Genome-Wide Characterization of the Phosphofructokinase Gene Family in Arabidopsis thaliana and Functional Analysis of AtPFK2 in Stress Tolerance
Abstract
1. Introduction
2. Results
2.1. Identification of the Phosphofructokinase (PFK) Gene Family in Arabidopsis thaliana
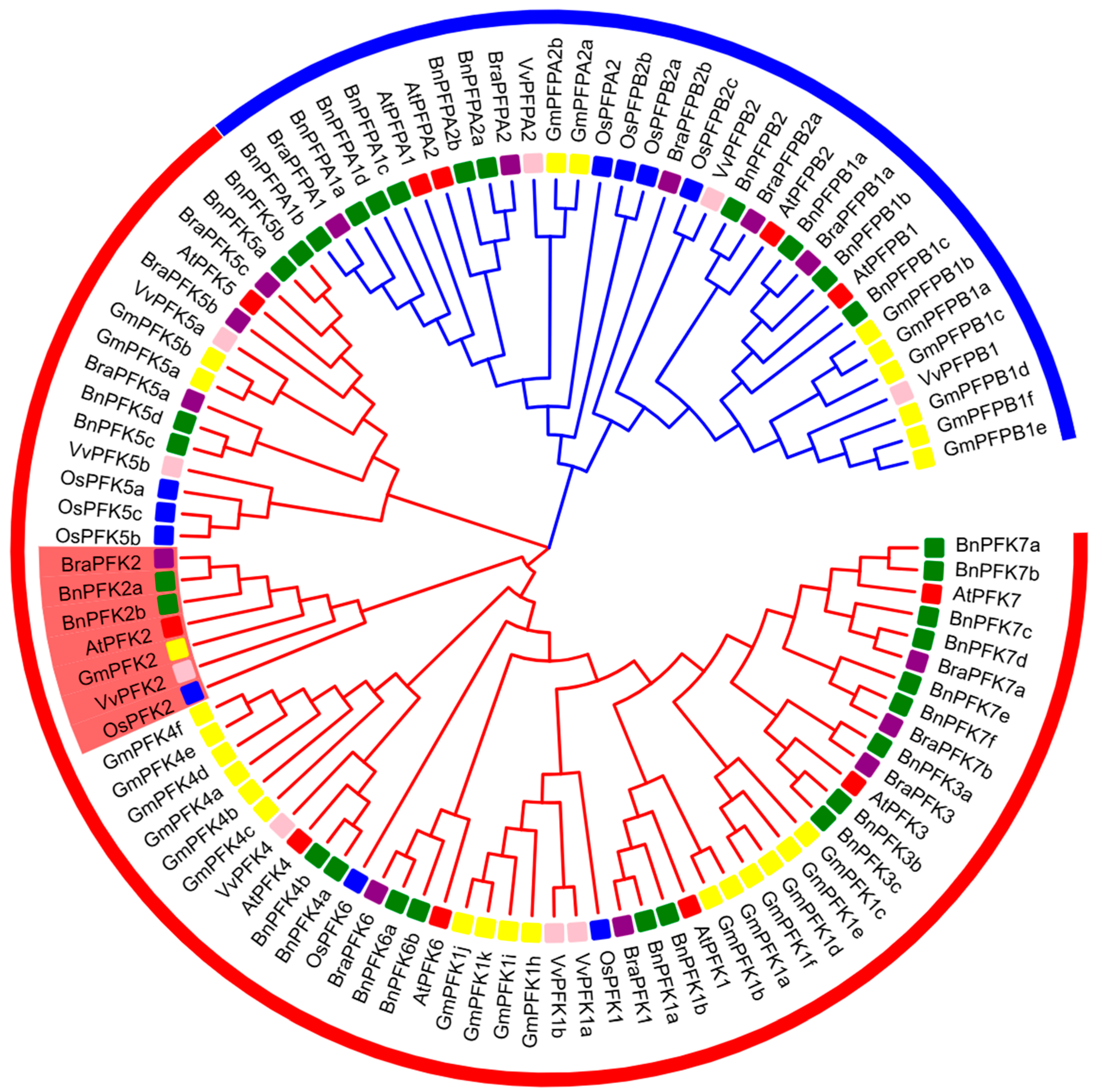
2.2. Phylogenetic Analysis of PFK Proteins in Arabidopsis thaliana
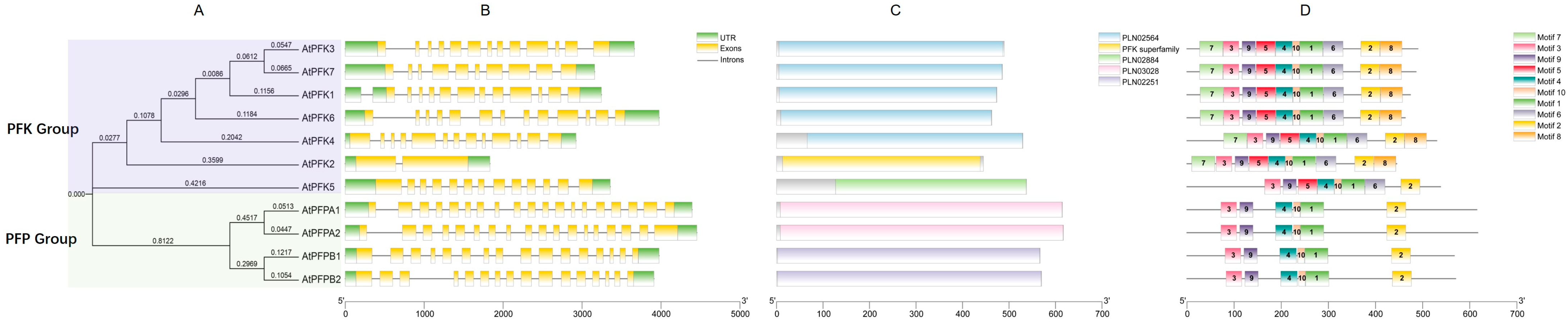
2.3. Motif and Gene Structure Analysis of the PFK Gene Family in Arabidopsis thaliana
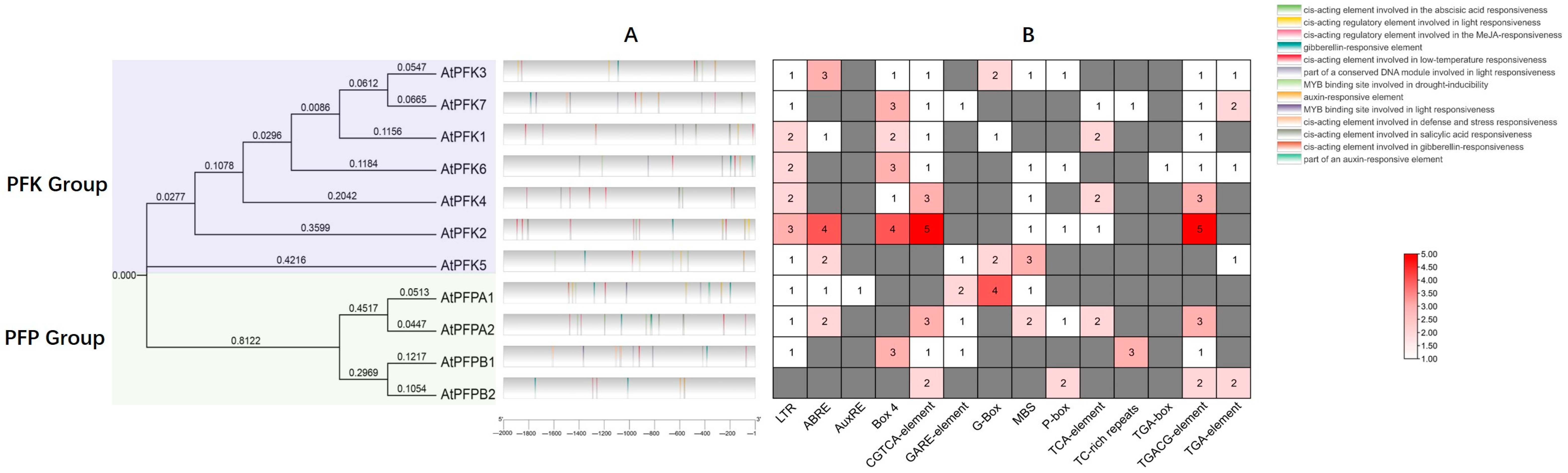
2.4. cis-Acting Element Analysis in the Promoter Regions of PFK Genes in Arabidopsis thaliana
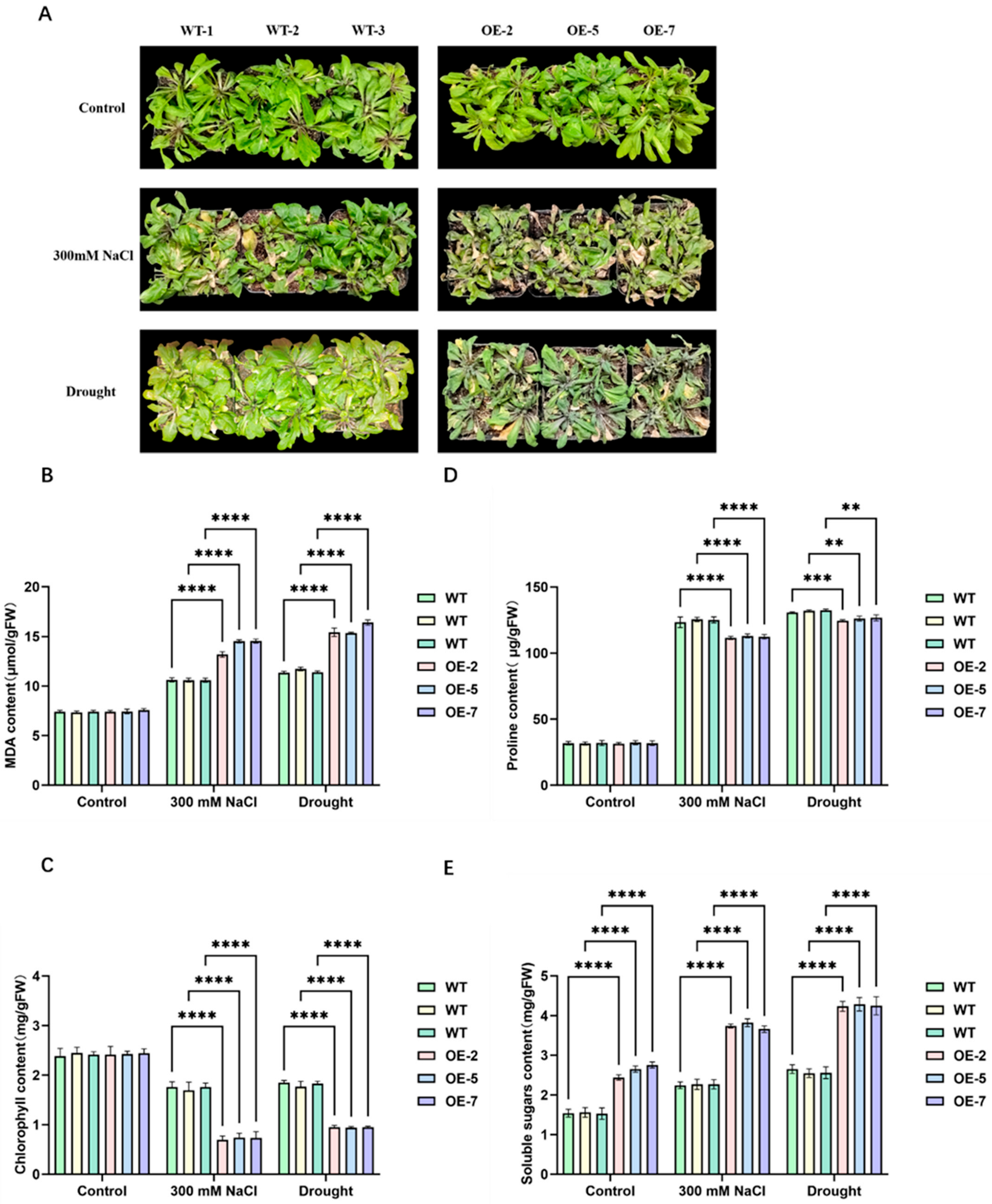
2.5. AtPFK2 Acts as a Negative Regulator of Salt and Drought Stress Tolerance in Arabidopsis thaliana
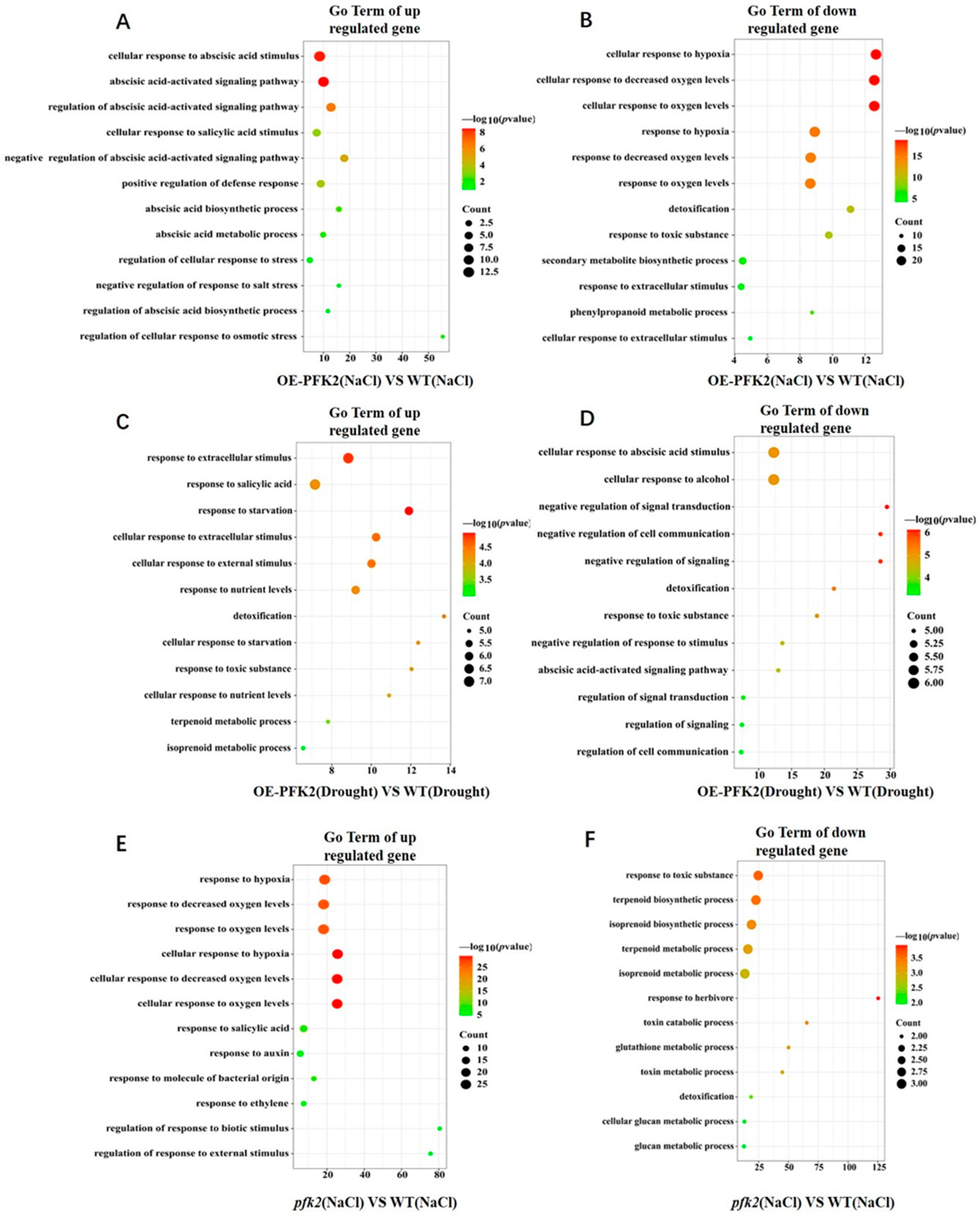
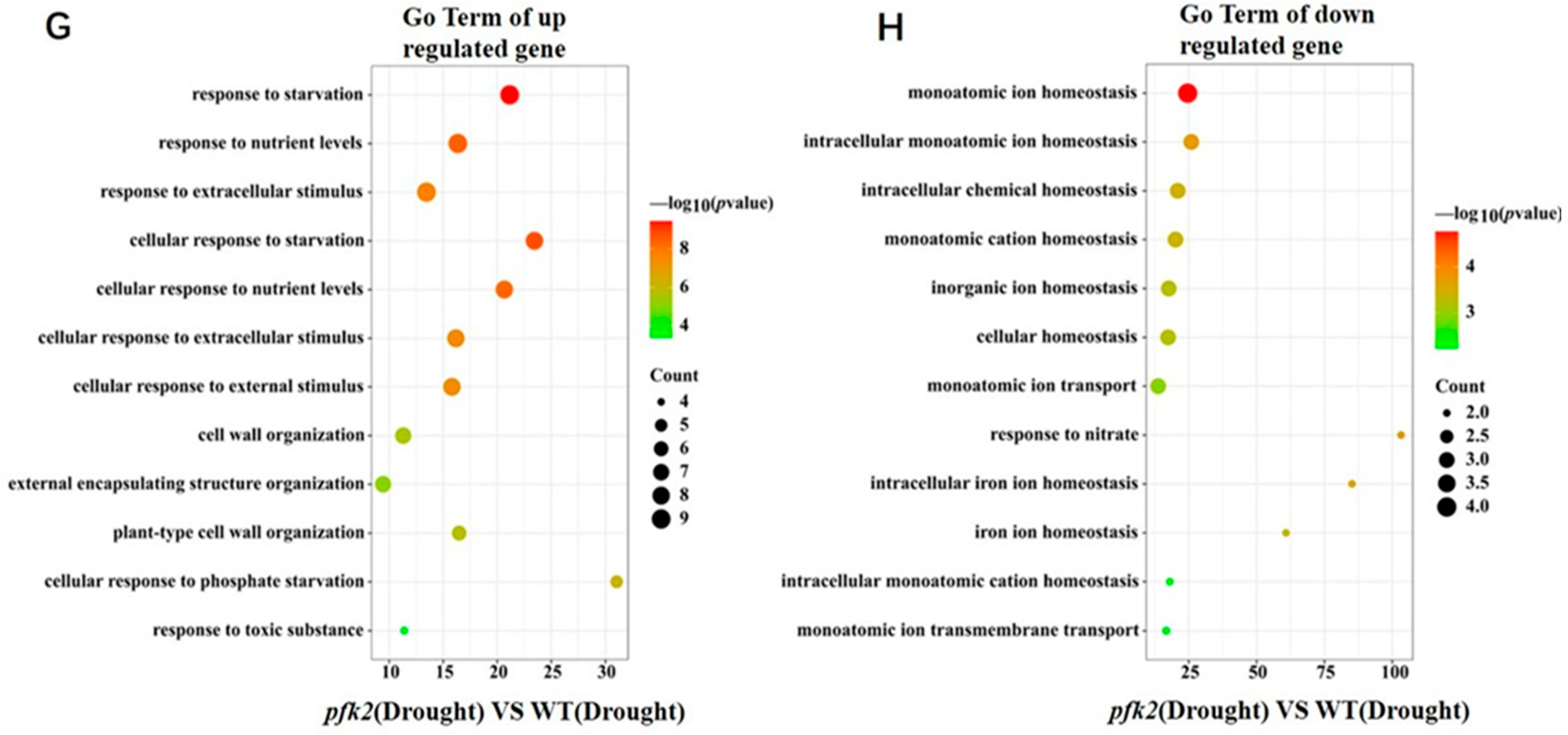
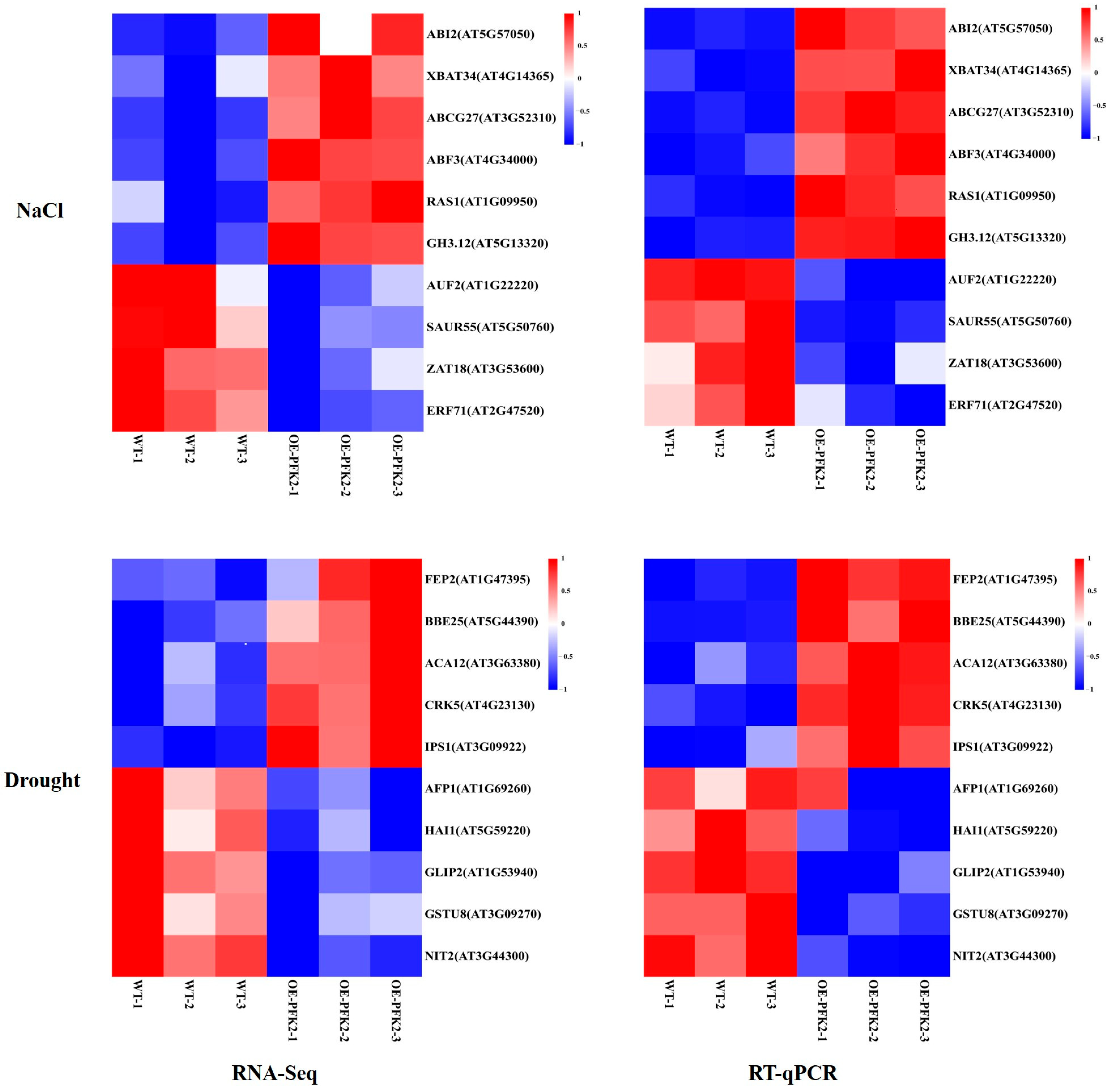
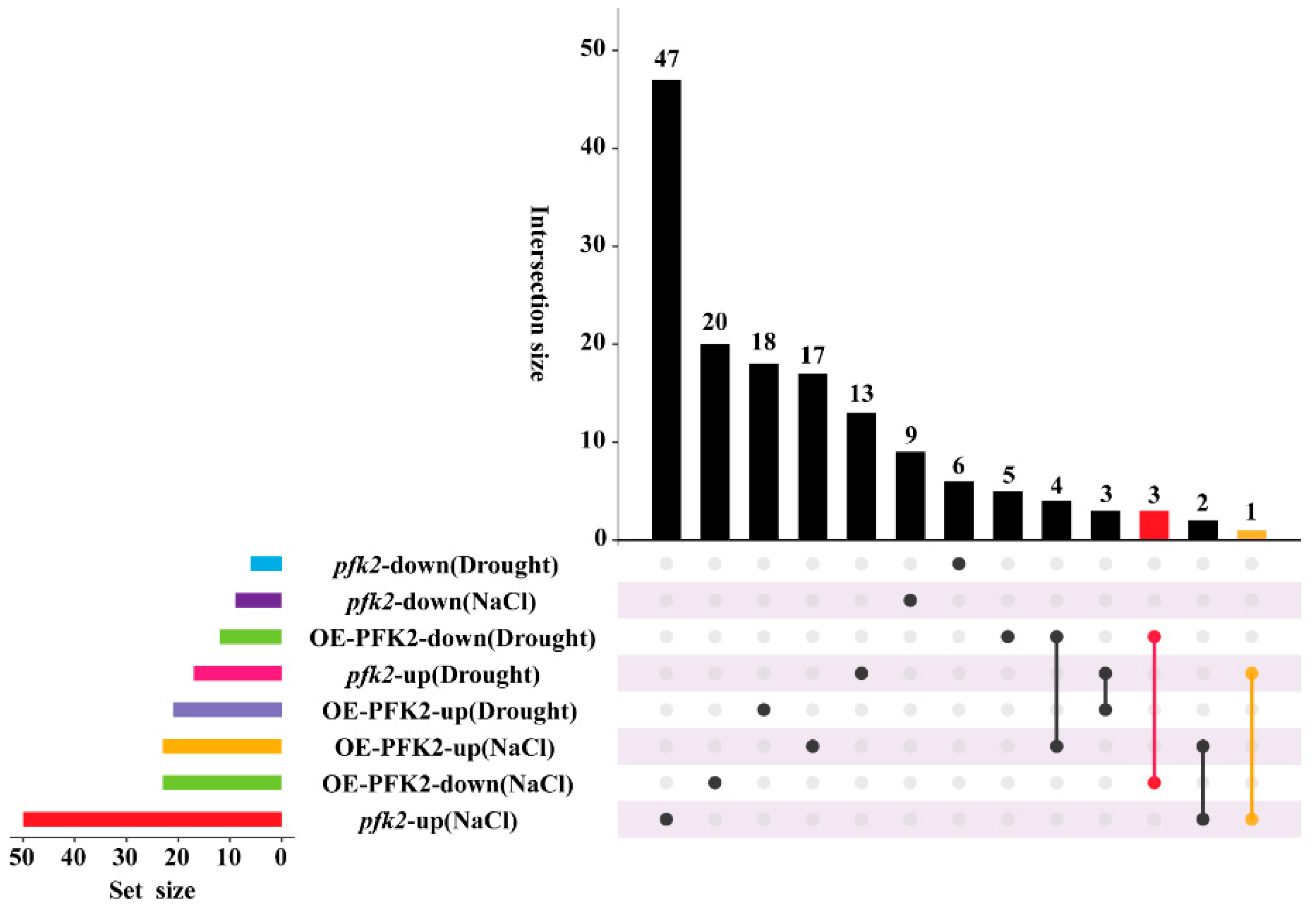
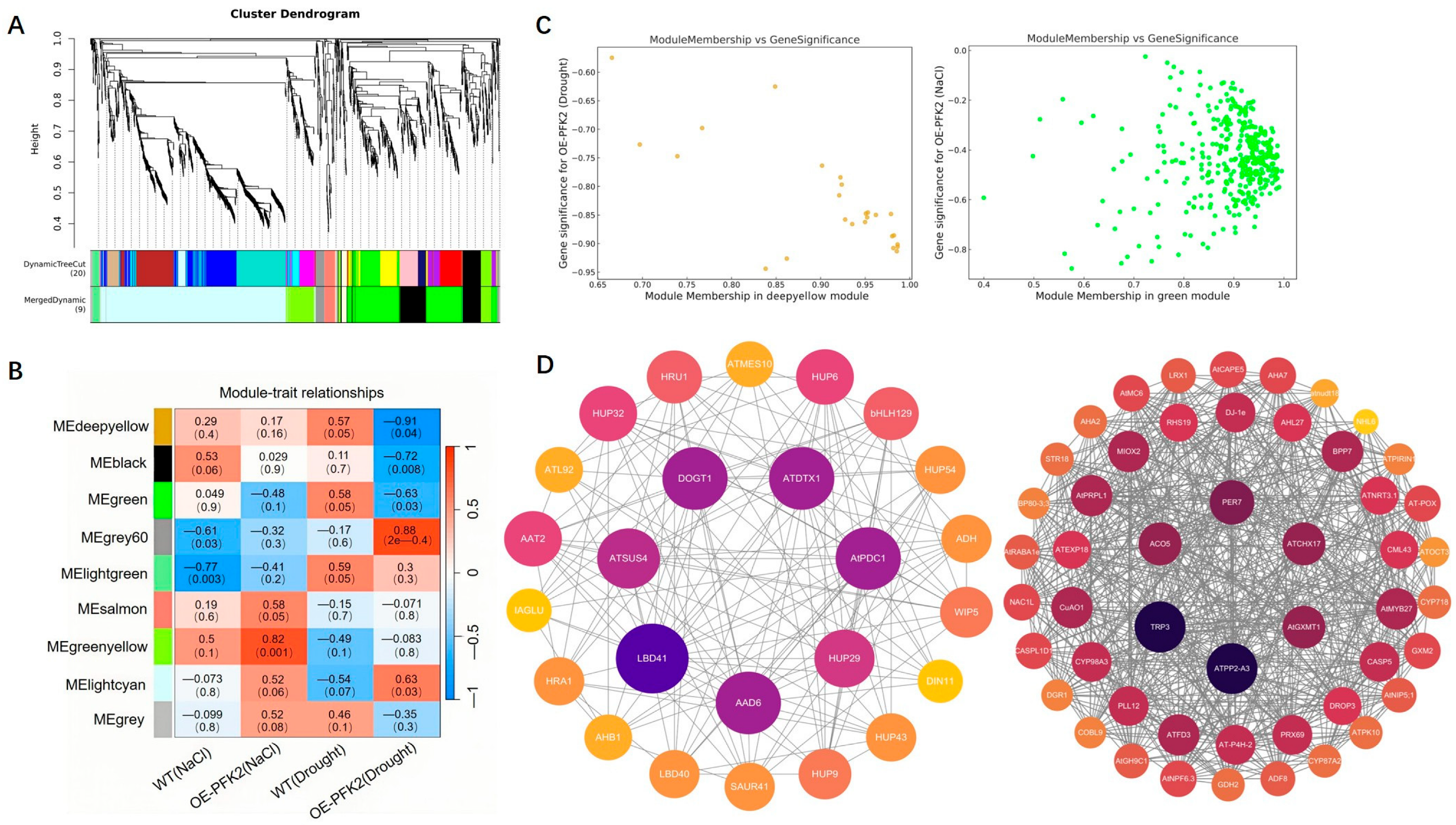
2.6. Transcriptomic Analysis of AtPFK2 in Response to Salt and Drought Stresses and the Protein–Protein Interaction Network of Core Genes
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification and Sequence Analysis
4.2. Prediction of Physiochemical Properties and Subcellular Localization
4.3. Phylogenetic Analysis
4.4. Conserved Motif, Gene Structure, and cis-Element Analysis
4.5. Construction of AtPFK2-Overexpression Vector and Genetic Transformation in Arabidopsis
4.6. RNA-Seq and qRT-PCR Analysis
4.7. Weighted Gene Co-Expression Network Analysis (WGCNA)
4.8. Abiotic Stress Tolerance Assays
4.9. Determination of Proline, MDA, Chlorophyll, and Soluble Sugar Contents
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arunachalam, E.; Keber, F.C.; Law, R.C.; Kumar, C.K.; Shen, Y.; Park, J.O.; Wühr, M.; Needleman, D.J. Robustness of mitochondrial biogenesis and respiration explain aerobic glycolysis. bioRxiv 2024. [Google Scholar]
- Khanna, M. Plant Metabolism during Water Deficit Stress: A Review. Agric. Rev. 2024, 45, 448–455. [Google Scholar] [CrossRef]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrange-ments and regulatory networks. J. Exp. Bot. 2012, 63, 1593. [Google Scholar] [CrossRef] [PubMed]
- Baena-González, E.; Sheen, J. Convergent energy and stress signaling. Trends Plant Sci. 2008, 13, 474–482. [Google Scholar] [CrossRef]
- Dumont, S.; Rivoal, J. Consequences of oxidative stress on plant glycolytic and respiratory metabolism. Front. Plant Sci. 2019, 10, 166. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Parvin, K.; Bhuiyan, T.F.; Fujita, M. Regulation of ROS metabolism in plants under environmental stress: A review of recent experimental evidence. Int. J. Mol. Sci. 2020, 21, 8695. [Google Scholar] [CrossRef]
- Gupta, B.; Huang, B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef]
- Aoki, M.; Kaku, K.; Inoue, H.; Matsutani, A.; Kaneko, T. Tolbutamide inhibits cAMP-dependent phosphorylation of liver 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase. Diabetes 1992, 41, 334–338. [Google Scholar] [CrossRef]
- Rasal, K.D.; Iquebal, M.A.; Dixit, S.; Vasam, M.; Sundaray, J.K. Revealing alteration in the hepatic glucose metabolism of genetically improved carp, jayanti rohu labeo rohita fed a high carbohydrate diet using transcriptome sequencing. Int. J. Mol. Sci. 2020, 21, 8180. [Google Scholar] [CrossRef]
- Kleszcz, R.; Paluszczak, J.; Belka, M.; Krajka-Kuniak, V. PRI-724 and IWP-O1 wnt signaling pathway inhibitors modulate the expression of glycolytic enzymes in tongue cancer cell lines. Curr. Issues Mol. Biol. 2023, 45, 9579–9592. [Google Scholar] [CrossRef]
- Ferreira, J.C.; Villanueva, A.J.; Fadl, S.; Adem, K.A.; Cinviz, Z.N.; Nedyalkova, L.; Cardoso, T.H.S.; Andrade, M.E.; Saksena, N.K.; Sensoy, O. Residues in the fructose-binding pocket are required for ketohexokinase-A activity. J. Biol. Chem. 2024, 300, 16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.L.; Zhu, H.; Chen, C.Y.; Shang, N.N.; Sheng, L.X.; Yu, J.Q. The function of an apple ATP-dependent Phosphofructokinase gene MdPFK5 in regulating salt stress. Physiol. Plant. 2024, 176, e13931. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Zhao, Q.; Chen, L.; Yao, X.; Zhang, W.; Zhang, B.; Xie, F. Effect of Drought Stress on Sugar Metabolism in Leaves and Roots of Soybean Seedlings. Plant. Physiol. Biochem. 2020, 146, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Cui, K.; Xu, A.; Nie, L.; Huang, J.; Peng, S. Drought Stress Condition Increases Root to Shoot Ratio via Alteration of Carbohydrate Partitioning and Enzymatic Activity in Rice Seedlings. Acta Physiol. Plant. 2015, 37, 1–11. [Google Scholar] [CrossRef]
- Mustroph, A.; Sonnewald, U.; Biemelt, S. Characterisation of the ATP-dependent phosphofructokinase gene family from Arab. thaliana. FEBS Lett. 2007, 581, 2401–2410. [Google Scholar] [CrossRef]
- Kelly, G.J.; Latzko, E. Evidence for phosphofructokinase in chloroplasts. Nature 1975, 256, 429–430. [Google Scholar] [CrossRef]
- Kelly, G.J.; Latzko, E. Chloroplast phosphofructokinase: I. Proof of phosphofructokinase activity in chloroplasts. Plant Physiol. 1977, 60, 290–294. [Google Scholar] [CrossRef]
- Kelly, G.J.; Latzko, E. Chloroplast phosphofructokinase: II. Partial purification, kinetic and regulatory properties. Plant Physiol. 1977, 60, 295–299. [Google Scholar] [CrossRef]
- Hausler, R.E.; Holtum, J.A.M.; Latzko, E. Cytosolic ATP-dependent Phosphofructokinase from Spinach. Plant Physiol. 1987, 84, 205–207. [Google Scholar] [CrossRef]
- Hausler, R.E.; Holtum, J.A.M.; Latzko, E. Cytosolic phosphofructokinase from spinach leaves: I. Purification, characteristics, and regulation. Plant Physiol. 1989, 90, 1498–1505. [Google Scholar] [CrossRef][Green Version]
- Hausler, R.E.; Holtum, J.A.M.; Latzko, E. Cytosolic Phosphofructokinase from Spinach Leaves: II. Affinity for Mg2+ and Nucleoside Phosphates. Plant Physiol. 1989, 90, 1506–1512. [Google Scholar] [CrossRef] [PubMed]
- Carnal, W.; Black, C.C. Pyrophosphate-dependent 6-phosphofructokinase, a new glycolytic enzyme in pineapple leaves. Biochem. Biophys. Res. Commun. 1979, 86, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Takeshige, K.; Tazawa, M. Determination of the inorganic pyrophosphate level and its subcellular localization in Chara corallina. J. Biol. Chem. 1989, 264, 3262. [Google Scholar] [CrossRef]
- Lyu, J.; Jin, N.; Meng, X.; Jin, L.; Wang, S.Y.; Xiao, X.M.; Liu, Z.C.; Tang, Z.Q.; Yu, J.H. Exogenous silicon alleviates the adverse effects of cinnamic acid-induced autotoxicity stress on cucumber seedling growth. Front. Plant Sci. 2022, 13, 968514. [Google Scholar] [CrossRef]
- Zhao, J.F.; Yu, A.L.; Du, Y.W.; Wang, G.H.; Zhao, G.Y.; Wang, X.D.; Zhang, W.Z.; Cheng, K.; Liu, X.; Wang, Z.H.; et al. Foxtail millet (Setaria italica (L.) P. Beauv) CIPKs are responsive to ABA and abiotic stresses. PLoS ONE 2019, 14, e0225091. [Google Scholar] [CrossRef]
- Winkler, C.; Delvos, B.; Martin, W.; Henze, K. Purification, microsequencing and cloning of spinach ATP-dependent phosphofructokinase link sequence and function for the plant enzyme. FEBS J. 2007, 274, 429–438. [Google Scholar] [CrossRef]
- Lin, W.X.; Wu, L.K.; Lin, S.; Zhang, A.J.; Zhou, M.M.; Lin, R.; Wang, H.B.; Chen, J.; Zhang, Z.X.; Lin, R.Y. Metaproteomic analysis of ratoon sugarcane rhizospheric soil. BMC Microbiol. 2013, 13, 1–13. [Google Scholar] [CrossRef]
- Enomoto, T.; Ohyama, H.; Kodama, M. Purification and characterization of pyrophosphate: D-fructose 6-phosphate 1-phosphotransferase from rice seedlings. Biosci. Biotechnol. Biochem. 1992, 56, 251–255. [Google Scholar] [CrossRef]
- Mertens, E. Pyrophosphate-dependent phosphofructokinase, an anaerobic glycolytic enzyme? FEBS Lett. 1991, 285, 1–5. [Google Scholar] [CrossRef]
- Mertens, E.; Larondelle, Y.; Hers, H.G. Induction of pyrophosphate: Fructose 6-phosphate 1-phosphotransferase by anoxia in rice seedlings. Plant Physiol. 1990, 93, 584–587. [Google Scholar] [CrossRef]
- Klepikova, A.V.; Kasianov, A.S.; Gerasimov, E.S.; Logacheva, M.D.; Penin, A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016, 88, 1058–1070. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.H.; Dong, C.; Liu, T.F.; Shi, Y.; Wang, H.D.; Tao, Z.; Liang, Y.; Lian, J.Z. Improved functional expression of cytochrome P450s in Saccharomyces cerevisiae through screening a cDNA library from Arab. thaliana. Front. Bioeng. Biotechnol. 2021, 9, 764851. [Google Scholar]
- Nabi, R.B.S.; Tayade, R.; Deshmukh, R.; Hussain, A.; Shahid, M.; Adhikari, A.; AbuQamar, F.S.; Yun, B.W. The stress-induced gene AtDUF569 positively regulates salt stress responses in Arabidopsis thaliana. BMC Plant Biol. 2025, 25, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Long, G.S.; Hussen, M.; Dench, J. Stéphane Aris-Brosou. Identifying genetic determinants of complex phenotypes from whole genome sequence data. bioRxiv 2019. [Google Scholar]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef]
- Jaina, M.; Sara, C.; Lowri, W.; Matloob, Q.; Gustavoa, S.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.; et al. Pfam: The protein families database in 2021. Nucleic. Acids. Res. 2021, 49, D412–D419. [Google Scholar]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Huala, E. The Arabidopsis information resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- He, S.C.; Xu, L.; Wu, W.H.; Zhang, J.J.; Hao, Z.D.; Lu, L.; Shi, J.S.; Chen, J.H. The identification and expression analysis of the liriodendron chinense F-box gene family. Plants 2024, 13, 171. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, N.N.; Song, W.L.; Yin, G.J.; Qin, Y.J.; Yan, Y.M.; Hu, Y.K. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef]
- Zhang, F.F.; Liu, Y.X.; Liu, F.; Yang, J.; Sohail, A.; Lu, C.K.; Xu, P. Genome-wide characterization and analysis of rice DUF247 gene family. BMC Genom. 2024, 25, 613. [Google Scholar] [CrossRef]
- Roulin, A.; Auer, P.L.; Libault, M.; Schlueter, J.; Farmer, A.; May, G.; Stacey, G.; Doerge, R.W.; Jackson, S.A. The fate of duplicated genes in a polyploid plant genome. Plant J. 2013, 73, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.W.; Wang, P.G.; Jia, H.C.; Wu, T.T.; Yuan, S.; Jiang, B.J.; Sun, S.; Zhang, Y.X.; Wang, L.W.; Han, T.F. Haplotype Analysis of GmSGF14 Gene Family Reveals Its Roles in Photoperiodic Flowering and Regional Adaptation of Soybean. Int. J. Mol. Sci. 2023, 24, 9436. [Google Scholar] [CrossRef]
- Fan, C.M.; Xu, W.; Wang, Y.W.; Hu, R.B.; Zhang, X.M.; Chen, J.X.; Fu, Y.F.; Zhang, T. Genome-wide expression analysis of soybean MADS genes showing potential function in the seed development. PLoS ONE 2013, 8, e62288. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zagnitko, O.; Rodionova, I.; Osterman, A.; Godzik, A. The FGGY carbohydrate kinase family: Insights into the evolution of functional specificities. PLOS Comput. Biol. 2011, 7, 1–13. [Google Scholar] [CrossRef]
- Huang, X.L.; Tian, T.; Chen, J.Z.; Wang, D.; Tong, B.L.; Liu, J.M. Transcriptome analysis of Cinnamomum migao seed germination in medicinal plants of southwest China. BMC Plant Biol. 2021, 21, 270. [Google Scholar] [CrossRef]
- Wang, H.T.; Zhou, X.S.; Liu, C.C.; Li, W.X.; Guo, W.Z. Suppression of GhGLU19 encoding β-1,3-glucanase promotes seed germination in cotton. BMC Plant Biol. 2022, 22, 1–15. [Google Scholar] [CrossRef]
- Lourkisti, R.; Antoine, S.; Pailly, O.; Luro, F.; Gibon, Y.; Oustric, J.; Berti, L. GABA shunt pathway is stimulated in response to early defoliation-induced carbohydrate limitation in mandarin fruits. Heliyon 2023, 9, e15573. [Google Scholar] [CrossRef]
- Lynch, E.M.; Hansen, H.; Salay, L.; Cooper, M.; Timr, S.; Kollman, J.M.; Kollman, J.M. Structural basis for allosteric regulation of human phosphofructokinase-1. Nat. Commun. 2024, 15, 7323. [Google Scholar] [CrossRef]
- Strater, N.; Marek, S.; Kuettner, E.B.; Kloos, M.; Keim, A.; Bruser, A.; Kirchberger, J.; Schoneberg, T. Molecular architecture and structural basis of allosteric regulation of eukaryotic phosphofructokinases. Faseb J. 2011, 25, 89–98. [Google Scholar] [CrossRef]
- Siebers, B.; Hensel, R. Pyrophosphate-dependent phosphofructokinase from Thermoproteus tenax. Methods Enzymol. 2001, 331, 54–62. [Google Scholar]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef] [PubMed]
- Kempa, S.; Krasensky, J.; Santo, S.D.; Kopka, J.; Jonak, C. A central role of abscisic acid in stress-regulated carbohydrate metabolism. PLoS ONE 2008, 3, e3935. [Google Scholar] [CrossRef] [PubMed]
- Ruan, L. Sucrose Metabolism: Gateway to Diverse Carbon Useand Sugar Signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef]
- Rook, F.; Hadingham, S.A.; Li, Y.H.; Bevan, M.W. Sugar and aba response pathways and the control of gene expression. Plant Cell Environ. 2006, 29, 426–434. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, J.; Khan, M.; Wang, Y.; Xiao, W.; Fang, T.; Qu, J.; Xiao, P.; Li, C.L.; Liu, J.H. Transcription factors ABF4 and ABR1 synergistically regulate amylase-mediated starch catabolism in drought tolerance. Plant Physiol. 2023, 191, 591–609. [Google Scholar] [CrossRef]
- Lee, J.M.; Hammarén, H.M.; Savitski, M.M.; Baek, S.H. Control of protein stability by post-translational modifications. Nat. Commun. 2023, 14, 201. [Google Scholar] [CrossRef]
- Hu, Y.Q.; Xu, W.X.; Chen, L. Post-translational modifications and the reprogramming of tumor metabolism. Discov. Oncol. 2025, 16, 929. [Google Scholar] [CrossRef]
- Jansson, S. The light-harvesting chlorophyll a/b-binding proteins. Biochim. Et. Biophys. Acta 1994, 1184, 1–19. [Google Scholar] [CrossRef]
- Verslues, P.E.; Sharma, S. Proline metabolism and its implications for plant-environment interaction. Arab. Book 2010, 8, e0140. [Google Scholar] [CrossRef]
- Herbers, K.; Meuwly, P.; Jean-Pierre Métraux Sonnewald, U. Salicylic acid-independent induction of pathogenesis-related protein transcripts by sugars is dependent on leaf developmental stage. FEBS Lett. 1996, 397, 239–244. [Google Scholar] [CrossRef]
- Sakaki, T.; Kondo, N.; Sugahara, K. Breakdown of photosynthetic pigments and lipids in spinach leaves with ozone fumigation: Role of active oxygens. Physiol. Plant. 2010, 59, 28–34. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Breusegem, F.V. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Thalmann, M.; Pazmino, D.; Seung, D.; Horrer, D.; Nigro, A.; Meier, T.; Ling, K.K.; Pfeifhofer, H.W.; Zeeman, S.C.; Santelia, D. Regulation of leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell 2016, 28, 1860. [Google Scholar] [CrossRef] [PubMed]
- Wim, V.D.E.; El-Esawe, S.K. Sucrose signaling pathways leading to fructan and anthocyanin accumulation: A dual function in abiotic and biotic stress responses? Environ. Exp. Bot. 2014, 108, 4–13. [Google Scholar]
- Petrillo, E.; Herz, M.A.G.; Barta, A.; Kalyna, M.; Kornblihtt, A.R. Let there be light: Regulation of gene expression in plants. RNA Biol. 2014, 11, 1215–1220. [Google Scholar] [CrossRef]
- Karam, M.A.; Abd-Elgawad, M.E.; Ali, R.M. Differential gene expression of salt-stressed Peganum harmala L. Genet. Eng. Biotechnol. J. 2016, 14, 319–326. [Google Scholar] [CrossRef]
- Mustroph, A.; Stock, J.; Hess, N.; Aldous, S.; Grimm, B. Characterization of the phosphofructokinase gene family in rice and its expression under oxygen deficiency stress. Front. Plant Sci. 2013, 4, 125. [Google Scholar] [CrossRef]
- Rognoni, S.; Teng, S.; Arru, L.; Smeekens, S.C.M.; Perata, P. Sugar effects on early seedling development in Arabidopsis. Plant Growth Regul. 2007, 52, 217–228. [Google Scholar] [CrossRef]
- Gong, M.; Chen, B.O.; Li, Z.G.; Guo, L.H. Heat-shock-induced cross adaptation to heat, chilling, drought and salt stress in maize seedlings and involvement of H2O2. J. Plant Physiol. 2001, 158, 1125–1130. [Google Scholar] [CrossRef]
- Yang, L.; Ji, W.; Zhu, Y.; Gao, P.; Li, Y.; Cai, H.; Bai, X.; Guo, D. GsCBRLK, a calcium/calmodulin-binding receptor-like kinase, is a positive regulator of plant tolerance to salt and ABA stress. J. Exp. Bot. 2010, 61, 2519–2533. [Google Scholar] [CrossRef]
- Yu, S.; Cao, L.; Zhou, C.M.; Zhang, T.Q.; Lian, H.; Sun, Y.; Wu, J.Q.; Huang, J.; Wang, G.D.; Wang, J.W. Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. eLife 2013, 2, e00269. [Google Scholar] [CrossRef] [PubMed]
- Jeroen, L.; Johannes, H.; Sjef, S. Sugar signals and the control of plant growth and development. J. Exp. Bot. 2014, 65, 799–807. [Google Scholar]
- Obata, T.; Fernie, A. The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef]
- Fernie, A.R.; Stitt, M. On the discordance of metabolomics with proteomics and transcriptomics: Coping with increasing complexity in logic, chemistry, and network interactions. Plant Physiol. 2012, 158, 1139–1145. [Google Scholar] [CrossRef]
- Liu, X.; Hou, X. Antagonistic regulation of ABA and GA in metabolism and signaling pathways. Front. Plant Sci. 2018, 9, 251. [Google Scholar] [CrossRef]
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Zulfiqar, F.; Akram, N.A.; Ashraf, M. Osmoprotection in plants under abiotic stresses: New insights into a classical phenomenon. Planta 2020, 251, 3. [Google Scholar] [CrossRef]
- El-Esawi, M. Regulation of genes and transcriptional factors involved in plant responses to abiotic stress. In Plant Life Under Changing Environment; Academic Press: Cambridge, MA, USA, 2020; pp. 825–833. [Google Scholar]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Vanholme, R.; Storme, V.; Vanholme, B.; Sundin, L.; Christensen, J.H.; Goeminne, G.; Halpin, C.; Rohde, A.; Morreel, K.; Boerjan, W. A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 2012, 24, 3506–3529. [Google Scholar] [CrossRef]
- Pauwels, L. Jasmonate signalling in Arabidopsis thaliana. Ph.D. Thesis, Ghent University, Gent, Belgium, 2009. [Google Scholar]
- Pieterse, C.M.; Leon-Reyes, A.; Van der Ent, S.; Van Wees, S.C. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef]
- Sudhir, K.; Glen, S.; Koichiro, T. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar]
- Powell, H.R.; Islam, S.A.; David, A.; Sternberg, M.J.E. Phyre2.2: A Community Resource for Template-based Protein Structure Prediction. J. Mol. Biol. 2025, 437, 168960. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Song, J.F.; Sheng, S.A.; Wang, D.J.; Wang, T.T.; Wang, N.; Chen, A.R.; Wang, L.X.; Peng, Y.X.; Ma, Y.H.; et al. Genome-wide characterization of Solanum tuberosum UGT gene family and functional analysis of StUGT178 in salt tolerance. BMC Genom. 2024, 25, 1206. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.H.; Lercher, M.J.; Hu, S.N.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. Issue 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Lesco, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1999, 16, 735–743. [Google Scholar] [CrossRef]
- Kim, J.H.; Cho, A.Y.; Lim, S.D.; Jang, C.S. Mutation of a RING E3 ligase, OsDIRH2, enhances drought tolerance in rice with low stomata density. Physiol. Plant. 2024, 176, 1–16. [Google Scholar] [CrossRef]

| Gene Name | Gene ID | Chromosome Localization | Number of Amino Acid | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity |
|---|---|---|---|---|---|---|---|---|
| AtPFK1 | AT4G29220 | Chr4 | 473 | 51,991.32 | 7.22 | 36.21 | 86.55 | −0.20 |
| AtPFK2 | AT5G47810 | Chr5 | 444 | 49,182.14 | 6.63 | 29.20 | 87.61 | −0.19 |
| AtPFK3 | AT4G26270 | Chr4 | 489 | 53,666.37 | 6.61 | 40.42 | 89.04 | −0.19 |
| AtPFK4 | AT5G61580 | Chr5 | 530 | 58,467.08 | 8.46 | 34.97 | 90.64 | −0.17 |
| AtPFK5 | AT2G22480 | Chr2 | 537 | 58,614.75 | 6.81 | 39.46 | 90.04 | −0.14 |
| AtPFK6 | AT4G32840 | Chr4 | 462 | 50,787.97 | 6.61 | 33.17 | 87.97 | −0.17 |
| AtPFK7 | AT5G56630 | Chr5 | 485 | 53,482.03 | 6.87 | 40.24 | 85.96 | −0.28 |
| AtPFPA1 | AT1G20950 | Chr1 | 614 | 67,119.18 | 6.53 | 40.62 | 94.38 | −0.08 |
| AtPFPA2 | AT1G76550 | Chr1 | 617 | 67,558.52 | 6.81 | 42.07 | 91.99 | −0.12 |
| AtPFPB1 | AT1G12000 | Chr1 | 566 | 61,459.46 | 5.80 | 37.17 | 88.07 | −0.18 |
| AtPFPB2 | AT4G04040 | Chr4 | 569 | 62,741.61 | 5.44 | 36.77 | 83.95 | −0.27 |
| Species | Accession (Ensembl Plants) | Name | Accession (UniProt) |
|---|---|---|---|
| Arabidopsis thaliana | AT4g29220 | AtPFK1 | Q9M0F9 |
| Arabidopsis thaliana | AT5g47810 | AtPFK2 | Q9FIK0 |
| Arabidopsis thaliana | AT4g26270 | AtPFK3 | Q94AA4 |
| Arabidopsis thaliana | AT5g61580 | AtPFK4 | Q9FKG3 |
| Arabidopsis thaliana | AT2g22480 | AtPFK5 | Q8VYN6 |
| Arabidopsis thaliana | AT4g32840 | AtPFK6 | Q9M076 |
| Arabidopsis thaliana | AT5g56630 | AtPFK7 | Q9C5J7 |
| Arabidopsis thaliana | AT1g20950 | AtPFPA1 | Q9SYP2 |
| Arabidopsis thaliana | AT1g76550 | AtPFPA2 | Q9C9K3 |
| Arabidopsis thaliana | AT1g12000 | AtPFPB1 | Q8W4M5 |
| Arabidopsis thaliana | AT4g04040 | AtPFPB2 | F4JGR5 |
| Oryza sativa | Os01g0191700 | OsPFK1 | Q0JPZ1 |
| Oryza sativa | Os09g0479800 | OsPFK2 | Q652D3 |
| Oryza sativa | Os10g0405600 | OsPFK5a | A0A8J8YCT6 |
| Oryza sativa | Os08g0439000 | OsPFK5b | Q0J5F5 |
| Oryza sativa | Os09g0415800 | OsPFK5c | A0A0P0XM93 |
| Oryza sativa | Os05g0524400 | OsPFK6 | Q65X97 |
| Oryza sativa | Os06g0326400 | OsPFPA2 | B9FT08 |
| Oryza sativa | Os09g0298100 | OsPFPB2a | A0A0P0XL64 |
| Oryza sativa | Os08g0345700 | OsPFPB2b | Q84QT9 |
| Oryza sativa | Os06g0247500 | OsPFPB2c | A3BA88 |
| Glycine max | GLYMA_06G088600 | GmPFK1a | I1K9I0 |
| Glycine max | GLYMA_06G088600 | GmPFK1b | K7KU09 |
| Glycine max | GLYMA_08G031700 | GmPFK1c | I1KPV0 |
| Glycine max | GLYMA_07G126400 | GmPFK1d | I1KJS2 |
| Glycine max | GLYMA_07G126400 | GmPFK1e | I1KJS3 |
| Glycine max | GLYMA_07G126400 | GmPFK1f | I1KJS5 |
| Glycine max | GLYMA_08G199800 | GmPFK1h | I1KV07 |
| Glycine max | GLYMA_13G353400 | GmPFK1g | I1M5F6 |
| Glycine max | GLYMA_13G353400 | GmPFK1i | A0A0R0H5D7 |
| Glycine max | GLYMA_15G020900 | GmPFK1k | I1MCW1 |
| Glycine max | GLYMA_15G020900 | GmPFK1J | I1MCW0 |
| Glycine max | GLYMA_07G269500 | GmPFK2 | I1KNN0 |
| Glycine max | GLYMA_10G194300 | GmPFK4a | A0A0R0HVU7 |
| Glycine max | GLYMA_10G194300 | GmPFK4b | A0A0R0HVV2 |
| Glycine max | GLYMA_10G194300 | GmPFK4c | I1LCJ1 |
| Glycine max | GLYMA_10G194300 | GmPFK4d | A0A0R0I3J4 |
| Glycine max | GLYMA_10G194300 | GmPFK4e | A0A0R0I3C2 |
| Glycine max | GLYMA_10G194300 | GmPFK4f | K7LKD1 |
| Glycine max | GLYMA_08G280700 | GmPFK5a | I1KX93 |
| Glycine max | GLYMA_18G145500 | GmPFK5b | I1N1M0 |
| Glycine max | GLYMA_20G007400 | GmPFPA2a | I1ND14 |
| Glycine max | GLYMA_07G160500 | GmPFPA2b | I1KKN7 |
| Glycine max | GLYMA_09G007900 | GmPFPB1a | I1KZW7 |
| Glycine max | GLYMA_15G112300 | GmPFPB1b | I1MFL2 |
| Glycine max | GLYMA_15G112300 | GmPFPB1c | A0A0R0FZ92 |
| Glycine max | GLYMA_17G010100 | GmPFPB1d | K7MJC5 |
| Glycine max | GLYMA_07G263800 | GmPFPB1e | I1KNH4 |
| Glycine max | GLYMA_07G263800 | GmPFPB1f | I1KNH5 |
| Brassica napus | GSBRNA2T00091362001 | BnPFK1a | A0A078IGT7 |
| Brassica napus | GSBRNA2T00123079001 | BnPFK1b | NO |
| Brassica napus | GSBRNA2T00001926001 | BnPFK2a | A0A078FZE1 |
| Brassica napus | GSBRNA2T00128839001 | BnPFK2b | A0A816MMS9 |
| Brassica napus | GSBRNA2T00082589001 | BnPFK3a | A0A078JTU5 |
| Brassica napus | GSBRNA2T00149578001 | BnPFK3b | A0A816RM02 |
| Brassica napus | GSBRNA2T00157466001 | BnPFK3c | A0A816RM02 |
| Brassica napus | GSBRNA2T00113526001 | BnPFK4a | A0A816W200 |
| Brassica napus | GSBRNA2T00021520001 | BnPFK4b | A0A078GCL9 |
| Brassica napus | GSBRNA2T00127444001 | BnPFK5a | A0A816UWW4 |
| Brassica napus | GSBRNA2T00078189001 | BnPFK5b | A0A078FKS0 |
| Brassica napus | GSBRNA2T00007196001 | BnPFK5c | A0A078IRE3 |
| Brassica napus | GSBRNA2T00150980001 | BnPFK5d | A0A817AB30 |
| Brassica napus | GSBRNA2T00121159001 | BnPFK6a | NO |
| Brassica napus | GSBRNA2T00130627001 | BnPFK6b | A0A816R9K6 |
| Brassica napus | GSBRNA2T00073694001 | BnPFK7a | A0A078HTA9 |
| Brassica napus | GSBRNA2T00012921001 | BnPFK7b | A0A078G2H5 |
| Brassica napus | GSBRNA2T00052829001 | BnPFK7c | A0A078H5N0 |
| Brassica napus | GSBRNA2T00053002001 | BnPFK7d | A0A078H7L8 |
| Brassica napus | GSBRNA2T00060751001 | BnPFK7e | A0A078FF53 |
| Brassica napus | GSBRNA2T00148613001 | BnPFK7f | A0A816V1T9 |
| Brassica napus | GSBRNA2T00058643001 | BnPFPA1a | A0A078JNQ8 |
| Brassica napus | GSBRNA2T00059752001 | BnPFPA1b | A0A078JHX5 |
| Brassica napus | GSBRNA2T00102813001 | BnPFPA1c | NO |
| Brassica napus | GSBRNA2T00055023001 | BnPFPA1d | A0A078H4Z3 |
| Brassica napus | GSBRNA2T00146592001 | BnPFPA2a | A0A816ZCQ6 |
| Brassica napus | GSBRNA2T00147797001 | BnPFPA2b | NO |
| Brassica napus | GSBRNA2T00105468001 | BnPFPB1a | NO |
| Brassica napus | GSBRNA2T00044584001 | BnPFPB1b | A0A078GX24 |
| Brassica napus | GSBRNA2T00057050001 | BnPFPB1c | A0A078HAW1 |
| Brassica napus | GSBRNA2T00157791001 | BnPFPB2 | NO |
| Brassica rapa | Bra011089 | BraPFK1 | M4D3N5 |
| Brassica rapa | Bra024914 | BraPFK2 | M4E808 |
| Brassica rapa | Bra026452 | BraPFK3 | A0A398AM83 |
| Brassica rapa | Bra010637 | BraPFK5a | M4D2D7 |
| Brassica rapa | Bra030394 | BraPFK5b | M4ENM4 |
| Brassica rapa | Bra038519 | BraPFK5c | M4FBQ3 |
| Brassica rapa | Bra011387 | BraPFK6 | M4D4I3 |
| Brassica rapa | Bra002801 | BraPFK7a | M4CF19 |
| Brassica rapa | Bra006864 | BraPFK7b | M4CRM3 |
| Brassica rapa | Bra025858 | BraPFPA1 | A0A397Z4U1 |
| Brassica rapa | Bra015734 | BraPFPA2 | M4DGV8 |
| Brassica rapa | Bra019733 | BraPFPB1 | M4DT90 |
| Brassica rapa | Bra029482 | BraPFPB2a | M4EL15 |
| Brassica rapa | Bra016799 | BraPFPB2b | M4DJX1 |
| Vitis vinifera | Vitvi11g00237 | VvPFK1a | A0A438DRZ9 |
| Vitis vinifera | Vitvi04g00040 | VvPFK1b | D7STJ8 |
| Vitis vinifera | Vitvi10g00212 | VvPFK2 | A0A438D403 |
| Vitis vinifera | Vitvi16g00381 | VvPFK4 | D7U799 |
| Vitis vinifera | Vitvi07g01462 | VvPFK5a | NO |
| Vitis vinifera | Vitvi14g01938 | VvPFK5b | A0A438D7F2 |
| Vitis vinifera | Vitvi18g00037 | VvPFPA2 | F6I6W5 |
| Vitis vinifera | Vitvi10g00129 | VvPFPB1 | D7TR81 |
| Vitis vinifera | Vitvi12g00427 | VvPFPB2 | D7TED0 |
| Sequence | Motif | E-Value | Entry Accession | Description |
|---|---|---|---|---|
| RRAGPRQKVYFEPDEVKACIVTCGGLCPGLNTVI | 1 | 2.70 × 10−17 | IPR035966 | Phosphofructokinase superfamily |
| RRAGPRQKVYFEPDEVKACIVTCGGLCPGLNTVI | 1 | 1.28 × 10−15 | IPR035966 | Phosphofructokinase superfamily |
| RRAGPRQKVYFEPDEVKACIVTCGGLCPGLNTVI | 1 | 5.70 × 10−26 | IPR050929 | Phosphofructokinase type A |
| RRAGPRQKVYFEPDEVKACIVTCGGLCPGLNTVI | 2 | 6.40 × 10−18 | IPR050929 | Phosphofructokinase type A |
| RRAGPRQKVYFEPDEVKACIVTCGGLCPGLNTVI | 2 | 6.54 × 10−12 | IPR035966 | Phosphofructokinase superfamily |
| RRAGPRQKVYFEPDEVKACIVTCGGLCPGLNTVI | 3 | 6.90 × 10−16 | IPR050929 | Phosphofructokinase type A |
| RRAGPRQKVYFEPDEVKACIVTCGGLCPGLNTVI | 3 | 8.37 × 10−10 | IPR035966 | Phosphofructokinase superfamily |
| RRAGPRQKVYFEPDEVKACIVTCGGLCPGLNTVI | 3 | 2.00 × 10−6 | - | - |
| GGDGTQKGAAAIFEEIRRRKLKVAVVGIPKTIDNDI | 4 | 1.70 × 10−13 | - | - |
| GGDGTQKGAAAIFEEIRRRKLKVAVVGIPKTIDNDI | 4 | 1.10 × 10−14 | IPR050929 | Phosphofructokinase type A |
| GGDGTQKGAAAIFEEIRRRKLKVAVVGIPKTIDNDI | 4 | 3.79 × 10−13 | IPR035966 | Phosphofructokinase superfamily |
| GLVNGRHTYIPFNRITEKQNKVVITDRMWARLLSSTNQPSF | 5 | 2.60 × 10−12 | IPR050929 | Phosphofructokinase type A |
| KVVNDIHKRGGTILGTSRGGHDTSKIVDSIQDRGINQVYII | 6 | 2.20 × 10−11 | - | - |
| KVVNDIHKRGGTILGTSRGGHDTSKIVDSIQDRGINQVYII | 6 | 3.90 × 10−16 | IPR050929 | Phosphofructokinase type A |
| KVVNDIHKRGGTILGTSRGGHDTSKIVDSIQDRGINQVYII | 6 | 1.70 × 10−8 | IPR035966 | Phosphofructokinase superfamily |
| KVVNDIHKRGGTILGTSRGGHDTSKIVDSIQDRGINQVYII | 6 | - | - | - |
| SPFYLEGKGGLFEFIEKRLKENGHMVIVIAEGAGQDLVAKSME | 7 | 4.19 × 10−5 | IPR035966 | Phosphofructokinase superfamily |
| SPFYLEGKGGLFEFIEKRLKENGHMVIVIAEGAGQDLVAKSME | 7 | 3.30 × 10−6 | IPR035966 | Phosphofructokinase superfamily |
| SPFYLEGKGGLFEFIEKRLKENGHMVIVIAEGAGQDLVAKSME | 7 | 6.60 × 10−15 | IPR050929 | Phosphofructokinase type A |
| VPHLSDYLPDLPTYPNPLQDNPAYSVVKQYFVDADDTVPQKIVVHKDSPR | 8 | 5.60 × 10−14 | IPR050929 | Phosphofructokinase type A |
| Gene | ID | cis-Elements | Starting Position | Termination Position |
|---|---|---|---|---|
| AtPFK1 | AT4G29220 | LTR | −1828 | −1822 |
| AtPFK1 | AT4G29220 | CGTCA-motif | −1690 | −1685 |
| AtPFK1 | AT4G29220 | TGACG-motif | −1690 | −1685 |
| AtPFK1 | AT4G29220 | TATC-box | −1270 | −1263 |
| AtPFK1 | AT4G29220 | Box 4 | −636 | −630 |
| AtPFK1 | AT4G29220 | Box 4 | −576 | −570 |
| AtPFK1 | AT4G29220 | TCA-element | −476 | −467 |
| AtPFK1 | AT4G29220 | TCA-element | −207 | −198 |
| AtPFK1 | AT4G29220 | ABRE | −139 | −134 |
| AtPFK1 | AT4G29220 | G-box | −139 | −133 |
| AtPFK1 | AT4G29220 | LTR | −22 | −16 |
| AtPFK2 | AT5G47810 | LTR | −1896 | −1890 |
| AtPFK2 | AT5G47810 | LTR | −1854 | −1848 |
| AtPFK2 | AT5G47810 | TCA-element | −1811 | −1802 |
| AtPFK2 | AT5G47810 | CGTCA-motif | −1473 | −1468 |
| AtPFK2 | AT5G47810 | TGACG-motif | −1473 | −1468 |
| AtPFK2 | AT5G47810 | CGTCA-motif | −1470 | −1465 |
| AtPFK2 | AT5G47810 | TGACG-motif | −1470 | −1465 |
| AtPFK2 | AT5G47810 | CGTCA-motif | −970 | −965 |
| AtPFK2 | AT5G47810 | TGACG-motif | −970 | −965 |
| AtPFK2 | AT5G47810 | MBS | −949 | −943 |
| AtPFK2 | AT5G47810 | CGTCA-motif | −922 | −917 |
| AtPFK2 | AT5G47810 | TGACG-motif | −922 | −917 |
| AtPFK2 | AT5G47810 | P-box | −659 | −652 |
| AtPFK2 | AT5G47810 | ABRE | −262 | −257 |
| AtPFK2 | AT5G47810 | G-Box | −262 | −256 |
| AtPFK2 | AT5G47810 | LTR | −235 | −229 |
| AtPFK2 | AT5G47810 | CGTCA-motif | −85 | −85 |
| AtPFK2 | AT5G47810 | TGACG-motif | −85 | −80 |
| AtPFK2 | AT5G47810 | G-box | −84 | −78 |
| AtPFK2 | AT5G47810 | ABRE | −83 | −78 |
| AtPFK2 | AT5G47810 | G-box | −56 | −46 |
| AtPFK2 | AT5G47810 | ABRE | −55 | −46 |
| AtPFK2 | AT5G47810 | ABRE | −53 | −48 |
| AtPFK2 | AT5G47810 | G-box | −53 | −47 |
| AtPFK3 | AT4G26270 | ABRE | −1887 | −1882 |
| AtPFK3 | AT4G26270 | G-Box | −1887 | −1881 |
| AtPFK3 | AT4G26270 | CGTCA-motif | −1860 | −1855 |
| AtPFK3 | AT4G26270 | TGACG-motif | −1860 | −1855 |
| AtPFK3 | AT4G26270 | ABRE | −1165 | −1160 |
| AtPFK3 | AT4G26270 | G-Box | −1165 | −1159 |
| AtPFK3 | AT4G26270 | P-box | −1093 | −1086 |
| AtPFK3 | AT4G26270 | LTR | −486 | −480 |
| AtPFK3 | AT4G26270 | ABRE | −470 | −463 |
| AtPFK3 | AT4G26270 | Box 4 | −464 | −458 |
| AtPFK3 | AT4G26270 | MBS | −423 | −417 |
| AtPFK3 | AT4G26270 | TGA-element | −321 | −315 |
| AtPFK4 | AT5G61580 | CGTCA-motif | −1818 | −1813 |
| AtPFK4 | AT5G61580 | TGACG-motif | −1818 | −1813 |
| AtPFK4 | AT5G61580 | Box 4 | −1546 | −1540 |
| AtPFK4 | AT5G61580 | CGTCA-motif | −1476 | −1471 |
| AtPFK4 | AT5G61580 | TGACG-motif | −1476 | −1471 |
| AtPFK4 | AT5G61580 | LTR | −1320 | −1314 |
| AtPFK4 | AT5G61580 | LTR | −1191 | −1185 |
| AtPFK4 | AT5G61580 | TCA-element | −609 | −600 |
| AtPFK4 | AT5G61580 | MBS | −581 | −575 |
| AtPFK4 | AT5G61580 | CGTCA-motif | −190 | −185 |
| AtPFK4 | AT5G61580 | TGACG-motif | −190 | −185 |
| AtPFK4 | AT5G61580 | TCA-element | −173 | −164 |
| AtPFK5 | AT2G22480 | MBS | −1594 | −1588 |
| AtPFK5 | AT2G22480 | GARE-motif | −1357 | −1350 |
| AtPFK5 | AT2G22480 | LTR | −979 | −973 |
| AtPFK5 | AT2G22480 | ABRE | −921 | −916 |
| AtPFK5 | AT2G22480 | G-box | −921 | −915 |
| AtPFK5 | AT2G22480 | MBS | −657 | −651 |
| AtPFK5 | AT2G22480 | ABRE | −593 | −588 |
| AtPFK5 | AT2G22480 | G-box | −593 | −587 |
| AtPFK5 | AT2G22480 | MBS | −538 | −532 |
| AtPFK5 | AT2G22480 | TGA-element | −94 | −88 |
| AtPFK6 | AT4G32840 | Box 4 | −1399 | −1393 |
| AtPFK6 | AT4G32840 | MBS | −1220 | −1214 |
| AtPFK6 | AT4G32840 | Box 4 | −854 | −848 |
| AtPFK6 | AT4G32840 | LTR | −659 | −653 |
| AtPFK6 | AT4G32840 | Box 4 | −265 | −259 |
| AtPFK6 | AT4G32840 | P-box | −198 | −191 |
| AtPFK6 | AT4G32840 | LTR | −164 | −158 |
| AtPFK6 | AT4G32840 | TGA-element | −123 | −117 |
| AtPFK6 | AT4G32840 | TGA-box | −28 | −20 |
| AtPFK6 | AT4G32840 | CGTCA-motif | −25 | −20 |
| AtPFK6 | AT4G32840 | TGACG-motif | −25 | −20 |
| AtPFK7 | AT5G56630 | GARE-motif | −1788 | −1781 |
| AtPFK7 | AT5G56630 | MRE | −1744 | −1737 |
| AtPFK7 | AT5G56630 | TC-rich repeats | −1500 | −1491 |
| AtPFK7 | AT5G56630 | Box 4 | −1474 | −1468 |
| AtPFK7 | AT5G56630 | Box 4 | −1095 | −1089 |
| AtPFK7 | AT5G56630 | LTR | −955 | −949 |
| AtPFK7 | AT5G56630 | TGA-element | −908 | −902 |
| AtPFK7 | AT5G56630 | TGA-element | −771 | −765 |
| AtPFK7 | AT5G56630 | Box 4 | −427 | −421 |
| AtPFK7 | AT5G56630 | CGTCA-motif | −321 | −316 |
| AtPFK7 | AT5G56630 | TGACG-motif | −321 | −316 |
| AtPFK7 | AT5G56630 | TCA-element | −110 | −101 |
| AtPFPα1 | AT1G20950 | TATC-box | −1487 | −1480 |
| AtPFPα1 | AT1G20950 | G-box | −1454 | −1448 |
| AtPFPα1 | AT1G20950 | MBS | −1430 | −1424 |
| AtPFPα1 | AT1G20950 | GARE-motif | −1283 | −1276 |
| AtPFPα1 | AT1G20950 | LTR | −1195 | −1189 |
| AtPFPα1 | AT1G20950 | MRE | −1026 | −1019 |
| AtPFPα1 | AT1G20950 | G-box | −552 | −546 |
| AtPFPα1 | AT1G20950 | ATC-motif | −438 | −430 |
| AtPFPα1 | AT1G20950 | AuxRE | −371 | −360 |
| AtPFPα1 | AT1G20950 | G-box | −272 | −264 |
| AtPFPα1 | AT1G20950 | G-Box | −270 | −264 |
| AtPFPα1 | AT1G20950 | ABRE | −269 | −264 |
| AtPFPα1 | AT1G20950 | GARE-motif | −201 | −194 |
| AtPFPα2 | AT1G76550 | CGTCA-motif | −1477 | −1472 |
| AtPFPα2 | AT1G76550 | TGACG-motif | −1477 | −1472 |
| AtPFPα2 | AT1G76550 | MBS | −1415 | −1409 |
| AtPFPα2 | AT1G76550 | CGTCA-motif | −1387 | −1382 |
| AtPFPα2 | AT1G76550 | TGACG-motif | −1387 | −1382 |
| AtPFPα2 | AT1G76550 | ABRE | −1198 | −1193 |
| AtPFPα2 | AT1G76550 | P-box | −1066 | −1059 |
| AtPFPα2 | AT1G76550 | MBS | −870 | −864 |
| AtPFPα2 | AT1G76550 | ABRE | −834 | −827 |
| AtPFPα2 | AT1G76550 | GARE-motif | −828 | −821 |
| AtPFPα2 | AT1G76550 | TCA-element | −769 | −760 |
| AtPFPα2 | AT1G76550 | TCA-element | −575 | −566 |
| AtPFPα2 | AT1G76550 | LTR | −253 | −247 |
| AtPFPα2 | AT1G76550 | CGTCA-motif | −80 | −75 |
| AtPFPα2 | AT1G76550 | TGACG-motif | −80 | −75 |
| AtPFPβ1 | AT1G12000 | TC-rich repeats | −1613 | −1604 |
| AtPFPβ1 | AT1G12000 | MRE | −1369 | −1362 |
| AtPFPβ1 | AT1G12000 | TC-rich repeats | −1111 | −1102 |
| AtPFPβ1 | AT1G12000 | TC-rich repeats | −1075 | −1066 |
| AtPFPβ1 | AT1G12000 | Box 4 | −973 | −967 |
| AtPFPβ1 | AT1G12000 | LTR | −925 | −919 |
| AtPFPβ1 | AT1G12000 | Box 4 | −818 | −812 |
| AtPFPβ1 | AT1G12000 | Box 4 | −421 | −415 |
| AtPFPβ1 | AT1G12000 | GARE-motif | −387 | −380 |
| AtPFPβ1 | AT1G12000 | CGTCA-motif | −75 | −70 |
| AtPFPβ1 | AT1G12000 | TGACG-motif | −75 | −70 |
| AtPFPβ2 | AT4G04040 | P-box | −1752 | −1745 |
| AtPFPβ2 | AT4G04040 | CGTCA-motif | −1295 | −1290 |
| AtPFPβ2 | AT4G04040 | TGACG-motif | −1295 | −1290 |
| AtPFPβ2 | AT4G04040 | CGTCA-motif | −1262 | −1257 |
| AtPFPβ2 | AT4G04040 | TGACG-motif | −1262 | −1257 |
| AtPFPβ2 | AT4G04040 | P-box | −1016 | −1009 |
| AtPFPβ2 | AT4G04040 | TGA-element | −599 | −593 |
| AtPFPβ2 | AT4G04040 | TGA-element | −566 | −560 |
| Module | Name | Gene ID | TFs Family | KEGG Pathway | Uniprot Annotation |
|---|---|---|---|---|---|
| Deepyellow | LBD41 | AT3G02550 | LBD | NO | LOB domain-containing protein 41 |
| Green | TRP3 | AT3G54640 | NO | Glycine, serine and threonine metabolism | Indole-3-glycerol-phosphate lyase |
| Green | PP2-A3 | AT2G26820 | IAR | NO | Immune-associated nucleotide-binding protein 1 |
| Black | SAUR10 | AT2G18010 | NO | Plant hormone signal transduction | Protein SMALL AUXIN UP-REGULATED RNA 10 |
| Black | IAA6 | AT1G52830 | Aux/IAA | Plant hormone signal transduction | Auxin-responsive protein IAA6 |
| Turquoise | JAZ1 | AT1G19180 | JAZ | Plant hormone signal transduction | Jasmonate ZIM domain-containing protein 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Gou, J.; Tang, Y.; Wei, Y.; Zhang, R. Genome-Wide Characterization of the Phosphofructokinase Gene Family in Arabidopsis thaliana and Functional Analysis of AtPFK2 in Stress Tolerance. Int. J. Mol. Sci. 2025, 26, 6828. https://doi.org/10.3390/ijms26146828
Liu S, Gou J, Tang Y, Wei Y, Zhang R. Genome-Wide Characterization of the Phosphofructokinase Gene Family in Arabidopsis thaliana and Functional Analysis of AtPFK2 in Stress Tolerance. International Journal of Molecular Sciences. 2025; 26(14):6828. https://doi.org/10.3390/ijms26146828
Chicago/Turabian StyleLiu, Siyu, Jiheng Gou, Yunni Tang, Yunxiao Wei, and Rui Zhang. 2025. "Genome-Wide Characterization of the Phosphofructokinase Gene Family in Arabidopsis thaliana and Functional Analysis of AtPFK2 in Stress Tolerance" International Journal of Molecular Sciences 26, no. 14: 6828. https://doi.org/10.3390/ijms26146828
APA StyleLiu, S., Gou, J., Tang, Y., Wei, Y., & Zhang, R. (2025). Genome-Wide Characterization of the Phosphofructokinase Gene Family in Arabidopsis thaliana and Functional Analysis of AtPFK2 in Stress Tolerance. International Journal of Molecular Sciences, 26(14), 6828. https://doi.org/10.3390/ijms26146828






