Comparative Analysis of Differentially Expressed Long Non-Coding RNA in Pre- and Postmenopausal Fibroids
Abstract
1. Introduction
2. Results
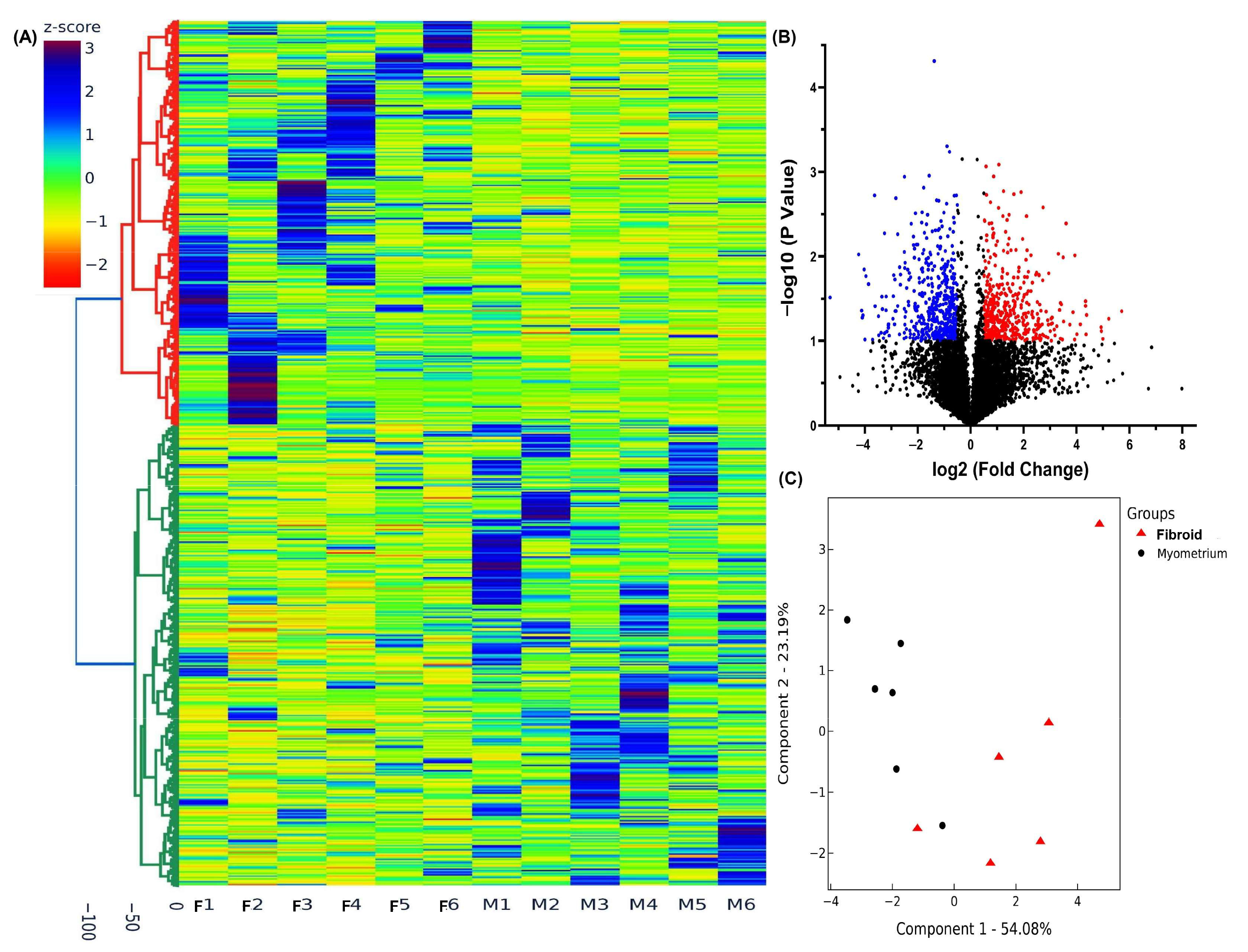
2.1. High-Throughput Sequencing Analysis of lncRNA Expression in Pre- and Postmenopausal Group
2.2. Validation of lncRNA Expression by qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Myometrium and Fibroid Tissues Collection
4.2. MED12 Mutation Analysis
4.3. RNA Sequencing and Bioinformatic Analysis
4.4. Quantitative RT-PCR
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stewart, E.A.; Cookson, C.L.; Gandolfo, R.A.; Schulze-Rath, R. Epidemiology of uterine fibroids: A systematic review. BJOG 2017, 124, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- De La Cruz, M.S.; Buchanan, E.M. Uterine Fibroids: Diagnosis and Treatment. Am. Fam. Physician 2017, 95, 100–107. [Google Scholar] [PubMed]
- Chuang, T.D.; Ton, N.; Rysling, S.; Boos, D.; Khorram, O. The Effect of Race/Ethnicity and MED12 Mutation on the Expression of Long Non-Coding RNAs in Uterine Leiomyoma and Myometrium. Int. J. Mol. Sci. 2024, 25, 1307. [Google Scholar] [CrossRef] [PubMed]
- Borahay, M.A.; Asoglu, M.R.; Mas, A.; Adam, S.; Kilic, G.S.; Al-Hendy, A. Estrogen Receptors and Signaling in Fibroids: Role in Pathobiology and Therapeutic Implications. Reprod. Sci. 2017, 24, 1235–1244. [Google Scholar] [CrossRef] [PubMed]
- Maruo, T.; Matsuo, H.; Samoto, T.; Shimomura, Y.; Kurachi, O.; Gao, Z.; Wang, Y.; Spitz, I.M.; Johansson, E. Effects of progesterone on uterine leiomyoma growth and apoptosis. Steroids 2000, 65, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Reis, F.M.; Bloise, E.; Ortiga-Carvalho, T.M. Hormones and pathogenesis of uterine fibroids. Best Pract. Res. Clin. Obstet. Gynaecol. 2016, 34, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Micić, J.; Macura, M.; Andjić, M.; Ivanović, K.; Dotlić, J.; Micić, D.D.; Arsenijević, V.; Stojnić, J.; Bila, J.; Babić, S.; et al. Currently Available Treatment Modalities for Uterine Fibroids. Medicina 2024, 60, 868. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Wang, D.; Wang, J.; Yu, W.; Yang, J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells 2019, 8, 1015. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, X.; Chen, S.; Zhang, S. Long noncoding RNAs: Fine-tuners hidden in the cancer signaling network. Cell Death Discov. 2021, 7, 283. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Zhao, Y.; Li, Q.; Jin, S.; Liu, Z. LncRNAs in tumor metabolic reprogramming and tumor microenvironment remodeling. Front. Immunol. 2024, 15, 1467151. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Rehan, A.; Khorram, O. Functional role of the long noncoding RNA X-inactive specific transcript in leiomyoma pathogenesis. Fertil. Steril. 2021, 115, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Ton, N.; Rysling, S.; Khorram, O. The in vivo effects of knockdown of long non-coding RNA XIST on fibroid growth and gene expression. FASEB J. 2024, 38, e70140. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Quintanilla, D.; Boos, D.; Khorram, O. Long Noncoding RNA MIAT Modulates the Extracellular Matrix Deposition in Leiomyomas by Sponging MiR-29 Family. Endocrinology 2021, 162, bqab186. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Ton, N.; Manrique, N.; Rysling, S.; Khorram, O. Targeting the long non-coding RNA MIAT for the treatment of fibroids in an animal model. Clin. Sci. 2024, 138, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Zhou, H.; Sun, Y.; Shen, B.; Chou, D. Long non-coding ribonucleic acid H19 and ten-eleven translocation enzyme 1 messenger RNA expression levels in uterine fibroids may predict their postoperative recurrence. Clinics 2021, 76, e2671. [Google Scholar] [CrossRef] [PubMed]
- Cao, T.; Jiang, Y.; Wang, Z.; Zhang, N.; Al-Hendy, A.; Mamillapalli, R.; Kallen, A.N.; Kodaman, P.; Taylor, H.S.; Li, D.; et al. H19 lncRNA identified as a master regulator of genes that drive uterine leiomyomas. Oncogene 2019, 38, 5356–5366. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Ton, N.; Rysling, S.; Khorram, O. The Functional Role of the Long Non-Coding RNA LINCMD1 in Leiomyoma Pathogenesis. Int. J. Mol. Sci. 2024, 25, 11539. [Google Scholar] [CrossRef] [PubMed]
- Naciri, I.; Andrade-Ludena, M.D.; Yang, Y.; Kong, M.; Sun, S. An emerging link between lncRNAs and cancer sex dimorphism. Hum. Genet. 2024, 143, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wesevich, V.; Chen, Z.; Zhang, D.; Kallen, A.N. Emerging roles for noncoding RNAs in female sex steroids and reproductive disease. Mol. Cell. Endocrinol. 2020, 518, 110875. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Quintanilla, D.; Boos, D.; Khorram, O. Differential Expression of Super-Enhancer-Associated Long Non-coding RNAs in Uterine Leiomyomas. Reprod. Sci. 2022, 29, 2960–2976. [Google Scholar] [CrossRef] [PubMed]
- George, J.W.; Fan, H.; Johnson, B.; Carpenter, T.J.; Foy, K.K.; Chatterjee, A.; Patterson, A.L.; Koeman, J.; Adams, M.; Madaj, Z.B.; et al. Integrated Epigenome, Exome, and Transcriptome Analyses Reveal Molecular Subtypes and Homeotic Transformation in Uterine Fibroids. Cell Rep. 2019, 29, 4069–4085.e6. [Google Scholar] [CrossRef] [PubMed]
- Benassayag, C.; Leroy, M.J.; Rigourd, V.; Robert, B.; Honoré, J.C.; Mignot, T.M.; Vacher-Lavenu, M.C.; Chapron, C.; Ferré, F. Estrogen receptors (ERalpha/ERbeta) in normal and pathological growth of the human myometrium: Pregnancy and leiomyoma. Am. J. Physiol. 1999, 276, E1112–E1118. [Google Scholar] [PubMed]
- Shozu, M.; Murakami, K.; Inoue, M. Aromatase and leiomyoma of the uterus. Semin. Reprod. Med. 2004, 22, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Gao, J.; Quintanilla, D.; McSwiggin, H.; Boos, D.; Yan, W.; Khorram, O. Differential Expression of MED12-Associated Coding RNA Transcripts in Uterine Leiomyomas. Int. J. Mol. Sci. 2023, 24, 3742. [Google Scholar] [CrossRef] [PubMed]
- Kasai, T.; Shozu, M.; Murakami, K.; Segawa, T.; Shinohara, K.; Nomura, K.; Inoue, M. Increased expression of type I 17beta-hydroxysteroid dehydrogenase enhances in situ production of estradiol in uterine leiomyoma. J. Clin. Endocrinol. Metab. 2004, 89, 5661–5668. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Mi, H.N.; Chai, L.Y.; Wang, H.N. Effects of progesterone receptor on proliferation of uterine leiomyoma cells. J. Biol. Regul. Homeost. Agents 2019, 33, 1685–1693. [Google Scholar] [CrossRef] [PubMed]
- Voronin, D.; Sotnikova, N.; Rukavishnikov, K.; Malyshkina, A.; Nagornii, S.; Antsiferova, Y. Differential regulatory effect of progesterone on the proliferation and apoptosis of uterine leiomyoma tissue explants and primary leiomyoma cell cultures. JBRA Assist. Reprod. 2021, 25, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Tsigkou, A.; Reis, F.M.; Lee, M.H.; Jiang, B.; Tosti, C.; Centini, G.; Shen, F.R.; Chen, Y.G.; Petraglia, F. Increased progesterone receptor expression in uterine leiomyoma: Correlation with age, number of leiomyomas, and clinical symptoms. Fertil. Steril. 2015, 104, 170–175.e1. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Ton, N.; Rysling, S.; Quintanilla, D.; Boos, D.; Gao, J.; McSwiggin, H.; Yan, W.; Khorram, O. The Influence of Race/Ethnicity on the Transcriptomic Landscape of Uterine Fibroids. Int. J. Mol. Sci. 2023, 24, 13441. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikov, V.Y.; Antonets, D.V.; Gulyaeva, L.F. The search of CAR, AhR, ESRs binding sites in promoters of intronic and intergenic microRNAs. J. Bioinform. Comput. Biol. 2018, 16, 1750029. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, L.; Ravo, M.; Nassa, G.; Tarallo, R.; De Filippo, M.R.; Giurato, G.; Cirillo, F.; Stellato, C.; Silvestro, S.; Cantarella, C.; et al. Effects of oestrogen on microRNA expression in hormone-responsive breast cancer cells. Horm. Cancer 2012, 3, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Bhat-Nakshatri, P.; Wang, G.; Collins, N.R.; Thomson, M.J.; Geistlinger, T.R.; Carroll, J.S.; Brown, M.; Hammond, S.; Srour, E.F.; Liu, Y.; et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009, 37, 4850–4861. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yu, W.; Tang, D.; Zhou, Y.; Bi, M.; Wang, H.; Zheng, Y.; Chen, M.; Li, L.; Xu, X.; et al. Epigenomics-based identification of oestrogen-regulated long noncoding RNAs in ER+ breast cancer. RNA Biol. 2020, 17, 1590–1602. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Ohta, T.; Oki, S. ChIP-Atlas 3.0: A data-mining suite to explore chromosome architecture together with large-scale regulome data. Nucleic Acids Res. 2024, 52, W45–W53. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.P.; Luo, H.; Matur, M.; Eshelman, M.A.; Hamamoto, K.; Sharma, A.; Lesperance, J.; Huang, S. A coordinated function of lncRNA HOTTIP and miRNA-196b underpinning leukemogenesis by targeting FAS signaling. Oncogene 2022, 41, 718–731. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Shi, C.; Deng, L.; Fu, W.; Zhang, J. LncRNA HOTTIP facilitated tumor growth via stimulating the hnRNPA2B1/DKK1/Wnt/β-catenin regulatory axis in hepatocellular carcinoma. Genes Dis. 2024, 11, 101013. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Ji, F. lncRNA HOTTIP facilitates osteosarcoma cell migration, invasion and epithelial-mesenchymal transition by forming a positive feedback loop with c-Myc. Oncol. Lett. 2019, 18, 1649–1656. [Google Scholar] [CrossRef] [PubMed]
- Tamang, S.; Acharya, V.; Roy, D.; Sharma, R.; Aryaa, A.; Sharma, U.; Khandelwal, A.; Prakash, H.; Vasquez, K.M.; Jain, A. SNHG12: An LncRNA as a Potential Therapeutic Target and Biomarker for Human Cancer. Front. Oncol. 2019, 9, 901. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Liu, Y.; Liu, H.; Chen, W.; Fan, H.N.; Zhang, J.; Zhu, J.S. The long non-coding RNA SNHG12 promotes gastric cancer by activating the phosphatidylinositol 3-kinase/AKT pathway. Aging 2019, 11, 10902–10922. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Xu, W.; Cai, Y.; Guo, C.; Zhou, G.; Yuan, C. CASC15: A Tumor-Associated Long Non-Coding RNA. Curr. Pharm. Des. 2021, 27, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xiong, Y.; Jiang, K.; Xin, B.; Jiang, T.; Wei, R.; Zou, Y.; Tan, H.; Jiang, T.; Yang, A.; et al. Hypoxia-sensitive long noncoding RNA CASC15 promotes lung tumorigenesis by regulating the SOX4/β-catenin axis. J. Exp. Clin. Cancer Res. 2021, 40, 12. [Google Scholar] [CrossRef] [PubMed]
- Mahboobeh, Z.; Pegah, M.; Fatemeh, S.; Elham, K.; Hanieh, A.; Milad, R.; Mohammad, S. lncRNA ZEB2-AS1: A promising biomarker in human cancers. IUBMB Life 2020, 72, 1891–1899. [Google Scholar] [CrossRef] [PubMed]
- Tanwar, P.S.; Lee, H.J.; Zhang, L.; Zukerberg, L.R.; Taketo, M.M.; Rueda, B.R.; Teixeira, J.M. Constitutive activation of Beta-catenin in uterine stroma and smooth muscle leads to the development of mesenchymal tumors in mice. Biol. Reprod. 2009, 81, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Li, Z.; Song, E.; Hu, P.; Yang, Q.; Hu, Y.; Liu, H.; Jin, A. LncRNA HOTTIP enhances human osteogenic BMSCs differentiation via interaction with WDR5 and activation of Wnt/β-catenin signalling pathway. Biochem. Biophys. Res. Commun. 2020, 524, 1037–1043. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Tang, Y.; Tang, J.; Liu, Z.; Wang, X. Downregulation of lncRNA HOTTIP Suppresses the Proliferation, Migration, and Invasion of Oral Tongue Squamous Cell Carcinoma by Regulation of HMGA2-Mediated Wnt/β-Catenin Pathway. Cancer Biother. Radiopharm. 2020, 35, 720–730. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Shoorei, H.; Hussen, B.M.; Abdullah, S.R.; Poornajaf, Y.; Taheri, M.; Samsami, M. LncRNA SNHG12: A budding star in human diseases. Pathol. Res. Pract. 2023, 251, 154897. [Google Scholar] [CrossRef] [PubMed]
- Daly, D.C.; Walters, C.A.; Prior, J.C.; Kuslis, S.T.; Chapitis, J.; Andreoli, J.; Riddick, D.H. Prolactin production from proliferative phase leiomyoma. Am. J. Obstet. Gynecol. 1984, 148, 1059–1063. [Google Scholar] [CrossRef] [PubMed]
- Nowak, R.A.; Rein, M.S.; Heffner, L.J.; Friedman, A.J.; Tashjian, A.H., Jr. Production of prolactin by smooth muscle cells cultured from human uterine fibroid tumors. J. Clin. Endocrinol. Metab. 1993, 76, 1308–1313. [Google Scholar] [PubMed]
- Nohara, A.; Ohmichi, M.; Koike, K.; Jikihara, H.; Kimura, A.; Masuhara, K.; Ikegami, H.; Inoue, M.; Miyake, A.; Murata, Y. Prolactin stimulates mitogen-activated protein kinase in human leiomyoma cells. Biochem. Biophys. Res. Commun. 1997, 238, 473–477. [Google Scholar] [CrossRef] [PubMed]
- DiMauro, A.; Seger, C.; Minor, B.; Amitrano, A.M.; Okeke, I.; Taya, M.; Rackow, A.R.; Kumar, D.; Kottman, R.M.; Bhagavath, B.; et al. Prolactin is Expressed in Uterine Leiomyomas and Promotes Signaling and Fibrosis in Myometrial Cells. Reprod. Sci. 2022, 29, 2525–2535. [Google Scholar] [CrossRef] [PubMed]
- do Amaral, V.C.; Carvalho, K.C.; Maciel, G.A.; Simoncini, T.; da Silva, P.L.; Marcondes, R.R.; Soares, J.M., Jr.; Baracat, E.C. The progesterone and estrogen modify the uterine prolactin and prolactin receptor expression of hyperprolactinemic mice. Gynecol. Endocrinol. 2014, 31, 148–151. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Khorram, O. Expression Profiling of lncRNAs, miRNAs, and mRNAs and Their Differential Expression in Leiomyoma Using Next-Generation RNA Sequencing. Reprod. Sci. 2018, 25, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Xie, Y.; Yan, W.; Khorram, O. Next-generation sequencing reveals differentially expressed small noncoding RNAs in uterine leiomyoma. Fertil. Steril. 2018, 109, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Ton, N.; Rysling, S.; Khorram, O. In Vivo Effects of Bay 11-7082 on Fibroid Growth and Gene Expression: A Preclinical Study. Cells 2024, 13, 1091. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Ton, N.; Rysling, S.; Baghdasarian, D.; Khorram, O. Differential Expression of Small Non-Coding RNAs in Uterine Leiomyomas. Int. J. Mol. Sci. 2025, 26, 1688. [Google Scholar] [CrossRef] [PubMed]
- de Sena Brandine, G.; Smith, A.D. Falco: High-speed FastQC emulation for quality control of sequencing data. F1000Research 2019, 8, 1874. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Delhomme, N.; Padioleau, I.; Furlong, E.E.; Steinmetz, L.M. easyRNASeq: A bioconductor package for processing RNA-Seq data. Bioinformatics 2012, 28, 2532–2533. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, A.; Duitama, C.; Metge, F.; Rosskopp, D.; Boucas, J. Flaski, Flaski (3.12.2); Zenodo: Geneva, Switzerland, 2021. [Google Scholar]
- Li, Z.; Zhang, Y.; Fang, J.; Xu, Z.; Zhang, H.; Mao, M.; Chen, Y.; Zhang, L.; Pian, C. NcPath: A novel platform for visualization and enrichment analysis of human non-coding RNA and KEGG signaling pathways. Bioinformatics 2023, 39, btac812. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Ton, N.; Rysling, S.; Quintanilla, D.; Boos, D.; Khorram, O. Therapeutic effects of in vivo administration of an inhibitor of tryptophan 2,3-dioxygenase (680c91) for the treatment of fibroids: A preclinical study. Fertil. Steril. 2024, 121, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Almeida, T.A.; Quispe-Ricalde, A.; Montes de Oca, F.; Foronda, P.; Hernandez, M.M. A high-throughput open-array qPCR gene panel to identify housekeeping genes suitable for myometrium and leiomyoma expression analysis. Gynecol. Oncol. 2014, 134, 138–143. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chuang, T.-D.; Rysling, S.; Ton, N.; Baghdasarian, D.; Khorram, O. Comparative Analysis of Differentially Expressed Long Non-Coding RNA in Pre- and Postmenopausal Fibroids. Int. J. Mol. Sci. 2025, 26, 6798. https://doi.org/10.3390/ijms26146798
Chuang T-D, Rysling S, Ton N, Baghdasarian D, Khorram O. Comparative Analysis of Differentially Expressed Long Non-Coding RNA in Pre- and Postmenopausal Fibroids. International Journal of Molecular Sciences. 2025; 26(14):6798. https://doi.org/10.3390/ijms26146798
Chicago/Turabian StyleChuang, Tsai-Der, Shawn Rysling, Nhu Ton, Daniel Baghdasarian, and Omid Khorram. 2025. "Comparative Analysis of Differentially Expressed Long Non-Coding RNA in Pre- and Postmenopausal Fibroids" International Journal of Molecular Sciences 26, no. 14: 6798. https://doi.org/10.3390/ijms26146798
APA StyleChuang, T.-D., Rysling, S., Ton, N., Baghdasarian, D., & Khorram, O. (2025). Comparative Analysis of Differentially Expressed Long Non-Coding RNA in Pre- and Postmenopausal Fibroids. International Journal of Molecular Sciences, 26(14), 6798. https://doi.org/10.3390/ijms26146798




