Genomic Insights into Vaccinium spp. Endophytes B. halotolerans and B. velezensis and Their Antimicrobial Potential
Abstract
1. Introduction
2. Results
2.1. Antagonistic Activity and Nutritional Preferences Test In Vitro
2.2. Genetic Characteristics
2.3. Horizontal Gene Transfer
3. Discussion
4. Materials and Methods
4.1. Microorganisms
4.2. Antagonism Evaluation In Vitro
4.3. Nutritional Preferences Test In Vitro
4.4. Genomic DNA Extraction, Whole-Genome Sequencing, Annotation
4.5. Comparative Genomic and Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial endophyte colonization and distribution within plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef] [PubMed]
- Tran, C.; Cock, I.E.; Chen, X.; Feng, Y. Antimicrobial Bacillus: Metabolites and their mode of action. Antibiotics 2022, 11, 88. [Google Scholar] [CrossRef]
- Kolytaitė, A.; Vaitiekūnaitė, D.; Antanynienė, R.; Baniulis, D.; Frercks, B. Monilinia fructigena-suppressing and plant growth-promoting endophytic Pseudomonas spp. bacteria isolated from plum. Microorganisms 2022, 10, 2402. [Google Scholar] [CrossRef]
- Su, Z.; Liu, G.; Liu, X.; Li, S.; Lu, X.; Wang, P.; Zhao, W.; Zhang, X.; Dong, L.; Qu, Y.; et al. Functional analyses of the Bacillus velezensis HMB26553 genome provide evidence that its genes are potentially related to the promotion of plant growth and prevention of cotton Rhizoctonia damping-off. Cells 2023, 12, 1301. [Google Scholar] [CrossRef] [PubMed]
- Kolytaitė, A.; Mažeikienė, I.; Kurgonaitė, M.; Raklevičiūtė, S.; Paškevičiūtė, G.; Frercks, B. Unlocking nature’s microbial defenders: Genetic mechanisms and potential against Monilinia spp. pathogens. Microorganisms 2025, 13, 818. [Google Scholar] [CrossRef] [PubMed]
- Islam, T.; Danishuddin; Tamanna, N.T.; Matin, M.N.; Barai, H.R.; Haque, M.A. Resistance mechanisms of plant pathogenic fungi to fungicide, environmental impacts of fungicides, and sustainable solutions. Plants 2024, 13, 2737. [Google Scholar] [CrossRef]
- Morales-Cedeño, L.R.; Orozco-Mosqueda, M.C.; Loeza-Lara, P.D.; Parra-Cota, F.I.; De Los Santos-Villalobos, S.; Santoyo, G. Plant growth-promoting bacterial endophytes as biocontrol agents of pre- and post-harvest diseases: Fundamentals, methods of application and future perspectives. Microbiol. Res. 2021, 242, 126612. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Hartmann, A.; Gao, X.; Borriss, R. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42—A review. Front. Microbiol. 2015, 6, 780. [Google Scholar] [CrossRef]
- Zhou, Y.; Ma, X. Genomic insights into biocontrol mechanisms of endophytic Bacillus velezensis against fungal diseases. Biol. Control. 2022, 173, 104986. [Google Scholar] [CrossRef]
- Iqbal, S.; Begum, F.; Rabaan, A.A.; Aljeldah, M.; Al Shammari, B.R.; Alawfi, A.; Alshengeti, A.; Sulaiman, T.; Khan, A. Classification and Multifaceted Potential of Secondary Metabolites Produced by Bacillus subtilis Group: A Comprehensive Review. Molecules 2023, 28, 927. [Google Scholar] [CrossRef]
- Zhong, X.; Jin, Y.; Ren, H.; Hong, T.; Zheng, J.; Fan, W.; Hong, J.; Chen, Z.; Wang, A.; Lu, H.; et al. Research progress of Bacillus velezensis in plant disease resistance and growth promotion. Front. Ind. Microbiol. 2024, 2, 1442980. [Google Scholar] [CrossRef]
- Taulavuori, K.; Laine, E.; Taulavuori, E. Experimental studies on Vaccinium myrtillus and Vaccinium vitis-idaea in relation to air pollution and global change at northern high latitudes: A review. Environ. Exp. Bot. 2013, 87, 191–196. [Google Scholar] [CrossRef]
- Zhou, C.; Zhang, T.; Duan, J.; Wu, M. Endophytic bacteria isolated from blueberry (Vaccinium spp.) show potential as biocontrol agents against Monilinia vaccinii-corymbosi. Biol. Control. 2020, 143, 104188. [Google Scholar] [CrossRef]
- Karlowski, W.M.; Varshney, D.; Zielezinski, A. Taxonomically restricted genes in Bacillus may form clusters of homologs and can be traced to a large reservoir of noncoding sequences. Genome Biol. Evol. 2023, 15, evad023. [Google Scholar] [CrossRef]
- Kalkreuter, E.; Pan, G.; Cepeda, A.J.; Shen, B. Targeting Bacterial Genomes for Natural Product Discovery. Trends Pharmacol. Sci. 2020, 41, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Langille, M.G.I. Current and Promising Approaches to Identify Horizontal Gene Transfer Events in Metagenomes. Genome Biol. Evol. 2019, 11, 2750–2766. [Google Scholar] [CrossRef]
- Mažeikienė, I.; Frercks, B.; Burokienė, D.; Mačionienė, I.; Šalaševičienė, A. Endophytic community composition and genetic-enzymatic features of cultivable bacteria in Vaccinium myrtillus L. in forests of the Baltic-Nordic region. Forests 2021, 12, 1647. [Google Scholar] [CrossRef]
- Vaitiekūnaitė, D.; Bružaitė, I.; Snitka, V. Endophytes from blueberry (Vaccinium) fruit: Characterization of yeast and bacteria via label-free surface-enhanced Raman spectroscopy (SERS). Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 275, 121158. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef]
- Danevčič, T.; Dragoš, A.; Spacapan, M.; Stefanic, P.; Dogsa, I.; Mandic-Mulec, I. Surfactin facilitates horizontal gene transfer in Bacillus subtilis. Front. Microbiol. 2021, 12, 657407. [Google Scholar] [CrossRef]
- Wang, X.; Leptihn, S. Defense and anti-defense mechanisms of bacteria and bacteriophages. J. Zhejiang Univ. Sci. B 2024, 25, 181–196. [Google Scholar] [CrossRef] [PubMed]
- Drożdżyński, P.; Rutkowska, N.; Rodziewicz, M.; Marchut-Mikołajczyk, O. Bioactive Compounds Produced by Endophytic Bacteria and Their Plant Hosts—An Insight into the World of Chosen Herbaceous Ruderal Plants in Central Europe. Molecules 2024, 29, 4456. [Google Scholar] [CrossRef] [PubMed]
- Al-Hawamdeh, F.; Ayad, J.Y.; Alananbeh, K.M.; Akash, M.W. Bacterial endophytes and their contributions to alleviating drought and salinity stresses in wheat: A systematic review of physiological mechanisms. Agriculture 2024, 14, 769. [Google Scholar] [CrossRef]
- Alshaikh, S.A.; El-Banna, T.; Sonbol, F.; Farghali, M.H. Correlation between antimicrobial resistance, biofilm formation, and virulence determinants in uropathogenic Escherichia coli from Egyptian hospital. Ann. Clin. Microbiol. Antimicrob. 2024, 23, 20. [Google Scholar] [CrossRef]
- Golnari, M.; Bahrami, N.; Milanian, Z.; Khorasgani, M.R.; Asadollahi, M.A.; Shafiei, R.; Fatemi, S.S.-A. Isolation and characterization of novel Bacillus strains with superior probiotic potential: Comparative analysis and safety evaluation. Sci. Rep. 2024, 14, 1457. [Google Scholar] [CrossRef]
- Kang, K.; Niu, Z.; Zhang, W.; Wei, S.; Lv, Y.; Hu, Y. Antagonistic strain Bacillus halotolerans Jk-25 mediates the biocontrol of wheat common root rot caused by Bipolaris sorokiniana. Plants 2023, 12, 828. [Google Scholar] [CrossRef]
- Do, T.Q.; Nguyen, T.T.; Dinh, V.M. Application of endophytic bacterium Bacillus velezensis BTR11 to control bacterial leaf blight disease and promote rice growth. Egypt. J. Biol. Pest Control 2023, 33, 97. [Google Scholar] [CrossRef]
- Rabbee, M.F.; Baek, K.H. Antimicrobial activities of lipopeptides and polyketides of Bacillus velezensis for agricultural applications. Molecules 2020, 25, 4973. [Google Scholar] [CrossRef]
- Liu, X.; Zhu, H.; Biswas, S.; Cheng, Y.Q. Improved production of cytotoxic thailanstatins A and D through metabolic engineering of Burkholderia thailandensis MSMB43 and pilot scale fermentation. Synth. Syst. Biotechnol. 2016, 1, 34–38. [Google Scholar] [CrossRef]
- Wei, Z.; Shan, C.; Zhang, L.; Ge, D.; Wang, Y.; Xia, X.; Liu, X.; Zhou, J. A novel subtilin-like lantibiotics subtilin JS-4 produced by Bacillus subtilis JS-4, and its antibacterial mechanism against Listeria monocytogenes. LWT 2021, 142, 110993. [Google Scholar] [CrossRef]
- Alajlani, M.M. Characterization of subtilosin gene in wild type Bacillus spp. and possible physiological role. Sci. Rep. 2022, 12, 10521. [Google Scholar] [CrossRef]
- Glick, B.R. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef] [PubMed]
- Vurukonda, S.S.K.P.; Vardharajula, S.; Shrivastava, M.; SkZ, A. Enhancement of drought stress tolerance in crops by plant growth-promoting rhizobacteria. Microbiol. Res. 2016, 184, 13–24. [Google Scholar] [CrossRef]
- Ahmad, H.M.; Fiaz, S.; Hafeez, S.; Zahra, S.; Shah, A.N.; Gul, B.; Aziz, O.; Mahmood-Ur-Rahman; Fakhar, A.; Rafique, M.; et al. Plant growth-promoting rhizobacteria eliminate the effect of drought stress in plants: A review. Front. Plant Sci. 2022, 13, 875774. [Google Scholar] [CrossRef]
- Liu, K.; Deng, F.; Zeng, F.; Chen, Z.H.; Qin, Y.; Chen, G. Plant growth-promoting rhizobacteria improve drought tolerance of crops: A review. Plant Growth Regul. 2025, 105, 567–581. [Google Scholar] [CrossRef]
- Ryu, C.M.; Farag, M.A.; Hu, C.H.; Reddy, M.S.; Wei, H.X.; Pare, P.W.; Kloepper, J.W. Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 4927–4932. [Google Scholar] [CrossRef] [PubMed]
- Brinsmade, S.R.; Alexander, E.L.; Livny, J.; Stettner, A.I.; Segrè, D.; Rhee, K.Y.; Sonenshein, A.L. Hierarchical expression of genes controlled by the Bacillus subtilis global regulatory protein CodY. Proc. Natl. Acad. Sci. USA 2014, 111, 8227–8232. [Google Scholar] [CrossRef]
- Rasiukevičiūtė, N.; Morkeliūnė, A.; Mažeikienė, I.; Lanauskas, J.; Valiuškaitė, A. Alternative Plant Protection Strategies Using Bacteria and Thyme to Improve Strawberry (cv. Elsanta) Yield and Quality. Plants 2025, 14, 1827. [Google Scholar] [CrossRef]
- Tiwari, P.; Bae, H. Horizontal gene transfer and endophytes: An implication for the acquisition of novel traits. Plants 2020, 9, 305. [Google Scholar] [CrossRef]
- Frost, L.S.; Leplae, R.; Summers, A.O.; Toussaint, A. Mobile genetic elements: The agents of open source evolution. Nat. Rev. Microbiol. 2005, 3, 722–732. [Google Scholar] [CrossRef]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Slomka, S.; Françoise, I.; Hornung, G.; Asraf, O.; Biniashvili, T.; Pilpel, Y.; Dahan, O. Experimental evolution of Bacillus subtilis reveals the evolutionary dynamics of horizontal gene transfer and suggests adaptive and neutral effects. Genetics 2020, 216, 543–558. [Google Scholar] [CrossRef]
- Nguyen, M.P.; Lehosmaa, K.; Martz, F.; Koskimäki, J.J.; Toth, K.; Ahonen, S.H.K.; Häggman, H.; Pirttilä, A.M. Dynamics of fungal endophytic communities in bilberry (Vaccinium myrtillus L.) fruits through development is shaped by host phenolic compounds. FEMS Microbiol. Ecol. 2025, 101, fiae168. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Zdobnov, E.M. BUSCO: Assessing genomic data quality and beyond. Curr. Protoc. 2021, 1, e323. [Google Scholar] [CrossRef] [PubMed]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Vader, L.; Szenei, J.; Reitz, Z.L.; Augustijn, H.E.; Cediel-Becerra, J.D.D.; de Crécy-Lagard, V.; Koetsier, R.A.; Williams, S.E.; et al. antiSMASH 8.0: Extended gene cluster detection capabilities and analyses of chemistry, enzymology, and regulation. Nucleic Acids Res. 2025, 53, W32–W38. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Ferrer Florensa, A.; Almagro Armenteros, J.J.; Kaas, R.S.; Clausen, P.T.; Nielsen, H.; Rost, B.; Aarestrup, F.M. Whole-genome prediction of bacterial pathogenic capacity on novel bacteria using protein language models, with PathogenFinder2. bioRxiv 2025, 648497. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Vernikos, G.S.; Parkhill, J. Interpolated variable order motifs for identification of horizontally acquired DNA: Revisiting the Salmonella pathogenicity islands. Bioinformatics 2006, 22, 2196–2203. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
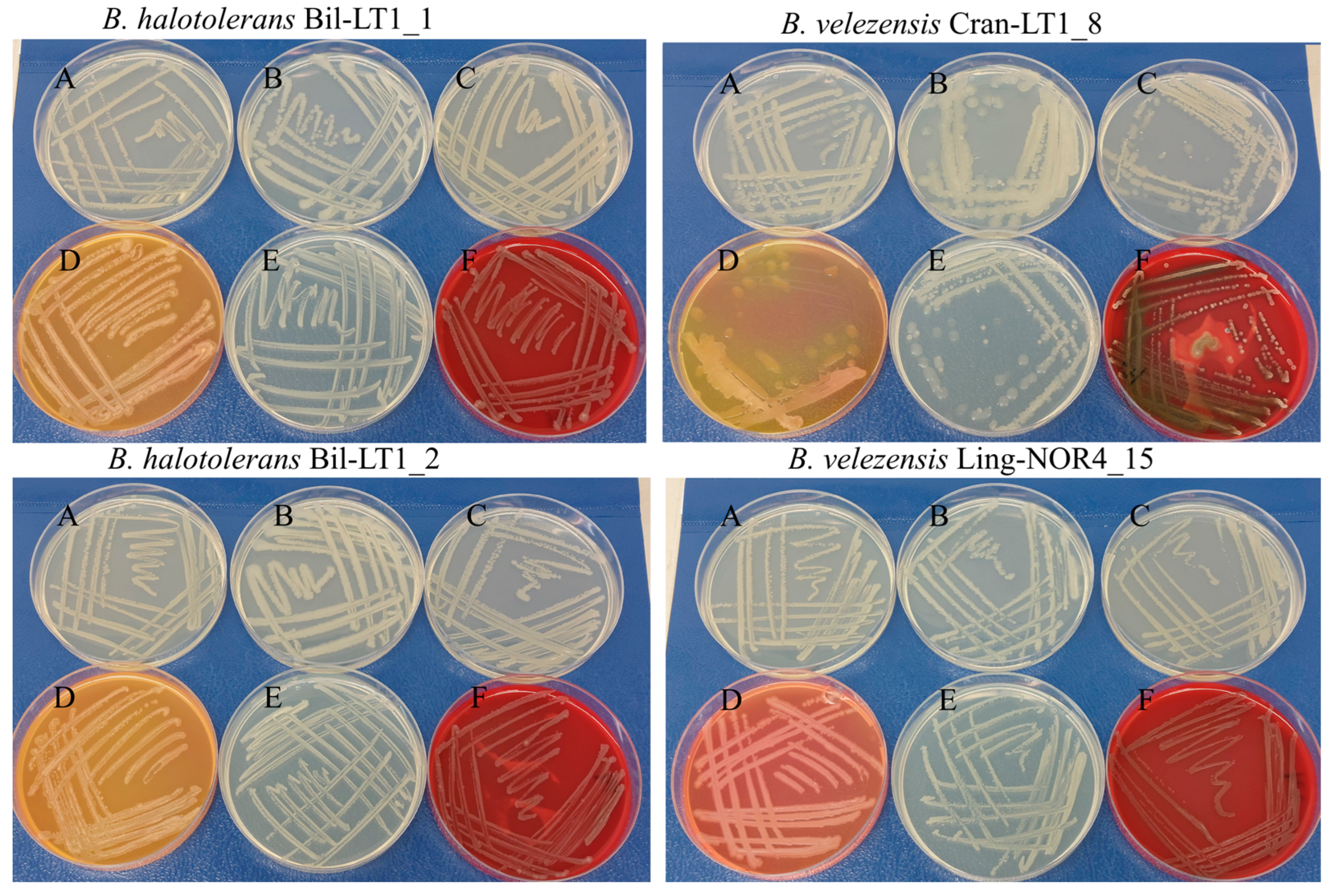
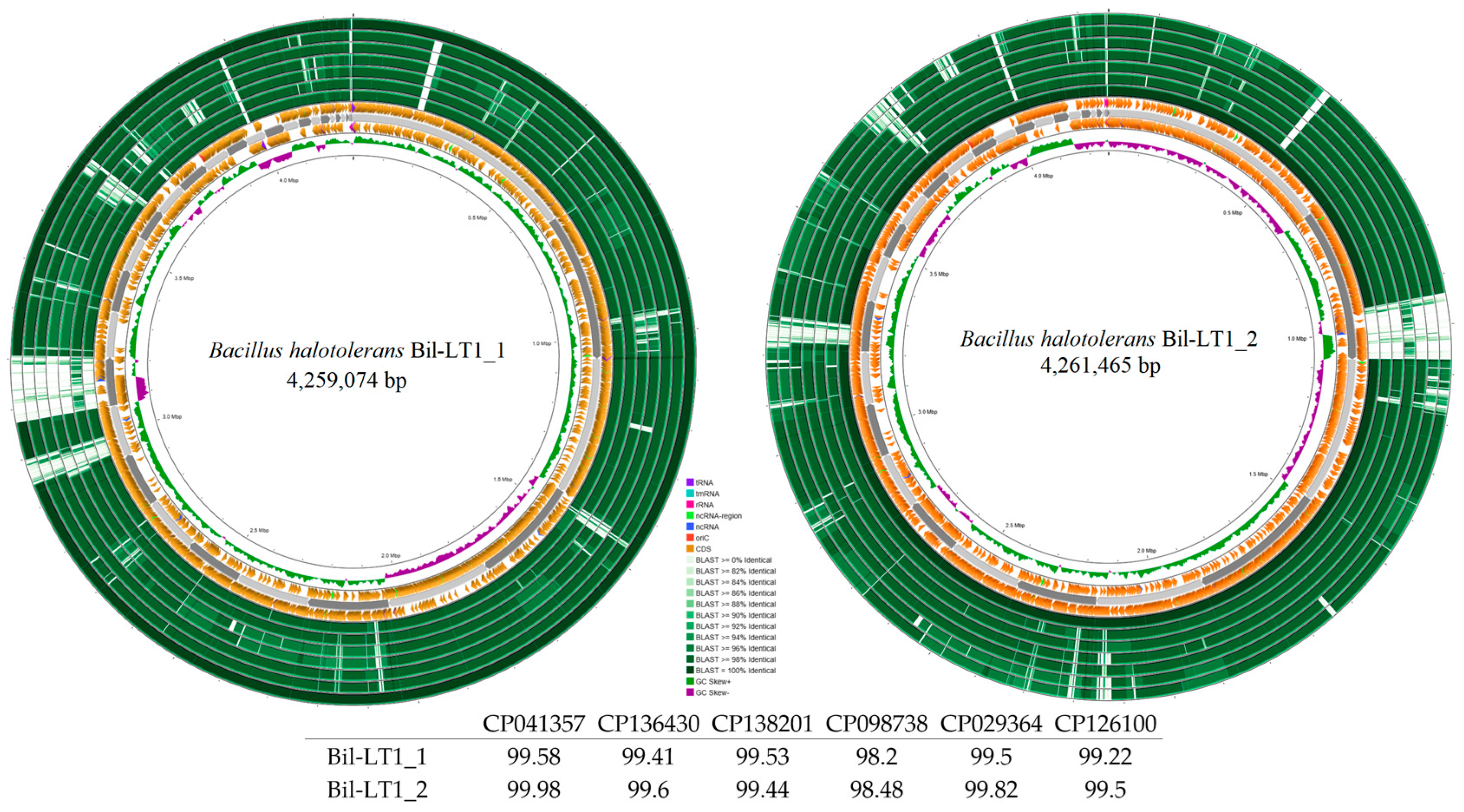

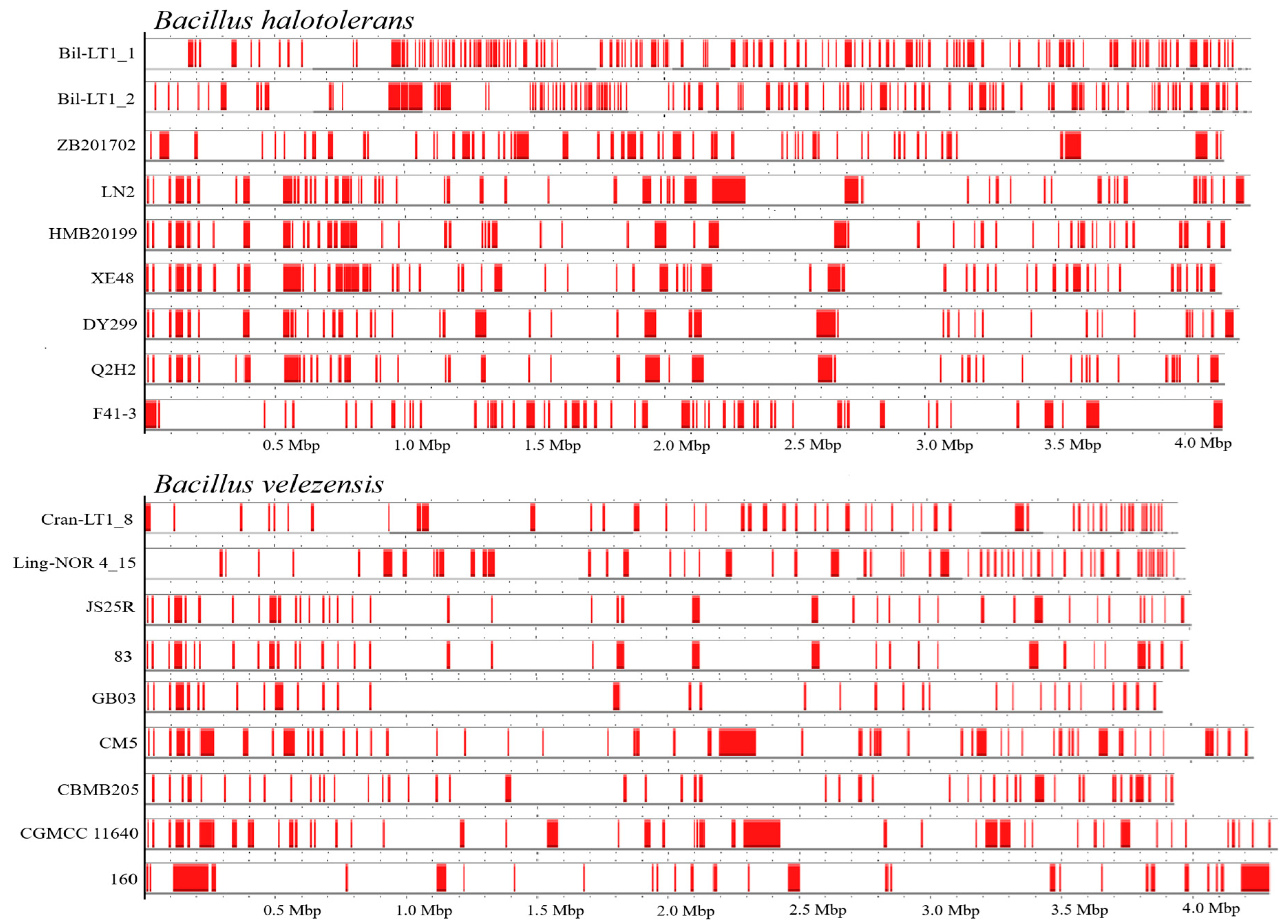
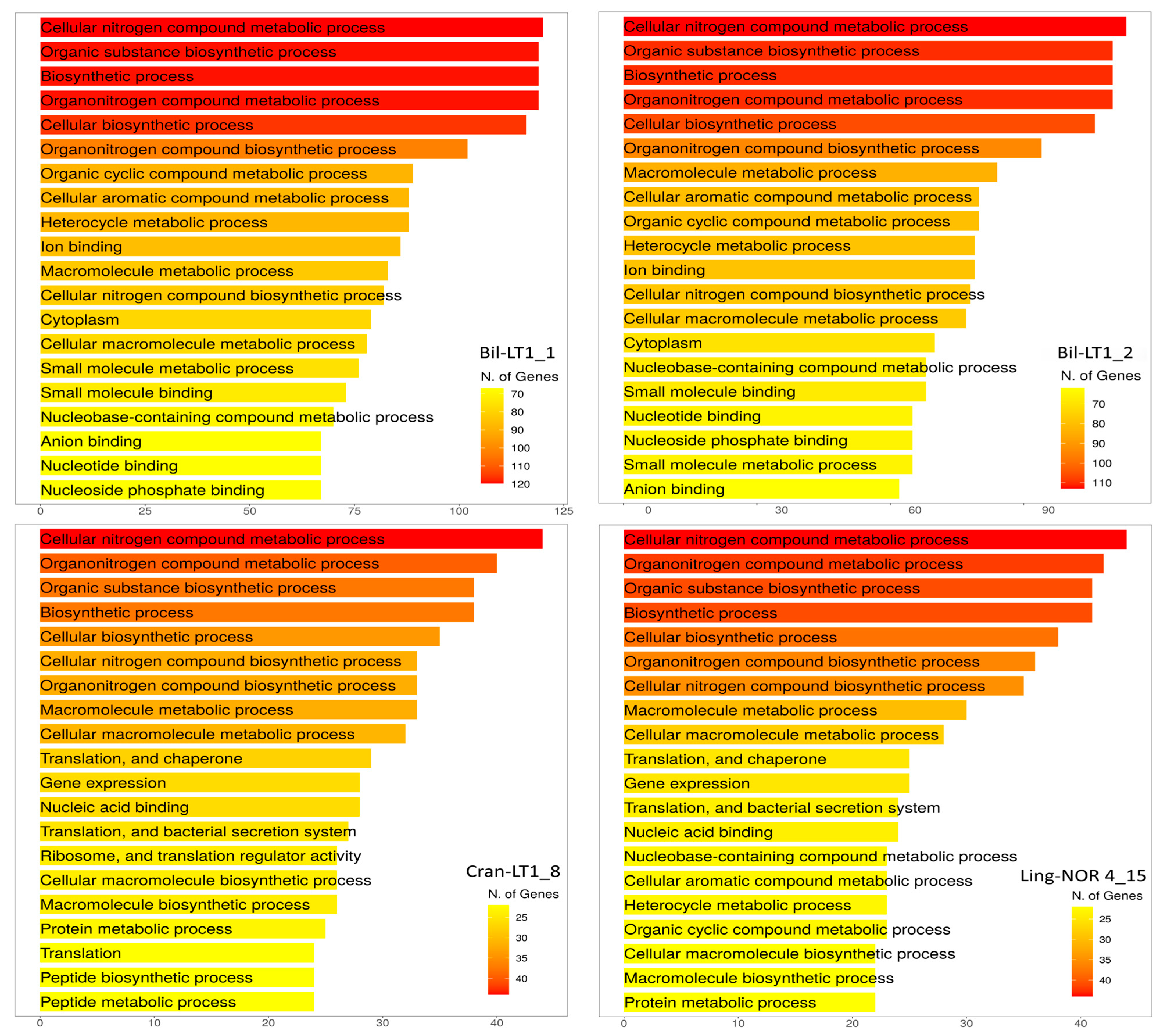
| Strain | Bil-LT1_1 | Bil-LT1_2 | Cran-LT1_8 | Ling-NOR4_15 |
|---|---|---|---|---|
| Antifungal Activity, Growth Suppression (%) | ||||
| B. cinerea T11B_BRA_189 | 52.4 ± 1.7 | 69.1 ± 1.5 | 79.7 ± 1.1 | 48.5 ± 0.6 |
| F. culmorum LT_1V23b | 23.3 ± 0.7 | 19.2 ± 0.7 | 26.9 ± 0.5 | 29.7 ± 0.9 |
| M. fructicola CBS 101512 | 50.6 ± 1.5 | 56.2 ± 0.3 | 59.4 ± 1.7 | 56.4 ± 0.8 |
| M. frutigena MFS01 | 68.7 ± 1.8 | 99.1 ± 0.0 | 91.7 ± 0.3 | 77.5 ± 0.6 |
| M. laxa CBS 489.50 | 61.2 ± 1.0 | 59.7 ± 1.1 | 84.8 ± 0.5 | 68.8 ± 1.5 |
| Antibacterial Activity, Inhibition Zone (mm) | ||||
| B. cereus ATCC 11778 | 10.0 ± 0.1 | 10.0 ± 0.1 | 16.0 ± 0.1 | 16.0 ± 0.1 |
| E. coli ATCC 25922 | N/A | N/A | 12.0 ± 0.1 | N/A |
| L. monocytogenes ATCC 13932 | 11.0 ± 0.2 | 12.0 ± 0.2 | 16.0 ± 0.1 | 16.0 ± 0.1 |
| S. enterica subsp. enterica serovar Typhimurium ATCC 13932 | N/A | N/A | 14.0 ± 0.1 | 11.0 ± 0.0 |
| S. aureus subsp. Aureus ATCC 25923 | 14.0 ± 0.3 | 11.0 ± 0.1 | 18.0 ± 0.3 | 14.0 ± 0.1 |
| Growth on media * | Good on all tested media (LB, PCA, TSA, NA, MYP) | Dense colonies on PCA; good on other media | Large, slightly mucoid colonies on PCA; low density on TSA; poor on NA | Uniform growth on NA; pink hue on MYP |
| Hemolysis on SBA | Beta | Beta | Alpha/Beta | Beta |
| Mannitol fermentation on MYP | Positive (orange/yellow shift) | Positive (orange/yellow shift) | Weak/partial (yellowish; inhibited growth) | Negative (pink hue) |
| Pigment production | Yellow on TSA | None observed | None observed | None observed |
| Isolated B. halotolerans | Isolated B. velezensis | |||
|---|---|---|---|---|
| Bil-LT1_1 | Bil-LT1_2 | Cran-LT1_8 | Ling-NOR4_15 | |
| SAMN36292479 | SAMN36292480 | SAMN36292481 | SAMN36292482 | |
| Statistics of Sequencing Data | ||||
| Raw reads | 4,383,886 | 4,967,478 | 4,499,288 | 4,914,838 |
| Raw data (G) | 1.1 | 1.2 | 1.1 | 1.2 |
| Clean data (G) | 1 | 1 | 1 | 1.1 |
| Effective (%)/Error (%) | 91.01/0.04 | 85.39/0.03 | 85.18/0.04 | 86.38/0.04 |
| GC (%) | 43.3 | 43.3 | 46.3 | 46.3 |
| Assembly Information | ||||
| Total reads | 4,259,074 | 4,261,465 | 4,170,815 | 4,593,628 |
| Number of contigs | 39 | 34 | 28 | 21 |
| Largest contig (bp) | 646,884 | 646,885 | 940,386 | 941,931 |
| Mapping Statistics to the Reference Genomes | ||||
| CP029364 (NCBI) | CP119675 (NCBI) | |||
| Average nucleotide identity (ANI), % | 99.50 | 99.82 | 98.01 | 97.95 |
| Annotation Information | ||||
| Bakta annotation | 4472 | 4472 | 3962 | 4025 |
| CDSs | 4313 | 4313 | 3805 | 3897 |
| tRNAs; tmRNAs; rRNAs | 62; 1; 4 | 62; 1; 4 | 73; 1; 3 | 77; 1; 3 |
| ncRNAs | 28 | 28 | 19 | 21 |
| ncRNA regions | 62 | 62 | 59 | 60 |
| Proteins with KEGG pathway assignments | 2457 | 2458 | 2315 | 2317 |
| Specialty Genes | ||||
| Antibiotic resistance (CARD) | 13 | 13 | 7 | 7 |
| Virulence factors (VFBDs) | 39 | 39 | 38 | 38 |
| Biosynthetic gene clusters (BGCs) | 14 | 14 | 20 | 21 |
| Biosynthesis of antibiotics | 5 | 5 | 3 | 2 |
| Bacterial Pathogenic Capacity Prediction | ||||
| Mean (std) | 0.0942 (0.0204) | 0.0825 (0.0181) | 0.0514 (0.0163) | 0.0511 (0.0191) |
| Bacteria Strain | Clusters for Secondary Metabolites Synthesis |
|---|---|
| Bil-LT1_1, Bil-LT1_2 Cran-LT1_8, Ling-NOR 4_15 | plipastatin, bacillaene, teichuronic acid, chejuenolide A/chejuenolide B, K53 capsular polysaccharide, bacillomycin D, surfactin, dipeptide aldehydes, fengycin, bacillibactin, bacilysin |
| Bil-LT1_1, Bil-LT1_2 | thailanstatin A, subtilosin A |
| Cran-LT1_8, Ling-NOR 4_15 | butirosin A/butirosin B, molybdenum cofactor, difficidin, macrolactin H |
| Ling-NOR 4_15 | plantazolicin |
| Cran-LT1_8 | mersacidin, rhizomide A/rhizomide B/rhizomide C |
| Function | B. halotolerans | B. velezensis | ||
|---|---|---|---|---|
| Bil-LT1_1 | Bil-LT1_2 | Cran-LT1_8 | Ling-NOR 4_15 | |
| Genes | ||||
| Phosphate metabolism | pstB3, pstA | |||
| pstS | - | pstS | pstS | |
| Nitrogen metabolism | nasE, nasD, rocG | |||
| Nitrogen fixation | nifS, salA, sufU | |||
| Siderophore | fhuD, pchA | |||
| - | fhuG | - | - | |
| Phytohormone production | trpA, trpB, trpF, trpC, trpD, trpE, amiE, aldH | |||
| Hydrolase | amyE, eglS, gmuD, ganB | |||
| Chitinase activity | sleL, ydhD | |||
| Biofilm | tasA, bslA | |||
| bslB | bslB | - | bslB | |
| 2,3-butanediol | alsD, ilvK, ilvE, ilvA, ilvD, ilvC, ilvH, ilvB | |||
| Access. No. in NCBI and Strain Name | HGTEs Size Range bp | No of HGTEs | Information About Isolate | Genome Size, bp | |
|---|---|---|---|---|---|
| Bacillus halotolerans | |||||
| SAMN36292479 | Bil-LT1_1 | 5000–37,501 | 123 | Bilberry leaves, Lithuania | 4,259,074 |
| SAMN36292480 | Bil-LT1_2 | 5000–52,501 | 107 | Bilberry leaves, Lithuania | 4,261,465 |
| CP029364 | ZB201702 | 5131–63,099 | 61 | Drought and salt rhizosphere soil of maize, China: Inner Mongolia | 4,154,245 |
| CP126100 | LN2 | 4055–128,846 | 58 | Rhizosphere soil, China: Longnan, Gansu Province | 4,252,134 |
| CP110264 | HMB20199 | 4640–47,395 | 54 | Plant, against Pseudoperonospora cubensis Rostow, China: Baoding | 4,175,808 |
| CP098738 | XE48 | 6651–66,933 | 62 | Marine low temperature surface sediment, West Indian Ocean | 4,140,100 |
| CP138201 | DY299 | 5023–74,450 | 46 | Soil, China: Inner Mongolia | 4,208,811 |
| CP136430 | Q2H2 | 5970–56,767 | 49 | Endophytic bacteria of potato root with strong antagonisticactivity, China: Inner Mongolia | 4,155,130 |
| CP041357 | F41-3 | 5001–50,720 | 55 | Flower of Chiness redbud, South Korea: Jeon-buk | 4,144,458 |
| Bacillus velezensis | |||||
| SAMN36292481 | Cran-LT1_8 | 5001–35,001 | 50 | Cranberry leaves, Lithuania | 3,958,789 |
| SAMN36292482 | Ling-NOR 4_15 | 5001–35,001 | 56 | Lingonberry leaves, Norway | 3,987,686 |
| CP009679 | JS25R | 3864–33,478 | 41 | Spikelet of wheat heads, with biocontrol activity China: Beijing | 4,006,002 |
| CP119675 | 160 | 5156–135,917 | 28 | Rhizosphere soil of corn crops, control agent of corn head smut, Mexico | 4,296,610 |
| CP011937 | CBMB205 | 4737–36,609 | 48 | Rice rhizosphere soil, South Korea: Cheongwon | 3,929,792 |
| CP026610 | CGMCC 11640 | 5090–142,965 | 50 | Bamboo forest soil, against Botryosphaeria dothidea, China:Tianmu Mountain | 4,322,979 |
| CP168028 | CM5 | 3808–140,634 | 56 | Peat casing layer, biocontrol in the mushroom microcosm Spain: Rioja | 4,240,819 |
| CP049904 | GB03 | 4777–34,819 | 33 | Biological Control Product Kodiak TM, Korea | 3,894,540 |
| CP034203 | 83 | 3879–36,447 | 35 | Biological Control Product Fungifree AB™, Korea | 3,997,902 |
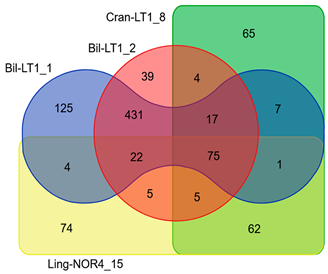 | List Strains | Total Number of CDS | Number of Annotated Genes | Antibiotic Resistance Genes | Virulence Factors |
| Bil-LT1_1 | 1155 | 682 | satA_Bs, rphC | 14 * | |
| Bil-LT1_2 | 1095 | 598 | satA_Bs, rphC | 13 ** | |
| Cran-LT1_8 | 525 | 236 | rphC, tet(L), satA_Bs | cheY | |
| Ling-NOR4_15 | 605 | 248 | satA_Bs, tet(L), clbA, rphC | dhbF, mbtH |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mažeikienė, I.; Frercks, B.; Kurgonaitė, M.; Rasiukevičiūtė, N.; Mačionienė, I. Genomic Insights into Vaccinium spp. Endophytes B. halotolerans and B. velezensis and Their Antimicrobial Potential. Int. J. Mol. Sci. 2025, 26, 6677. https://doi.org/10.3390/ijms26146677
Mažeikienė I, Frercks B, Kurgonaitė M, Rasiukevičiūtė N, Mačionienė I. Genomic Insights into Vaccinium spp. Endophytes B. halotolerans and B. velezensis and Their Antimicrobial Potential. International Journal of Molecular Sciences. 2025; 26(14):6677. https://doi.org/10.3390/ijms26146677
Chicago/Turabian StyleMažeikienė, Ingrida, Birutė Frercks, Monika Kurgonaitė, Neringa Rasiukevičiūtė, and Irena Mačionienė. 2025. "Genomic Insights into Vaccinium spp. Endophytes B. halotolerans and B. velezensis and Their Antimicrobial Potential" International Journal of Molecular Sciences 26, no. 14: 6677. https://doi.org/10.3390/ijms26146677
APA StyleMažeikienė, I., Frercks, B., Kurgonaitė, M., Rasiukevičiūtė, N., & Mačionienė, I. (2025). Genomic Insights into Vaccinium spp. Endophytes B. halotolerans and B. velezensis and Their Antimicrobial Potential. International Journal of Molecular Sciences, 26(14), 6677. https://doi.org/10.3390/ijms26146677








