Gray-Horse Melanoma—A Wolf in Sheep’s Clothing
Abstract
1. Introduction
2. Results
2.1. RNAseq Reveals Deregulated Gene Transcription That Notably Affects ECM-Associated Pathways in ghM
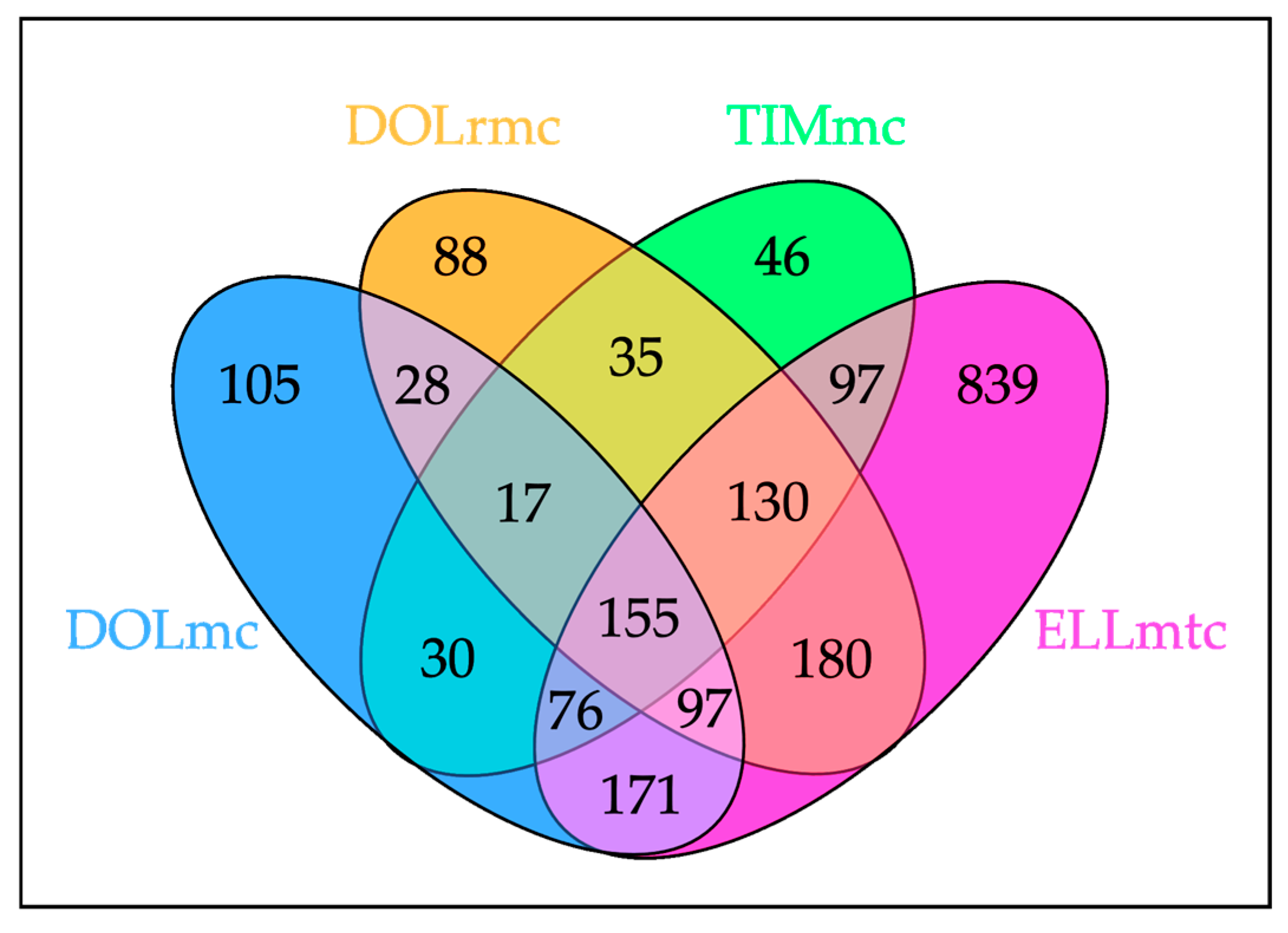
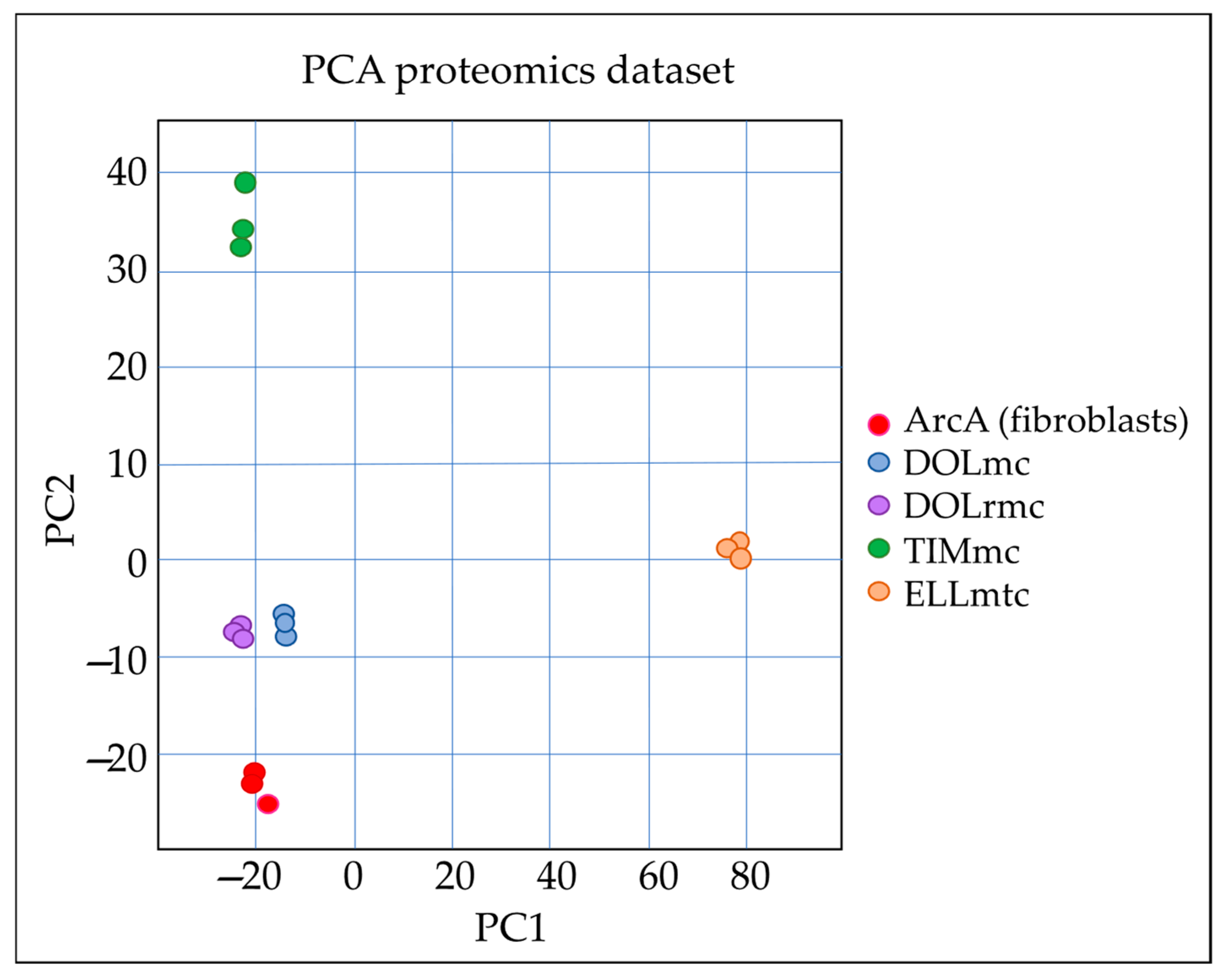
2.2. Proteome Analysis Unambiguously Identifies ghM as a Malignant Disease
3. Discussion
4. Materials and Methods
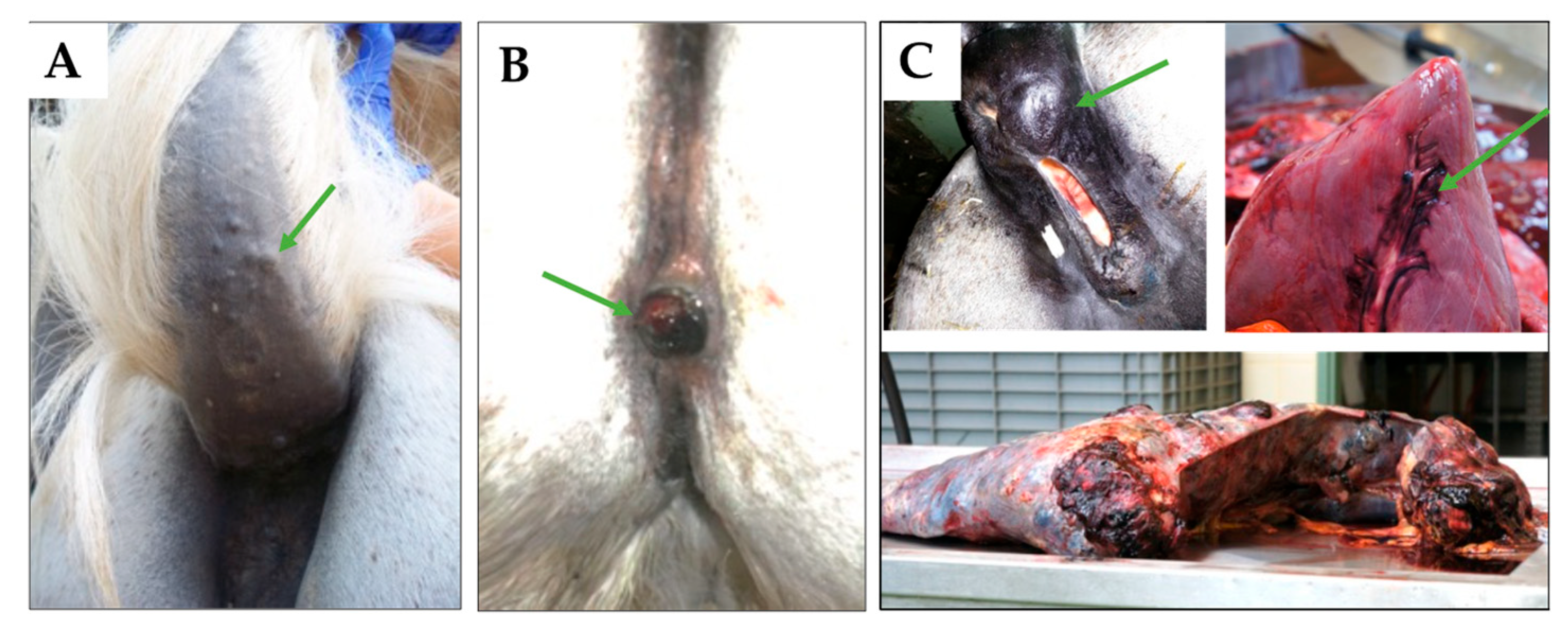
4.1. Sample Material
4.2. Transcriptome Analysis in Gray-Horse Melanoma Versus Intact Skin
4.3. Proteome Analysis in ghM Cells Versus Fibroblasts
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MM | malignant melanoma |
| hMM | human malignant melanoma |
| ghM | gray-horse melanoma |
| VM | vasculogenic mimicry |
| RNAseq | RNA sequencing |
| FDR | false discovery rate |
| RPKM | reads per kilobase million |
References
- Berwick, M.; Buller, D.B.; Cust, A.; Gallagher, R.; Lee, T.K.; Meyskens, F.; Pandey, S.; Thomas, N.E.; Veierod, M.B.; Ward, S. Melanoma Epidemiology and Prevention. Cancer Treat. Res. 2016, 167, 17–49. [Google Scholar] [CrossRef]
- Butler, A.P.; Trono, D.; Beard, R.; Fraijo, R.; Nairn, R.S. Melanoma susceptibility and cell cycle genes in Xiphophorus hybrids. Mol. Carcinog. 2007, 46, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.; Wanke, R.; Distl, O. Inheritance of melanocytic lesions and their association with the white colour phenotype in miniature swine. J. Anim. Breed. Genet. 2001, 118, 275–283. [Google Scholar] [CrossRef]
- Bourneuf, E. The MeLiM Minipig: An Original Spontaneous Model to Explore Cutaneous Melanoma Genetic Basis. Front. Genet. 2017, 8, 146. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.W.; Miller, W.H.J. Melanoma. In Equine Dermatology, 2nd ed.; Elsevier Saunders: Maryland Heights, MO, USA, 2011; pp. 506–508. [Google Scholar]
- Seltenhammer, M.H.; Simhofer, H.; Scherzer, S.; Zechner, R.; Curik, I.; Solkner, J.; Brandt, S.M.; Jansen, B.; Pehamberger, H.; Eisenmenger, E. Equine melanoma in a population of 296 grey Lipizzaner horses. Equine Vet. J. 2003, 35, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Valentine, B.A. Equine melanocytic tumors: A retrospective study of 53 horses (1988 to 1991). J. Vet. Intern. Med. 1995, 9, 291–297. [Google Scholar] [CrossRef]
- Rosengren Pielberg, G.; Golovko, A.; Sundstrom, E.; Curik, I.; Lennartsson, J.; Seltenhammer, M.H.; Druml, T.; Binns, M.; Fitzsimmons, C.; Lindgren, G.; et al. A cis-acting regulatory mutation causes premature hair graying and susceptibility to melanoma in the horse. Nat. Genet. 2008, 40, 1004–1009. [Google Scholar] [CrossRef]
- Sundstrom, E.; Imsland, F.; Mikko, S.; Wade, C.; Sigurdsson, S.; Pielberg, G.R.; Golovko, A.; Curik, I.; Seltenhammer, M.H.; Solkner, J.; et al. Copy number expansion of the STX17 duplication in melanoma tissue from Grey horses. BMC Genom. 2012, 13, 365. [Google Scholar] [CrossRef]
- Curik, I.; Druml, T.; Seltenhammer, M.; Sundstrom, E.; Pielberg, G.R.; Andersson, L.; Solkner, J. Complex inheritance of melanoma and pigmentation of coat and skin in Grey horses. PLoS Genet. 2013, 9, e1003248. [Google Scholar] [CrossRef]
- Hollis, A.R. Equine Melanoma Updates. Vet. Clin. N. Am. Equine Pract. 2024, 40, 431–439. [Google Scholar] [CrossRef]
- Brodesser, D.M.; Kummer, S.; Eichberger, J.A.; Schlangen, K.; Corteggio, A.; Borzacchiello, G.; Bertram, C.A.; Brandt, S.; Pratscher, B. Deregulation of Metalloproteinase Expression in Gray Horse Melanoma Ex Vivo and In Vitro. Cells 2024, 13, 956. [Google Scholar] [CrossRef] [PubMed]
- Pozniak, J.; Marine, J.C. Decoding melanoma’s cellular mosaic to unlock immunotherapy potential. Trends Cell Biol. 2025. [Google Scholar] [CrossRef]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef]
- Pearlman, R.L.; Montes de Oca, M.K.; Pal, H.C.; Afaq, F. Potential therapeutic targets of epithelial-mesenchymal transition in melanoma. Cancer Lett. 2017, 391, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef]
- Xiao, D.; Barry, S.; Kmetz, D.; Egger, M.; Pan, J.; Rai, S.N.; Qu, J.; McMasters, K.M.; Hao, H. Melanoma cell-derived exosomes promote epithelial-mesenchymal transition in primary melanocytes through paracrine/autocrine signaling in the tumor microenvironment. Cancer Lett. 2016, 376, 318–327. [Google Scholar] [CrossRef]
- Kumar, D.; Gorain, M.; Kundu, G.; Kundu, G.C. Therapeutic implications of cellular and molecular biology of cancer stem cells in melanoma. Mol. Cancer 2017, 16, 7. [Google Scholar] [CrossRef]
- Marzagalli, M.; Raimondi, M.; Fontana, F.; Montagnani Marelli, M.; Moretti, R.M.; Limonta, P. Cellular and molecular biology of cancer stem cells in melanoma: Possible therapeutic implications. Semin. Cancer Biol. 2019, 59, 221–235. [Google Scholar] [CrossRef]
- Maniotis, A.J.; Folberg, R.; Hess, A.; Seftor, E.A.; Gardner, L.M.; Pe’er, J.; Trent, J.M.; Meltzer, P.S.; Hendrix, M.J. Vascular channel formation by human melanoma cells in vivo and in vitro: Vasculogenic mimicry. Am. J. Pathol. 1999, 155, 739–752. [Google Scholar] [CrossRef]
- Hendrix, M.J.; Seftor, E.A.; Hess, A.R.; Seftor, R.E. Vasculogenic mimicry and tumour-cell plasticity: Lessons from melanoma. Nat. Rev. Cancer 2003, 3, 411–421. [Google Scholar] [CrossRef]
- Hendrix, M.J.; Seftor, E.A.; Seftor, R.E.; Chao, J.T.; Chien, D.S.; Chu, Y.W. Tumor cell vascular mimicry: Novel targeting opportunity in melanoma. Pharmacol. Ther. 2016, 159, 83–92. [Google Scholar] [CrossRef]
- Lugassy, C.; Zadran, S.; Bentolila, L.A.; Wadehra, M.; Prakash, R.; Carmichael, S.T.; Kleinman, H.K.; Peault, B.; Larue, L.; Barnhill, R.L. Angiotropism, pericytic mimicry and extravascular migratory metastasis in melanoma: An alternative to intravascular cancer dissemination. Cancer Microenviron. 2014, 7, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.P.; Liao, Y.D.; Mai, D.M.; Xie, P.; Qiang, Y.Y.; Zheng, L.S.; Wang, M.Y.; Mei, Y.; Meng, D.F.; Xu, L.; et al. Tumor vasculogenic mimicry predicts poor prognosis in cancer patients: A meta-analysis. Angiogenesis 2016, 19, 191–200. [Google Scholar] [CrossRef]
- Long, G.V.; Swetter, S.M.; Menzies, A.M.; Gershenwald, J.E.; Scolyer, R.A. Cutaneous melanoma. Lancet 2023, 402, 485–502. [Google Scholar] [CrossRef] [PubMed]
- Seltenhammer, M.H.; Heere-Ress, E.; Brandt, S.; Druml, T.; Jansen, B.; Pehamberger, H.; Niebauer, G.W. Comparative histopathology of grey-horse-melanoma and human malignant melanoma. Pigment Cell Res. 2004, 17, 674–681. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, Q.; Zhang, N.; Du, X.; Xu, G.; Yan, X. CD146, from a melanoma cell adhesion molecule to a signaling receptor. Signal Transduct. Target. Ther. 2020, 5, 148. [Google Scholar] [CrossRef] [PubMed]
- Cannon, A.C.; Uribe-Alvarez, C.; Chernoff, J. RAC1 as a Therapeutic Target in Malignant Melanoma. Trends Cancer 2020, 6, 478–488. [Google Scholar] [CrossRef]
- Acevedo, A.; Gonzalez-Billault, C. Crosstalk between Rac1-mediated actin regulation and ROS production. Free Radic. Biol. Med. 2018, 116, 101–113. [Google Scholar] [CrossRef]
- Cannon, C.M.; Trembley, J.H.; Kren, B.T.; Unger, G.M.; O’Sullivan, M.G.; Cornax, I.; Modiano, J.F.; Ahmed, K. Therapeutic Targeting of Protein Kinase CK2 Gene Expression in Feline Oral Squamous Cell Carcinoma: A Naturally Occurring Large-Animal Model of Head and Neck Cancer. Hum. Gene Ther. Clin. Dev. 2017, 28, 80–86. [Google Scholar] [CrossRef]
- Shaverdashvili, K.; Wong, P.; Ma, J.; Zhang, K.; Osman, I.; Bedogni, B. MT1-MMP modulates melanoma cell dissemination and metastasis through activation of MMP2 and RAC1. Pigment Cell Melanoma Res. 2014, 27, 287–296. [Google Scholar] [CrossRef]
- Zamolo, G.; Grahovac, M.; Zauhar, G.; Vucinic, D.; Kovac, L.; Brajenic, N.; Grahovac, B. Matrix metalloproteinases MMP-1, MMP-2, and MMP-13 are overexpressed in primary nodular melanoma. J. Cutan. Pathol. 2020, 47, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Zigrino, P.; Kuhn, I.; Bauerle, T.; Zamek, J.; Fox, J.W.; Neumann, S.; Licht, A.; Schorpp-Kistner, M.; Angel, P.; Mauch, C. Stromal expression of MMP-13 is required for melanoma invasion and metastasis. J. Investig. Dermatol. 2009, 129, 2686–2693. [Google Scholar] [CrossRef]
- Moro, N.; Mauch, C.; Zigrino, P. Metalloproteinases in melanoma. Eur. J. Cell Biol. 2014, 93, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Weinberg, R.A. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev. Cell 2008, 14, 818–829. [Google Scholar] [CrossRef]
- Hodis, E.; Watson, I.R.; Kryukov, G.V.; Arold, S.T.; Imielinski, M.; Theurillat, J.P.; Nickerson, E.; Auclair, D.; Li, L.; Place, C.; et al. A landscape of driver mutations in melanoma. Cell 2012, 150, 251–263. [Google Scholar] [CrossRef]
- Yeh, I.; Bastian, B.C. Melanoma pathology: New approaches and classification. Br. J. Dermatol. 2021, 185, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Tschandl, P.; Berghoff, A.S.; Preusser, M.; Burgstaller-Muehlbacher, S.; Pehamberger, H.; Okamoto, I.; Kittler, H. NRAS and BRAF mutations in melanoma-associated nevi and uninvolved nevi. PLoS ONE 2013, 8, e69639. [Google Scholar] [CrossRef]
- Petiot, A.; Conti, F.J.; Grose, R.; Revest, J.M.; Hodivala-Dilke, K.M.; Dickson, C. A crucial role for Fgfr2-IIIb signalling in epidermal development and hair follicle patterning. Development 2003, 130, 5493–5501. [Google Scholar] [CrossRef]
- Hertzler-Schaefer, K.; Mathew, G.; Somani, A.K.; Tholpady, S.; Kadakia, M.P.; Chen, Y.; Spandau, D.F.; Zhang, X. Pten loss induces autocrine FGF signaling to promote skin tumorigenesis. Cell Rep. 2014, 6, 818–826. [Google Scholar] [CrossRef]
- Grose, R.; Fantl, V.; Werner, S.; Chioni, A.M.; Jarosz, M.; Rudling, R.; Cross, B.; Hart, I.R.; Dickson, C. The role of fibroblast growth factor receptor 2b in skin homeostasis and cancer development. EMBO J. 2007, 26, 1268–1278. [Google Scholar] [CrossRef] [PubMed]
- Goyal, L.; Saha, S.K.; Liu, L.Y.; Siravegna, G.; Leshchiner, I.; Ahronian, L.G.; Lennerz, J.K.; Vu, P.; Deshpande, V.; Kambadakone, A.; et al. Polyclonal Secondary FGFR2 Mutations Drive Acquired Resistance to FGFR Inhibition in Patients with FGFR2 Fusion-Positive Cholangiocarcinoma. Cancer Discov. 2017, 7, 252–263. [Google Scholar] [CrossRef]
- Gartside, M.G.; Chen, H.; Ibrahimi, O.A.; Byron, S.A.; Curtis, A.V.; Wellens, C.L.; Bengston, A.; Yudt, L.M.; Eliseenkova, A.V.; Ma, J.; et al. Loss-of-function fibroblast growth factor receptor-2 mutations in melanoma. Mol. Cancer Res. 2009, 7, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Cismas, S.; Pasca, S.; Crudden, C.; Trocoli Drakensjo, I.; Suleymanova, N.; Zhang, S.; Gebhard, B.; Song, D.; Neo, S.; Shibano, T.; et al. Competing Engagement of beta-arrestin Isoforms Balances IGF1R/p53 Signaling and Controls Melanoma Cell Chemotherapeutic Responsiveness. Mol. Cancer Res. 2023, 21, 1288–1302. [Google Scholar] [CrossRef]
- Lee, J.T.; Brafford, P.; Herlyn, M. Unraveling the mysteries of IGF-1 signaling in melanoma. J. Investig. Dermatol. 2008, 128, 1358–1360. [Google Scholar] [CrossRef]
- Davies, M.A. The role of the PI3K-AKT pathway in melanoma. Cancer J. 2012, 18, 142–147. [Google Scholar] [CrossRef]
- Brodesser, D.M.; Schlangen, K.; Burgstaller, J.; Brandt, S.; Pratscher, B. Expression signature of grey horse melanoma. In Proceedings of the ESVONC, European Society of Veterinary Oncology Annual Congress, Lyon, France, 18–22 April 2017. [Google Scholar]
- Wade, C.M.; Giulotto, E.; Sigurdsson, S.; Zoli, M.; Gnerre, S.; Imsland, F.; Lear, T.L.; Adelson, D.L.; Bailey, E.; Bellone, R.R.; et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 2009, 326, 865–867. [Google Scholar] [CrossRef]
- Li, R.; Yu, C.; Li, Y.; Lam, T.W.; Yiu, S.M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Drai, D.; Elmer, G.; Kafkafi, N.; Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001, 125, 279–284. [Google Scholar] [CrossRef]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Forner, O.; Marin-Garcia, P.; Arnau, V.; D’Eustachio, P.; Stein, L.; Hermjakob, H. Reactome pathway analysis: A high-performance in-memory approach. BMC Bioinform. 2017, 18, 142. [Google Scholar] [CrossRef]
- Hainisch, E.K.; Jindra, C.; Reicher, P.; Miglinci, L.; Brodesser, D.M.; Brandt, S. Bovine Papillomavirus Type 1 or 2 Virion-Infected Primary Fibroblasts Constitute a Near-Natural Equine Sarcoid Model. Viruses 2022, 14, 2658. [Google Scholar] [CrossRef] [PubMed]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
| Transcription | Genes | Definition |
|---|---|---|
| Upregulated | MAGE D2 | Melanoma-associated antigen D2 |
| MITF | Microphthalmia-associated transcription factor | |
| PMEL17 (SILV, gp100) | Premelanosome protein | |
| TYR | Tyrosinase | |
| TYRP1 | Tyrosinase related protein 1 | |
| Downregulated | MCAM (CD146; MUC18) | Melanoma cell adhesion molecule |
| S100A6 | S100 Calcium Binding Protein A6; Calcyclin | |
| S100A7 | S100 Calcium Binding Protein A7; Psoriasin |
| Pathway Name | Reactome Entities | Reactions | ||||
|---|---|---|---|---|---|---|
| Found | Ratio | p-Value | FDR | Found | Ratio | |
| RAC1 GTPase cycle | 54/191 | 0.012 | 4.76 × 10−6 | 0.006 | 4/6 | 3.93 × 10−4 |
| ECM organization | 84/350 | 0.0022 | 8.06 × 10−6 | 0.006 | 256/330 | 0.022 |
| RHO GTPase cycle | 104/460 | 0.029 | 8.76 × 10−6 | 0.006 | 52/91 | 0.006 |
| RHOA GTPase cycle | 44/154 | 0.01 | 2.70 × 10−5 | 0.014 | 4/6 | 3.93 × 10−4 |
| Integrin cell surface interaction | 29/86 | 0.005 | 3.87 × 10−5 | 0.016 | 44/55 | 0.004 |
| ECM proteoglycans | 27/79 | 0.005 | 5.63 × 10−5 | 0.019 | 20/23 | 0.002 |
| Activation of gene expression by SREBF | 24/71 | 0.005 | 1.65 × 10−4 | 0.049 | 42/42 | 0.003 |
| GLI proteins bind promoters of Hh-responsive genes to promote transcription | 7/8 | 5.07 × 10−4 | 1.99 × 10−4 | 0.052 | 4/4 | 2.62 × 10−4 |
| Non-Integrin membrane-ECM interactions | 26/83 | 0.005 | 2.89 × 10−4 | 0.058 | 19/33 | 0.002 |
| AP−2 family regulates transcription of growth factors and their receptors | 11/21 | 0.001 | 3.15 × 10−4 | 0.058 | 9/18 | 0.001 |
| TRP channels | 14/32 | 0.002 | 3.20 × 10−4 | 0.058 | 1/7 | 4.59 × 10−4 |
| EGR2- and SOX10-mediated initiation of Schwann cell myelination | 16/40 | 0.003 | 3.32 × 10−4 | 0.058 | 26/27 | 0.002 |
| Formation of the cornified envelope | 37/138 | 0.009 | 3.71 × 10−4 | 0.059 | 17/27 | 0.002 |
| Syndecan interactions | 13/29 | 0.002 | 4.12 × 10−4 | 0.061 | 9/15 | 9.83 × 10−4 |
| Kinesins | 22/68 | 0.004 | 5.36 × 10−4 | 0.074 | 11/14 | 9.17 × 10−4 |
| Degradation of the ECM | 38/148 | 0.009 | 6.88 × 10−4 | 0.085 | 80/105 | 0.007 |
| RND3 GTPase cycle | 16/43 | 0.003 | 7.12 × 10−4 | 0.085 | 1/2 | 1.31 × 10−4 |
| Defective B3GALTL causes PpS | 15/39 | 0.002 | 7.43 × 10−4 | 0.085 | 1/1 | 6.55 × 10−5 |
| Assembly of collagen fibrils and other multimeric structures | 21/67 | 0.004 | 0.001 | 0.114 | 23/26 | 0.002 |
| O-glycosylation of TSR domain-containing proteins | 15/41 | 0.003 | 0.001 | 0.122 | 2/2 | 1.31 × 10−4 |
| Type I hemidesmosome assembly | 7/11 | 6.98 × 10−4 | 0.001 | 0.122 | 6/6 | 3.93 × 10−4 |
| CDC42 GTPase cycle | 39/159 | 0.01 | 0.001 | 0.122 | 4/6 | 3.93 × 10−4 |
| EPH-ephrin-mediated repulsion of cells | 18/55 | 0.003 | 0.001 | 0.122 | 9/9 | 5.90 × 10−4 |
| ERBB2 activates PTK6 signaling | 9/18 | 0.001 | 0.001 | 0.122 | 2/2 | 1.31 × 10−4 |
| Signaling by RHO GTPases | 133/708 | 0.045 | 0.002 | 0.127 | 113/203 | 0.013 |
| DOLmc | DOLrmc | TIMmc | ELLmtc |
|---|---|---|---|
| MCAM | MCAM | ||
| MAGE D2 | |||
| MLANA | |||
| PMEL | |||
| S100A1 | S100A1 | ||
| S100A10 | |||
| S100A11 | S100A11 | ||
| S100A4 | S100A4 | S100A4 | |
| S100A6 | S100A6 | S100A6 | S100A6 |
| TYR | |||
| TYRP |
| Pathway Name | Entities | Reactions | ||||
|---|---|---|---|---|---|---|
| Found | Ratio | p-Value | FDR | Found | Ratio | |
| HSF1-dependent transactivation | 8/59 | 0.002 | 3.93 × 10−7 | 6.42 × 10−4 | 5/8 | 5.24 × 10−4 |
| Attenuation phase | 7/47 | 0.002 | 1.14 × 10−6 | 9.34 × 10−4 | 3/5 | 3.28 × 10−4 |
| NFE2L2 regulates pentose phosphate pathway genes | 4/23 | 9.52 × 10−4 | 1.35 × 10−4 | 0.074 | 4/8 | 5.24 × 10−4 |
| Orc1 removal from chromatin | 5/64 | 0.003 | 7.74 × 10−4 | 0.211 | 3/4 | 2.62 × 10−4 |
| Signaling by BRAF and RAF1 fusions | 5/73 | 0.003 | 0.001 | 0.322 | 5/5 | 3.28 × 10−4 |
| RHOF GTPase cycle | 4/46 | 0.002 | 0.002 | 0.363 | 1/3 | 1.97 × 10−4 |
| Regulation of HSF1-mediated heat shock response | 13/302 | 0.013 | 0.002 | 0.378 | 11/14 | 9.17 × 10−4 |
| NFE2L2 regulating TCA cycle genes | 2/7 | 2.90 × 10−4 | 0.003 | 0.455 | 2/4 | 2.62 × 10−4 |
| RHOD GTPase cycle | 4/57 | 0.002 | 0.004 | 0.566 | 1/9 | 5.90 × 10−4 |
| Cellular response to heat stress | 14/391 | 0.016 | 0.004 | 0.591 | 17/29 | 0.002 |
| RAC3 GTPase cycle | 5/100 | 0.004 | 0.005 | 0.591 | 1/6 | 3.93 × 10−4 |
| DNA strand elongation | 5/101 | 0.004 | 0.005 | 0.591 | 7/15 | 9.83 × 10−4 |
| RSK activation | 2/11 | 4.55 × 10−4 | 0.007 | 0.665 | 4/4 | 2.62 × 10−4 |
| Oncogenic MAPK signaling | 5/112 | 0.005 | 0.008 | 0.665 | 30/46 | 0.003 |
| Signaling by moderate kinase activity BRAF mutants | 4/73 | 0.003 | 0.009 | 0.665 | 5/7 | 4.59 × 10−4 |
| Signaling downstream of RAS mutants | 4/73 | 0.003 | 0.009 | 0.665 | 5/7 | 4.59 × 10−4 |
| Paradoxical activation of RAF signaling by kinase inactive BRAF | 4/73 | 0.003 | 0.009 | 0.665 | 5/7 | 4.59 × 10−4 |
| Activation of ATR in response to replication stress | 5/73 | 0.003 | 0.009 | 0.665 | 5/9 | 5.90 × 10−4 |
| Signaling by RAS mutants | 4/73 | 0.003 | 0.009 | 0.665 | 5/9 | 5.90 × 10−4 |
| CREB1 phosphorylation through NMDA receptor-mediated activation of RAS signaling | 3/39 | 0.002 | 0.009 | 0.669 | 7/7 | 4.59 × 10−4 |
| Cluster 1 | Cluster 2 | Cluster 3 |
|---|---|---|
| Signaling by FGFR Signaling by FGFR2 SHC-mediated cascade: FGFR2 FRS-mediated FGFR2 signaling Signaling by FGFR2 IIIa TM Downstream signaling of FGFR2 FGFR2 mutant receptor activation | Insulin receptor signaling cascade IRS-mediated signaling IRS-related events triggered by IGF1R IGF1R signaling cascade Signaling by IGF1R AP-2 family regulates transcription of growth factors and their receptors | PI5P, PP2A and IER3 regulate PI3K/AKT signaling Negative regulation of PI3K/AKT network Constitutive signaling by aberrant PI3K in cancer |
| Code | Breed | Sex | Age | Short Disease Description | Samples Collected |
|---|---|---|---|---|---|
| ARI | Warmblood | G | 22 | Four melanomas, thoracic region | One tumor |
| DIM | Trakehner mix | G | 14 | Multiple melanomas in the anogenital, parotidal, ocular, and labial regions | One anogenital tumor, |
| Intact skin | |||||
| DOL | Icelandic horse | G | 14 | Multiple anogenital melanomas and a recurrent lesion after surgical excision | One anogenital tumor |
| Recurrent tumor | |||||
| ELL | Trakehner mix | M | 13 | Large perianal tumor, metastases (spleen, liver, heart) | Liver metastasis |
| GYN | Shagya Arabian | M | >20 | Two encapsulated melanomas under the tail root, maximum diameter < 2 cm | One tumor |
| Intact skin | |||||
| MAN | Lipizzan horse | M | 14 | Two encapsulated melanomas under the tail root | One tumor |
| MAK | Arabian horse | S | 4 | Single small, encapsulated melanoma, inner thigh | Tumor |
| TIM | Warmblood | M | 15 | Single encapsulated melanoma under the tail root | Tumor |
| HAC | PRE | S | 13 | Melanoma-free | Intact skin |
| ARC | GRP | G | 19 | Melanoma-free | Intact skin |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brodesser, D.M.; Schlangen, K.; Rodríguez-Rojas, A.; Kuropka, B.; Doulidis, P.G.; Brandt, S.; Pratscher, B. Gray-Horse Melanoma—A Wolf in Sheep’s Clothing. Int. J. Mol. Sci. 2025, 26, 6620. https://doi.org/10.3390/ijms26146620
Brodesser DM, Schlangen K, Rodríguez-Rojas A, Kuropka B, Doulidis PG, Brandt S, Pratscher B. Gray-Horse Melanoma—A Wolf in Sheep’s Clothing. International Journal of Molecular Sciences. 2025; 26(14):6620. https://doi.org/10.3390/ijms26146620
Chicago/Turabian StyleBrodesser, Daniela M., Karin Schlangen, Alexandro Rodríguez-Rojas, Benno Kuropka, Pavlos G. Doulidis, Sabine Brandt, and Barbara Pratscher. 2025. "Gray-Horse Melanoma—A Wolf in Sheep’s Clothing" International Journal of Molecular Sciences 26, no. 14: 6620. https://doi.org/10.3390/ijms26146620
APA StyleBrodesser, D. M., Schlangen, K., Rodríguez-Rojas, A., Kuropka, B., Doulidis, P. G., Brandt, S., & Pratscher, B. (2025). Gray-Horse Melanoma—A Wolf in Sheep’s Clothing. International Journal of Molecular Sciences, 26(14), 6620. https://doi.org/10.3390/ijms26146620





