DNA-Barcoding for Cultivar Identification and Intraspecific Diversity Analysis of Agricultural Crops
Abstract
1. Introduction
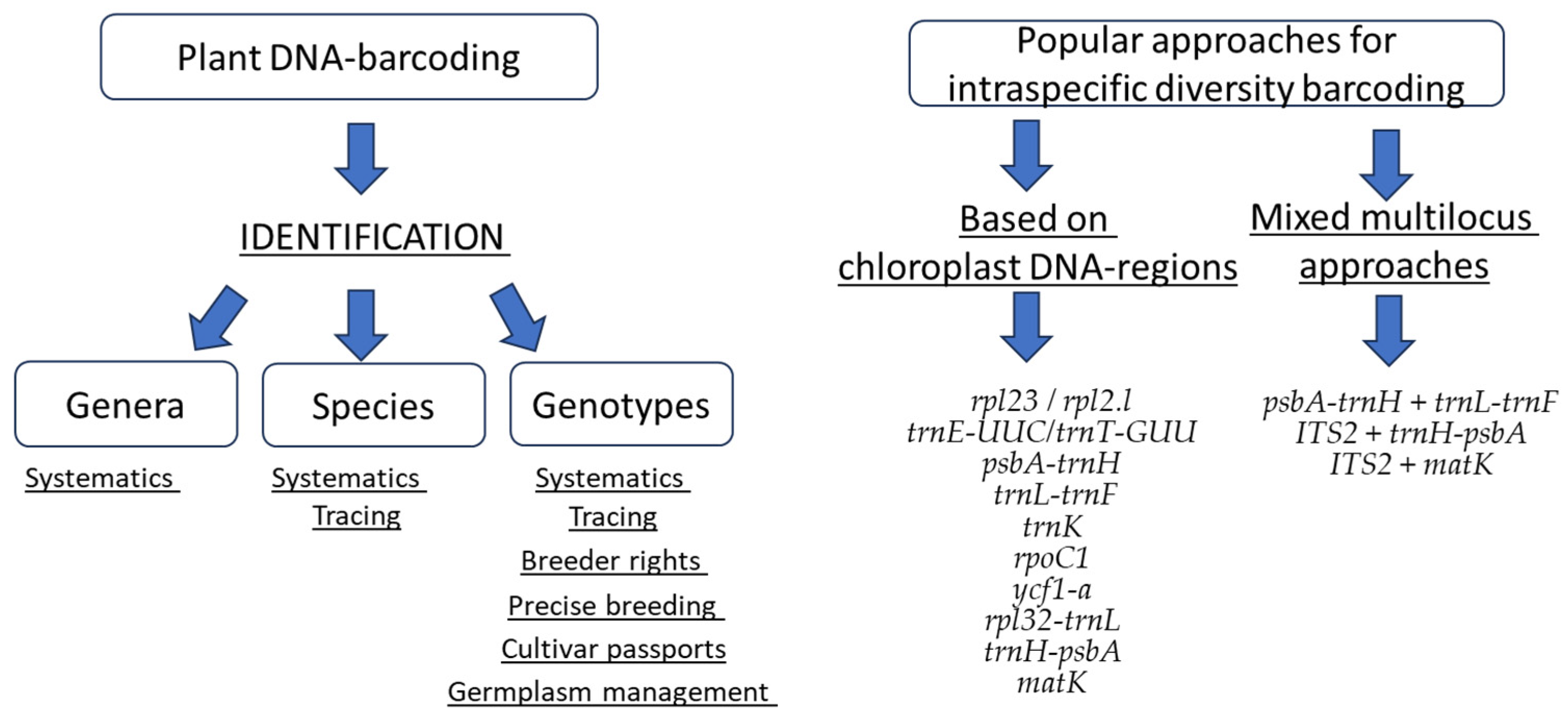
2. Selection of an Appropriate Loci
2.1. Mitochondrial DNA for Cultivars Identification
2.2. Chloroplast DNA for Cultivars Identification
2.3. Nuclear DNA for Cultivars Identification
2.4. Multilocus DNA Barcoding Systems for Cultivars Identification
3. Selection of Primers
4. Strategies of Amplicon Analysis
4.1. Restriction Analysis
4.2. High Resolution Melting for DNA Barcoding of Cultivars
4.3. Sanger Sequencing for Amplicon Analysis
4.4. Next Generation Sequencing (NGS) for Cultivars Identification
4.4.1. Pooled Amplicon Sequencing for Cultivar Identification
4.4.2. Genotyping-by-Sequencing for Cultivars Identification
4.4.3. RAD-Seq Approach for Cultivar Identification
4.4.4. Oxford Nanopore Technologies for Cultivars Identification
4.4.5. Application of Whole-Genome Resequencing as Super-Barcode
5. Bioinformatic Tools for Barcoding Data Analysis
6. Barcoding Databases and Libraries
7. Conclusions
Supplementary Materials
Funding
Data Availability Statement
Conflicts of Interest
References
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; de Waard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Jones, L.; Twyford, A.D.; Ford, C.R.; Rich, T.C.G.; Davies, H.; Forrest, L.L.; Hart, M.L.; McHaffie, H.; Brown, M.R.; Hollingsworth, P.M.; et al. Barcode UK: A complete DNA barcoding resource for the flowering plants and conifers of the United Kingdom. Mol. Ecol. Resour. 2021, 21, 2050–2062. [Google Scholar] [CrossRef]
- Xiang, X.G.; Hu, H.; Wang, W.; Jin, X.H. DNA barcoding of the recently evolved genus Holcoglossum (Orchidaceae: Aeridinae): A test of DNA barcode candidates. Mol. Ecol. Resour. 2011, 11, 1012–1021. [Google Scholar] [CrossRef] [PubMed]
- Abubakar, B.M.; Salleh, F.M.; Omar, M.S.S.; Wagiran, A. DNA Barcoding and Chromatography Fingerprints for the Authentication of Botanicals in Herbal Medicinal Products. Evid.-Based Complement. Altern. Med. 2017, 2017, 1352948. [Google Scholar] [CrossRef] [PubMed]
- Shneer, V.S.; Rodionov, A.V. DNA barcodes of plants. Adv. Mod. Biol. 2018, 138, 531–537. [Google Scholar] [CrossRef]
- Shekhovtsov, S.V.; Shekhovtsova, I.N.; Peltek, S.E. DNA-barcoding: Methods and approaches. Adv. Mod. Biol. 2019, 139, 211–220. [Google Scholar] [CrossRef]
- Savina, N.V.; Kubrak, S.V.; Milko, L.V.; Kilchevsky, A.V.; Nikitina, E.V.; Tozhibaev, K.S. DNA-barcoding as a tool for environmental monitoring and assessment of species diversity of rare plant species. Mol. Appl. Genet. 2020, 29, 25–36. [Google Scholar]
- Shadrin, D.M. DNA-barcoding: Areas of application. Genetics 2021, 57, 478–488. [Google Scholar] [CrossRef]
- Musou-Yahada, A.; Honjoh, K.I.; Yamamoto, K.; Miyamoto, T.; Ohta, H. Utilization of single nucleotide polymorphism-based allele-specific PCR to identify Shiikuwasha (Citrus depressa Hayata) and Calamondin (Citrus madurensis Lour.) in processed juice. Food Sci. Technol. Res. 2019, 25, 19–27. [Google Scholar] [CrossRef]
- Nazar, N.; Saxena, A.; Sebastian, A.; Slater, A.; Sundaresan, V.; Sgamma, T. Integrating DNA Barcoding Within an Orthogonal Approach for Herbal Product Authentication: A Narrative Review. Phytochem. Anal. 2024, 36, 7–29. [Google Scholar] [CrossRef]
- DeSalle, R.; Goldstein, P. Review and Interpretation of Trends in DNA Barcoding. Front. Ecol. Evol. 2019, 7, 302. [Google Scholar] [CrossRef]
- Kress, W.J.; Wurdack, K.J.; Zimmer, E.A.; Weigt, L.A.; Janzen, D.H. Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. USA 2005, 102, 8369–8374. [Google Scholar] [CrossRef] [PubMed]
- Hamza, H.; Villa, S.; Torre, S.; Marchesini, A.; Benabderrahim, M.A.; Rejili, M.; Sebastiani, F. Whole mitochondrial and chloroplast genome sequencing of Tunisian date palm cultivars: Diversity and evolutionary relationships. BMC Genom. 2023, 24, 772. [Google Scholar] [CrossRef] [PubMed]
- Park, H.S.; Jayakodi, M.; Lee, S.H.; Jeon, J.H.; Lee, H.O.; Park, J.Y.; Moon, B.C.; Kim, C.K.; Wing, R.A.; Newmaster, S.G.; et al. Mitochondrial plastid DNA can cause DNA barcoding paradox in plants. Sci. Rep. 2020, 10, 6112. [Google Scholar] [CrossRef] [PubMed]
- Ying, Z.; Awais, M.; Akter, R.; Xu, F.; Baik, S.; Jung, D.; Yang, D.C.; Kwak, G.-Y.; Wenying, Y. Discrimination of Panax ginseng from counterfeits using single nucleotide polymorphism: A focused review. Front. Plant Sci. 2022, 13, 903306. [Google Scholar] [CrossRef]
- Xiao, S.; Xu, P.; Deng, Y.; Dai, X.; Zhao, L.; Heider, B.; Zhang, A.; Zhou, Z.; Cao, Q. Comparative analysis of chloroplast genomes of cultivars and wild species of sweetpotato (Ipomoea batatas [L.] Lam). BMC Genom. 2021, 22, 262. [Google Scholar] [CrossRef]
- Ekram, A.E.; Hamilton, R.; Campbell, M.; Plett, C.; Kose, S.H.; Russell, J.M.; Stevenson, J.; Coolen, M.J.L. A 1 Ma Record of Climate-induced Vegetation Changes Using Sed ADNA and Pollen in A Biodiversity Hotspot: Lake Towuti, Sulawesi, Indonesia. In Proceedings of the EGU General Assembly 2021, Online, 19–30 April 2021; p. EGU21-4003. [Google Scholar] [CrossRef]
- Bibi, A.; Marountas, J.; Kouklinos, Y.; Kafetzopoulos, D.; Lefort, F.; Roubelakis-Angelakis, K. Revitalization of The Greek Vitis Database: A Multimedia Web-backed Genetic Database for Germplasm Management of Vitis Resources in Greece. J. Wine Res. 2021, 32, 1–10. [Google Scholar] [CrossRef]
- Dong, W.; Cheng, T.; Li, C.; Xu, C.; Long, P.; Chen, C.; Zhou, S. Discriminating plants using the DNA barcode rbcLb: An appraisal based on a large data set. Mol. Ecol. Resour. 2013, 14, 336–343. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef]
- CBOL Plant Working Group. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef]
- Haider, N.; Wilkinson, M.J. A Set of Plastid DNA-Specific Universal Primers for Flowering Plants. Russ. J. Genet. 2011, 47, 1066–1077. [Google Scholar] [CrossRef]
- Corvalán, L.C.J.; de Melo-Ximenes, A.A.; Carvalho, L.R.; e Silva-Neto, C.M.; Diniz-Filho, J.A.F.; de C Telles, M.P.; Nunes, R. Is There a Key Primer for Amplification of Core Land Plant DNA Barcode Regions (rbcL and matK)? Ecol. Evol. 2025, 15, e70961. [Google Scholar] [CrossRef] [PubMed]
- China Plant BOL Group; Li, D.Z.; Gao, L.M.; Li, H.T.; Wang, H.; Ge, X.J.; Liu, J.Q.; Chen, Z.D.; Zhou, S.L.; Chen, S.L.; et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Xu, C.; Li, C.; Sun, J.; Zuo, Y.; Shi, S.; Cheng, T.; Guo, J.; Zhou, S. ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 2015, 5, 8348. [Google Scholar] [CrossRef] [PubMed]
- Samarina, L.S.; Koninskaya, N.G.; Shkhalakhova, R.M.; Simonyan, T.A.; Tsaturyan, G.A.; Shurkina, E.S.; Kulyan, R.V.; Omarova, Z.M.; Tutberidze, T.V.; Ryndin, A.V.; et al. The potential of universal primers for barcoding of subtropical crops: Actinidia, Feijoa, Citrus, and Tea. Int. J. Mol. Sci. 2025; submitted. [Google Scholar]
- Dong, W.; Liu, J.; Yu, J.; Wang, L.; Zhou, S. Highly Variable Chloroplast Markers for Evaluating Plant Phylogeny at Low Taxonomic Levels and for DNA Barcoding. PLoS ONE 2012, 7, e35071. [Google Scholar] [CrossRef]
- Galimberti, A.; Labra, M.; Sandionigi, A.; Bruno, A.; Mezzasalma, V.; De Mattia, F. DNA Barcoding for Minor Crops and Food Traceability. Adv. Agric. 2014, 2014, 831875. [Google Scholar] [CrossRef]
- Sayed, H.A.; Mostafa, S.; Haggag, I.M.; Hassan, N.A. DNA Barcoding of Prunus Species Collection Conserved in the National Gene Bank of Egypt. Mol. Biotechnol. 2023, 65, 410–418. [Google Scholar] [CrossRef]
- Pang, X.; Liu, C.; Shi, L.; Liu, R.; Liang, D.; Li, H.; Cherny, S.S.; Chen, S. Utility of the trnH-psbA Intergenic Spacer Region and Its Combinations as Plant DNA Barcodes: A Meta-Analysis. PLoS ONE 2012, 7, e48833. [Google Scholar] [CrossRef]
- Feng, S.; Jiao, K.; Zhu, Y.; Wang, H.; Jiang, M.; Wang, H. Molecular identification of species of Physalis (Solanaceae) using a candidate DNA barcode: The chloroplast psbA-trnH intergenic region. Genome 2018, 61, 15–20. [Google Scholar] [CrossRef]
- Awad, M.; Fahmy, R.M.; Mosa, K.A.; Helmy, M.; El-Feky, F.A. Identification of effective DNA barcodes for Triticum plants through chloroplast genome-wide analysis. Comput. Biol. Chem. 2017, 71, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Jena, B.; Pati, K.; Nedunchezhiyan, M.; Giri, A.K.; Acharya, V. Application of DNA Barcode for Cultivar Identification in Tuber Crops. Biotica Res. Today 2022, 4, 606–608. [Google Scholar]
- Badr, A.; Elsherif, N.; Aly, S.; Ibrahim, S.; Ibrahim, M. Genetic Diversity among Selected Medicago sativa Cultivars Using Inter-Retrotransposon-Amplified Polymorphism, Chloroplast DNA Barcodes and Morpho-Agronomic Trait Analyses. Plants 2020, 9, 995. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhu, C.; Wang, S.; Wang, F.; Sun, Z. Identification of three cultivated varieties of Scutellaria baicalensis using the complete chloroplast genome as a super-barcode. Sci. Rep. 2023, 13, 5602. [Google Scholar] [CrossRef]
- Ma, Y.-P.; Zhao, L.; Zhang, W.-J.; Zhang, Y.-H.; Xing, X.; Duan, X.-X.; Hu, J.; Harris, A.J.; Liu, P.-L.; Dai, S.-L.; et al. Origins of cultivars of Chrysanthemum-Evidence from the chloroplast genome and nuclear LFY gene. J. Syst. Evol. 2020, 58, 925–944. [Google Scholar] [CrossRef]
- Al-Andal, A. Unraveling the genetic diversity and evolutionary lineages of Catharanthus roseus cultivars through plastome analysis and DNA barcoding. Crop Pasture Sci. 2025, 76, CP24363. [Google Scholar] [CrossRef]
- Attia, O.A.; Ismail, A.I.; El Dessoky, S.D.; Bandar, S.A. Using of DNA-Barcoding, SCoT and SDS-PAGE Protein to Assess Soma-Clonal Variation in Micro-Propagated Fig (Ficus carica L.) Plant. Pak. J. Biol. Sci. 2022, 25, 415–425. [Google Scholar] [CrossRef]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation Of The ITS2 Region As A Novel DNA Barcode For Identifying Medicinal Plant Species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef]
- Li, R.; Dao, Z. Identification of Meconopsis Species By A DNA Barcode Sequence: The Nuclear Internal Transcribed Spacer (ITS) Region of Ribosomal Deoxyribonucleic Acid (DNA). Afr. J. Biotechnol. 2011, 10, 1802–1807. [Google Scholar] [CrossRef]
- Dissanayake, U.H.K.; Senevirathna, R.W.K.M.; Ranaweera, L.T.; Wijesundara, W.W.M.U.K.; Jayarathne, H.S.M.; Weebadde, C.K.; Sooriyapathirana, S.D.S.S. Characterization of Cassava (Manihot Esculenta Crantz) Cultivars in Sri Lanka Using Morphological, Molecular and Organoleptic Parameters. Trop. Agric. Res. 2019, 30, 51–70. [Google Scholar] [CrossRef]
- Tatlises, M.B.; Hasançebi, S. Identification of Lens Cultivars in Market By Molecular Tools: DNA Barcoding and SSRs. Trak. Univ. J. Nat. Sci. 2023, 24, 91–100. [Google Scholar] [CrossRef]
- Dhivya, S.; Ashutosh, S.; Gowtham, I.; Baskar, V.; Harini, A.B.; Mukunthakumar, S.; Sathishkumar, R. Molecular Identification and Evolutionary Relationships Between The Subspecies of Musa By DNA Barcodes. BMC Genom. 2020, 21, 659. [Google Scholar] [CrossRef]
- Hidayat, T.; Abdullah, F.I.; Kuppusamy, C.; Samad, A.A.; Wagiran, A. Molecular Identification of Malaysian Pineapple Cultivar Based on Internal Transcribed Spacer Region. APCBEE Procedia 2012, 4, 146–151. [Google Scholar] [CrossRef]
- Hürkan, K. Employing Barcode High-Resolution Melting Technique for Authentication of Apricot Cultivars. J. Agric. Sci.-Tarim Bilim. Derg. 2022, 28, 251–258. [Google Scholar] [CrossRef]
- Meghana, B.N.; Reshma, S.V. DNA Barcoding of Geographical Indication Tagged Byadagi Chilli and Its Cultivars Using ITS2, MatK and RbcL Coding Sequences. Mol. Biol. Rep. 2025, 52, 286. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.P.; Do, K.T. Identification of Mango (Mangifera indica L.) cultivars in the Mekong Delta using ISSR markers and DNA barcodes. J. Appl. Biol. Biotechnol. 2025, 13, 68–75. [Google Scholar] [CrossRef]
- Alzahrani, O.F.; Dguimi, H.M.; Alshaharni, M.O.; Albalawi, D.; Zaoui, S. Employing plant DNA barcodes for pomegranate species identification in Al-Baha Region, Saudi Arabia. J. Umm Al-Qura Univ. Appl. Sci. 2024, 10, 136–144. [Google Scholar] [CrossRef]
- Castro, C.; Hernandez, A.; Alvarado, L.; Flores, D. DNA Barcodes in Fig Cultivars (Ficus carica L.) Using ITS Regions of Ribosomal DNA, the psbA-trnH Spacer and the matK Coding Sequence. Am. J. Plant Sci. 2015, 6, 95–102. [Google Scholar] [CrossRef]
- Kjærandsen, J. Current State of DNA Barcoding of Sciaroidea (Diptera)—Highlighting the Need to Build the Reference Library. Insects 2022, 13, 147. [Google Scholar] [CrossRef]
- Ho, V.T.; Tu, N.T.; Nguyen, T.A. DNA fingerprinting and molecular characterization of mango (Mangifera spp.) cultivars in Vietnam using ITS DNA barcode. Bulg. J. Agric. Sci. 2021, 27, 128. [Google Scholar]
- Gogoi, B.; Wann, S.B.; Saikia, S.P. DNA barcodes for delineating Clerodendrum species of North East India. Sci. Rep. 2020, 10, 13490. [Google Scholar] [CrossRef]
- Wang, J.; Yan, Z.; Zhong, P.; Shen, Z.; Yang, G.; Ma, L. Screening of universal DNA barcodes for identifying grass species of Gramineae. Front. Plant Sci. 2022, 13, 998863. [Google Scholar] [CrossRef] [PubMed]
- Raskoti, B.B.; Ale, R. DNA barcoding of medicinal orchids in Asia. Sci. Rep. 2021, 11, 23651. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Choudhary, K.; Shelja, A.; Singh, H.; Kumar, A.; Bagga, P. Molecular Approaches for Authentication and Identification of Medicinal Plants. Plant Mol. Biol. Rep. 2025. [Google Scholar] [CrossRef]
- Said, E.M.; Hassan, M.E. DNA barcodes in Egyptian olive cultivars (Olea europaea L.) using the rbcL and matK coding sequences. J. Crop Sci. Biotechnol. 2023, 26, 447–456. [Google Scholar] [CrossRef]
- Boudadi, I.; Lachheb, M.; Merzougui, S.E.; Lachguer, K.; Serghini, M.A. Exploring genetic variation in saffron (Crocus sativus L.) accessions through two-locus DNA barcoding. Ind. Crops Prod. 2024, 220, 119177. [Google Scholar] [CrossRef]
- Abouseada, H.H.; Mohamed, A.-S.H.; Teleb, S.S.; Bard, A.; Tantawy, M.E.; Ibrahim, S.D.; Ellmouni, F.Y.; Ibrahim, M. Genetic diversity analysis in wheat cultivars using SCoT and ISSR markers, chloroplast DNA barcoding and grain SEM. BMC Plant Biol. 2023, 23, 193. [Google Scholar] [CrossRef]
- Zhang, M.; Zhai, X.; He, L.; Wang, Z.; Cao, H.; Wang, P.; Ren, W.; Ma, W. Morphological description and DNA barcoding research of nine Syringa species. Front. Genet. 2025, 16, 1544062. [Google Scholar] [CrossRef]
- Chen, Y.; Zhu, X.; Loukopoulos, P.; Weston, L.; Albrecht, D.E.; Quinn, J.C. Genotypic identification of Panicum spp. in New South Wales, Australia using DNA barcoding. Sci. Rep. 2021, 11, 16055. [Google Scholar] [CrossRef]
- Aljuaid, B.S.; Ismail, I.A.; Attia, A.O.; Dessoky, D.S. Genetic Stability of in vitro Propagated Grapevine (Vitis vinifera L.) cv. Al-Bayadi. J. Agric. Crops 2022, 8, 12–19. [Google Scholar] [CrossRef]
- Letsiou, S.; Madesis, P.; Vasdekis, E.; Montemurro, C.; Grigoriou, M.E.; Skavdis, G.; Moussis, V.; Koutelidakis, A.E.; Tzakos, A.G. DNA Barcoding as a Plant Identification Method. Appl. Sci. 2024, 14, 1415. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, J.; Zhang, Y.; Su, H.; Qiu, X.; Gong, L.; Huang, J.; Bai, J.; Huang, Z.; Xu, W. Chemical and genetic discrimination of commercial Guangchenpi (Citrus reticulata ‘Chachi’) by using UPLC-QTOF-MS/MS based metabolomics and DNA barcoding approaches. RSC Adv. 2019, 9, 23373–23381. [Google Scholar] [CrossRef]
- Fomina, N.A.; Antonova, O.Y.; Chukhina, I.G.; Gimaeva, E.A.; Stashevski, Z.; Gavrilenko, T.A. Nomenclatural Standards and Genetic Passports of Potato Cultivars Bred By The Tatar Research Institute of Agriculture «Kazan Scientific Center of The Russian Academy of Sciences». Plant Biotechnol. Breed. 2020, 3, 55–67. [Google Scholar] [CrossRef]
- Kress, W.J. Plant DNA barcodes: Applications today and in the future. J. Syst. Evol. 2017, 55, 291–307. [Google Scholar] [CrossRef]
- Barcaccia, G.; Palumbo, F.; Scariolo, F.; Vannozzi, A.; Borin, M.; Bona, S. Potentials and Challenges of Genomics for Breeding Cannabis Cultivars. Front. Plant Sci. 2020, 11, 573299. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Xu, C.; Lei, L.; Li, C.; Zhang, Y.; Zhou, S. Barcoding the kingdom Plantae: New PCR primers for ITS regions of plants with improved universality and specificity. Mol. Ecol. Resour. 2016, 16, 138–149. [Google Scholar] [CrossRef]
- de Vere, N.; Rich, T.C.; Ford, C.R.; Trinder, S.A.; Long, C.; Moore, C.W.; Satterthwaite, D.; Davies, H.; Allainguillaume, J.; Ronca, S.; et al. DNA barcoding the native flowering plants and conifers of Wales. PLoS ONE 2012, 7, e37945. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L.; Jones, F.A.; Swenson, N.G.; Perez, R.; Sanjur, O.; Bermingham, E. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proc. Natl. Acad. Sci. USA 2009, 106, 18621–18626. [Google Scholar] [CrossRef]
- Fazekas, A.J.; Burgess, K.S.; Kesanakurti, P.R.; Graham, S.W.; Newmaster, S.G.; Husband, B.C.; Percy, D.M.; Hajibabaei, M.; Barrett, S.C.H. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE 2008, 3, e2802. [Google Scholar] [CrossRef]
- Fay, M.F.; Swensen, S.M.; Chase, M.W. Taxonomic affinities of Medusagyne oppositifolia (Medusagynaceae). Kew Bull. 1997, 52, 111–120. [Google Scholar] [CrossRef]
- Ford, C.S.; Ayres, K.L.; Toomey, N.; Haider, N.; Van Alphen Stahl, J.; Kelly, L.J.; Wikström, N.; Hollingsworth, P.M.; Duff, R.J.; Hoot, S.B.; et al. Selection of candidate coding DNA barcoding regions for use on land plants. Bot. J. Linn. Soc. 2009, 159, 1–11. [Google Scholar] [CrossRef]
- de Vere, N.; Rich, T.C.; Trinder, S.A.; Long, C. DNA barcoding for plants. In Plant Genotyping: Methods and Protocols; Batley, J., Ed.; Humana Press: New York, NY, USA, 2015; pp. 101–118. [Google Scholar] [CrossRef]
- Cuénoud, P.; Savolainen, V.; Chatrou, L.W.; Powell, M.; Grayer, R.J.; Chase, M.W. Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am. J. Bot. 2002, 89, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hu, Y.; He, M.; Zhang, B.; Wu, W.; Cai, P.; Huo, D.; Hong, Y. Comparative chloroplast genomes: Insights into the evolution of the chloroplast genome of Camellia sinensis and the phylogeny of Camellia. BMC Genom. 2021, 22, 138. [Google Scholar] [CrossRef] [PubMed]
- Wolff, K.; Zietkiewicz, E.; Hofstra, H. Identification of chrysanthemum cultivars and stability of DNA fingerprint patterns. Theor. Appl. Genet. 1995, 91, 439–447. [Google Scholar] [CrossRef]
- Caetano-Anollés, G.; Bassam, B.J.; Gresshoff, P.M. Enhanced detection of polymorphic DNA by multiple arbitrary amplicon profiling of endonuclease-digested DNA: Identification of markers tightly linked to the supernodulation locus in soybean. Molec. Gen. Genet. 1993, 241, 57–64. [Google Scholar] [CrossRef]
- Herten, K.; Hestand, M.S.; Vermeesch, J.R.; Van Houdt, J.K.J. GBSX: A toolkit for experimental design and demultiplexing genotyping by sequencing experiments. BMC Bioinform. 2015, 16, 73. [Google Scholar] [CrossRef]
- Muleo, R.; Colao, M.C.; Miano, D.; Cirilli, M.; Intrieri, M.C.; Baldoni, L.; Rugini, E. Mutation scanning and genotyping by high-resolution DNA melting analysis in olive germplasm. Genome 2009, 52, 252–260. [Google Scholar] [CrossRef][Green Version]
- Jaakola, L.; Suokas, M.; Häggman, H. Novel approaches based on DNA barcoding and high-resolution melting of amplicons for authenticity analyses of berry species. Food Chem. 2010, 123, 494–500. [Google Scholar] [CrossRef]
- Bosmali, I.; Ganopoulos, I.; Madesis, P.; Tsaftaris, A. Microsatellite and DNA-barcode regions typing combined with high resolution melting (HRM) analysis for food forensic uses: A case study on lentils (Lens culinaris). Food Res. Int. 2012, 46, 141–147. [Google Scholar] [CrossRef]
- Madesis, P.; Ganopoulos, I.; Anagnostis, A.; Tsaftaris, A. The application of Bar-HRM (Barcode DNA-High Resolution Melting) analysis for authenticity testing and quantitative detection of bean crops (Leguminosae) without prior DNA purification. Food Control 2012, 25, 576–582. [Google Scholar] [CrossRef]
- Zeng, H.; Huang, B.; Xu, L.; Wu, Y. Banana Classification Using Sanger Sequencing of the Ribosomal DNA Internal Transcribed Spacer (ITS) Region. Plants 2024, 13, 2173. [Google Scholar] [CrossRef]
- Fernandes, T.J.R.; Amaral, J.S.; Mafra, I. DNA Barcode Markers Applied to Seafood Authentication: An Updated Review. Crit. Rev. Food Sci. Nutr. 2020, 61, 3904–3935. [Google Scholar] [CrossRef] [PubMed]
- Richardson, M.A.; Nenadic, N.; Wingfield, M.; McDougall, C. The Development of Multiplex PCR As-says for The Rapid Identification of Multiple Saccostrea Species, and Their Practical Applications in Restoration and Aquaculture. BMC Ecol. Evol. 2024, 24, 67. [Google Scholar] [CrossRef]
- Pandit, R.; Travadi, T.; Sharma, S.; Joshi, C.; Joshi, M. DNA Meta-barcoding Using rbcL Based Mini-barcode Revealed Presence of Unspecified Plant Species in Ayurvedic Polyherbal Formulations. Phytochem. Anal. 2021, 32, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Shokralla, S.; Gibson, J.F.; Nikbakht, H.; Janzen, D.H.; Hallwachs, W.; Hajibabaei, M. Next-generation DNA barcoding: Using next-generation sequencing to enhance and accelerate DNA barcode capture from single specimens. Mol. Ecol. Resour. 2014, 14, 892–901. [Google Scholar] [CrossRef]
- Antil, S.; Abraham, J.S.; Sripoorna, S.; Maurya, S.; Dagar, J.; Makhija, S.; Bhagat, P.; Gupta, R.; Sood, U.; Lal, R.; et al. DNA Barcoding, An Effective Tool for Species Identification: A Review. Mol. Biol. Rep. 2023, 50, 761–775. [Google Scholar] [CrossRef]
- Jia, Y.; Zhao, S.; Guo, W.; Peng, L.; Zhao, F.; Wang, L.; Fan, G.; Zhu, Y.; Xu, D.; Liu, G.; et al. Sequencing Introduced False Positive Rare Taxa Lead to Biased Microbial Community Diversity, Assembly, and Interaction Interpretation in Amplicon Studies. Environ. Microbiome 2022, 17, 43. [Google Scholar] [CrossRef]
- Urra, C.; Sanhueza, D.; Pavez, C.; Tapia, P.; Núñez-Lillo, G.; Minio, A.; Miossec, M.; Blanco-Herrera, F.; Gainza, F.; Castro, A.; et al. Identification of Grapevine Clones Via High-throughput Amplicon Sequencing: A Proof-of-concept Study. G3 Genes Genomes Genet. 2023, 13, jkad145. [Google Scholar] [CrossRef]
- Hawliczek, A.; Bolibok, L.; Tofil, K.; Borzęcka, E.; Jankowicz-Cieślak, J.; Gawroński, P.; Kral, A.; Till, B.J.; Bolibok-Brągoszewska, H. Deep sampling and pooled amplicon sequencing reveals hidden genic variation in heterogeneous rye accessions. BMC Genom. 2020, 21, 845. [Google Scholar] [CrossRef]
- Samarina, L.; Fedorina, J.; Kuzmina, D.; Malyukova, L.; Manakhova, K.; Kovalenko, T.; Matskiv, A.; Xia, E.; Tong, W.; Zhang, Z.; et al. Analysis of Functional Single-Nucleotide Polymorphisms (SNPs) and Leaf Quality in Tea Collection under Nitrogen-Deficient Conditions. Int. J. Mol. Sci. 2023, 24, 14538. [Google Scholar] [CrossRef]
- Whitford, W.; Hawkins, V.; Moodley, K.; Grant, M.J.; Lehnert, K.; Snell, R.G.; Jacobsen, J.C. Optimised multiplex amplicon sequencing for mutation identification using the MinION nanopore sequencer. bioRxiv 2021. [Google Scholar] [CrossRef]
- Whitford, W.; Hawkins, V.; Moodley, K.S.; Grant, M.J.; Lehnert, K.; Snell, R.G.; Jacobsen, J.C. Proof of concept for multiplex amplicon sequencing for mutation identification using the MinION nanopore sequencer. Sci. Rep. 2022, 12, 8572. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, A.; Steindl, S.; Kirchner, S.; Schwahofer, P.; Haring, E.; Szucsich, N.; Kruckenhauser, L.; Kapun, M. AmpliPiper: A versatile amplicon-seq analysis tool for multilocus DNA barcoding. bioRxiv 2024. [Google Scholar] [CrossRef]
- Uitdewilligen, J.G.; Wolters, A.M.; D’hoop, B.B.; Borm, T.J.; Visser, R.G.; van Eck, H.J. Correction: A next-generation sequencing method for genotyping-by-sequencing of highly heterozygous autotetraploid potato. PLoS ONE 2015, 10, e0141940. [Google Scholar] [CrossRef] [PubMed]
- Niimi, E.; Fujii, H.; Ohta, S.; Iwakura, T.; Endo, T.; Shimada, T. Development of Acid Citrus Cultivar Identification System By SNP Markers. Hortic. Res. 2022, 21, 111–122. [Google Scholar] [CrossRef]
- Park, J.Y.; Jang, Y.J.; Jung, J.K.; Shim, E.J.; Sim, S.C.; Chung, S.M.; Lee, G.P. Development of SNP Markers for The Identification of Commercial Korean Watermelon Cultivars Using Fluidigm Genotyping Analysis. Korean J. Hortic. Sci. Technol. 2022, 40, 75–84. [Google Scholar] [CrossRef]
- Meng, J.; Zhang, Y.; Wei, Y.; Li, R.; Li, Z.; Zhong, C. Identification of Commercial Cultivars in the Tabebuia Alliance Using Genotyping-by-Sequencing. Forests 2023, 14, 271. [Google Scholar] [CrossRef]
- Scariolo, F.; Palumbo, F.; Vannozzi, A.; Sacilotto, G.B.; Gazzola, M.; Barcaccia, G. Genotyping Analysis by RAD-Seq Reads Is Useful to Assess the Genetic Identity and Relationships of Breeding Lines in Lavender Species Aimed at Managing Plant Variety Protection. Genes 2021, 12, 1656. [Google Scholar] [CrossRef]
- Yu, Q.; Ling, Y.; Xiong, Y.; Zhao, W.; Xiong, Y.; Dong, Z.; Yang, J.; Zhao, J.; Zhang, X.; Ma, X. RAD-seq as an effective strategy for heterogenous variety identification in plants–a case study in Italian ryegrass (Lolium multiflorum). BMC Plant Biol. 2022, 22, 231. [Google Scholar] [CrossRef]
- Shen, Y.; Yao, G.; Li, Y.; Tian, X.; Li, S.; Wang, N.; Zhang, C.; Wang, F.; Ma, Y. RAD-seq data reveals robust phylogeny and morphological evolutionary history of Rhododendron. Hortic. Plant J. 2024, 10, 866–878. [Google Scholar] [CrossRef]
- Li, H.; Ikram, M.; Xia, Y.; Li, R.; Yuan, Q.; Zhao, W.; Siddique, K.H.M.; Guo, P. Genome-wide identification and development of InDel markers in tobacco (Nicotiana tabacum L.) using RAD-seq. Physiol. Mol. Biol. Plants 2022, 28, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Casanova, A.; Maroso, F.; Blanco, A.; Hermida, M.; Ríos, N.; García, G.; Manuzzi, A.; Zane, L.; Verissimo, A.; García-Marín, J.L.; et al. Low impact of different SNP panels from two building-loci pipelines on RAD-Seq population genomic metrics: Case study on five diverse aquatic species. BMC Genom. 2021, 22, 150. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Guo, P.; Xu, Q.; Du, W. Cotton Seed Cultivar Identification Based on the Fusion of Spectral and Textural Features. PLoS ONE 2024, 19, e0303219. [Google Scholar] [CrossRef] [PubMed]
- Blazakis, K.N.; Stupichev, D.; Kosma, M.; El Chami, M.A.H.; Apodiakou, A.; Kostelenos, G.; Kalaitzis, P. Discrimination of 14 olive cultivars using morphological analysis and machine learning algorithms. Front. Plant Sci. 2024, 15, 1441737. [Google Scholar] [CrossRef]
- Wahyuni, D.; Dwiyanti, F.G.; Pratama, R.; Majiidu, M.; Rachmat, H.H.; Siregar, I.Z. Chloroplast Genome Draft of Dryobalanops aromatica Generated Using Oxford Nanopore Technology and Its Potential Application for Phylogenetic Study. Forests 2021, 12, 1515. [Google Scholar] [CrossRef]
- Aury, J.M.; Engelen, S.; Istace, B.; Monat, C.; Lasserre-Zuber, P.; Belser, C.; Cruaud, C.; Rimbert, H.; Leroy, P.; Arribatet, S.; et al. Long-read and chromosome-scale assembly of the hexaploid wheat genome achieves high resolution for research and breeding. GigaScience 2021, 11, giac034. [Google Scholar] [CrossRef]
- Song, B.; Ning, W.; Wei, D.; Jiang, M.; Zhu, K.; Wang, X.; Edwards, D.; Odeny, D.A.; Cheng, S. Plant genome resequencing and population genomics: Current status and future prospects. Mol. Plant 2023, 16, 1252–1268. [Google Scholar] [CrossRef]
- Wee, Y.; Bhyan, S.B.; Liu, Y.; Lu, J.; Li, X.; Zhao, M. The bioinformatics tools for the genome assembly and analysis based on third-generation sequencing. Brief. Funct. Genomics 2019, 18, 1–12. [Google Scholar] [CrossRef]
- Duan, Y.; Wang, Y.; Ding, W.; Wang, C.; Meng, L.; Meng, J.; Chen, N.; Liu, Y.; Xing, S. Comparative and phylogenetic analysis of the chloroplast genomes of four commonly used medicinal cultivars of Chrysanthemums morifolium. BMC Plant Biol. 2024, 24, 992. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, H.; Xu, W.; Wan, X.; Zhu, K. Comparative analysis of chloroplast genome of Lonicera japonica cv. Damaohua. Open Life Sci. 2024, 19, 20220984. [Google Scholar] [CrossRef]
- Lee, D.J.; Kim, C.K.; Lee, T.H.; Lee, S.J.; Moon, D.G.; Kwon, Y.H.; Cho, J.Y. The complete chloroplast genome sequence of economical standard tea plant, Camellia sinensis L. cultivar Sangmok, in Korea. Mitochondrial DNA B Resour. 2020, 5, 2835–2836. [Google Scholar] [CrossRef]
- Vieira, M.B.; Faustino, M.V.; Lourenço, T.F.; Oliveira, M.M. DNA-Based Tools to Certify Authenticity of Rice Varieties—An Overview. Foods 2022, 11, 258. [Google Scholar] [CrossRef]
- Wang, X.; Gussarova, G.; Ruhsam, M.; de Vere, N.; Metherell, C.; Hollingsworth, P.M.; Twyford, A.D. DNA barcoding British Euphrasia reveals deeply divergent polyploids but lack of species-level resolution. AoB PLANTS 2018, 10, ply026. [Google Scholar] [CrossRef]
- Steinke, D.; Vences, M.; Salzburger, W.; Meyer, A. TaxI: A software tool for DNA barcoding using distance methods. Philos. Trans. R. Soc. B 2005, 360, 1975–1980. [Google Scholar] [CrossRef] [PubMed]
- Hakimzadeh, A.; Asbun, A.A.; Albanese, D.; Bernard, M.; Buchner, D.; Callahan, B.; Caporaso, J.G.; Curd, E.; Djemiel, C.; Durling, M.B.; et al. A pile of pipelines: An overview of the bioinformatics software for metabarcoding data analyses. Mol. Ecol. Resour. 2024, 24, e13847. [Google Scholar] [CrossRef] [PubMed]
- Zinger, L.; Lionnet, C.; Benoiston, A.-S.; Donald, J.; Mercier, C.; Boyer, F. metabaR: An R package for the evaluation and improvement of DNA metabarcoding data quality. Methods Ecol. Evol. 2021, 12, 586–592. [Google Scholar] [CrossRef]
- Stucky, B.J. SeqTrace: A graphical tool for rapidly processing DNA sequencing chromatograms. J. Biomol. Tech. 2012, 23, 90–93. [Google Scholar] [CrossRef]
- Oliveira, R.R.M.; Nunes, G.L.; de Lima, T.G.L.; Oliveira, G.; Alves, R. PIPEBAR and OverlapPER: Tools for a fast and accurate DNA barcoding analysis and paired-end assembly. BMC Bioinform. 2018, 19, 297. [Google Scholar] [CrossRef]
- Williams, J.; Nash, B.; Ghiban, C.; Khalfan, M.; Hilgert, U.; Lauter, S.; Yang, C.; Micklos, D.A. Analysis of DNA Barcodes Using DNA Subway. In DNA Barcoding. Methods in Molecular Biology; DeSalle, R., Ed.; Humana Press: New York, NY, USA, 2024; Volume 2744, pp. 551–560. [Google Scholar] [CrossRef]
- Han, R.; Qi, J.; Xue, Y.; Sun, X.; Zhang, F.; Gao, X.; Li, G. HycDemux: A hybrid unsupervised approach for accurate barcoded sample demultiplexing in nanopore sequencing. Genome Biol. 2023, 24, 222. [Google Scholar] [CrossRef]
- Papetti, D.M.; Spolaor, S.; Nazari, I.; Tirelli, A.; Leonardi, T.; Caprioli, C.; Besozzi, D.; Vlachou, T.; Pelicci, P.G.; Cazzaniga, P.; et al. Barcode Demultiplexing of Nanopore Sequencing Raw Signals by Unsupervised Machine Learning. Front. Bioinform. 2023, 3, 1067113. [Google Scholar] [CrossRef]
- Sintsova, A.; Ruscheweyh, H.J.; Field, C.M.; Feer, L.; Nguyen, B.D.; Daniel, B.; Hardt, W.D.; Vorholt, J.A.; Sunagawa, S. mBARq: A versatile and user-friendly framework for the analysis of DNA barcodes from transposon insertion libraries, knockout mutants, and isogenic strain populations. Bioinformatics 2024, 40, btae078. [Google Scholar] [CrossRef]
- Sahlin, K.; Lim, M.C.W.; Prost, S. NGSpeciesID: DNA barcode and amplicon consensus generation from long-read sequencing data. Ecol. Evol. 2021, 11, 1392–1398. [Google Scholar] [CrossRef] [PubMed]
- Rafeie, M.; Vafaee, F.; Faridani, O.R. Medlib: A feature-rich C/C++ library for exact alignment of nanopore sequences using multiple edit distance. bioRxiv 2025. [Google Scholar] [CrossRef]
- Vasiljevic, N.; Lim, M.; Humble, E.; Seah, A.; Kratzer, A.; Morf, N.V.; Prost, S.; Ogden, R. Developmental Validation of Oxford Nanopore Technology MinION Sequence Data and the NGSpeciesID Bioinformatic Pipeline for Forensic Genetic Species Identification. Forensic Sci. Int. Genet. 2021, 53, 102493. [Google Scholar] [CrossRef] [PubMed]
- Porter, T.M.; Hajibabaei, M. Profile hidden Markov model sequence analysis can help remove putative pseudogenes from DNA barcoding and metabarcoding datasets. BMC Bioinform. 2021, 22, 256. [Google Scholar] [CrossRef]
- Narayanan, R.; DeGroat, W.; Peker, E.; Zeeshan, S.; Ahmed, Z. VAREANT: A Bioinformatics Application for Gene Variant Reduction and Annotation. Bioinform. Adv. 2025, 5, vbae210. [Google Scholar] [CrossRef]
- Sarhan, M.S.; Filosi, M.; Maixner, F.; Fuchsberger, C. Taxonize-gb: A tool for filtering GenBank non-redundant databases based on taxonomy. bioRxiv 2024. [Google Scholar] [CrossRef]
- Seoane, P.; Diaz-Martinez, L.; Viguera, E.; Claros, M.G.; Grande-Perez, A. QuasiFlow: A bioinformatic tool for genetic variability analysis from next generation sequencing data. bioRxiv 2022. [Google Scholar] [CrossRef]
- Fearnley, L.G.; Bennett, M.F.; Bahlo, M. Ultrafast, alignment-free detection of repeat expansions in next-generation DNA and RNA sequencing data. bioRxiv 2021. [Google Scholar] [CrossRef]
- Koh, E.; Sunil, R.S.; Lam, H.Y.I.; Mutwil, M. Confronting the data deluge: How artificial intelligence can be used in the study of plant stress. Comput. Struct. Biotechnol. J. 2024, 23, 3454–3466. [Google Scholar] [CrossRef]
- Salvi, D.; Berrilli, E.; D’Alessandro, P.; Biondi, M. Sharpening the DNA barcoding tool through a posteriori taxonomic validation: The case of Longitarsus flea beetles (Coleoptera: Chrysomelidae). PLoS ONE 2020, 15, e0233573. [Google Scholar] [CrossRef]
- Yu, J.; Wu, X.; Liu, C.; Newmaster, S.; Ragupathy, S.; Kress, W.J. Progress in the use of DNA barcodes in the identification and classification of medicinal plants. Ecotoxicol. Environ. Saf. 2021, 208, 111691. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Wei, C.; Chan, D.; Agda, J.; Ballesteros-Mejia, L.; Ait Boutou, H.; Boutou, H.A.; El Bastami, Z.M.; Ma, E.; Manjunathet, R.; et al. BOLD v4: A Centralized Bioinformatics Platform for DNA-Based Biodiversity Data. In DNA Barcoding: Methods and Protocols; Springer: New York, NY, USA, 2024; pp. 403–441. [Google Scholar]
- Jin, L.; Shi, H.Y.; Li, T.; Zhao, N.; Xu, Y.; Xiao, T.W.; Song, F.; Ma, C.X.; Li, Q.M.; Lin, L.X.; et al. A DNA barcode library for woody plants in tropical and subtropical China. Sci. Data 2023, 10, 819. [Google Scholar] [CrossRef]
- Song, F.; Deng, Y.F.; Yan, H.F.; Lin, Z.L.; Delgado, A.; Trinidad, H.; Gonzales-Arce, P.; Riva, S.; Cano-Echevarría, A.; Ramos, E.; et al. Flora diversity survey and establishment of a plant DNA barcode database of Lomas ecosystems in Peru. Sci. Data 2023, 10, 294. [Google Scholar] [CrossRef]
- Grant, D.M.; Brodnicke, O.B.; Evankow, A.M.; Ferreira, A.O.; Fontes, J.T.; Hansen, A.K.; Jensen, M.R.; Kalaycı, T.E.; Leeper, A.; Patil, S.K.; et al. The Future of DNA Barcoding: Reflections from Early Career Researchers. Diversity 2021, 13, 313. [Google Scholar] [CrossRef]
- Bartolo, A.G.; Zammit, G.; Peters, A.F.; Küpper, F.C. The current state of DNA barcoding of macroalgae in the Mediterranean Sea: Presently lacking but urgently required. Bot. Mar. 2020, 63, 253–272. [Google Scholar] [CrossRef]
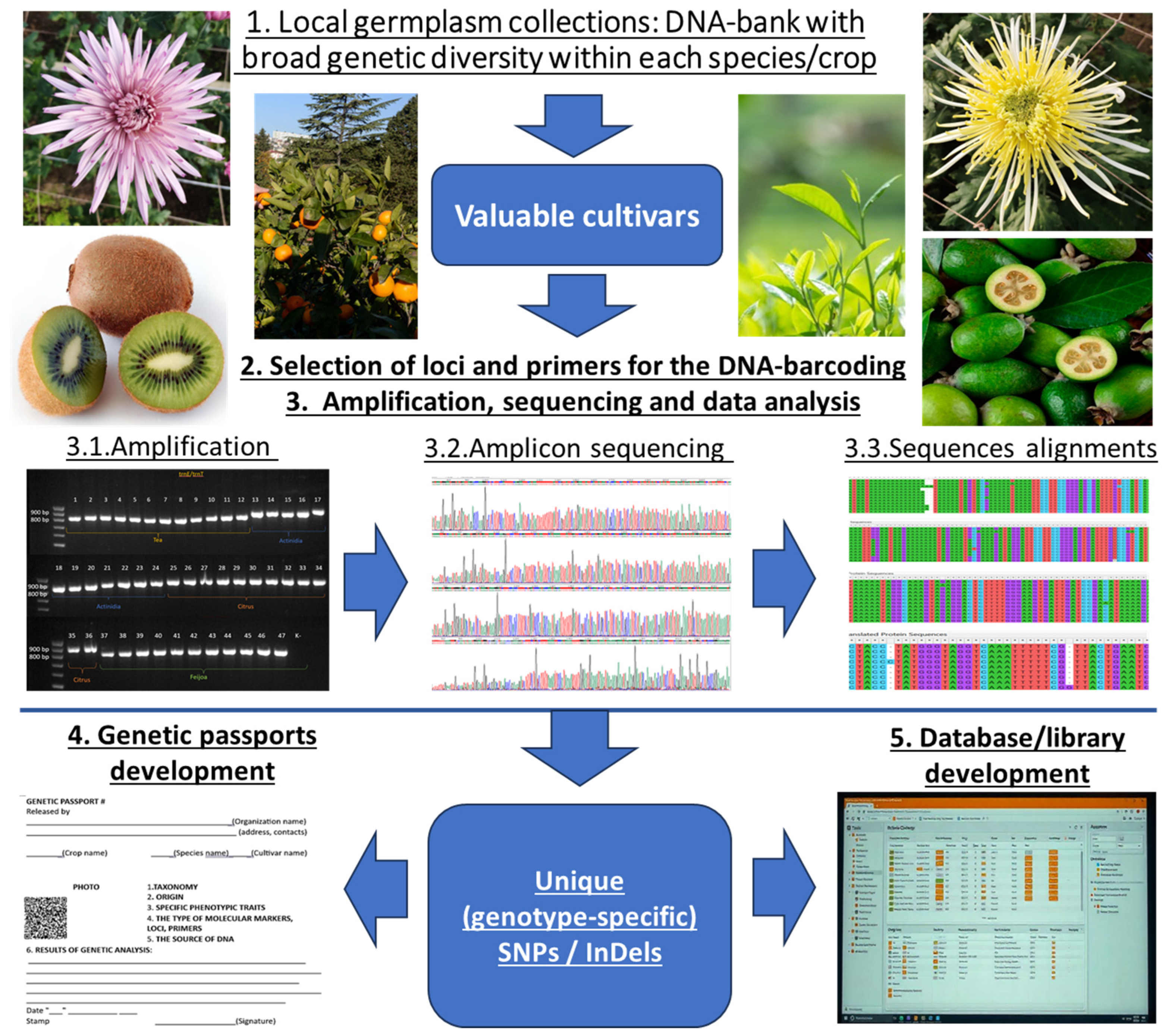
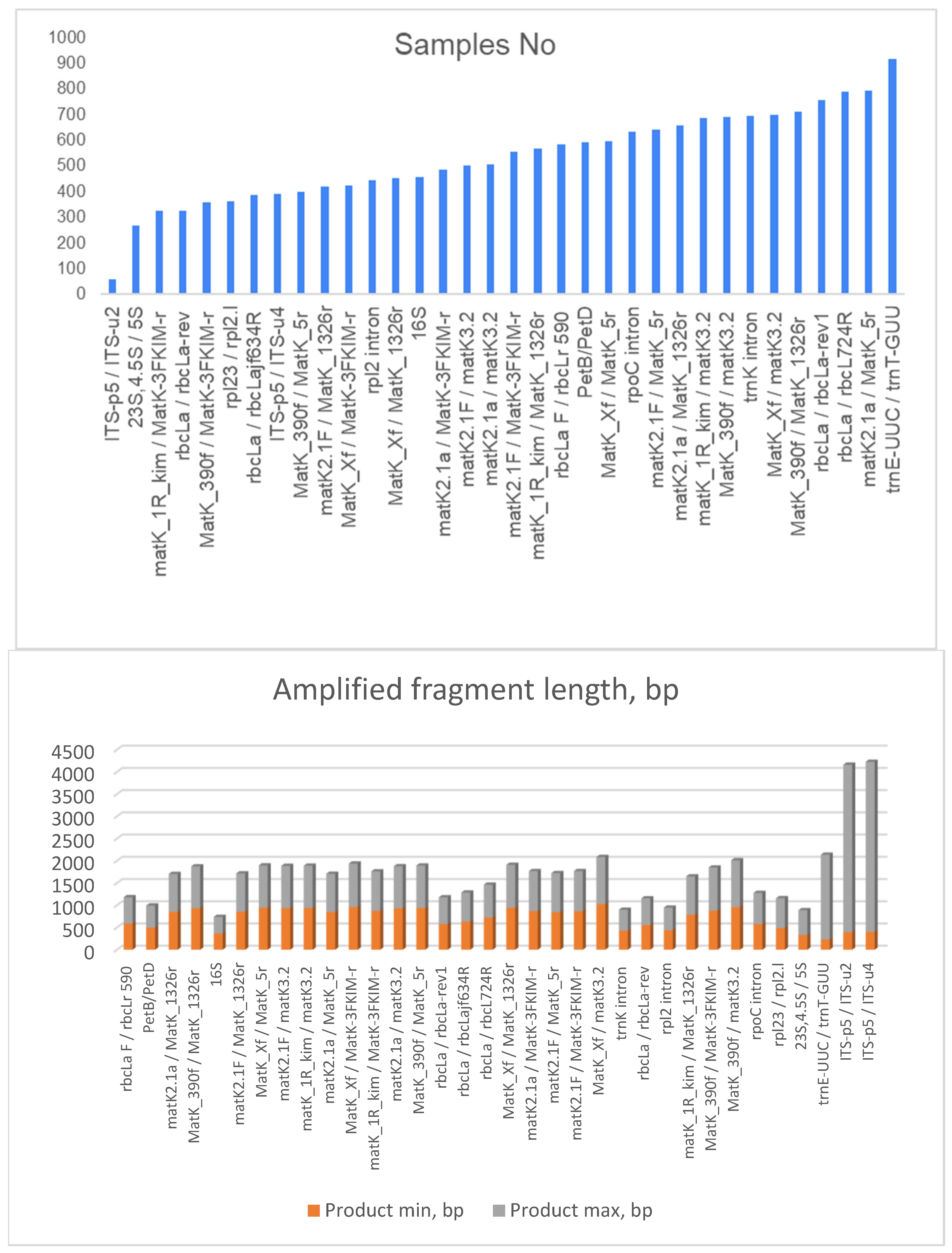
| Primer Code | Forward/Reverse | Sequence 5′-3′ | Reference |
|---|---|---|---|
| rbcLa-F | F | ATGTCACCACAAACAGAGACTAAAGC | [20] |
| rbcLr590 | R | AGTCCACCGCGTAGACATTCAT | [68] |
| rbcLa-rev | R | GTAAAATCAAGTCCACCRCG | [69] |
| rbcLajf634R | R | GAAACGGTCTCTCCAACGCAT | [70] |
| rbcL724R | R | TCGCATGTACCTGCAGTAGC | [71] |
| matK2.1a | F | ATCCATCTGGAAATCTTAGTTC | [72] |
| matK2.1F | F | CCTATCCATCTGGAAATCTTAG | [72] |
| matK_1R_kim | F | ACCCAGTCCATCTGGAAATCTTGGTCC | [73] |
| MatK_390f | F | CGATCTATTCATTCAATATTTC | [74] |
| MatK_Xf | F | TAATTTACGATCAATTCATTC | [72] |
| MatK-3FKIM-r | R | CGTACAGTACTTTTGTGTTTACGAG | [73] |
| MatK_1326r | R | TCTAGCACACGAAAGTCGAAGT | [74] |
| MatK_5r | R | GTTCTAGCACAAGAAAGTCG | [72] |
| matK3.2 | R | CTTCCTCTGTAAAGAATTC | [72] |
| 23S,4.5S/5S | F | TCTCCTCCGACTTCCCTAG | [22] |
| 23S,4.5S/5S | R | ACCATGAACGAGGAAAGGC | [22] |
| 16S | F | ATTGCGTCGTTGTGCCTGG | [22] |
| 16S | R | GATACGTTGTTAGGTGCTCC | [22] |
| petB/petD | F | TAGGGGGAATTACACTTAC | [22] |
| petB/petD | R | CATTAACATGAATACGGCAG | [22] |
| rpl23/rpl2.l | F | GAAGAAGCTTGTACAGTTTGG | [22] |
| rpl23/rpl2.l | R | TTTACTTACGGCGACGAAG | [22] |
| rpl2 intron | F | ATTGAGTTCAGTAGTTCCTC | [22] |
| rpl2 intron | R | CCAAACTGTACAAGCTTCTTC | [22] |
| rpoC1 intron | F | GAGTAACATGAAGCTCAG | [22] |
| rpoC1 intron | R | GTTTCCTTTCATCCGGCT | [22] |
| trnK intron | F | GTCTACATCATCGGTAGAG | [22] |
| trnK intron | R | CAACCCAATCGCTCTTTTG | [22] |
| trnE-UUC/trnT-GUU | F | TCCTGAACCACTAGACGATG | [75] |
| trnE-UUC/trnT-GUU | R | ATGGCGTTACTCTACCACTG | [75] |
| ITS-p5 | F | CCTTATCAYTTAGAGGAAGGAG | [67] |
| ITS-p3 | F | YGACTCTCGGCAACGGATA | [67] |
| ITS-u4 | R | RGTTTCTTTTCCTCCGCTTA | [67] |
| ITS-u2 | R | GCGTTCAAAGAYTCGATGRTTC | [67] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samarina, L.S.; Koninskaya, N.G.; Shkhalakhova, R.M.; Simonyan, T.A.; Kuzmina, D.O. DNA-Barcoding for Cultivar Identification and Intraspecific Diversity Analysis of Agricultural Crops. Int. J. Mol. Sci. 2025, 26, 6808. https://doi.org/10.3390/ijms26146808
Samarina LS, Koninskaya NG, Shkhalakhova RM, Simonyan TA, Kuzmina DO. DNA-Barcoding for Cultivar Identification and Intraspecific Diversity Analysis of Agricultural Crops. International Journal of Molecular Sciences. 2025; 26(14):6808. https://doi.org/10.3390/ijms26146808
Chicago/Turabian StyleSamarina, Lidiia S., Natalia G. Koninskaya, Ruset M. Shkhalakhova, Taisiya A. Simonyan, and Daria O. Kuzmina. 2025. "DNA-Barcoding for Cultivar Identification and Intraspecific Diversity Analysis of Agricultural Crops" International Journal of Molecular Sciences 26, no. 14: 6808. https://doi.org/10.3390/ijms26146808
APA StyleSamarina, L. S., Koninskaya, N. G., Shkhalakhova, R. M., Simonyan, T. A., & Kuzmina, D. O. (2025). DNA-Barcoding for Cultivar Identification and Intraspecific Diversity Analysis of Agricultural Crops. International Journal of Molecular Sciences, 26(14), 6808. https://doi.org/10.3390/ijms26146808






