Alternative Lengthening of Telomeres: The Need for ATRX Mutations Is Lineage-Dependent
Abstract
1. Introduction
2. Results
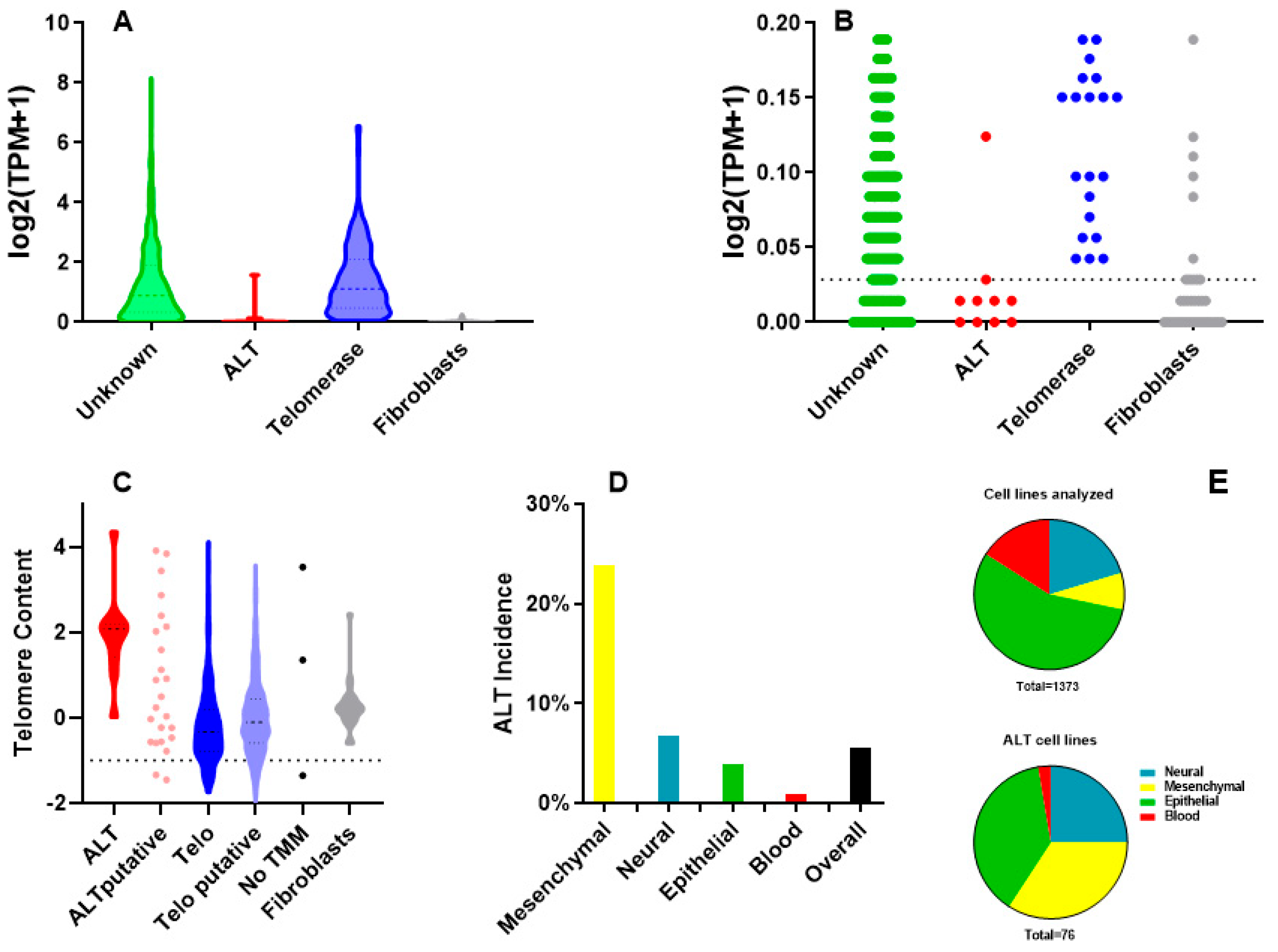
2.1. Identification of ALT Cell Lines
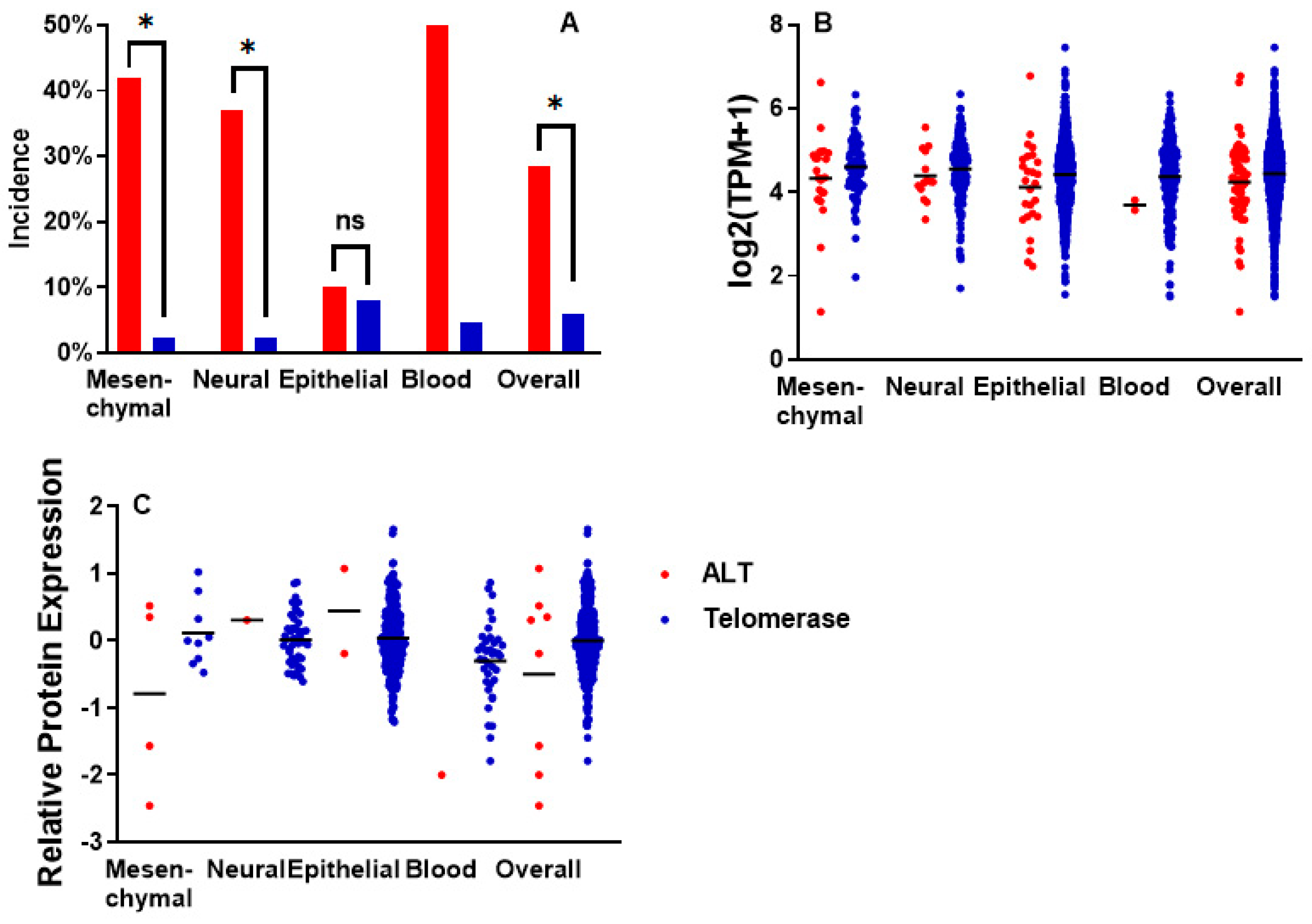
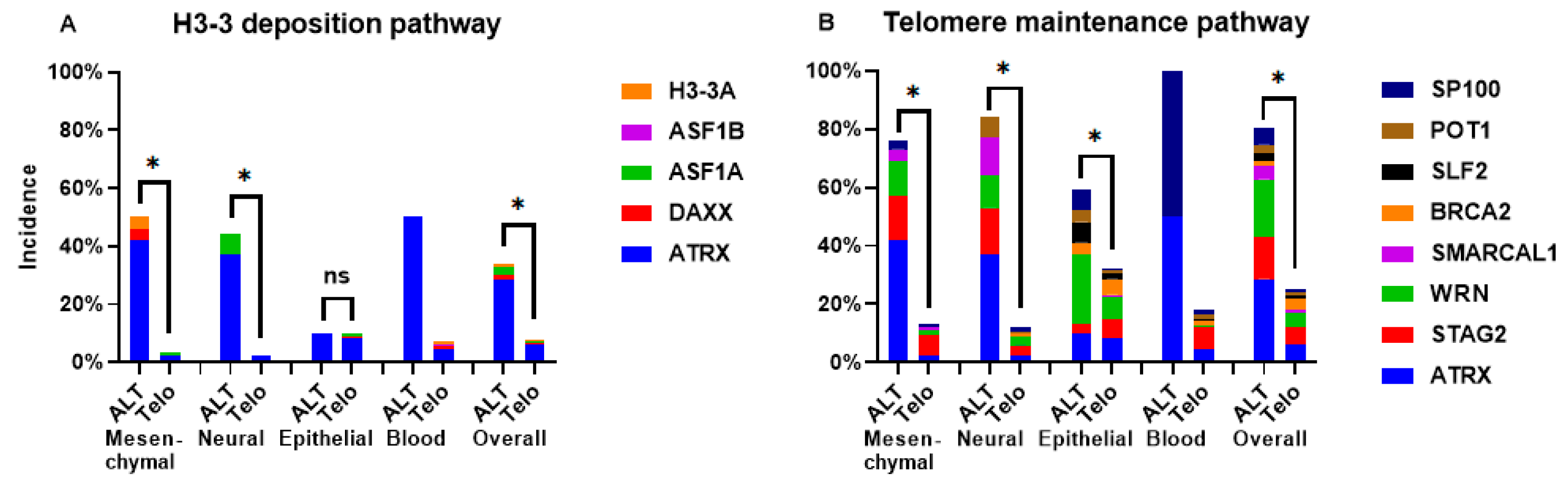
2.2. Incidence of ATRX Alterations and ATRX Expression in ALT Cell Lines
2.3. Incidence of Mutations in ALT Cell Lines
3. Discussion
4. Material and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yuan, X.; Larsson, C.; Xu, D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: Old actors and new players. Oncogene 2019, 38, 6172–6183. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, R.J.; Greenberg, R.A. Mechanisms of Alternative Lengthening of Telomeres. Cold Spring Harb. Perspect. Biol. 2025, 17, a041690. [Google Scholar] [CrossRef] [PubMed]
- Clatterbuck Soper, S.F.; Meltzer, P.S. ATRX/DAXX: Guarding the Genome against the Hazards of ALT. Genes 2023, 14, 790. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Yu, D.; Guo, J.; Hu, J.; Zhao, Y.; Jiang, C.; Meng, X.; Cai, J.; Zhao, Y. From pathology to therapy: A comprehensive review of ATRX mutation related molecular functions and disorders. Mutat. Res. Rev. Mutat. Res. 2025, 795, 108537. [Google Scholar] [CrossRef] [PubMed]
- Lewis, P.W.; Elsaesser, S.J.; Noh, K.M.; Stadler, S.C.; Allis, C.D. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. USA 2010, 107, 14075–14080. [Google Scholar] [CrossRef] [PubMed]
- Kovatcheva, M.; Liao, W.; Klein, M.E.; Robine, N.; Geiger, H.; Crago, A.M.; Dickson, M.A.; Tap, W.D.; Singer, S.; Koff, A. ATRX is a regulator of therapy induced senescence in human cells. Nat. Commun. 2017, 8, 386. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.W.C.; Ghosal, G.; Wang, W.; Shen, X.; Wang, J.; Li, L.; Chen, J. Alpha thalassemia/mental retardation syndrome X-linked gene product ATRX is required for proper replication restart and cellular resistance to replication stress. J. Biol. Chem. 2013, 288, 6342–6350. [Google Scholar] [CrossRef] [PubMed]
- Blasco, M.A. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007, 8, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Udroiu, I.; Marinaccio, J.; Sgura, A. Effects of p53 and ATRX inhibition on telomeric recombination in aging fibroblasts. Front. Oncol. 2024, 14, 1322438. [Google Scholar] [CrossRef] [PubMed]
- Lovejoy, C.A.; Li, W.; Reisenweber, S.; Thongthip, S.; Bruno, J.; de Lange, T.; De, S.; Petrini, J.H.J.; Sung, P.A.; Jasin, M.; et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012, 8, e1002772. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M.; Englezou, A.; Dalla-Pozza, L.; Dunham, M.A.; Reddel, R.R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997, 3, 1271–1274. [Google Scholar] [CrossRef] [PubMed]
- Heaphy, C.M.; Subhawong, A.P.; Hong, S.M.; Goggins, M.G.; Montgomery, E.A.; Gabrielson, E.; Netto, G.J.; Epstein, J.I.; Lotan, T.L.; Westra, W.H.; et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am. J. Pathol. 2011, 179, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a Cancer Dependency Map. Cell 2017, 170, 564–576. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, D., Jr.; Watters, A.K.; To, J.T.; Young, M.W.; Muratori, J.; Wilkoff, M.H.; Abraham, R.G.; Plummer, M.M.; Zhang, D. ALT Positivity in Human Cancers: Prevalence and Clinical Insights. Cancers 2021, 13, 2384. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.Q.; Zhong, Z.H.; Henson, J.D.; Neumann, A.A.; Chang, A.C.M.; Reddel, R.R. Suppression of alternative lengthening of telomeres by Sp100-mediated sequestration of the MRE11/RAD50/NBS1 complex. Mol. Cell. Biol. 2005, 25, 2708–2721. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Multani, A.S.; He, H.; Cosme-Blanco, W.; Deng, Y.; Deng, J.M.; Bachilo, O.; Pathak, S.; Tahara, H.; Bailey, S.M.; et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 2006, 126, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Furuichi, Y.; Ide, T.; Goto, M. Involvement of WRN helicase in immortalization and tumorigenesis by the telomeric crisis pathway. Oncol. Lett. 2011, 2, 609–611. [Google Scholar] [CrossRef] [PubMed]
- Daniloski, Z.; Smith, S. Loss of tumor suppressor STAG2 promotes telomere recombination and extends the replicative lifespan of normal human cells. Cancer Res. 2017, 77, 5530–5542. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.; Lee, J.J.; Min, J.; Hwang, K.; Park, S.G.; Kim, E.H.; Kim, B.C.; Bhak, J.; Lee, H. Brca2 abrogation engages with the alternative lengthening of telomeres via break-induced replication. FEBS J. 2019, 286, 1841–1858. [Google Scholar] [CrossRef] [PubMed]
- Scott, W.A.; Dhanji, E.Z.; Dyakov, B.J.A.; Dreseris, E.S.; Asa, J.S.; Grange, L.J.; Mirceta, M.; Pearson, C.E.; Stewart, G.S.; Gingras, A.-C.; et al. ATRX proximal protein associations boast roles beyond histone deposition. PLoS Genet. 2021, 17, e1009909. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, C.; Diplas, B.H.; Brown, A.; Strickland, L.M.; Yao, H.; Ling, J.; E McLendon, R.; Keir, S.T.; Ashley, D.M.; et al. Cancer-associated SMARCAL1 loss-of-function mutations promote alternative lengthening of telomeres and tumorigenesis in telomerase-negative glioblastoma cells. Neuro-Oncol. 2023, 25, 1563–1575. [Google Scholar] [CrossRef] [PubMed]
- Flynn, R.L.; Centore, R.C.; O’sUllivan, R.J.; Rai, R.; Tse, A.; Songyang, Z.; Chang, S.; Karlseder, J.; Zou, L. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 2011, 471, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Raghunandan, M.; Geelen, D.; Majerova, E.; Decottignies, A. NHP2 downregulation counteracts hTR-mediated activation of the DNA damage response at ALT telomeres. EMBO J. 2021, 40, e106336. [Google Scholar] [CrossRef] [PubMed]
- Udroiu, I.; Marinaccio, J.; Sgura, A. Many functions of telomerase components: Certainties, doubts, and inconsistencies. Int. J. Mol. Sci. 2022, 23, 15189. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Wang, Z.J.; Yi, B.Q.; Ma, H.C.; Xu, H.M. hRad21 overexpresses and localizes to the ALT-associated promyelocytic leukemia body in ALT cells. Cancer Biol. Ther. 2010, 9, 978–983. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Rossi, M.L.; Singh, D.K.; Dunn, C.; Ramamoorthy, M.; Croteau, D.L.; Liu, Y.; Bohr, V.A. RECQL4, the protein mutated in Rothmund-Thomson syndrome, functions in telomere maintenance. J. Biol. Chem. 2012, 287, 196–209. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; O’Rourke, J.J.; Sobinoff, A.P.; Allen, J.A.M.; Nelson, C.B.; Tomlinson, C.G.; Lee, M.; Reddel, R.R.; Deans, A.J.; Pickett, H.A. The FANCM-BLM-TOP3A-RMI complex suppresses alternative lengthening of telomeres (ALT). Nat. Commun. 2019, 10, 2252. [Google Scholar] [CrossRef] [PubMed]
- de Nonneville, A.; Salas, S.; Bertucci, F.; Sobinoff, A.P.; Adélaïde, J.; Guille, A.; Finetti, P.; Noble, J.R.; Churikov, D.; Chaffanet, M.; et al. TOP3A amplification and ATRX inactivation are mutually exclusive events in pediatric osteosarcomas using ALT. EMBO Mol. Med. 2022, 14, e15859. [Google Scholar] [CrossRef] [PubMed]
- Henson, J.D.; Reddel, R.R. Assaying and investigating Alternative Lengthening of Telomeres activity in human cells and cancers. FEBS Lett. 2010, 584, 3800–3811. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, A.S.; Dagg, R.A.; Choi, L.M.R.; Shay, J.W.; Reynolds, C.P.; Lau, L.M. Alternative lengthening of telomeres in neuroblastoma cell lines is associated with a lack of MYCN genomic amplification and with p53 pathway aberrations. J. Neuro-Oncol. 2014, 119, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Flørenes, V.A.; Mælandsmo, G.M.; Forus, A.; Andreassen, Å.; Myklebost, O.; Fodstad, Ø. MDM2 gene amplification and transcript levels in human sarcomas: Relationship to TP53 gene status. JNCI J. Natl. Cancer Inst. 1994, 86, 1297–1302. [Google Scholar] [CrossRef] [PubMed]
- Landers, J.E.; Cassel, S.L.; George, D.L. Translational enhancement of mdm2 oncogene expression in human tumor cells containing a stabilized wild-type p53 protein. Cancer Res. 1997, 57, 3562–3568. [Google Scholar] [PubMed]
- Kleiblova, P.; Shaltiel, I.A.; Benada, J.; Sevcik, J.; Pecháčková, S.; Pohlreich, P.; Voest, E.E.; Dundr, P.; Bartek, J.; Kleibl, Z.; et al. Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint. J. Cell Biol. 2013, 201, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Udroiu, I.; Marinaccio, J.; Goffi, R.S.; Micheli, E.; Sgura, A. Specificity and sensitivity of ALT-associated markers in cancer cells. FEBS Lett. 2025, 599, 989–1005. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.; Bernier, C.; Kaiser, B.; Fournier, S.; Li, L.; Desjardins, J.; Skeldon, A.; Rimkunas, V.; Veloso, A.; Young, J.T.; et al. Guiding ATR and PARP inhibitor combinations with chemogenomic screens. Cell Rep. 2022, 40, 111081. [Google Scholar] [CrossRef] [PubMed]
- Udroiu, I.; Sgura, A. Alternative lengthening of telomeres and chromatin status. Genes 2019, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Hoang, S.M.; O’Sullivan, R.J. Alternative Lengthening of Telomeres: Building bridges to connect chromosome ends. Trends Cancer 2020, 6, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Behjati, S.; Tarpey, P.S.; Presneau, N.; Scheipl, S.; Pillay, N.; Van Loo, P.; Wedge, D.C.; Cooke, S.L.; Gundem, G.; Davies, H.; et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat. Genet. 2013, 45, 1479–1482. [Google Scholar] [CrossRef] [PubMed]
- Forsyth, R.G.; De Boeck, G.; Bekaert, S.; De Meyer, T.; Taminiau, A.H.; Uyttendaele, D.; Roels, H.; Praet, M.M.; Hogendoorn, P.C. Telomere biology in giant cell tumour of bone. J. Pathol. 2008, 214, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Laud, P.R.; Multani, A.S.; Bailey, S.M.; Wu, L.; Ma, J.; Kingsley, C.; Lebel, M.; Pathak, S.; DePinho, R.A.; Chang, S. Elevated telomere-telomere recombination in WRN-deficient, telomere dysfunctional cells promotes escape from senescence and engagement of the ALT pathway. Genes Dev. 2005, 19, 2560–2570. [Google Scholar] [CrossRef] [PubMed]
- Napier, C.E.; Huschtscha, L.I.; Harvey, A.; Bower, K.; Noble, J.R.; Hendrickson, E.A.; Reddel, R.R. ATRX represses alternative lengthening of telomeres. Oncotarget 2015, 6, 16543. [Google Scholar] [CrossRef] [PubMed]
- Bérubé, N.G. ATRX in chromatin assembly and genome architecture during development and disease. Biochem. Cell Biol. 2011, 89, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Kernohan, K.D.; Jiang, Y.; Tremblay, D.C.; Bonvissuto, A.C.; Eubanks, J.H.; Mann, M.R.; Bérubé, N.G. ATRX partners with cohesin and MeCP2 and contributes to developmental silencing of imprinted genes in the brain. Dev. Cell 2010, 18, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Voon, H.P.; Hughes, J.R.; Rode, C.; De La Rosa-Velazquez, I.A.; Jenuwein, T.; Feil, R.; Higgs, D.R.; Gibbons, R.J. ATRX plays a key role in maintaining silencing at interstitial heterochromatic loci and imprinted genes. Cell Rep. 2015, 11, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Kernohan, K.D.; Vernimmen, D.; Gloor, G.B.; Bérubé, N.G. Analysis of neonatal brain lacking ATRX or MeCP2 reveals changes in nucleosome density, CTCF binding and chromatin looping. Nucleic Acids Res. 2014, 42, 8356–8368. [Google Scholar] [CrossRef] [PubMed]
- Bieluszewska, A.; Wulfridge, P.; Doherty, J.; Ren, W.; Sarma, K. ATRX histone binding and helicase activities have distinct roles in neuronal differentiation. Nucleic Acids Res. 2022, 50, 9162–9174. [Google Scholar] [CrossRef] [PubMed]
- Huh, M.S.; O’dEa, T.P.; Ouazia, D.; McKay, B.C.; Parise, G.; Parks, R.J.; Rudnicki, M.A.; Picketts, D.J. Compromised genomic integrity impedes muscle growth after Atrx inactivation. J. Clin. Investig. 2012, 122, 4412–4423. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-T.; Jiang, M.-M.; Leynes, C.; Adeyeye, M.; Majano, C.F.; Ibrahim, B.; Polak, U.; Hung, G.; Jin, Z.; Lanza, D.G.; et al. ATRX silences Cartpt expression in osteoblastic cells during skeletal development. J. Clin. Investig. 2025, 135, e163587. [Google Scholar] [CrossRef] [PubMed]
- Deguchi, K.; Clewing, J.M.; Elizondo, L.I.; Hirano, R.; Huang, C.; Choi, K.; Sloan, E.A.; Lücke, T.; Marwedel, K.M.; Powell, R.D.; et al. Neurologic phenotype of Schimke immuno-osseous dysplasia and neurodevelopmental expression of SMARCAL1. J. Neuropathol. Exp. Neurol. 2008, 67, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Beatty, A.; Rubin, A.M.; Wada, H.; Heidinger, B.; Hood, W.R.; Schwartz, T.S. Postnatal expression of IGF2 is the norm in amniote vertebrates. Proc. R. Soc. B 2022, 289, 20212278. [Google Scholar] [CrossRef] [PubMed]
- Matouk, I.J.; DeGroot, N.; Mezan, S.; Ayesh, S.; Abu-lail, R.; Hochberg, A.; Galun, E. The H19 non-coding RNA is essential for human tumor growth. PLoS ONE 2007, 2, e845. [Google Scholar] [CrossRef] [PubMed]
- Barsyte-Lovejoy, D.; Lau, S.K.; Boutros, P.C.; Khosravi, F.; Jurisica, I.; Andrulis, I.L.; Tsao, M.S.; Penn, L.Z. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006, 66, 5330–5337. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Yao, F.; Yin, M.; Liao, Y.; Li, K.; Li, L.; Xiao, X.; Guo, J.; Hu, F.; Feng, H. Anti-senescent effects of long non-coding RNA H19 on human dermal fibroblast cells through impairing microRNA-296-5p-dependent inhibition of IGF2. Cell. Signal. 2022, 94, 110327. [Google Scholar] [CrossRef] [PubMed]
- Fu, V.X.; Schwarze, S.R.; Kenowski, M.L.; LeBlanc, S.; Svaren, J.; Jarrard, D.F. A loss of insulin-like growth factor-2 imprinting is modulated by CCCTC-binding factor down-regulation at senescence in human epithelial cells. J. Biol. Chem. 2004, 279, 52218–52226. [Google Scholar] [CrossRef] [PubMed]
- Akter, J.; Katai, Y.; Sultana, P.; Takenobu, H.; Haruta, M.; Sugino, R.P.; Mukae, K.; Satoh, S.; Wada, T.; Ohira, M.; et al. Loss of p53 suppresses replication stress-induced DNA damage in ATRX-deficient neuroblastoma. Oncogenesis 2021, 10, 73. [Google Scholar] [CrossRef] [PubMed]
- Udroiu, I.; Marinaccio, J.; Sgura, A. Inhibition of p53 and ATRX increases telomeric recombination in primary fibroblasts. FEBS Open Bio 2023, 13, 1683–1698. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Mangino, M.; Aviv, A.; UK10KConsortium Spector, T.; Durbin, R. Estimating telomere length from whole genome sequence data. Nucleic Acids Res. 2014, 42, e75. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Ghandi, M.; Huang, F.W. Integrated evaluation of telomerase activation and telomere maintenance across cancer cell lines. eLife 2021, 10, e66198. [Google Scholar] [CrossRef] [PubMed]
- Abid, H.Z.; McCaffrey, J.; Raseley, K.; Young, E.; Lassahn, K.; Varapula, D.; Riethman, H.; Xiao, M. Single-molecule analysis of subtelomeres and telomeres in Alternative Lengthening of Telomeres (ALT) cells. BMC Genom. 2020, 21, 485. [Google Scholar] [CrossRef] [PubMed]
- Alawi, F.; Lin, P.; Ziober, B.; Patel, R. Dyskerin expression correlates with active proliferation independently of telomerase. Head Neck 2010, 33, 1041. [Google Scholar] [CrossRef] [PubMed]
- Atri, S.; Nasoohi, N.; Hodjat, M. Azacitidine, as a DNMT inhibitor decreases hTERT gene expression and telomerase activity more effective compared with HDAC inhibitor in human head and neck squamous cell carcinoma cell lines. Curr. Mol. Pharmacol. 2021, 14, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Binz, N.; Shalaby, T.; Rivera, P.; Shin-Ya, K.; Grotzer, M.A. Telomerase inhibition, telomere shortening, cell growth suppression and induction of apoptosis by telomestatin in childhood neuroblastoma cells. Eur. J. Cancer 2005, 41, 2873–2881. [Google Scholar] [CrossRef] [PubMed]
- Bojovic, B.; Crowe, D.L. Resistance to telomerase inhibition by human squamous cell carcinoma cell lines. Int. J. Oncol. 2011, 38, 1175–1181. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bovée, J.V.; van den Broek, L.J.; Cleton-Jansen, A.M.; Hogendoorn, P.C. Chondrosarcoma is not characterized by detectable telomerase activity. J. Pathol. 2001, 193, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Brosnan-Cashman, J.A.; Yuan, M.; Graham, M.K.; Rizzo, A.J.; Myers, K.M.; Davis, C.; Zhang, R.; Esopi, D.M.; Raabe, E.H.; Eberhart, C.G.; et al. ATRX loss induces multiple hallmarks of the alternative lengthening of telomeres (ALT) phenotype in human glioma cell lines in a cell line-specific manner. PLoS ONE 2018, 13, e0204159. [Google Scholar] [CrossRef] [PubMed]
- Counter, C.M.; Hahn, W.C.; Wei, W.; Caddle, S.D.; Beijersbergen, R.L.; Lansdorp, P.M.; Sedivy, J.M.; Weinberg, R.A. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc. Natl. Acad. Sci. USA 1998, 95, 14723–14728. [Google Scholar] [CrossRef] [PubMed]
- Diplas, B.H.; He, X.; Brosnan-Cashman, J.A.; Liu, H.; Chen, L.H.; Wang, Z.; Moure, C.J.; Killela, P.J.; Loriaux, D.B.; Lipp, E.S.; et al. The genomic landscape of TERT promoter wildtype-IDH wildtype glioblastoma. Nat. Commun. 2018, 9, 2087. [Google Scholar] [CrossRef] [PubMed]
- Flynn, R.L.; Cox, K.E.; Jeitany, M.; Wakimoto, H.; Bryll, A.R.; Ganem, N.J.; Bersani, F.; Pineda, J.R.; Suvà, M.L.; Benes, C.H.; et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science 2015, 347, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, M.; Kamma, H.; Wu, W.; Yano, Y.; Homma, S.; Satoh, H. Alternative lengthening of telomeres in the human adrenocortical carcinoma cell line H295R. Int. J. Oncol. 2006, 29, 445–451. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Glaessl, A.; Bosserhoff, A.K.; Buettner, R.; Hohenleutner, U.; Landthaler, M.; Stolz, W. Increase in telomerase activity during progression of melanocytic cells from melanocytic naevi to malignant melanomas. Arch. Dermatol. Res. 1999, 291, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Gordon, K.E.; Ireland, H.; Roberts, M.; Steeghs, K.; McCaul, J.A.; MacDonald, D.G.; Parkinson, E.K. High levels of telomere dysfunction bestow a selective disadvantage during the progression of human oral squamous cell carcinoma. Cancer Res. 2003, 63, 458–467. [Google Scholar] [PubMed]
- Hou, M.; Xu, D.; Bjorkholm, M.; Gruber, A. Real-time quantitative telomeric repeat amplification protocol assay for the detection of telomerase activity. Clin. Chem. 2001, 47, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Bobb, D.; Lu, Y.; He, J.; Dome, J.S. Effect of telomerase inhibition on preclinical models of malignant rhabdoid tumor. Cancer Genet. 2014, 207, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Isaka, K.; Nishi, H.; Sagawa, Y.; Nakada, T.; Osakabe, Y.; Serizawa, H.; Ebihara, Y.; Takayama, M. Establishment of a new human cell line (EN) with TP53 mutation derived from endometrial carcinoma. Cancer Genet. Cytogenet. 2003, 141, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Khaw, A.K.; Silasudjana, M.; Banerjee, B.; Suzuki, M.; Baskar, R.; Hande, M.P. Inhibition of telomerase activity and human telomerase reverse transcriptase gene expression by histone deacetylase inhibitor in human brain cancer cells. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2007, 625, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.C.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Kleideiter, E.; Schwab, M.; Friedrich, U.; Koscielniak, E.; Schäfer, B.W.; Klotz, U. Telomerase activity in cell lines of pediatric soft tissue sarcomas. Pediatr. Res. 2003, 54, 718–723. [Google Scholar] [CrossRef] [PubMed]
- Kyo, S.; Kanaya, T.; Ishikawa, H.; Ueno, H.; Inoue, M. Telomerase activity in gynecological tumors. Clin. Cancer Res. 1996, 2, 2023–2028. [Google Scholar] [PubMed]
- Lee, J.C.; Jong, H.S.; Yoo, C.G.; Han, S.K.; Shim, Y.S.; Kim, Y.W. Telomerase activity in lung cancer cell lines and tissues. Lung Cancer 1998, 21, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Lim, S.; Viani, M.A.; Sapp, M.; Lim, M.S. Down-regulation of telomerase activity in malignant lymphomas by radiation and chemotherapeutic agents. Am. J. Pathol. 2001, 159, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Urquidi, V.; Wong, J.; Kleeman, J.; Goodison, S. Telomerase reverse transcriptase promoter regulation during myogenic differentiation of human RD rhabdomyosarcoma cells. Mol. Cancer Res. 2003, 1, 739–746. [Google Scholar] [PubMed]
- Maellaro, E.; Pacenti, L.; Del Bello, B.; Valentini, M.A.; Mangiavacchi, P.; De Felice, C.; Rubegni, P.; Luzi, P.; Miracco, C. Different effects of interferon-α on melanoma cell lines: A study on telomerase reverse transcriptase, telomerase activity and apoptosis. Br. J. Dermatol. 2003, 148, 1115–1124. [Google Scholar] [CrossRef] [PubMed]
- Mason-Osann, E.; Dai, A.; Floro, J.; Lock, Y.J.; Reiss, M.; Gali, H.; Matschulat, A.; Labadorf, A.; Flynn, R.L. Identification of a novel gene fusion in ALT positive osteosarcoma. Oncotarget 2018, 9, 32868–32880. [Google Scholar] [CrossRef] [PubMed]
- Milas, M.; Yu, D.; Sun, D.; Pollock, R.E. Telomerase activity of sarcoma cell lines and fibroblasts is independent of p53 status. Clin. Cancer Res. 1998, 4, 1573–1579. [Google Scholar] [PubMed]
- Mochida, A.; Gotoh, E.; Senpuku, H.; Harada, S.; Kitamura, R.; Takahashi, T.; Yanagi, K. Telomere size and telomerase activity in Epstein-Barr virus (EBV)-positive and EBV-negative Burkitt’s lymphoma cell lines. Arch. Virol. 2005, 150, 2139–2150. [Google Scholar] [CrossRef] [PubMed]
- Nishi, H.; Ohyashiki, K.; Fujito, A.; Yahata, N.; Ohyashiki, J.H.; Isaka, K.; Takayama, M. Expression of telomerase subunits and localization of telomerase activation in hydatidiform mole. Placenta 1999, 20, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Panier, S.; Maric, M.; Hewitt, G.; Mason-Osann, E.; Gali, H.; Dai, A.; Labadorf, A.; Guervilly, J.-H.; Ruis, P.; Segura-Bayona, S.; et al. SLX4IP antagonizes promiscuous BLM activity during ALT maintenance. Mol. Cell 2019, 76, 27–43. [Google Scholar] [CrossRef] [PubMed]
- Parsch, D.; Brassat, U.; Brümmendorf, T.H.; Fellenberg, J. Consequences of telomerase inhibition by BIBR1532 on proliferation and chemosensitivity of chondrosarcoma cell lines. Cancer Investig. 2008, 26, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Rha, S.Y.; Jeung, H.C.; Yang, W.I.; Kim, J.J.; Oh, T.J.; An, S.W.; Chung, H.C. Alteration of hTERT full-length variant expression level showed different gene expression profiles and genomic copy number changes in breast cancer. Oncol. Rep. 2006, 15, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Saretzki, G.; Petersen, S.; Petersen, I.; Kölble, K.; von Zglinicki, T. hTERT gene dosage correlates with telomerase activity in human lung cancer cell lines. Cancer Lett. 2002, 176, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Maehara, N.; Mizumoto, K.; Nagai, E.; Yasoshima, T.; Hirata, K.; Tanaka, M. Telomerase activity of cultured human pancreatic carcinoma cell lines correlates with their potential for migration and invasion. Cancer 2001, 91, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Shervington, A.; Patel, R.; Lu, C.; Cruickshanks, N.; Lea, R.; Roberts, G.; Dawson, T.; Shervington, L. Telomerase subunits expression variation between biopsy samples and cell lines derived from malignant glioma. Brain Res. 2007, 1134, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Tahara, H.; Kuniyasu, H.; Yokozaki, H.; Yasui, W.; Shay, J.W.; Ide, T.; Tahara, E. Telomerase activity in preneoplastic and neoplastic gastric and colorectal lesions. Clin. Cancer Res. 1995, 1, 1245–1251. [Google Scholar] [PubMed]
- Terasaki, T.; Kyo, S.; Takakura, M.; Maida, Y.; Tsuchiya, H.; Tomita, K.; Inoue, M. Analysis of telomerase activity and telomere length in bone and soft tissue tumors. Oncol. Rep. 2004, 11, 1307–1311. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ulaner, G.A.; Hoffman, A.R.; Otero, J.; Huang, H.Y.; Zhao, Z.; Mazumdar, M.; Gorlick, R.; Meyers, P.; Healey, J.H.; Ladanyi, M. Divergent patterns of telomere maintenance mechanisms among human sarcomas: Sharply contrasting prevalence of the alternative lengthening of telomeres mechanism in Ewing’s sarcomas and osteosarcomas. Genes Chromosomes Cancer 2004, 41, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Villa, R.; Folini, M.; Perego, P.; Supino, R.; Setti, E.; Daidone, M.G.; Zunino, F.; Zaffaroni, N. Telomerase activity and telomere length in human ovarian cancer and melanoma cell lines: Correlation with sensitivity to DNA damaging agents. Int. J. Oncol. 2000, 16, 995–1002. [Google Scholar] [CrossRef] [PubMed]
- Wege, H.; Chui, M.S.; Le, H.T.; Tran, J.M.; Zern, M.A. SYBR Green real-time telomeric repeat amplification protocol for the rapid quantification of telomerase activity. Nucleic Acids Res. 2002, 31, e3. [Google Scholar] [CrossRef] [PubMed]
- Yu, E.Y.; Cheung, I.Y.; Feng, Y.; Rabie, M.O.; Roboz, G.J.; Guzman, M.L.; Cheung, N.-K.V.; Lue, N.F. Telomere trimming and DNA damage as signatures of high risk neuroblastoma. Neoplasia 2019, 21, 689–701. [Google Scholar] [CrossRef] [PubMed]


| Pathway | Genes | Alterations |
|---|---|---|
| ATRX | Frequent damaging alterations in mesenchymal (42%) and neural cells (37%), much rarer in epithelial ones (10%) | |
| Telomeric integrity | WRN | Damaging alterations in epithelial (24%), mesenchymal (12%) and neural cells (11%) |
| STAG2 | Damaging alterations in mesenchymal (15%) and neural cells (16%), much rarer in epithelial ones (3%) | |
| SMARCAL1 | Damaging alterations in neural cells (13%), much rarer in mesenchymal cells (4%), absent in epithelial ones | |
| H3-3 deposition | DAXX, ASF1A,
H3-3A | Quite rare damage mutations in mesenchymal and neural cells (<7%), none in epithelial ones |
| TOP3A | Amplified in 11% of cell lines | |
| Cell proliferation | RB1 | Damaging alterations in 19% of cell lines |
| MYC | Amplified in 31% of cell lines | |
| P53 | TP53 | Frequent damaging alterations (68%) |
| MDM2 | Amplified in 13% of cell lines |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Udroiu, I.; Sgura, A. Alternative Lengthening of Telomeres: The Need for ATRX Mutations Is Lineage-Dependent. Int. J. Mol. Sci. 2025, 26, 6765. https://doi.org/10.3390/ijms26146765
Udroiu I, Sgura A. Alternative Lengthening of Telomeres: The Need for ATRX Mutations Is Lineage-Dependent. International Journal of Molecular Sciences. 2025; 26(14):6765. https://doi.org/10.3390/ijms26146765
Chicago/Turabian StyleUdroiu, Ion, and Antonella Sgura. 2025. "Alternative Lengthening of Telomeres: The Need for ATRX Mutations Is Lineage-Dependent" International Journal of Molecular Sciences 26, no. 14: 6765. https://doi.org/10.3390/ijms26146765
APA StyleUdroiu, I., & Sgura, A. (2025). Alternative Lengthening of Telomeres: The Need for ATRX Mutations Is Lineage-Dependent. International Journal of Molecular Sciences, 26(14), 6765. https://doi.org/10.3390/ijms26146765






