | 0.978 | 0.975 | 0.955 | 1.000 | 0.978 | 1.000 |
| sgd | assoc | none | 0.975 | 1.000 | 0.978 | 0.975 | 0.961 | 0.988 | 0.978 | 0.990 |
| gandalf | embed_lgbm | lgbm | 0.972 | 0.998 | 0.969 | 0.971 | 0.975 | 0.976 | 0.969 | 0.973 |
| sgd | none | none | 0.970 | 1.000 | 0.973 | 0.969 | 0.952 | 0.988 | 0.973 | 0.990 |
| lr | none | none | 0.964 | 1.000 | 0.968 | 0.964 | 0.941 | 0.988 | 0.968 | 0.990 |
| gandalf | embed_linear | linear | 0.958 | 0.996 | 0.953 | 0.955 | 0.944 | 0.982 | 0.953 | 0.990 |
| rf | embed_linear | linear | 0.957 | 1.000 | 0.957 | 0.956 | 0.976 | 0.945 | 0.957 | 0.934 |
| rf | embed_lgbm | lgbm | 0.957 | 1.000 | 0.957 | 0.956 | 0.973 | 0.948 | 0.957 | 0.937 |
| rf | assoc | none | 0.957 | 1.000 | 0.957 | 0.956 | 0.973 | 0.948 | 0.957 | 0.937 |
| lr | assoc | none | 0.956 | 1.000 | 0.962 | 0.956 | 0.929 | 0.988 | 0.962 | 0.990 |
| rf | none | none | 0.954 | 1.000 | 0.953 | 0.952 | 0.973 | 0.942 | 0.953 | 0.930 |
| rf | pred | none | 0.954 | 0.999 | 0.953 | 0.952 | 0.973 | 0.942 | 0.953 | 0.930 |
| rf | wrap | none | 0.954 | 0.999 | 0.953 | 0.952 | 0.973 | 0.942 | 0.953 | 0.930 |
| knn | embed_lgbm | lgbm | 0.953 | 0.976 | 0.958 | 0.946 | 0.924 | 0.984 | 0.958 | 0.987 |
| gandalf | pred | none | 0.948 | 0.998 | 0.943 | 0.927 | 0.933 | 0.979 | 0.943 | 0.969 |
| knn | wrap | none | 0.927 | 0.938 | 0.933 | 0.923 | 0.929 | 0.960 | 0.933 | 0.950 |
| knn | pred | none | 0.924 | 0.937 | 0.927 | 0.915 | 0.894 | 0.955 | 0.927 | 0.956 |
| knn | embed_linear | linear | 0.916 | 0.950 | 0.917 | 0.915 | 0.881 | 0.959 | 0.917 | 0.956 |
| knn | none | none | 0.916 | 0.950 | 0.917 | 0.915 | 0.881 | 0.959 | 0.917 | 0.956 |
| knn | assoc | none | 0.916 | 0.950 | 0.917 | 0.915 | 0.881 | 0.959 | 0.917 | 0.956 |
| gandalf | none | none | 0.889 | 0.992 | 0.884 | 0.883 | 0.842 | 0.984 | 0.884 | 0.978 |
| gandalf | wrap | none | 0.869 | 0.990 | 0.879 | 0.860 | 0.861 | 0.964 | 0.879 | 0.967 |
| gandalf | assoc | none | 0.830 | 0.954 | 0.834 | 0.826 | 0.788 | 0.924 | 0.834 | 0.921 |
| dummy | embed_linear | linear | 0.443 | 0.500 | 0.500 | 0.307 | 0.429 | 0.452 | 0.500 | 0.400 |
| dummy | none | none | 0.443 | 0.500 | 0.500 | 0.307 | 0.429 | 0.452 | 0.500 | 0.400 |
| dummy | assoc | none | 0.443 | 0.500 | 0.500 | 0.307 | 0.429 | 0.452 | 0.500 | 0.400 |
| dummy | embed_lgbm | lgbm | 0.443 | 0.500 | 0.500 | 0.307 | 0.429 | 0.452 | 0.500 | 0.400 |
| dummy | pred | none | 0.443 | 0.500 | 0.500 | 0.307 | 0.429 | 0.452 | 0.500 | 0.400 |
| dummy | wrap | none | 0.443 | 0.500 | 0.500 | 0.307 | 0.429 | 0.452 | 0.500 | 0.400 |
| Feature | Score |
|---|---|
| SOD1N | 0.944 |
| pERKN | 0.994 |
| BDNFN_NAN | 0.994 |
| NR2AN | 0.964 |
| H3AcK18N_NAN | 0.983 |
| BCL2N_NAN | 0.961 |
| Model | Selection | Embed Selector | Acc | AUROC | Bal-Acc | F1 | NPV | PPV | Sens | Spec |
|---|---|---|---|---|---|---|---|---|---|---|
| sgd | embed_lgbm | lgbm | 0.877 | 0.893 | 0.876 | 0.873 | 0.850 | 0.917 | 0.876 | 0.891 |
| rf | none | none | 0.876 | 0.890 | 0.889 | 0.875 | 0.812 | 0.959 | 0.889 | 0.964 |
| rf | pred | none | 0.874 | 0.893 | 0.887 | 0.873 | 0.811 | 0.955 | 0.887 | 0.960 |
| rf | embed_linear | linear | 0.874 | 0.893 | 0.887 | 0.873 | 0.811 | 0.955 | 0.887 | 0.960 |
| knn | embed_lgbm | lgbm | 0.873 | 0.889 | 0.867 | 0.869 | 0.853 | 0.889 | 0.867 | 0.840 |
| rf | assoc | none | 0.872 | 0.875 | 0.883 | 0.871 | 0.811 | 0.952 | 0.883 | 0.953 |
| rf | wrap | none | 0.872 | 0.908 | 0.884 | 0.870 | 0.803 | 0.971 | 0.884 | 0.969 |
| lr | embed_lgbm | lgbm | 0.860 | 0.917 | 0.847 | 0.853 | 0.872 | 0.863 | 0.847 | 0.796 |
| gandalf | none | none | 0.825 | 0.903 | 0.771 | 0.773 | 0.914 | 0.810 | 0.771 | 0.596 |
| lgbm | wrap | none | 0.823 | 0.900 | 0.807 | 0.805 | 0.799 | 0.884 | 0.807 | 0.804 |
| gandalf | pred | none | 0.819 | 0.918 | 0.792 | 0.799 | 0.888 | 0.801 | 0.792 | 0.660 |
| gandalf | embed_lgbm | lgbm | 0.817 | 0.926 | 0.782 | 0.765 | 0.913 | 0.814 | 0.782 | 0.618 |
| lgbm | embed_lgbm | lgbm | 0.815 | 0.920 | 0.797 | 0.799 | 0.824 | 0.858 | 0.797 | 0.758 |
| rf | embed_lgbm | lgbm | 0.809 | 0.859 | 0.797 | 0.797 | 0.815 | 0.852 | 0.797 | 0.764 |
| lr | none | none | 0.808 | 0.894 | 0.794 | 0.798 | 0.778 | 0.850 | 0.794 | 0.762 |
| lr | embed_linear | linear | 0.806 | 0.891 | 0.792 | 0.796 | 0.778 | 0.847 | 0.792 | 0.758 |
| lgbm | pred | none | 0.806 | 0.878 | 0.801 | 0.800 | 0.769 | 0.850 | 0.801 | 0.789 |
| knn | wrap | none | 0.805 | 0.867 | 0.775 | 0.789 | 0.840 | 0.800 | 0.775 | 0.667 |
| sgd | embed_linear | linear | 0.803 | 0.821 | 0.800 | 0.798 | 0.775 | 0.844 | 0.800 | 0.787 |
| lr | assoc | none | 0.800 | 0.891 | 0.784 | 0.789 | 0.777 | 0.842 | 0.784 | 0.749 |
| sgd | wrap | none | 0.799 | 0.899 | 0.768 | 0.778 | 0.808 | 0.807 | 0.768 | 0.669 |
| lr | wrap | none | 0.799 | 0.904 | 0.771 | 0.781 | 0.808 | 0.805 | 0.771 | 0.673 |
| sgd | none | none | 0.799 | 0.898 | 0.787 | 0.790 | 0.778 | 0.839 | 0.787 | 0.760 |
| sgd | pred | none | 0.798 | 0.879 | 0.790 | 0.791 | 0.779 | 0.820 | 0.790 | 0.738 |
| lgbm | assoc | none | 0.797 | 0.878 | 0.784 | 0.787 | 0.784 | 0.839 | 0.784 | 0.758 |
| sgd | assoc | none | 0.796 | 0.808 | 0.782 | 0.786 | 0.775 | 0.835 | 0.782 | 0.751 |
| gandalf | assoc | none | 0.792 | 0.886 | 0.744 | 0.748 | 0.833 | 0.781 | 0.744 | 0.547 |
| lr | pred | none | 0.791 | 0.883 | 0.786 | 0.784 | 0.782 | 0.818 | 0.786 | 0.738 |
| lgbm | embed_linear | linear | 0.788 | 0.889 | 0.773 | 0.776 | 0.781 | 0.831 | 0.773 | 0.736 |
| lgbm | none | none | 0.783 | 0.888 | 0.764 | 0.768 | 0.790 | 0.818 | 0.764 | 0.709 |
| knn | pred | none | 0.746 | 0.785 | 0.733 | 0.729 | 0.741 | 0.774 | 0.733 | 0.649 |
| knn | embed_linear | linear | 0.745 | 0.800 | 0.722 | 0.730 | 0.732 | 0.764 | 0.722 | 0.629 |
| knn | assoc | none | 0.745 | 0.800 | 0.722 | 0.730 | 0.732 | 0.764 | 0.722 | 0.629 |
| knn | none | none | 0.745 | 0.800 | 0.722 | 0.730 | 0.732 | 0.764 | 0.722 | 0.629 |
| gandalf | wrap | none | 0.730 | 0.865 | 0.707 | 0.687 | 0.808 | 0.784 | 0.707 | 0.629 |
| dummy | embed_linear | linear | 0.611 | 0.500 | 0.500 | 0.378 | – | 0.611 | 0.500 | 0.000 |
| dummy | pred | none | 0.611 | 0.500 | 0.500 | 0.378 | – | 0.611 | 0.500 | 0.000 |
| dummy | none | none | 0.611 | 0.500 | 0.500 | 0.378 | – | 0.611 | 0.500 | 0.000 |
| dummy | assoc | none | 0.611 | 0.500 | 0.500 | 0.378 | – | 0.611 | 0.500 | 0.000 |
| dummy | embed_lgbm | lgbm | 0.611 | 0.500 | 0.500 | 0.378 | – | 0.611 | 0.500 | 0.000 |
| dummy | wrap | none | 0.611 | 0.500 | 0.500 | 0.378 | – | 0.611 | 0.500 | 0.000 |
| gandalf | embed_linear | linear | 0.589 | 0.696 | 0.539 | 0.435 | 0.358 | 0.638 | 0.539 | 0.311 |
| Feature | Score |
|---|---|
| CaNAN | 0.720 |
| SOD1N | 0.893 |
| pP70S6N | 0.906 |
| BADN_NAN | 0.912 |
| UbiquitinN | 0.916 |
| H3AcK18N | 0.918 |
| pRSKN | 0.914 |
| pPKCABN | 0.901 |
| NR2AN | 0.905 |
| pCASP9N | 0.903 |
| GSK3BN | 0.897 |
| pCAMKIIN | 0.862 |
| Measurement | Data | Description |
|---|---|---|
| Genotype | c | Control mouse |
| t | Trisomy mouse (Ts65Dn model) | |
| Treatment Type | m | Mouse injected with Memantine |
| s | Mouse injected with saline (control) | |
| Behavior | CS | Context-shock: Mice explored test chamber before shock |
| SC | Shock-context: Mice received shock before exploration | |
| Class (Target) | c-CS-s | Control mouse: Context-shock conditioning + saline |
| c-CS-m | Control mouse: Context-shock conditioning + memantine | |
| c-SC-s | Control mouse: Shock-context conditioning + saline | |
| c-SC-m | Control mouse: Shock-context conditioning + memantine | |
| t-CS-s | Ts65Dn mouse: Context-shock conditioning + saline | |
| t-CS-m | Ts65Dn mouse: Context-shock conditioning + memantine | |
| t-SC-s | Ts65Dn mouse: Shock-context conditioning + saline | |
| t-SC-m | Ts65Dn mouse: Shock-context conditioning + memantine |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X.; Gauthier, C.; Berger, D.; Cai, H.; Levman, J. Identifying Cortical Molecular Biomarkers Potentially Associated with Learning in Mice Using Artificial Intelligence. Int. J. Mol. Sci. 2025, 26, 6878. https://doi.org/10.3390/ijms26146878
Huang X, Gauthier C, Berger D, Cai H, Levman J. Identifying Cortical Molecular Biomarkers Potentially Associated with Learning in Mice Using Artificial Intelligence. International Journal of Molecular Sciences. 2025; 26(14):6878. https://doi.org/10.3390/ijms26146878
Chicago/Turabian StyleHuang, Xiyao, Carson Gauthier, Derek Berger, Hao Cai, and Jacob Levman. 2025. "Identifying Cortical Molecular Biomarkers Potentially Associated with Learning in Mice Using Artificial Intelligence" International Journal of Molecular Sciences 26, no. 14: 6878. https://doi.org/10.3390/ijms26146878
APA StyleHuang, X., Gauthier, C., Berger, D., Cai, H., & Levman, J. (2025). Identifying Cortical Molecular Biomarkers Potentially Associated with Learning in Mice Using Artificial Intelligence. International Journal of Molecular Sciences, 26(14), 6878. https://doi.org/10.3390/ijms26146878






