1. Introduction
Cutaneous adverse reactions (CARs) are the most prevalent side effects associated with the administration of Epidermal Growth Factor Receptor (EGFR) inhibitors, due to the pronounced expression of EGFR in the skin and its adnexal structures [
1]. Among these dermatological lesions, the most frequently encountered are encapsulated within the PRIDE syndrome—an acronym representing Papulopustules, Paronychia, Regulatory abnormalities of hair growth, Itching, and Dryness [
2]. The initiation and intensity of papulopustular eruptions have been demonstrated to positively correlate with both therapeutic response and overall survival [
3,
4]. Consequently, these eruptions are considered indicators of therapeutic efficacy and prognostic markers of clinical outcomes [
3,
5]. Scientific evidence suggests that dose escalation in patients exhibiting grade ≤1 toxicity may enhance response rates [
6]. Moreover, patients who experience a variety of CARs have superior survival outcomes [
7].
Papulopustular eruptions—commonly known as acneiform reactions—represent the predominant CARs, occurring with greater frequency in individuals receiving monoclonal antibodies such as cetuximab (80%) compared to those undergoing treatment with tyrosine kinase inhibitors like erlotinib (67%) [
8,
9]. Clinically, these lesions present as erythematous papules and follicular pustules devoid of comedones or cysts, primarily affecting seborrheic areas, and are frequently exacerbated in a cyclical manner after cetuximab infusions [
10]. While such eruptions have prognostic relevance, they may also considerably diminish quality of life, often demanding dose adjustments in the oncological treatment regimens [
3,
4,
5].
EGFR inhibition compromises epidermal barrier integrity by downregulating critical structural and antimicrobial constituents such as claudin-1, LL37, and RNase 7, thereby fostering colonization by
Staphylococcus aureus and potentially worsening dermatological lesions through toxin-mediated pathways [
11,
12,
13,
14,
15,
16].
S. aureus represents a prevalent pathogen in nosocomial infections and poses a significant threat to immunocompromised individuals [
17]. Its pathogenesis involves a complex interaction between host defense mechanisms and bacterial virulence determinants [
18]. These factors encompass cell wall-associated adhesins—such as protein A, clumping factors A and B, and proteins that bind to fibronectin, collagen, elastin, and fibrinogen—alongside soluble proteins and exotoxins, including hemolysins, coagulases, enterotoxins, exfoliative toxins, toxic shock syndrome toxin-1, and Panton–Valentine leukocidin [
19]. Adhesins and coagulases facilitate adherence to host cells and implanted devices, initiating a colonization process that may evolve into an infection [
19,
20].
The pathogenicity of
S. aureus is driven not only by antibiotic resistance and production of a diverse array of virulence factors, but also by its capacity to form biofilms [
21,
22]. Biofilms are highly organized and dynamic microbial aggregates encapsulated within a self-generated extracellular mucopolysaccharide matrix, which exhibits compositional variability across different colonies [
21,
22,
23]. This matrix, consisting of polysaccharides, lipids, nucleic acids, and proteins—derived from host or bacterial cell lysis or directly secreted—affords protection against antimicrobial agents and host immune responses [
23,
24,
25,
26]. It also hampers phagocytosis and attenuates local immune activity [
27,
28]. Furthermore, the close spatial arrangement within biofilms facilitates horizontal gene transfer, including the spread of antibiotic resistance genes [
29]. The formation of biofilms has been intimately associated with chronic infections, reduced therapeutic efficacy, and heightened resistance, to the degree that biofilms have been likened to “primitive multicellular organisms” [
30,
31,
32].
Given these patterns and the suspected role of microbial colonization—particularly with S. aureus—in the development and persistence of CARs, we conducted an observational, prospective investigation in the Oncology Departments of the Bucharest University Emergency Hospital, Elias University Emergency Hospital, and the Bucharest Oncology Institute, between May 2014 and January 2015, analyzing the phenotypic and genotypic characteristics of S. aureus strains isolated from papulopustular lesions and paronychia in patients undergoing EGFR inhibitor therapy. This study aimed to explore the link between bacterial virulence, antibiotic resistance, and the clinical severity of dermatologic toxicity. In Romania, data regarding bacterial colonization, antibiotic resistance profiles, or virulence traits in CARs is lacking. This is among the few prospective studies characterizing phenotypic and genotypic features of isolated strains and their resistance to antibiotics.
3. Discussion
The papulopustular eruption was reported in most patients, of whom the majority were undergoing treatment with cetuximab and only a few with erlotinib. Severe paronychia manifested in six patients receiving high-dose cetuximab, generally in conjunction with FOLFOX or FOLFIRI regimens. These lesions emerged after an eight-week treatment period as painful inflammation of the periungual folds, frequently exacerbated by the formation of abscesses or pyogenic granulomas. Although dose reductions were rare, Grade 2 papulopustular eruptions and paronychia significantly affected quality of life in 38% of the cohort.
Initially, the administration of systemic antibiotics was restricted to the period following the onset of CARs; however, emerging evidence supporting the prophylactic use of tetracyclines (200 mg/day), in conjunction with emollients and appropriate skin care, has illustrated a greater than 50% reduction in Grade 2 or lower CARs, thereby supporting their integration into standardized care protocols [
9,
33,
34]. Despite a rising incidence of tetracycline-resistant
S. aureus strains, doxycycline continues to exhibit clinical relevance due to its anti-inflammatory properties, which include the inhibition of matrix metalloproteinases [
35,
36]. CARs typically intensify during the initial month of treatment and subsequently achieve stabilization and improvement by the 6–8 week mark, underscoring the critical need for early prophylactic interventions [
37]. Although CAR severity has functioned as a pharmacodynamic indicator in clinical trials, the extensive implementation of prophylactic measures may potentially diminish its prognostic significance [
6]. In the context of clinical practice, dosing modifications for cetuximab are not predicated upon the grading of CARs [
6].
Microbiological evaluations revealed the presence of
S. aureus in 90% of patients exhibiting CARs. Additional pathogenic organisms included
S. marcescens, C. diversus K. oxytoca, P. aeruginosa, S. epidermidis, and
E. faecalis. Importantly, no instances of systemic infections were reported. Sampling methodologies were designed to ensure that the isolated pathogens accurately represented true infections rather than mere surface colonization [
38,
39]. Moreover, approximately one-third of patients included in the study carried nasal strains that were genetically and phenotypically congruent with those isolated from skin lesions, implying autoinoculation as a plausible mechanism. This finding underscores the significance of nasal colonization not only as a reservoir for
S. aureus but also as a modifiable risk factor that may contribute to the persistence or relapse of dermatologic toxicity. The high concordance between nasal and cutaneous isolates highlights the need for targeted screening and decolonization strategies—particularly in patients with recurrent or severe CARs—to improve therapeutic outcomes and reduce lesion chronicity.
Antibiotic susceptibility evaluations indicated elevated resistance rates among
S. aureus isolates, particularly concerning penicillins (85%) and tetracyclines (57%). All strains maintained susceptibility to fluoroquinolones, linezolid, vancomycin, and rifampicin. Genotypic analyses revealed the presence of SCC
mec type IV in roughly 20% of strains, suggesting a potential community-acquired origin [
40,
41]. The
mecA gene was identified in only 7% of isolates, which may be attributable to alternative resistance mechanisms or genetic loss during subculturing procedures [
42,
43,
44].
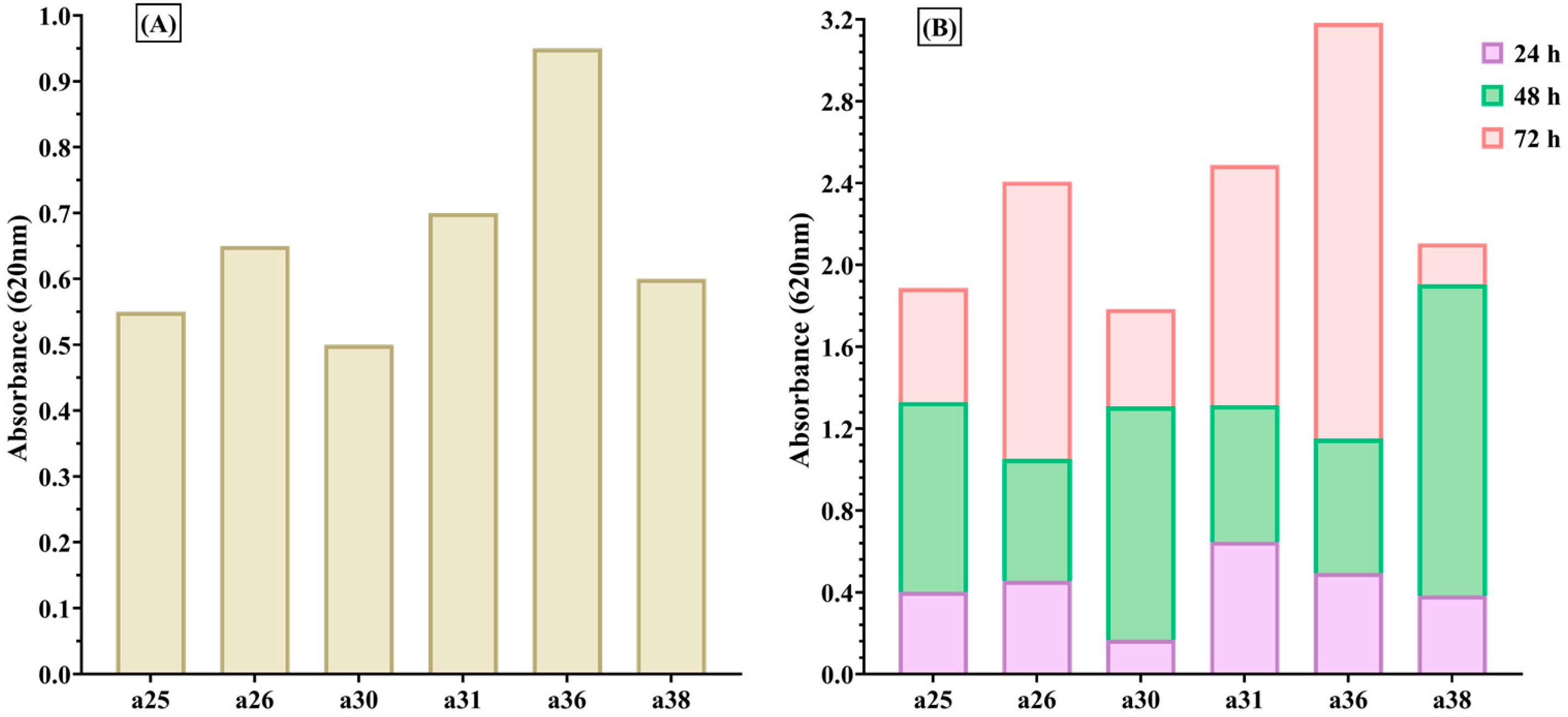
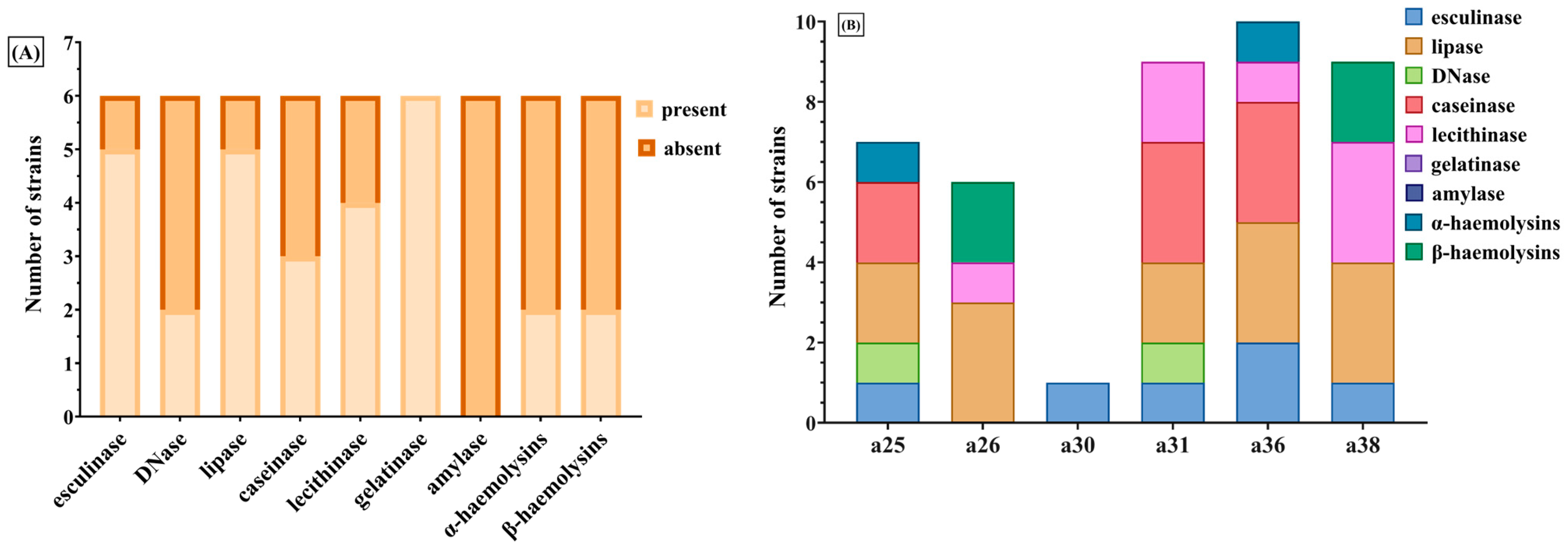
All isolates demonstrated the capacity for biofilm formation, with strains originating from paronychia and late-onset papulopustular eruptions exhibiting more pronounced biofilm production. This observation suggests a potential correlation with chronicity and resistance to treatment [
45,
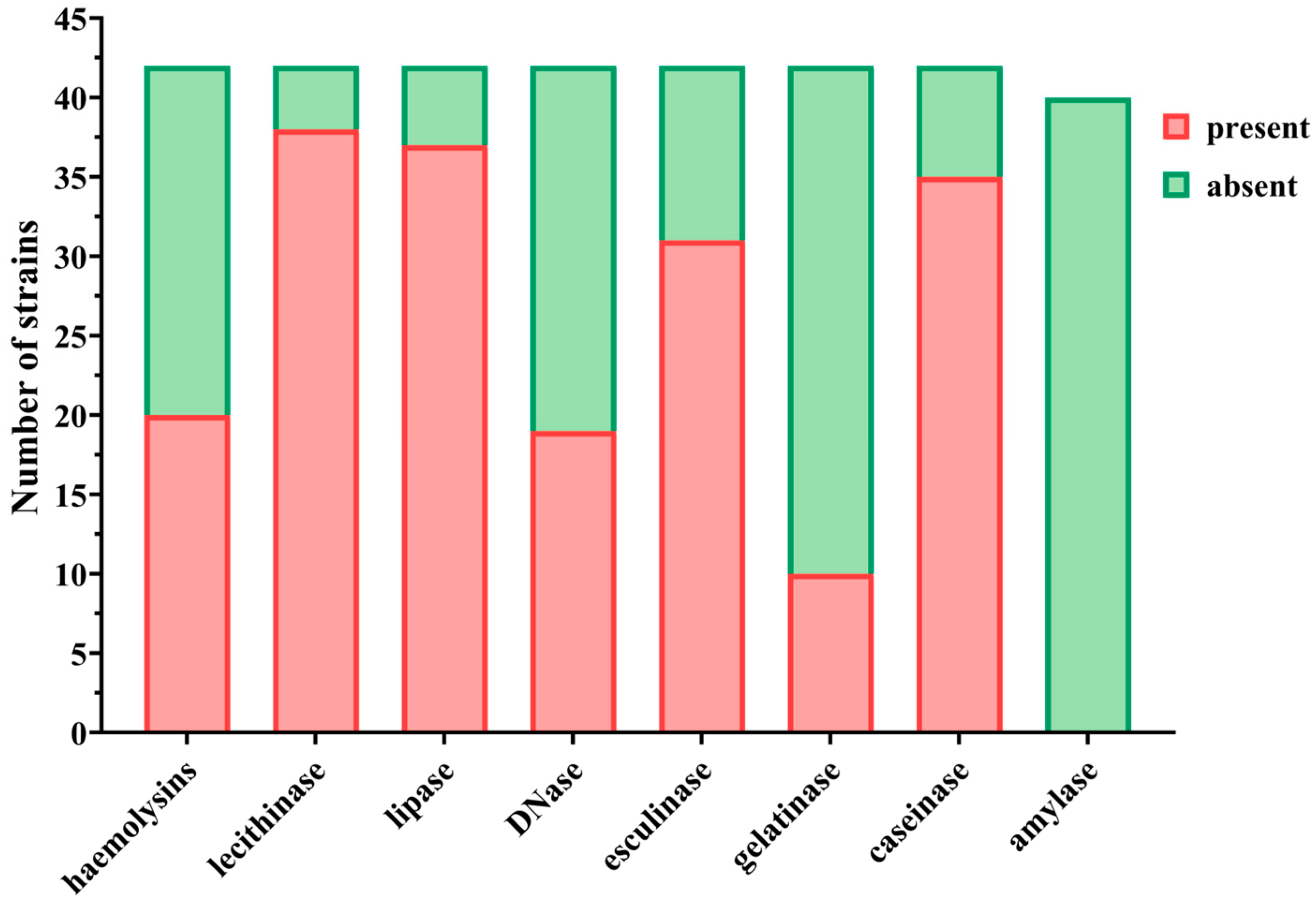
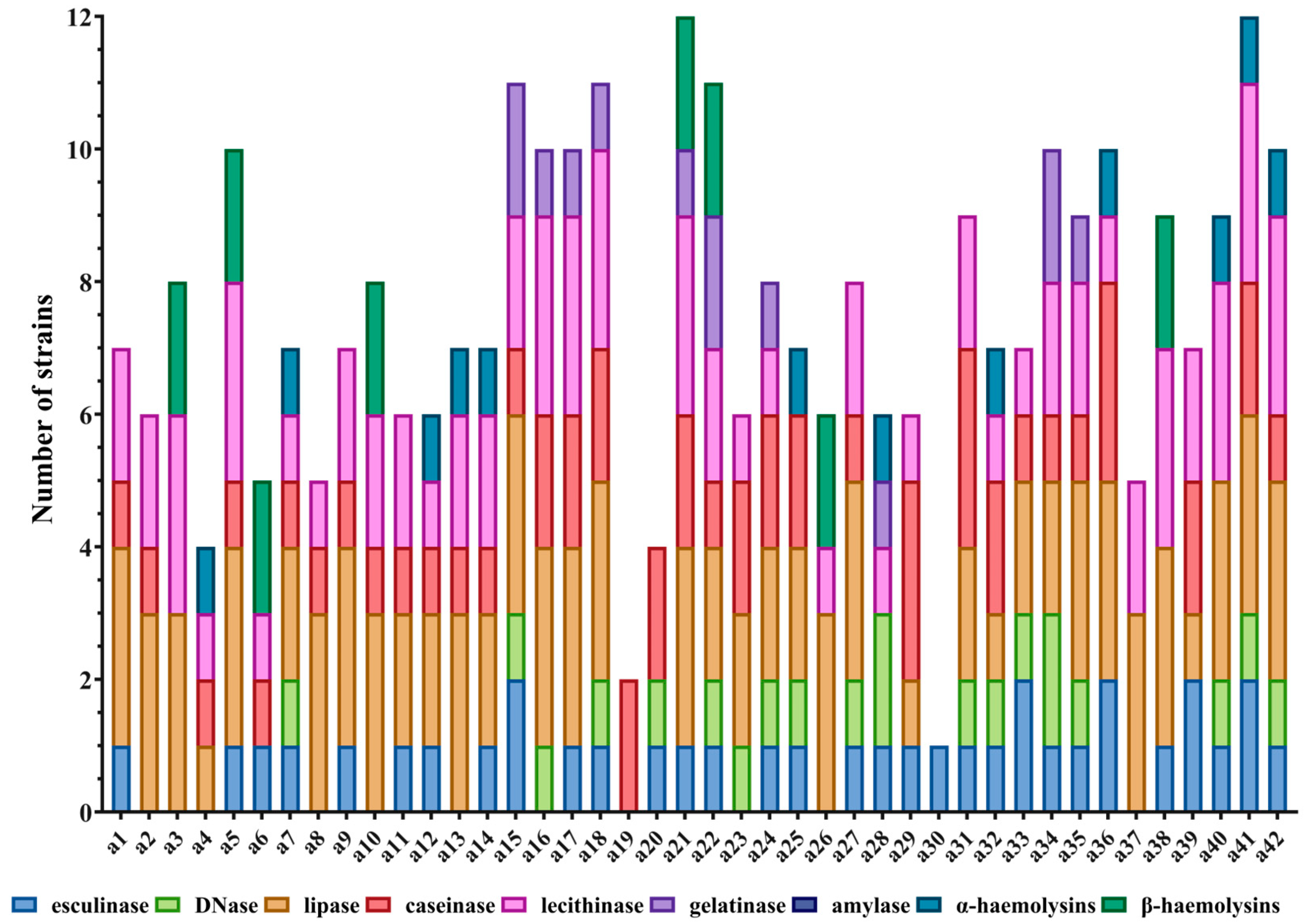
46]. Phenotypically, all strains displayed multiple virulence factors, with high prevalence rates of hemolysins, lecithinase, lipase, esculinase, and caseinase. Conversely, gelatinase was infrequently detected (23.8%), while DNase activity was recorded in 45.2% of strains.
Virulence index scores, derived from enzyme expression profiles, exhibited a weak yet statistically significant correlation with CTCAE-based grading of CARs severity. Among the evaluated factors, DNase and esculinase emerged as the most predictive, with an enzymatic score of 3 conferring an 18-fold elevated risk for the development of severe CARs, making it a potentially robust marker for early identification of high-risk patients. Principal component analysis did not reveal stronger associations beyond these individual enzymes. The association between the enzymatic score and the clinical severity of cutaneous adverse reactions suggests that virulence factor profiling could serve as a predictive tool in clinical practice. Given the ease of phenotypic assessment through standard microbiological techniques, incorporating such enzymatic profiling into diagnostic workflows could allow for timely, risk-adapted therapeutic strategies. However, prospective validation in larger cohorts is necessary to determine its reproducibility and clinical utility. Although we characterized both phenotypic and genotypic virulence features, we did not directly correlate gene presence with enzymatic activity. This is a limitation of the current study and should be addressed in future investigations using larger datasets and transcriptomic validation to determine whether gene expression consistently translates into functional protein activity.
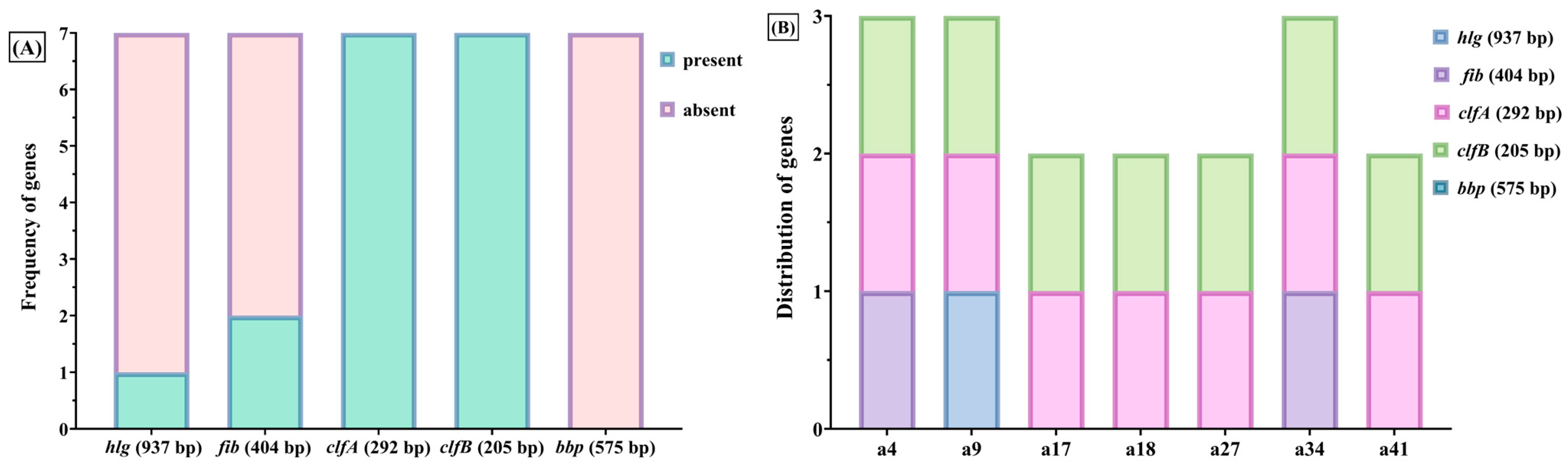
Genotypic profiling indicated a high expression of adhesion-related genes such as clfA and clfB (85.7%) and fib (~33%), while extracellular virulence genes, including hlg and bbp, were rarely identified. Notably, ebps, fnbB, and lpv were absent across all isolates. The dominance of cell wall-associated virulence genes may play a significant role in prolonging colonization and contributing to resistance against therapeutic interventions.
Protein A and the fibronectin-binding proteins A and B (encoded by
fnbA/fnbB) facilitate the adherence to fibronectin in fibrin clots or extracellular matrices of the host and have been associated with the pathogenesis of endocarditis [
19]. Clumping factors A and B (
clfA, clfB) promote the binding of fibrinogen—where
clfA specifically interacts with the γ-chain and
clfB with the α- and β-chains—thereby contributing to immune evasion and the persistence of chronic infections [
19,
47,
48]. Additionally, Protein A exhibits affinity for the Fc region of immunoglobulin G and von Willebrand factor, thereby further compromising host immune defenses [
49]. Strains that express
bbp, as well as collagen- and fibronectin-binding proteins, have been correlated with the pathogenesis of osteomyelitis [
50].
S. aureus augments tissue invasiveness and immune evasion through the secretion of α-, β-, and γ-hemolysins, along with Panton–Valentine leukocidin, which collectively form pores in host cell membranes and instigate cellular lysis [
51,
52]. While α-hemolysin specifically targets platelets and monocytes, PVL—encoded by
lpv—selectively affects leukocytes and has been implicated in necrotizing skin infections and methicillin-resistant
S. aureus pneumonia [
19,
53,
54]. Although γ-hemolysin, encoded by
hlg, was found to be expressed in less than 20% of our isolates, it is noteworthy that all strains demonstrated some degree of hemolytic activity [
19,
55].
Coagulase plays a significant role in immune evasion by forming staphylothrombin complexes with prothrombin, which facilitates localized fibrin clot formation [
19]. Lecithinase and lipase contribute to the degradation of phosphatidylcholine, thereby enhancing tissue invasion, while hemolysins promote iron acquisition through the breakdown of hemoglobin [
56,
57,
58]. Esculinase catalyzes the hydrolysis of esculin, resulting in the production of esculetol that chelates iron, a critical factor for the expression of virulence determinants [
56,
57]. Caseinase and gelatinase compromise the integrity of connective tissue, thereby enhancing the organism’s invasiveness, while DNase and amylase facilitate immune evasion and metabolic processes [
56,
57,
58,
59].
4. Materials and Methods
4.1. Patient Selection and Sample Processing
Fifty-five adult oncology patients who developed EGFR inhibitor–associated CARs were enrolled in our study. S. aureus identification was based on colony morphology, Gram staining, catalase and coagulase positivity, and confirmation via the VITEK® 2 Compact system (bioMérieux, Marcy-l’Étoile, France).
Only those with microbiologically confirmed S. aureus colonization or infection were selected for microbiological and molecular analysis. A total of 42 S. aureus strains were isolated from skin lesions and underwent phenotypic and genotypic characterization for antibiotic resistance and virulence.
4.2. Antibiotic Susceptibility Testing
Phenotypic antibiotic susceptibility was assessed using the Kirby–Bauer disk diffusion methodology on Mueller–Hinton agar in accordance with the established guidelines of the 2014 Clinical and Laboratory Standards Institute (CLSI) and subsequently verified through the automated VITEK 2 Compact system [
60,
61,
62]. The resistance profiles were classified within the framework of multidrug-resistant (MDR), extensively drug-resistant (XDR), and pandrug-resistant (PDR) bacteria, as delineated by Magiorakos et al. (2012) [
63].
Nevertheless, it is important to note that the classification was contingent upon the spectrum of antibiotics encompassed within the local testing panel available at the time of the investigation. Not all antimicrobial categories listed by Magiorakos et al. (2012) [
63] were subjected to testing; consequently, the MDR/XDR/PDR classification was employed within a constrained context and should be interpreted accordingly. This limitation has been acknowledged, and the terminology used is primarily intended to facilitate a comparative discourse regarding resistance patterns.
4.3. Biofilm Formation Assay
The biofilm-producing ability of
S. aureus isolates was evaluated using the standard microtiter plate assay. The isolates were cultivated in Tryptic Soy Broth (TSB) without glucose supplementation, adjusted to a turbidity equivalent to 0.5 McFarland standard (approximately 1.5 × 10
8 CFU/mL). A volume of 10 µL of this standardized inoculum was added to each well of a 96-well microtiter plate containing fresh TSB and incubated at 37 °C for 24, 48, and 72 h [
64,
65]. After incubation, wells were gently washed, stained with 0.1% crystal violet, and the dye retained by the adherent biofilm was solubilized using 33% acetic acid. The optical density was measured at 492 nm to quantify biofilm biomass [
22,
65].
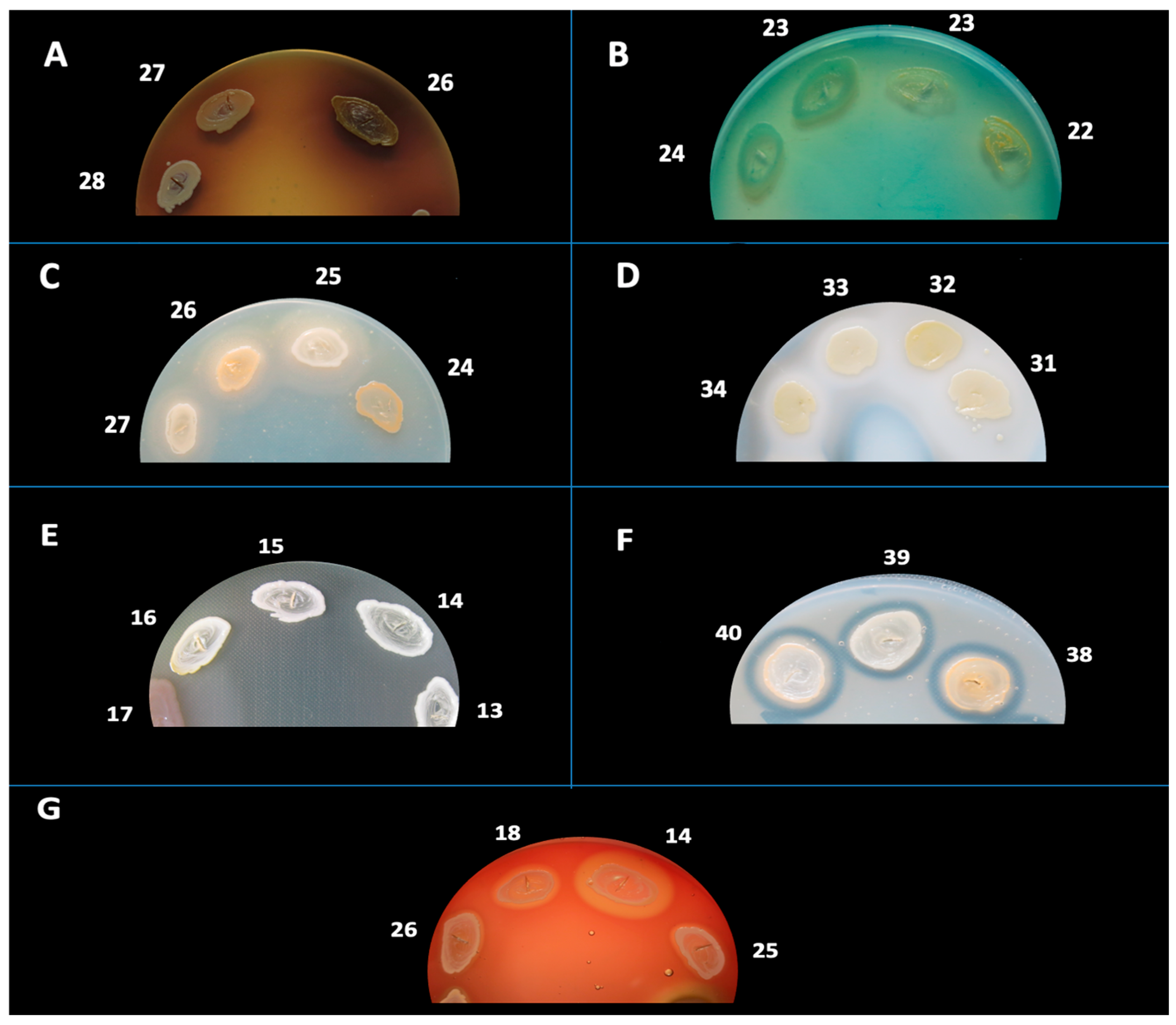
4.4. Phenotypic Detection of Soluble Virulence Factors
Phenotypic identification of soluble virulence determinants in
S. aureus was conducted through spot inoculation on nutrient agar media augmented with enzyme-specific substrates. The enzymatic virulence determinants subjected to evaluation included: hemolysins (α and β), amylase, caseinase, gelatinase, esculinase, DNase, lipase, and lecithinase. Reactions were interpreted based on the appearance of characteristic halos or precipitates as described in previous literature [
19,
54,
60].
Hemolysins were examined on 7% sheep blood agar, incubated at 37 °C for a duration of 24 h. Hemolysis was categorized into β-hemolysis (complete, transparent halo), α-hemolysis (partial, greenish halo), or γ-hemolysis (absence of hemolysis). In certain instances, plates were stored at 4 °C for a period of 30 min prior to analysis to augment visualization.
Lecithinase activity was evaluated on agar enriched with 2.5% egg yolk and incubated at 37 °C for a maximum of 7 days. A positive response was signified by an opaque zone (precipitation) resulting from diglyceride formation or a transparent halo due to lipid complex hydrolysis.
Lipase was assessed utilizing agar fortified with 1% sorbitol monooleate and incubated at 37 °C for a maximum of 7 days. The presence of a white, opaque precipitation zone signaled enzymatic degradation of monooleate and the subsequent formation of insoluble calcium oleate crystals.
DNase activity was identified on DNase agar augmented with methyl green, incubated at 37 °C for 24 h. The presence of a clear zone surrounding the colony indicated DNA hydrolysis.
Esculinase activity was evaluated using agar composed of 1% esculin and 1% ferric ammonium citrate. Following 24 h of incubation at 37 °C, the darkening of the medium indicated the production of esculetol and subsequent chelation of iron.
Caseinase activity was examined on agar containing 15% casein and incubated at 37 °C for 24 h. The manifestation of a precipitation zone indicated proteolytic breakdown of casein and the formation of insoluble paracaseinate.
Gelatinase was assessed on agar containing 3% gelatin. Plates were incubated at 37 °C for 24 h, and the emergence of a clear halo post-refrigeration confirmed enzymatic activity.
Amylase was evaluated on starch agar incubated at 37 °C for 24 h, followed by staining with Lugol’s iodine. A yellow halo surrounding colonies on a blue background indicated the hydrolysis of starch.
Each strain was evaluated in duplicate. Only consistently positive results were documented, and all assays were interpreted visually by trained personnel utilizing standardized criteria.
4.5. Genotypic Characterization of Virulence and Resistance Markers
Genomic DNA was extracted using an alkaline lysis method, and its concentration and purity were verified by agarose gel electrophoresis [
19]. Virulence genes—
bbp,
clfA,
clfB,
ebps,
fnbB,
fib,
lpv, and
hlg—were detected using multiplex polymerase chain reaction (PCR) based on the protocol by Cotar et al. [
19] (
Table 10,
Table 11 and
Table 12,
Supplementary Table S2). enotypic methicillin resistance profiling was performed through multiplex PCR targeting
S. aureus SCC
mec cassette types I–V, using primer sets and amplification protocols described by Milheiriço et al. and Zhang et al. [
66,
67] (
Table 10,
Table 11 and
Table 12). PCR amplification was performed with GoTaq
® Green Master Mix (Jena Bioscience, Jena, Germany), and reactions were carried out on a Corbett Thermal Cycler with specific cycling conditions for each gene group.
PCR amplicons were visualized by 1.5% agarose gel electrophoresis stained with ethidium bromide (10 µg/mL) under ultraviolet (UV) light and compared with 100 bp molecular weight markers (M-Bench Top 100 bp DNA Ladder, Promega, Madison, WI, USA).
4.6. Clinical Correlation
Clinical severity of skin reactions was graded using the common terminology criteria for adverse events CTCAE v4.03 scale and correlated with bacterial genotypic profiles, biofilm-forming ability, and expression of soluble virulence factors.
4.7. Statistical Analysis
Patient data were entered into an OpenOffice Calc database (version 4.1.1, Apache Software Foundation). Statistical analyses were conducted using SAS University Edition (SAS Institute Inc., Cary, NC, USA) and R version 3.3.0 (R Foundation for Statistical Computing, Vienna, Austria), with supplementary analyses performed in RStudio version 0.99.902. Several R packages were used, including asbio, boot, bootstrap, and arm.
Descriptive statistics included the calculation of absolute and relative frequencies for categorical variables. For continuous variables, central tendency and variability were assessed using the mean, median, standard deviation, variance, and interquartile range (IQR). Distribution characteristics such as skewness and kurtosis were also computed. The Shapiro–Wilk test was applied to evaluate normality, and graphical representations included histograms, density plots, normal probability plots, and boxplots.
Inferential analyses were selected based on the distribution of the data. For variables approximating a normal distribution, Welch’s two-sample t-test was used to compare groups. For variables deviating from Gaussian distribution, a bootstrap resampling approach was employed to assess differences in medians. Comparisons involving more than two patient groups were conducted using analysis of variance (ANOVA). Associations between continuous variables were examined using Spearman’s rank correlation coefficient (ρ). A p-value below 0.05 was considered statistically significant.
5. Conclusions
S. aureus was identified in 90% of papulopustular and paronychia lesions associated with EGFR inhibitor therapy, with numerous strains demonstrating multidrug resistance (MDR)—particularly to β-lactams and tetracyclines—and a pronounced capacity for biofilm formation, especially in lesions exhibiting chronic progression. Virulence profiling revealed the frequent expression of pore-forming enzymes, esculinase, and caseinase, while DNase and esculinase expression showed a significant correlation with clinical severity.
From a genotypic standpoint, adhesion-related virulence genes (clfA, clfB, fib) were frequently expressed, whereas genes encoding exotoxins (hlg, lpv) were less commonly detected, suggesting a virulence profile that favors colonization and persistence over acute toxicity.
Notably, approximately one-third of patients harbored nasal strains that were both genetically and phenotypically identical to those isolated from skin lesions, suggesting that nasal colonization may serve as a reservoir for autoinoculation and recurrence. This finding underscores the need for routine nasal screening and decolonization strategies in patients initiating EGFR inhibitor therapy, particularly those with recurrent or severe cutaneous adverse reactions (CARs).
The enzymatic score—especially when based on DNase and esculinase expression levels—showed strong predictive value for CAR severity and may serve as a promising clinical biomarker. Its simplicity and accessibility support further validation in prospective clinical trials.
Collectively, these findings highlight the pivotal role of S. aureus in the pathogenesis of CARs and support the integration of routine microbial sampling and antibiogram-guided therapy into standard management. Future research should explore molecular distinctions between early- and late-onset eruptions and validate enzymatic scoring as a tool for risk stratification and personalized therapeutic interventions.