Protein Functional Effector (pfe) Noncoding RNAS Are Identical to Fragments from Various Noncoding RNAs
Abstract
1. Introduction
2. Results
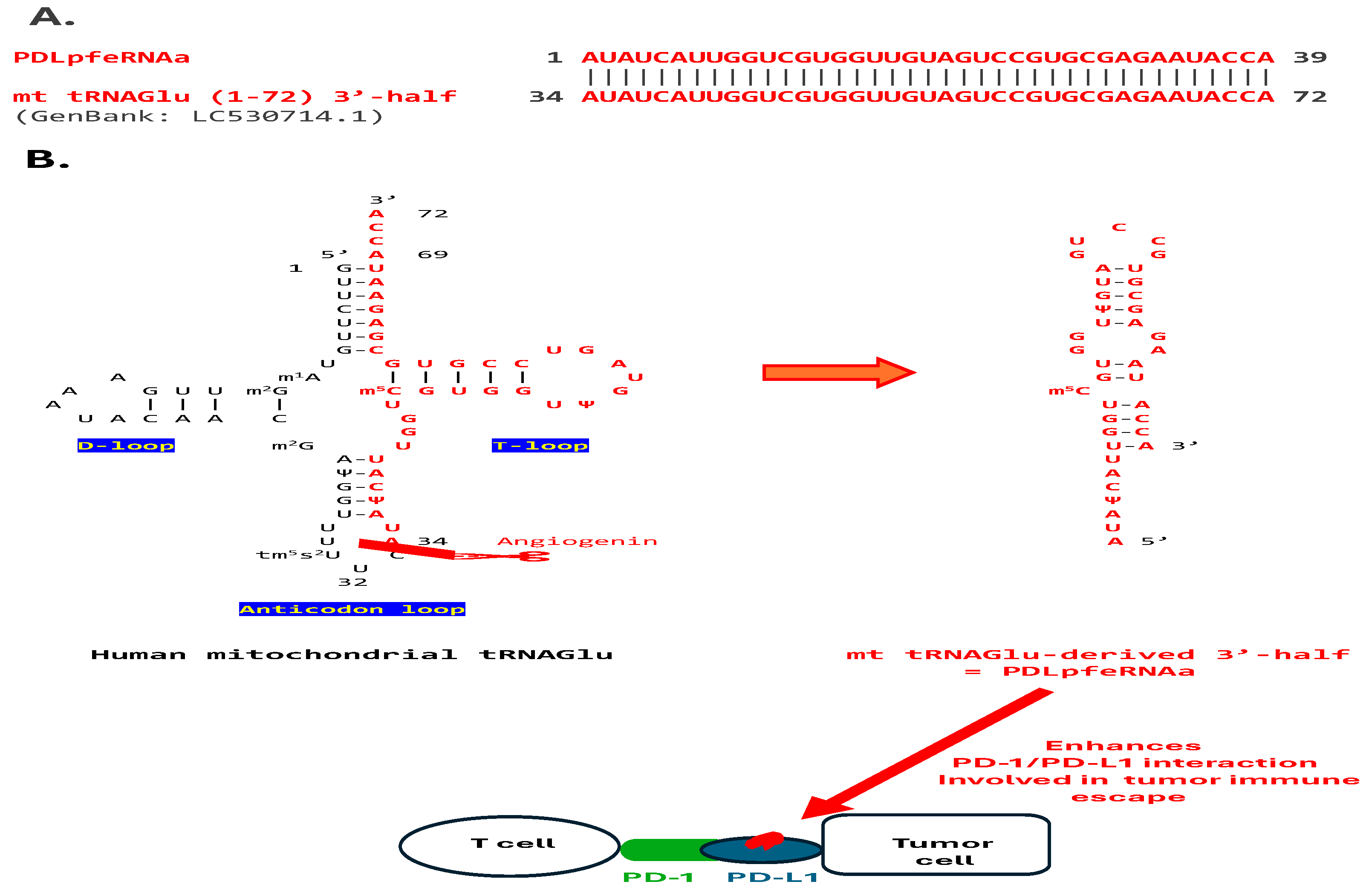
2.1. Analysis of pfeRNAs Related to PD-1/PD-L1 Interactions, Termed PDLpfeRNAs
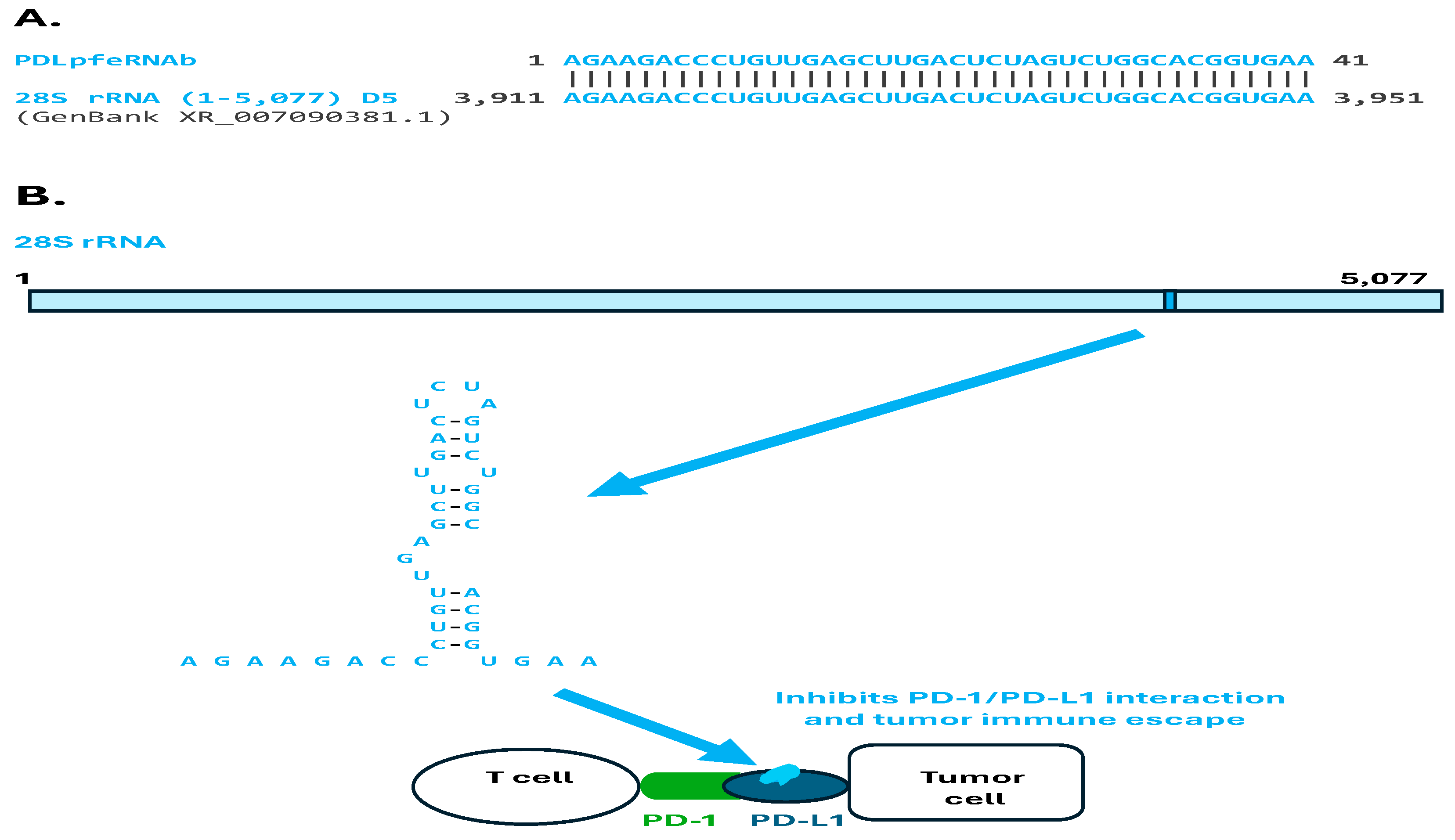
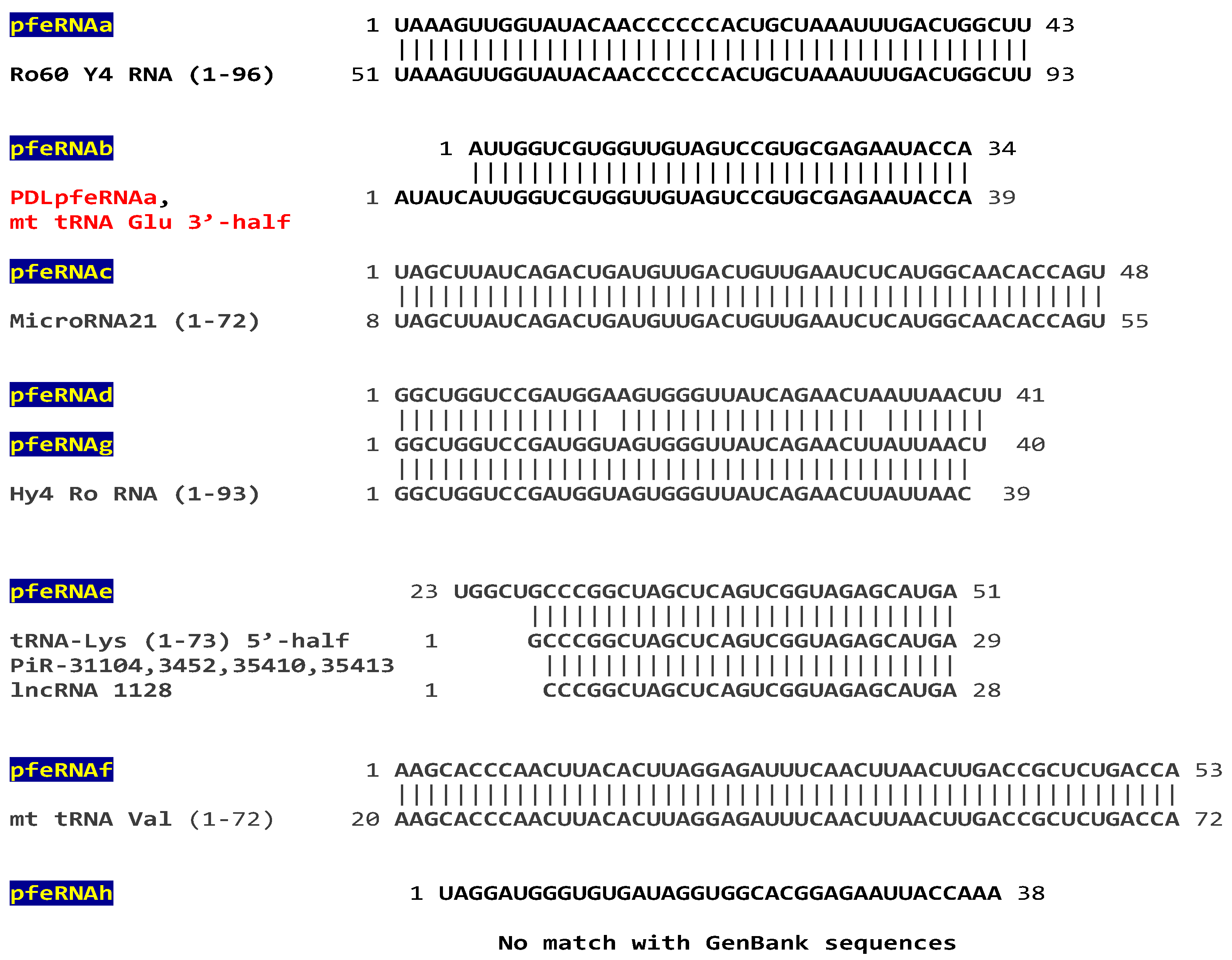
2.2. Analysis of Plasma pfeRNAs Related to Pulmonary Nodules: pfeRNAa-h
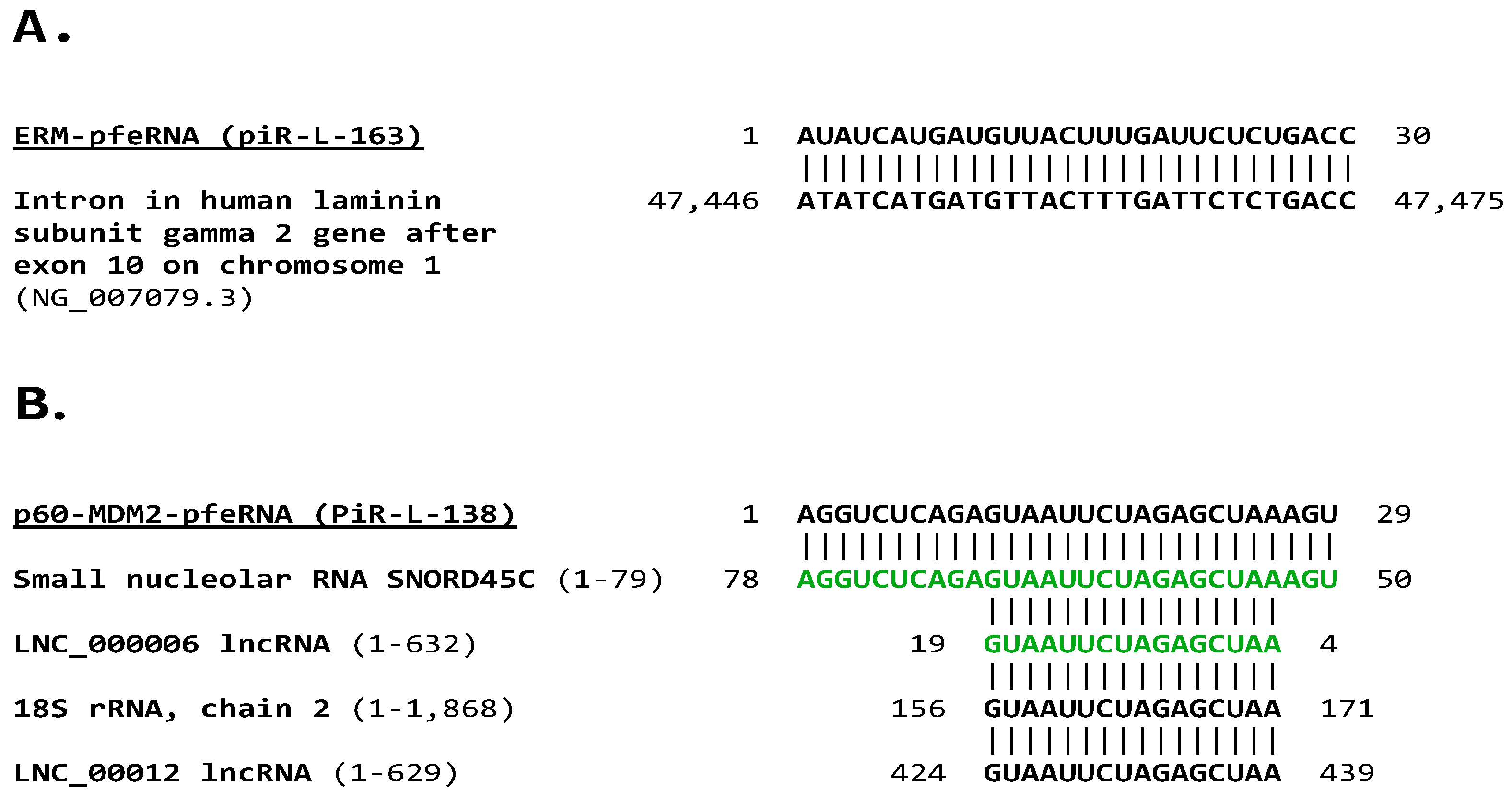
2.3. Analysis of Plasma pfeRNAs Related to p-ERM Proteins and p60-MDM2
3. Discussion
3.1. PD-L1-Binding pfeRNAs (PDLpfeRNAs a and b)
3.2. Pulmonary Nodule-Related pfeRNAs
3.3. ERM-pfeRNA and p-60-MDM2-pfeRNA
3.4. pfeRNAs, glycoRNAs, and Nicked tRNA Halves Among RNAs Affecting Cell Surface and Extracellular Protein Functions and PD-1/PD-L1 Interactions
3.5. 3′-end 2′-O-Methylation Is Also Present in ncRNAs Other than the Characterized pfeRNAs
4. Materials and Methods
4.1. Detection of Genomic Sequences Identical or Highly Similar to pfeRNAs, and Their Chromosomal Location for Pulmonary Nodule-Related pfeRNAs
4.2. Visualization of RNA Secondary Structures and Estimation of Their Minimum Free Energies
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mattick, J.S. Non-coding RNAs: The architects of eukaryotic complexity. EMBO Rep. 2001, 2, 986–991. [Google Scholar] [CrossRef] [PubMed]
- Haseltine, W.A.; Patarca, R. The RNA Revolution in the Central Molecular Biology Dogma Evolution. Int. J. Mol. Sci. 2024, 25, 12695. [Google Scholar] [CrossRef] [PubMed]
- Gerstein, M.B.; Bruce, C.; Rozowsky, J.S.; Zheng, D.; Du, J.; Korbel, J.O.; Emanuelsson, O.; Zhang, Z.D.; Weissman, S.; Snyder, M. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007, 17, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Rusk, N. Understanding noncoding RNAs. Nat. Methods 2015, 12, 35. [Google Scholar] [CrossRef]
- Chen, Z.; Sun, Y.; Yang, X.; Wu, Z.; Guo, K.; Niu, X.; Wang, Q.; Ruan, J.; Bu, W.; Gao, S. Two featured series of rRNA-derived RNA fragments (rRFs) constitute a novel class of small RNAs. PLoS ONE 2017, 12, e0176458. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Wilson, B.; Kumar, P.; Dutta, A. Noncanonical Roles of tRNAs: tRNA Fragments and Beyond. Annu. Rev. Genet. 2020, 54, 47–69. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Yong, J.; Liu, H.; Shi, Y.; Meinkoth, J.; Dreyfuss, G.; Yang, X. tRNA binds to cytochrome c and inhibits caspase activation. Mol. Cell 2010, 37, 668–678. [Google Scholar] [CrossRef] [PubMed]
- Read, D.F.; Waller, T.J.; Tse, E.; Southworth, D.R.; Engelke, D.R.; Smaldino, P.J. Aggregation of Mod5 is affected by tRNA binding with implications for tRNA gene-mediated silencing. FEBS Lett. 2017, 591, 1601–1610. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Gebetsberger, J.; Polacek, N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013, 10, 1798–1806. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Li, X.; Khan, F.A.; Yuan, C.; Pandupuspitasari, N.S.; Huang, C.; Sun, F.; Guan, K. tRNA modifications and tRNA-derived small RNAs: New insights of tRNA in human disease. Cell Biol. Toxicol. 2024, 40, 76. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Wang, Y.; Kumari, P.; Shetty, A.C.; Clark, D.; Gable, T.; MacKerell, A.D.; Ma, M.Z.; Weber, D.J.; Yang, A.J.; et al. A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells. Nat. Commun. 2015, 6, 7316. [Google Scholar] [CrossRef] [PubMed]
- Gable, T.; Wang, Y.; Clark, D.; Kumari, P.; Shetty, A.C.; Li, M.; Mei, Y. A phosphorylation-wide sncRNA screen reveals Protein Functional Effector sncRNAs (pfeRNAs) in human lung somatic cells. Cancer Lett. 2017, 396, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Brock, M.; Mei, Y. Protein functional effector sncRNAs (pfeRNAs) in lung cancer. Cancer Lett. 2017, 403, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gable, T.; Ma, M.Z.; Clark, D.; Zhao, J.; Zhang, Y.; Liu, W.; Mao, L.; Mei, Y. A piRNA-like Small RNA Induces Chemoresistance to Cisplatin-Based Therapy by Inhibiting Apoptosis in Lung Squamous Cell Carcinoma. Mol. Ther. Nucleic Acids 2017, 6, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Khan, H.; Shishikura, M.; Ishiyama, S.; Khan, A.; Orita, H.; Brock, M.V. pfeRNAs—A Novel Class of Small Non-coding RNAs With Real Translational Potential. J. Surg. Res. 2023, 284, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Gurau, A.; Yamauchi, S.; Ecoff, K.; Rodgers, K.P.; Eshleman, J.R.; Talbot, C.C., Jr.; Huang, P.; Choi, J.; Forde, P.M.; Anagnostou, V.; et al. PD-L1 pfeRNAs as blood-based predictors of treatment response of unresectable malignant pleural mesothelioma patients administered Durvalumab with cisplatin and pemetrexed as first-line therapy. Noncoding RNA Res. 2025, 12, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, Y.; Huang, H.; Fackche, N.; Rodgers, K.; Lee, B.; Nizam, W.; Khan, H.; Lu, Z.; Kong, X.; et al. A Cost-Effective and Non-Invasive pfeRNA-Based Test Differentiates Benign and Suspicious Pulmonary Nodules from Malignant Ones. Noncoding RNA 2021, 7, 80. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Yashiro, Y.; Kikuchi, I.; Ishigami, Y.; Saito, H.; Matsuzawa, I.; Okada, S.; Mito, M.; Iwasaki, S.; Ma, D.; et al. Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 2020, 11, 4269. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Feng, J.; Liu, Q.; Sun, F.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009, 583, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Boccitto, M.; Wolin, S.L. Ro60 and Y RNAs: Structure, functions, and roles in autoimmunity. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 133–152. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.H.; Tsao, C.J. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016, 5, 395–402. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, C.A.; Harley, J.B. A subset of hY RNAs is associated with erythrocyte Ro ribonucleoproteins. EMBO J. 1990, 9, 3683–3689. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lowe, T.M. GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009, 37, D93–D97. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lowe, T.M. GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016, 44, D184–D189. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. In Gene Prediction; Methods in Molecular Biology; Humana: New York, NY, USA, 2019; Volume 1962, pp. 1–14. [Google Scholar] [CrossRef]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006, 442, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef] [PubMed]
- Ribas, J.; Ni, X.; Castanares, M.; Liu, M.M.; Esopi, D.; Yegnasubramanian, S.; Rodriguez, R.; Mendell, J.T.; Lupold, S.E. A novel source for miR-21 expression through the alternative polyadenylation of VMP1 gene transcripts. Nucleic Acids Res. 2012, 40, 6821–6833. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Stanton, B.A. Transfer RNA-derived fragments, the underappreciated regulatory small RNAs in microbial pathogenesis. Front. Microbiol. 2021, 12, 687632. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yan, M.; Cao, Z.; Li, X.; Zhang, Y.; Shi, J.; Feng, G.H.; Peng, H.; Zhang, X.; Zhang, Y.; et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016, 351, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Gu, J.; Wang, M.; Zhang, J.; Chen, Y.; Jiang, P.; Zhu, T.; Zhang, X. Emerging roles of tRNA-derived fragments in cancer. Mol. Cancer 2023, 22, 30. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yu, Z.; Jiang, W.; Lyu, X.; Guo, A.; Sun, X.; Yang, Y.; Zhang, Y. tRNA and tsRNA: From Heterogeneity to Multifaceted Regulators. Biomolecules 2024, 14, 1340. [Google Scholar] [CrossRef] [PubMed]
- Loveland, A.B.; Koh, C.S.; Ganesan, R.; Jacobson, A.; Korostelev, A.A. Structural mechanism of angiogenin activation by the ribosome. Nature 2024, 630, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Grigoriev, A. Computational meta-analysis of ribosomal RNA fragments: Potential targets and interaction mechanisms. Nucleic Acids Res. 2021, 49, 4085–4103. [Google Scholar] [CrossRef] [PubMed]
- Butler, P.; Pascheto, I.; Lizzi, M.; St-Onge, R.; Lanner, C.; Guo, B.; Masilamani, T.; Pritzker, L.B.; Kovala, A.T.; Parissenti, A.M. RNA disruption is a widespread phenomenon associated with stress-induced cell death in tumour cells. Sci. Rep. 2023, 13, 1711. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Zhou, B.; Zhang, F.; Tu, Y.; Hu, Y.; Zhang, B.; Zhai, Q. Profiling and identification of small rDNA-derived RNAs and their potential biological functions. PLoS ONE 2013, 8, e56842. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.; Benmoussa, A.; Provost, P. Small Non-Coding RNAs Derived From Eukaryotic Ribosomal RNA. Noncoding RNA 2019, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Guglas, K.; Kołodziejczak, I.; Kolenda, T.; Kopczyńska, M.; Teresiak, A.; Sobocińska, J.; Bliźniak, R.; Lamperska, K. YRNAs and YRNA-Derived Fragments as New Players in Cancer Research and Their Potential Role in Diagnostics. Int. J. Mol. Sci. 2020, 21, 5682. [Google Scholar] [CrossRef] [PubMed]
- Valkov, N.; Das, S. Y RNAs: Biogenesis, Function and Implications for the Cardiovascular System. In Non-Coding RNAs in Cardiovascular Diseases; Advances in Experimental Medicine and Biology; Springer: Singapore, 2020; Volume 1229, pp. 327–342. [Google Scholar] [CrossRef]
- Telonis, A.G.; Loher, P.; Kirino, Y.; Rigoutsos, I. Nuclear and mitochondrial tRNA-lookalikes in the human genome. Front. Genet. 2014, 5, 344. [Google Scholar] [CrossRef] [PubMed]
- Bofill-De Ros, X.; Yang, A.; Gu, S. IsomiRs: Expanding the miRNA repression toolbox beyond the seed. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2020, 1863, 194373. [Google Scholar] [CrossRef] [PubMed]
- Morin, R.D.; O’Connor, M.D.; Griffith, M.; Kuchenbauer, F.; Delaney, A.; Prabhu, A.L.; Zhao, Y.; McDonald, H.; Zeng, T.; Hirst, M.; et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008, 18, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Rigoutsos, I.; Londin, E.; Kirino, Y. Short RNA regulators: The past, the present, the future, and implications for precision medicine and health disparities. Curr. Opin. Biotechnol. 2019, 58, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.W.; Evans, J.M.; Huang, S.; Mahoney, D.W.; Dukek, B.A.; Taylor, W.R.; Yab, T.C.; Smyrk, T.C.; Jen, J.; Kisiel, J.B.; et al. A Comprehensive Approach to Sequence-oriented IsomiR annotation (CASMIR): Demonstration with IsomiR profiling in colorectal neoplasia. BMC Genom. 2018, 19, 401. [Google Scholar] [CrossRef] [PubMed]
- McCall, M.N.; Kim, M.S.; Adil, M.; Patil, A.H.; Lu, Y.; Mitchell, C.J.; Leal-Rojas, P.; Xu, J.; Kumar, M.; Dawson, V.L.; et al. Toward the human cellular microRNAome. Genome Res. 2017, 27, 1769–1781. [Google Scholar] [CrossRef] [PubMed]
- Tomasello, L.; Distefano, R.; Nigita, G.; Croce, C.M. The MicroRNA Family Gets Wider: The IsomiRs Classification and Role. Front. Cell Dev. Biol. 2021, 9, 668648. [Google Scholar] [CrossRef] [PubMed]
- Loher, P.; Karathanasis, N.; Londin, E.F.; Bray, P.; Pliatsika, V.; Telonis, A.G.; Rigoutsos, I. IsoMiRmap: Fast, deterministic and exhaustive mining of isomiRs from short RNA-seq datasets. Bioinformatics 2021, 37, 1828–1838. [Google Scholar] [CrossRef] [PubMed]
- Telonis, A.G.; Loher, P.; Jing, Y.; Londin, E.; Rigoutsos, I. Beyond the one-locus-one-miRNA paradigm: microRNA isoforms enable deeper insights into breast cancer heterogeneity. Nucleic Acids Res. 2015, 43, 9158–9175. [Google Scholar] [CrossRef] [PubMed]
- Neilsen, C.T.; Goodall, G.J.; Bracken, C.P. IsomiRs—The overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012, 28, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Kumarswamy, R.; Volkmann, I.; Thum, T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011, 8, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Sheedy, F.J. Turning 21: Induction of miR-21 as a Key Switch in the Inflammatory Response. Front. Immunol. 2015, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The Promising Role of miR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. Nucleic Acids 2020, 20, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, R.; Vijakumaran, U.; Shanmuganantha, L.; Law, J.X.; Alias, E.; Ng, M.H. The Role and Mechanism of MicroRNA 21 in Osteogenesis: An Update. Int. J. Mol. Sci. 2023, 24, 11330. [Google Scholar] [CrossRef] [PubMed]
- Pop-Bica, C.; Pintea, S.; Magdo, L.; Cojocneanu, R.; Gulei, D.; Ferracin, M.; Berindan-Neagoe, I. The Clinical Utility of miR-21 and let-7 in Non-small Cell Lung Cancer (NSCLC). A Systematic Review and Meta-Analysis. Front. Oncol. 2020, 10, 516850. [Google Scholar] [CrossRef] [PubMed]
- Stark, V.A.; Facey, C.O.B.; Viswanathan, V.; Boman, B.M. The Role of miRNAs, miRNA Clusters, and isomiRs in Development of Cancer Stem Cell Populations in Colorectal Cancer. Int. J. Mol. Sci. 2021, 22, 1424. [Google Scholar] [CrossRef] [PubMed]
- Lo, T.F.; Tsai, W.C.; Chen, S.T. MicroRNA-21-3p, a berberine-induced miRNA, directly down-regulates human methionine adenosyltransferases 2A and 2B and inhibits hepatoma cell growth. PLoS ONE 2013, 8, e75628. [Google Scholar] [CrossRef] [PubMed]
- Meseguer, S. MicroRNAs and tRNA-Derived Small Fragments: Key Messengers in Nuclear—Mitochondrial Communication. Front. Mol. Biosci. 2021, 8, 643575. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Clark, D.; Mao, L. Novel dimensions of piRNAs in cancer. Cancer Lett. 2013, 336, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Ross, R.J.; Weiner, M.M.; Lin, H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature 2014, 505, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Hu, H.Y.; Jiang, X.; Maierhofer, V.; Neb, E.; He, L.; Hu, Y.; Hu, H.; Li, N.; Chen, W.; et al. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 2011, 39, 6596–6607. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Shi, J.; Liu, H.; Zhang, X.; Zhou, J.J.; Li, M.; Zhou, D.; Li, R.; Lv, J.; Wen, G.; et al. Peripheral blood non-canonical small non-coding RNAs as novel biomarkers in lung cancer. Mol. Cancer 2020, 19, 159. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhang, Y.; Zhou, T.; Chen, Q. tsRNAs: The Swiss Army Knife for Translational Regulation. Trends Biochem. Sci. 2019, 44, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Kosanam, H.; Prassas, I.; Chrystoja, C.C.; Soleas, I.; Chan, A.; Dimitromanolakis, A.; Blasutig, I.M.; Rückert, F.; Gruetzmann, R.; Pilarsky, C.; et al. Laminin, gamma 2 (LAMC2): A promising new putative pancreatic cancer biomarker identified by proteomic analysis of pancreatic adenocarcinoma tissues. Mol. Cell. Proteom. 2013, 12, 2820–2832. [Google Scholar] [CrossRef] [PubMed]
- Moon, Y.W.; Rao, G.; Kim, J.J.; Shim, H.S.; Park, K.S.; An, S.S.; Kim, B.; Steeg, P.S.; Sarfaraz, S.; Lee, L.C.; et al. LAMC2 enhances the metastatic potential of lung adenocarcinoma. Cell Death Differ. 2015, 22, 1341–1352. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Tian, J.; Lou, J.; Ke, J.; Li, L.; Li, J.; Yang, Y.; Gong, Y.; Zhu, Y.; Zhang, Y.; et al. A functional polymorphism in lnc-LAMC2-1:1 confers risk of colorectal cancer by affecting miRNA binding. Carcinogenesis 2016, 37, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.F.; Liu, J.; Cheng, J.; Wu, W.D.; Liu, X.Q. Silencing of LAMC2 Reverses Epithelial-Mesenchymal Transition and Inhibits Angiogenesis in Cholangiocarcinoma via Inactivation of the Epidermal Growth Factor Receptor Signaling Pathway. Am. J. Pathol. 2019, 189, 1637–1653. [Google Scholar] [CrossRef] [PubMed]
- Okada, Y.; Nishiwada, S.; Yamamura, K.; Sho, M.; Baba, H.; Takayama, T.; Goel, A. Identification of laminin γ2 as a prognostic and predictive biomarker for determining response to gemcitabine-based therapy in pancreatic ductal adenocarcinoma. Eur. J. Cancer 2021, 146, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Lu, Q.; Xu, B. hsa-miR-5580-3p inhibits oral cancer cell viability, proliferation and migration by suppressing LAMC2. Mol. Med. Rep. 2021, 23, 453. [Google Scholar] [CrossRef] [PubMed]
- Berndt, A.; Gaßler, N.; Franz, M. Invasion-Associated Reorganization of Laminin 332 in Oral Squamous Cell Carcinomas: The Role of the Laminin γ2 Chain in Tumor Biology, Diagnosis, and Therapy. Cancers 2022, 14, 4903. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ji, F.; Tan, Y.; Zhao, L.; Zhao, Y.; Liu, J.; Shao, L.; Shi, J.; Ye, M.; He, X.; et al. Oncogenic Roles of Laminin Subunit Gamma-2 in Intrahepatic Cholangiocarcinoma via Promoting EGFR Translation. Adv. Sci. 2024, 11, e2309010. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.L.; Miller, J.D.; Ying, S.Y. Intronic microRNA (miRNA). BioMed Res. Int. 2006, 2006, 26818. [Google Scholar] [CrossRef] [PubMed]
- Taft, R.J.; Glazov, E.A.; Lassmann, T.; Hayashizaki, Y.; Carninci, P.; Mattick, J.S. Small RNAs derived from snoRNAs. RNA 2009, 15, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Martens-Uzunova, E.S.; Olvedy, M.; Jenster, G. Beyond microRNA—Novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett. 2013, 340, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Falaleeva, M.; Stamm, S. Processing of snoRNAs as a new source of regulatory non-coding RNAs: snoRNA fragments form a new class of functional RNAs. Bioessays 2013, 35, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Liao, J.; Gao, L.; Shen, J.; Guarnera, M.A.; Zhan, M.; Fang, H.; Stass, S.A.; Jiang, F. Analysis of small nucleolar RNAs in sputum for lung cancer diagnosis. Oncotarget 2016, 7, 5131–5142. [Google Scholar] [CrossRef] [PubMed]
- Stępiński, D. The nucleolus, an ally, and an enemy of cancer cells. Histochem. Cell Biol. 2018, 150, 607–629. [Google Scholar] [CrossRef] [PubMed]
- Kiss-László, Z.; Henry, Y.; Bachellerie, J.P.; Caizergues-Ferrer, M.; Kiss, T. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell 1996, 85, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- Bachellerie, J.P.; Cavaillé, J. Guiding ribose methylation of rRNA. Trends Biochem. Sci. 1997, 22, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Bratkovič, T.; Rogelj, B. Biology and applications of small nucleolar RNAs. Cell Mol. Life Sci. 2011, 68, 3843–3851. [Google Scholar] [CrossRef] [PubMed]
- Falaleeva, M.; Pages, A.; Matuszek, Z.; Hidmi, S.; Agranat-Tamir, L.; Korotkov, K.; Nevo, Y.; Eyras, E.; Sperling, R.; Stamm, S. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 2016, 113, E1625–E1634. [Google Scholar] [CrossRef] [PubMed]
- Flynn, R.A.; Pedram, K.; Malaker, S.A.; Batista, P.J.; Smith, B.A.H.; Johnson, A.G.; George, B.M.; Majzoub, K.; Villalta, P.W.; Carette, J.E.; et al. Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell 2021, 184, 3109–3124.e22. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chai, P.; Till, N.A.; Hemberger, H.; Lebedenko, C.G.; Porat, J.; Watkins, C.P.; Caldwell, R.M.; George, B.M.; Perr, J.; et al. The modified RNA base acp3U is an attachment site for N-glycans in glycoRNA. Cell 2024, 187, 5228–5237.e12. [Google Scholar] [CrossRef] [PubMed]
- Nachtergaele, S.; Krishnan, Y. New Vistas for Cell-Surface GlycoRNAs. N. Engl. J. Med. 2021, 385, 658–660. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, W.; Pandey, V.; Pokharel, Y.R. Membrane linked RNA glycosylation as new trend to envision epi-transcriptome epoch. Cancer Gene Ther. 2023, 30, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Tang, W.; Torres, L.; Wang, X.; Ajaj, Y.; Zhu, L.; Luan, Y.; Zhou, H.; Wang, Y.; Zhang, D.; et al. Cell surface RNAs control neutrophil recruitment. Cell 2024, 187, 846–860.e17. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Guo, W.; Mou, Q.; Shao, X.; Lyu, M.; Garcia, V.; Kong, L.; Lewis, W.; Ward, C.; Yang, Z.; et al. Spatial imaging of glycoRNA in single cells with ARPLA. Nat. Biotechnol. 2024, 42, 608–616. [Google Scholar] [CrossRef] [PubMed]
- Köhn, M.; Pazaitis, N.; Hüttelmaier, S. Why YRNAs? About Versatile RNAs and Their Functions. Biomolecules 2013, 3, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Blanco, F.; Kalsi, J.; Isenberg, D.A. Analysis of antibodies to RNA in patients with systemic lupus erythematosus and other autoimmune rheumatic diseases. Clin. Exp. Immunol. 1991, 86, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Tang, X.; Xie, Y.; Cuffari, B.J.; Tang, C.; Cao, F.; Gao, X.; Meng, Z.; Noble, P.W.; Young, M.R.; et al. A lupus-derived autoantibody that binds to intracellular RNA activates cGAS-mediated tumor immunity and can deliver RNA into cells. Sci. Signal. 2025, 18, eadk3320. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Fan, X.; Zaleta-Rivera, K.; Nguyen, T.C.; Zhou, J.; Luo, Y.; Gao, J.; Fang, R.H.; Yan, Z.; Chen, Z.B.; et al. Natural display of nuclear-encoded RNA on the cell surface and its impact on cell interaction. Genome Biol. 2020, 21, 225. [Google Scholar] [CrossRef] [PubMed]
- Dellar, E.R.; Hill, C.; Melling, G.E.; Carter, D.R.F.; Baena-Lopez, L.A. Unpacking extracellular vesicles: RNA cargo loading and function. J. Extracell. Biol. 2022, 1, e40. [Google Scholar] [CrossRef] [PubMed]
- Tosar, J.P.; Witwer, K.; Cayota, A. Revisiting Extracellular RNA Release, Processing, and Function. Trends Biochem. Sci. 2021, 46, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Costa, B.; Li Calzi, M.; Castellano, M.; Blanco, V.; Cuevasanta, E.; Litvan, I.; Ivanov, P.; Witwer, K.; Cayota, A.; Tosar, J.P. Nicked tRNAs are stable reservoirs of tRNA halves in cells and biofluids. Proc. Natl. Acad. Sci. USA 2023, 120, e2216330120. [Google Scholar] [CrossRef] [PubMed]
- Costa, B.; Blanco, V.; Cayota, A.; Tosar, J.P. Methods for purification and characterization of nicked tRNAs. Methods Enzymol. 2025, 711, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Yang, Z.; Li, J.; Minakhina, S.; Yang, M.; Padgett, R.W.; Steward, R.; Chen, X. Methylation as a crucial step in plant microRNA biogenesis. Science 2005, 307, 932–955. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Sakaguchi, Y.; Suzuki, T.; Suzuki, T.; Siomi, H.; Siomi, M.C. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007, 21, 1603–1608. [Google Scholar] [CrossRef] [PubMed]
- Galloway, A.; Cowling, V.H. mRNA cap regulation in mammalian cell function and fate. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2019, 1862, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Mui Chan, C.; Zhou, C.; Brunzelle, J.S.; Huang, R.H. Structural and biochemical insights into 2′-O-methylation at the 3′-terminal nucleotide of RNA by Hen1. Proc. Natl. Acad. Sci. USA 2009, 106, 17699–17704. [Google Scholar] [CrossRef] [PubMed]
- Kurth, H.M.; Mochizuki, K. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA 2009, 15, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Maden, B.E. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 1990, 39, 241–303. [Google Scholar] [CrossRef] [PubMed]
- Krogh, N.; Kongsbak-Wismann, M.; Geisler, C.; Nielsen, H. Substoichiometric ribose methylations in spliceosomal snRNAs. Org. Biomol. Chem. 2017, 15, 8872–8876. [Google Scholar] [CrossRef] [PubMed]
- Marchand, V.; Pichot, F.; Thüring, K.; Ayadi, L.; Freund, I.; Dalpke, A.; Helm, M.; Motorin, Y. Next-Generation Sequencing-Based RiboMethSeq Protocol for Analysis of tRNA 2′-O-Methylation. Biomolecules 2017, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Höfler, S.; Carlomagno, T. Structural and functional roles of 2′-O-ribose methylations and their enzymatic machinery across multiple classes of RNAs. Curr. Opin. Struct. Biol. 2020, 65, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Khoshnevis, S.; Dreggors-Walker, R.E.; Marchand, V.; Motorin, Y.; Ghalei, H. Ribosomal RNA 2′-O-methylations regulate translation by impacting ribosome dynamics. Proc. Natl. Acad. Sci. USA 2022, 119, e2117334119. [Google Scholar] [CrossRef] [PubMed]
- Ayadi, L.; Galvanin, A.; Pichot, F.; Marchand, V.; Motorin, Y. RNA ribose methylation (2′-O-methylation): Occurrence, biosynthesis and biological functions. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2019, 1862, 253–269. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.I.; Pecot, C.V.; Holley, C.L. 2′-O-methylation (Nm) in RNA: Progress, challenges, and future directions. RNA 2024, 30, 570–582. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Cai, Y.; Yang, H.; Zhang, B.; Li, N.; Ren, G. PBOX-sRNA-seq uncovers novel features of miRNA modification and identifies selected 5′-tRNA fragments bearing 2′-O-modification. Nucleic Acids Res. 2024, 52, e65. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Kim, H.K.; Xu, J.; Jing, Y.; Kay, M.A. The 3′tsRNAs are aminoacylated: Implications for their biogenesis. PLoS Genet. 2021, 17, e1009675. [Google Scholar] [CrossRef] [PubMed]
- Monaco, P.L.; Marcel, V.; Diaz, J.J.; Catez, F. 2′-O-Methylation of Ribosomal RNA: Towards an Epitranscriptomic Control of Translation? Biomolecules 2018, 8, 106. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Kerpedjiev, P.; Hammer, S.; Hofacker, I.L. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics 2015, 31, 3377–3379. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Höner Zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Wadler, C.S.; Vanderpool, C.K. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl. Acad. Sci. USA 2007, 104, 20454–20459. [Google Scholar] [CrossRef] [PubMed]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Mattick, J.S. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput. Biol. 2008, 4, e1000176. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.Z.; Li, W.; Liu, Z.Y. Alternative role of noncoding RNAs: Coding and noncoding properties. J. Zhejiang Univ. Sci. B 2019, 20, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Keiler, K.C.; Ramadoss, N.S. Bifunctional transfer-messenger RNA. Biochimie 2011, 93, 1993–1997. [Google Scholar] [CrossRef] [PubMed]
- Leygue, E. Steroid receptor RNA activator (SRA1): Unusual bifaceted gene products with suspected relevance to breast cancer. Nucl. Recept. Signal. 2007, 5, e006. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; King, M.L. Xenopus VegT RNA is localized to the vegetal cortex during oogenesis and encodes a novel T-box transcription factor involved in mesodermal patterning. Development 1996, 122, 4119–4129. [Google Scholar] [CrossRef] [PubMed]
- Kloc, M.; Wilk, K.; Vargas, D.; Shirato, Y.; Bilinski, S.; Etkin, L.D. Potential structural role of non-coding and coding RNAs in the organization of the cytoskeleton at the vegetal cortex of Xenopusoocytes. Development 2005, 132, 3445–3457. [Google Scholar] [CrossRef] [PubMed]
- Jenny, A.; Hachet, O.; Závorszky, P.; Cyrklaff, A.; Weston, M.D.; Johnston, D.S.; Erdélyi, M.; Ephrussi, A. A translation-independent role of oskar RNA in early Drosophila oogenesis. Development 2006, 133, 2827–2833. [Google Scholar] [CrossRef] [PubMed]
- Gultyaev, A.P.; Roussis, A. Identification of conserved secondary structures and expansion segments in enod40 RNAs reveals new enod40 homologues in plants. Nucleic Acids Res. 2007, 35, 3144–3152. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Sampath, K. cncRNAs: Bi-functional RNAs with protein coding and non-coding functions. Semin. Cell Dev. Biol. 2015, 47–48, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Gimpel, M.; Preis, H.; Barth, E.; Gramzow, L.; Brantl, S. SR1—A small RNA with two remarkably conserved functions. Nucleic Acids Res. 2012, 40, 11659–11672. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, J.J.; Raina, M.; Zhong, A.; Storz, G. Dual-function Spot 42 RNA encodes a 15-amino acid protein that regulates the CRP transcription factor. Proc. Natl. Acad. Sci. USA 2022, 119, e2119866119. [Google Scholar] [CrossRef] [PubMed]
- Francastel, C.; Hubé, F. Coding or non-coding: Need they be exclusive? Biochimie 2011, 93, vi–vii. [Google Scholar] [CrossRef] [PubMed]
- Saraiya, A.A.; Wang, C.C. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008, 4, e1000224. [Google Scholar] [CrossRef] [PubMed]
- Ender, C.; Krek, A.; Friedländer, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A human snoRNA with microRNA-like functions. Mol. Cell 2008, 32, 519–528. [Google Scholar] [CrossRef] [PubMed]

| Feature | pfeRNAs | miRNAs, siRNAs, piRNAs, and Other ncRNAs |
|---|---|---|
| Length | 26–60 nts; longer than miRNAs, siRNAs, piRNAs, but shorter than pre-miRNAs, snoRNAs, snRNAs, and others | miRNAS and siRNAs: ~21–23 nts; piRNAs: ~24–31 nts; pre-miRNAs, snoRNAs, snRNAs: >60 nts |
| Database presence | According to previous publications: Not found in existing RNA databases; not predicted by current RNA prediction software. This publication: Matches in GenBank database with fragments of other ncRNAs | Well-annotated in databases like miRbase, piRNABank, etc. |
| 3′ end modification | 2′-O-methylation at the 3′ end, conferring stability against RNase degradation | miRNAs: some have 2′-O-methylation (mainly in plants); piRNAs: 2′-O-methylation; others vary |
| Cellular origin | Identified in somatic cells (not restricted to germline) | piRNAs: mainly germline; miRNAs, siRNAs: somatic and germline |
| Target interaction | Directly bind to specific proteins; regulate protein function without altering protein levels | miRNAs/siRNAs: bind to mRNA targets, regulate gene expression via degradation or translation inhibition; piRNAs: transposon silencing |
| Target specificity | One protein can be affected by more than one pfeRNA; a pfeRNA may recognize proteins with common phosphorylated sites | miRNAs/siRNAs: sequence-specific to mRNA; piRNAs: sequence-specific to transposons |
| Functional role | Modulate biological activities of target proteins (e.g., cell growth, immune escape) | miRNAs/siRNAs: posttranscriptional gene regulation; piRNAs: genome defense |
| Biogenesis & discovery | Not well understood; discovered via protein immunoprecipitation and deep sequencing | Biogenesis pathways are well-characterized for most other ncRNAs and their fragments |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patarca, R.; Haseltine, W.A. Protein Functional Effector (pfe) Noncoding RNAS Are Identical to Fragments from Various Noncoding RNAs. Int. J. Mol. Sci. 2025, 26, 6870. https://doi.org/10.3390/ijms26146870
Patarca R, Haseltine WA. Protein Functional Effector (pfe) Noncoding RNAS Are Identical to Fragments from Various Noncoding RNAs. International Journal of Molecular Sciences. 2025; 26(14):6870. https://doi.org/10.3390/ijms26146870
Chicago/Turabian StylePatarca, Roberto, and William A. Haseltine. 2025. "Protein Functional Effector (pfe) Noncoding RNAS Are Identical to Fragments from Various Noncoding RNAs" International Journal of Molecular Sciences 26, no. 14: 6870. https://doi.org/10.3390/ijms26146870
APA StylePatarca, R., & Haseltine, W. A. (2025). Protein Functional Effector (pfe) Noncoding RNAS Are Identical to Fragments from Various Noncoding RNAs. International Journal of Molecular Sciences, 26(14), 6870. https://doi.org/10.3390/ijms26146870








