Dirhodium Tetraacetate Binding to Lysozyme at Body Temperature
Abstract
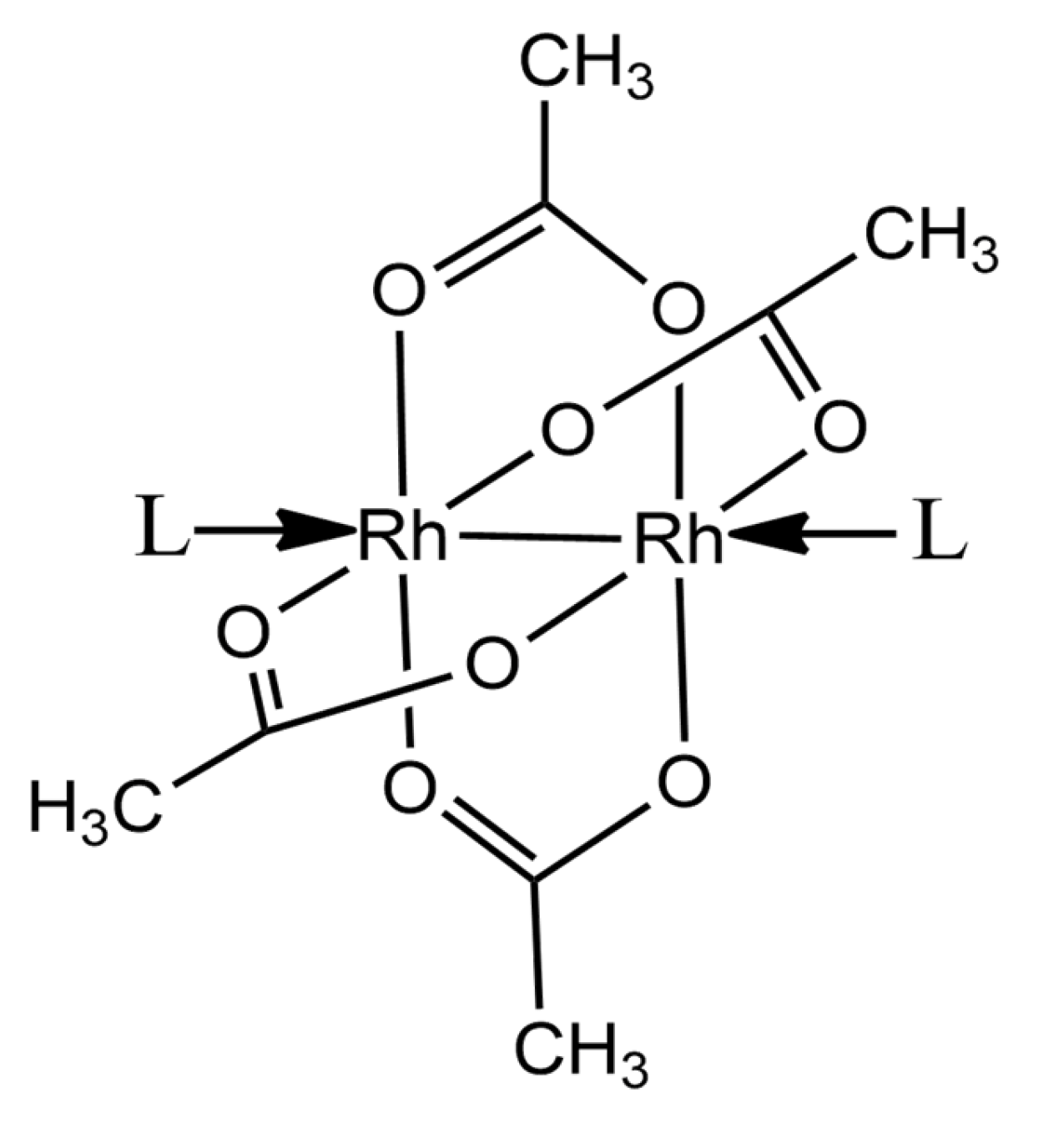
1. Introduction
2. Results and Discussion
2.1. Crystals of the Rh/HEWL Adduct at 310 K
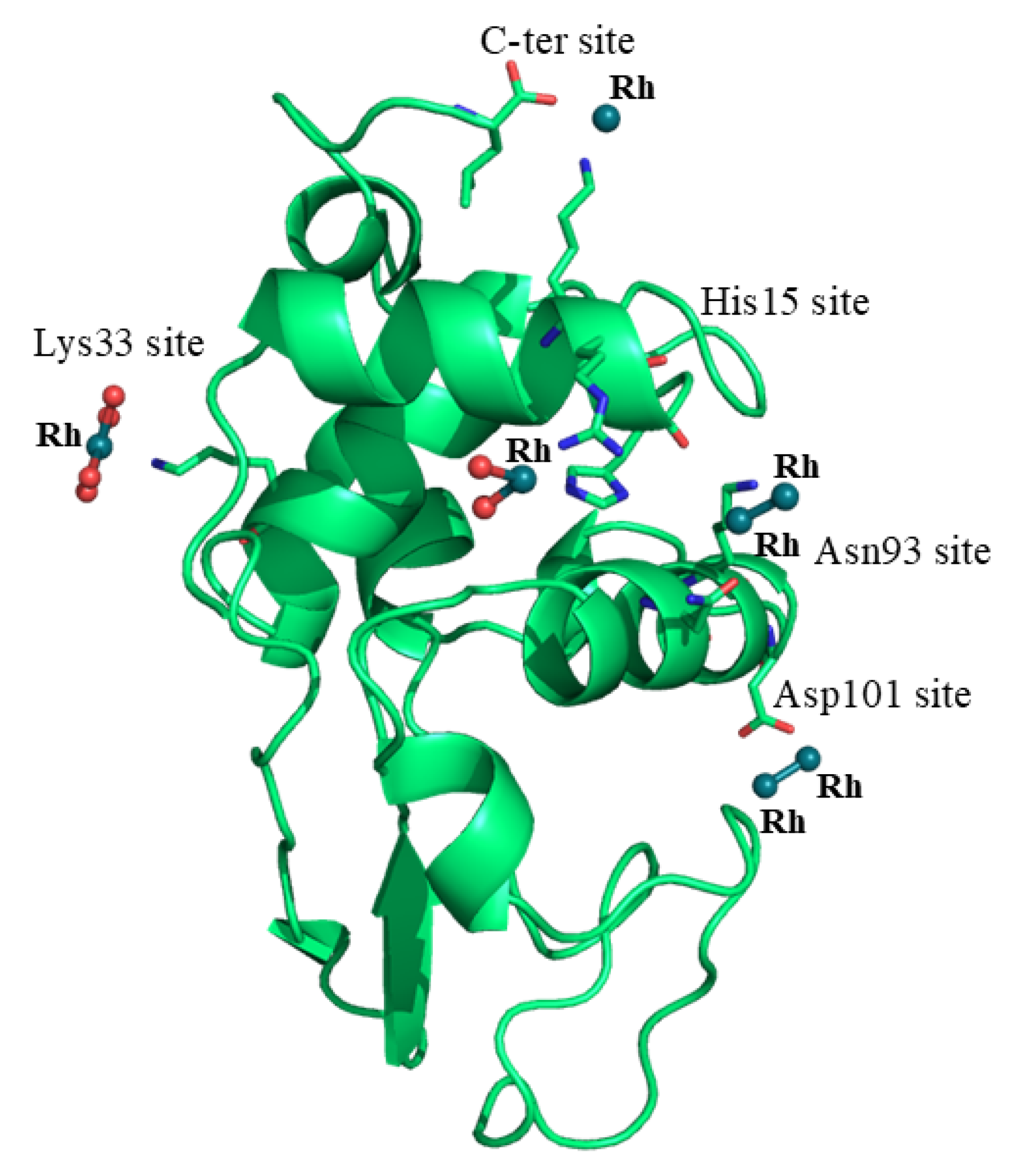
2.2. Structure of the Rh/HEWL Adduct at 310 K
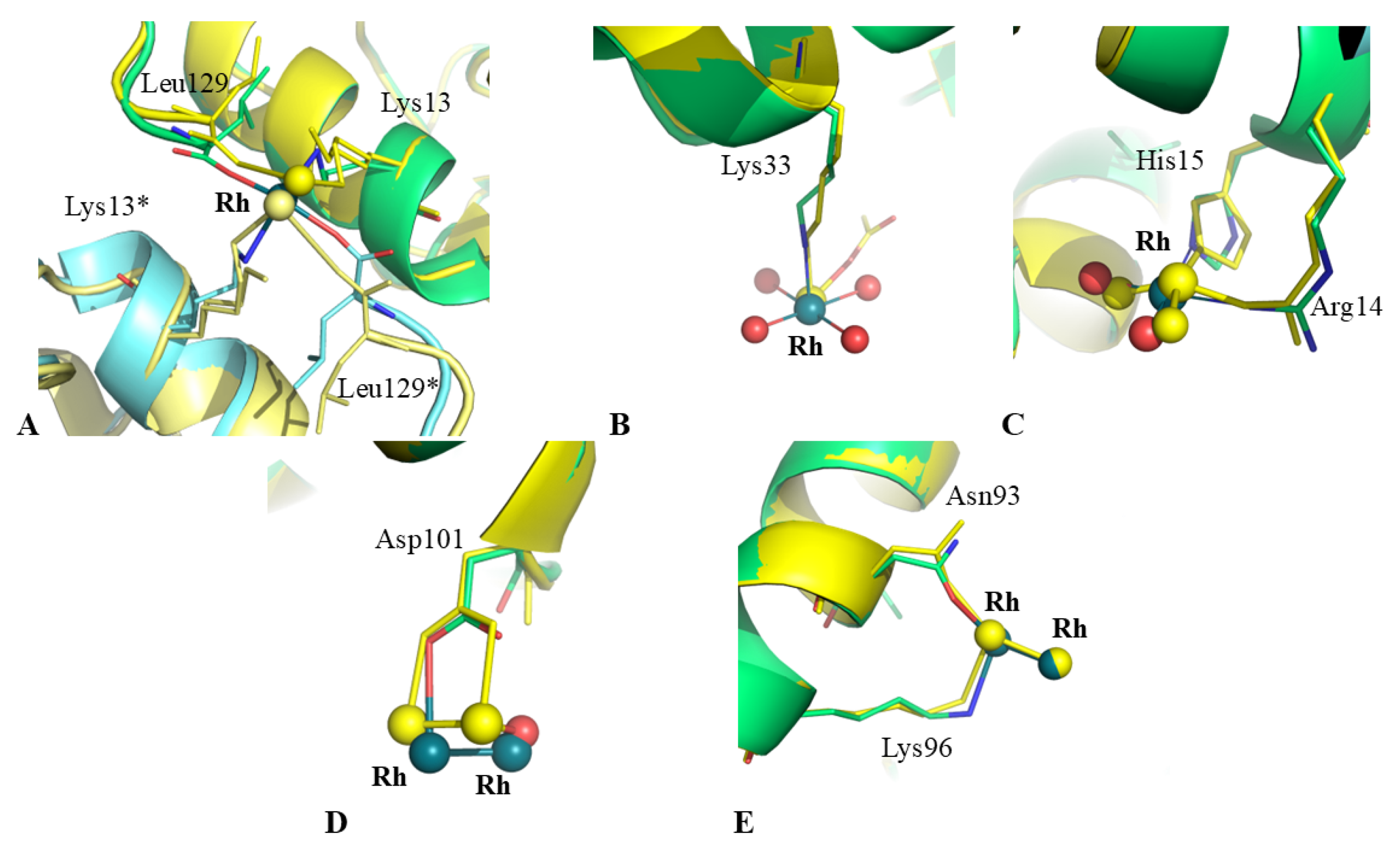
2.3. Structural Comparison with Data from Literature
3. Materials and Methods
3.1. Crystallization of HEWL and Formation of Rh/HEWL Adduct at 37 °C
3.2. Data Collection at 37 °C, Structure Solution and Refinement
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CD | Circular dichroism |
| HEWL | Hen egg white lysozyme |
| PDB | Protein Data Bank |
| R.m.s.d. | Root mean square deviation |
| RNase A | Bovine pancreatic ribonuclease |
References
- Thompson, M.C. Combining Temperature Perturbations with X-Ray Crystallography to Study Dynamic Macromolecules: A Thorough Discussion of Experimental Methods. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2023; Volume 688, pp. 255–305. ISBN 978-0-443-15926-8. [Google Scholar]
- Garman, E. “Cool” Crystals: Macromolecular Cryocrystallography and Radiation Damage. Curr. Opin. Struct. Biol. 2003, 13, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Garman, E.F.; Weik, M. Radiation Damage in Macromolecular Crystallography. In Protein Crystallography; Wlodawer, A., Dauter, Z., Jaskolski, M., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; Volume 1607, pp. 467–489. ISBN 978-1-4939-6998-2. [Google Scholar]
- Juers, D.H.; Matthews, B.W. Cryo-Cooling in Macromolecular Crystallography: Advantages, Disadvantages and Optimization. Quart. Rev. Biophys. 2004, 37, 105–119. [Google Scholar] [CrossRef]
- Doukov, T.; Herschlag, D.; Yabukarski, F. Instrumentation and Experimental Procedures for Robust Collection of X-Ray Diffraction Data from Protein Crystals across Physiological Temperatures. J. Appl. Crystallogr. 2020, 53, 1493–1501. [Google Scholar] [CrossRef] [PubMed]
- Aponte-Santamaría, C.; Fischer, G.; Båth, P.; Neutze, R.; De Groot, B.L. Temperature Dependence of Protein-Water Interactions in a Gated Yeast Aquaporin. Sci. Rep. 2017, 7, 4016. [Google Scholar] [CrossRef]
- Ferraro, G.; Garribba, E.; Merlino, A. Exploring Polyoxidovanadate–Protein Interaction. Trends Chem. 2025, 7, 3–6. [Google Scholar] [CrossRef]
- Hu, J.; Park, S.J.; Walter, T.; Orozco, I.J.; O.‘Dea, G.; Ye, X.; Du, J.; Lü, W. Physiological Temperature Drives TRPM4 Ligand Recognition and Gating. Nature 2024, 630, 509–515. [Google Scholar] [CrossRef]
- Schuller, M.; Correy, G.J.; Gahbauer, S.; Fearon, D.; Wu, T.; Díaz, R.E.; Young, I.D.; Carvalho Martins, L.; Smith, D.H.; Schulze-Gahmen, U.; et al. Fragment Binding to the Nsp3 Macrodomain of SARS-CoV-2 Identified through Crystallographic Screening and Computational Docking. Sci. Adv. 2021, 7, eabf8711. [Google Scholar] [CrossRef]
- Ebrahim, A.; Riley, B.T.; Kumaran, D.; Andi, B.; Fuchs, M.R.; McSweeney, S.; Keedy, D.A. The Temperature-Dependent Conformational Ensemble of SARS-CoV-2 Main Protease (Mpro). IUCrJ 2022, 9, 682–694. [Google Scholar] [CrossRef]
- Fischer, M. Macromolecular Room Temperature Crystallography. Quart. Rev. Biophys. 2021, 54, e1. [Google Scholar] [CrossRef]
- Jacobs, F.J.F.; Helliwell, J.R.; Brink, A. Body Temperature Protein X-Ray Crystallography at 37 °C: A Rhenium Protein Complex Seeking ac Physiological Condition Structure. Chem. Commun. 2024, 60, 14030–14033. [Google Scholar] [CrossRef]
- Tito, G.; Ferraro, G.; Garribba, E.; Merlino, A. Formation of Mixed-Valence Cage-Like Polyoxidovanadates at 37 °C Upon Reaction of VIV O(Acetylacetonato)2 with Lysozyme. Chem. –A Eur. J. 2025, 31, e202500488. [Google Scholar] [CrossRef]
- Merlino, A. Recent Advances in Protein Metalation: Structural Studies. Chem. Commun. 2021, 57, 1295–1307. [Google Scholar] [CrossRef]
- Marzo, T.; Ferraro, G.; Merlino, A.; Messori, L. Protein Metalation by Inorganic Anticancer Drugs. In Encyclopedia of Inorganic and Bioinorganic Chemistry; Scott, R.A., Ed.; Wiley: Hoboken, NJ, USA, 2020; pp. 1–17. ISBN 978-1-119-95143-8. [Google Scholar]
- Merlino, A.; Marzo, T.; Messori, L. Protein Metalation by Anticancer Metallodrugs: A Joint ESI MS and XRD Investigative Strategy. Chem. –A Eur. J. 2017, 23, 6942–6947. [Google Scholar] [CrossRef]
- Messori, L.; Merlino, A. Protein Metalation by Metal-Based Drugs: X-Ray Crystallography and Mass Spectrometry Studies. Chem. Commun. 2017, 53, 11622–11633. [Google Scholar] [CrossRef]
- Loreto, D.; Ferraro, G.; Merlino, A. Unusual Structural Features in the Adduct of Dirhodium Tetraacetate with Lysozyme. Int. J. Mol. Sci. 2021, 22, 1496. [Google Scholar] [CrossRef]
- Erck, A.; Rainen, L.; Whileyman, J.; Chang, I.-M.; Kimball, A.P.; Bear, J. Studies of Rhodium(II) Carboxylates as Potential Antitumor Agents. Exp. Biol. Med. 1974, 145, 1278–1283. [Google Scholar] [CrossRef]
- Bear, J.L.; Gray, H.B., Jr.; Rainen, L.; Chang, I.M.; Howard, R. Interaction of Rhodium (II) Carboxylates With Molecules of Biologic Importance. Cancer Chemother. Rep. 1975, 59, 611–620. [Google Scholar]
- Chang, I.; Woo, W.S. Effects of Rh2 (O2CC2H5) 4L2 on the Replication of Ehrlich Tumor Cells in Vivo. Korean Biochem. J. 1976, 9, 175–180. [Google Scholar]
- Loreto, D.; Maity, B.; Morita, T.; Nakamura, H.; Merlino, A.; Ueno, T. Cross-Linked Crystals of Dirhodium Tetraacetate/RNase A Adduct Can Be Used as Heterogeneous Catalysts. Inorg. Chem. 2023, 62, 7515–7524. [Google Scholar] [CrossRef]
- Hrdina, R. Dirhodium(II, II) Paddlewheel Complexes. Eur. J. Inorg. Chem. 2021, 2021, 501–528. [Google Scholar] [CrossRef]
- Chifotides, H.T.; Dunbar, K.R. Rhodium Compounds. In Multiple Bonds Between Metal Atoms; Cotton, F.A., Murillo, C.A., Walton, R.A., Eds.; Springer: New York, NY, USA, 2005; pp. 465–589. ISBN 978-0-387-25084-7. [Google Scholar]
- Cotton, F.A.; Murillo, C.A.; Walton, R.A. Multiple Bonds Between Metal Atoms; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Hope, H. Cryocrystallography of Biological Macromolecules: A Generally Applicable Method. Acta Crystallogr. B Struct. Sci. 1988, 44, 22–26. [Google Scholar] [CrossRef]
- Vaney, M.C.; Maignan, S.; Riès-Kautt, M.; Ducruix, A. High-Resolution Structure (1.33 Å) of a HEW Lysozyme Tetragonal Crystal Grown in the APCF Apparatus. Data and Structural Comparison with a Crystal Grown under Microgravity from SpaceHab-01 Mission. Acta Crystallogr. D Biol. Crystallogr. 1996, 52, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, F.J.F.; Helliwell, J.R.; Brink, A. Time-Series Analysis of Rhenium(I) Organometallic Covalent Binding to a Model Protein for Drug Development. IUCrJ 2024, 11, 359–373. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, G.; Pica, A.; Russo Krauss, I.; Pane, F.; Amoresano, A.; Merlino, A. Effect of Temperature on the Interaction of Cisplatin with the Model Protein Hen Egg White Lysozyme. J. Biol. Inorg. Chem. 2016, 21, 433–442. [Google Scholar] [CrossRef]
- Russo Krauss, I.; Ferraro, G.; Pica, A.; Márquez, J.A.; Helliwell, J.R.; Merlino, A. Principles and Methods Used to Grow and Optimize Crystals of Protein–Metallodrug Adducts, to Determine Metal Binding Sites and to Assign Metal Ligands. Metallomics 2017, 9, 1534–1547. [Google Scholar] [CrossRef]
- Vonrhein, C.; Flensburg, C.; Keller, P.; Sharff, A.; Smart, O.; Paciorek, W.; Womack, T.; Bricogne, G. Data Processing and Analysis with the autoPROC Toolbox. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 293–302. [Google Scholar] [CrossRef]
- Karplus, P.A.; Diederichs, K. Linking Crystallographic Model and Data Quality. Science 2012, 336, 1030–1033. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser Crystallographic Software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC 5 for the Refinement of Macromolecular Crystal Structures. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef]
- Willard, L. VADAR: A Web Server for Quantitative Evaluation of Protein Structure Quality. Nucleic. Acids. Res. 2003, 31, 3316–3319. [Google Scholar] [CrossRef] [PubMed]
- Shelley, K.L.; Dixon, T.P.E.; Brooks-Bartlett, J.C.; Garman, E.F. RABDAM: Quantifying Specific Radiation Damage in Individual Protein Crystal Structures. J. Appl. Crystallogr. 2018, 51, 552–559. [Google Scholar] [CrossRef] [PubMed]
- Shelley, K.L.; Garman, E.F. Quantifying and Comparing Radiation Damage in the Protein Data Bank. Nat. Commun. 2022, 13, 1314. [Google Scholar] [CrossRef] [PubMed]
- Merlino, A.; Russo Krauss, I.; Castellano, I.; Vendittis, E.D.; Rossi, B.; Conte, M.; Vergara, A.; Sica, F. Structure and Flexibility in Cold-Adapted Iron Superoxide Dismutases: The Case of the Enzyme Isolated from Pseudoalteromonas Haloplanktis. J. Struct. Biol. 2010, 172, 343–352. [Google Scholar] [CrossRef]
- Loreto, D.; Merlino, A. The Interaction of Rhodium Compounds with Proteins: A Structural Overview. Coord. Chem. Rev. 2021, 442, 213999. [Google Scholar] [CrossRef]
- Enriquez Garcia, A.; Jalilehvand, F.; Niksirat, P.; Gelfand, B.S. Methionine Binding to Dirhodium(II) Tetraacetate. Inorg. Chem. 2018, 57, 12787–12799. [Google Scholar] [CrossRef]
- Howard, R.A.; Spring, T.G.; Bear, J. L The Interaction of Rhodium (II) Carboxylates with Enzymes. Cancer Res. 1976, 36, 4402–4405. [Google Scholar]
- Loreto, D.; Esposito, A.; Demitri, N.; Guaragna, A.; Merlino, A. Reactivity of a Fluorine-Containing Dirhodium Tetracarboxylate Compound with Proteins. Dalton Trans. 2022, 51, 3695–3705. [Google Scholar] [CrossRef]
- Loreto, D.; Esposito, A.; Demitri, N.; Guaragna, A.; Merlino, A. Digging into Protein Metalation Differences Triggered by Fluorine Containing-Dirhodium Tetracarboxylate Analogues. Dalton Trans. 2022, 51, 7294–7304. [Google Scholar] [CrossRef]
- Loreto, D.; Fasulo, F.; Muñoz-García, A.B.; Pavone, M.; Merlino, A. Unexpected Imidazole Coordination to the Dirhodium Center in a Protein Environment: Insights from X-Ray Crystallography and Quantum Chemistry. Inorg. Chem. 2022, 61, 8402–8405. [Google Scholar] [CrossRef]
- Ferraro, G.; Pratesi, A.; Messori, L.; Merlino, A. Protein Interactions of Dirhodium Tetraacetate: A Structural Study. Dalton Trans. 2020, 49, 2412–2416. [Google Scholar] [CrossRef] [PubMed]
- Tolbatov, I.; Marrone, A. Reaction of Dirhodium and Diruthenium Paddlewheel Tetraacetate Complexes with Nucleophilic Protein Sites: A Computational Study. Inorganica Chim. Acta 2022, 530, 120684. [Google Scholar] [CrossRef]
- Tolbatov, I.; Umari, P.; Marrone, A. Diruthenium Paddlewheel Complexes Attacking Proteins: Axial versus Equatorial Coordination. Biomolecules 2024, 14, 530. [Google Scholar] [CrossRef] [PubMed]


| Crystal | |
|---|---|
| Temperature | 37 °C |
| Crystallization condition | 0.01 M HEPES pH 7.5 and 2.00 M sodium formate |
| Data collection | |
| Space group | P43212 |
| a = b (Å) | 80.43 |
| c (Å) | 36.14 |
| Resolution range (Å) | 56.87–2.12 (2.15–2.12) |
| Unique reflections | 6717 (342) |
| Completeness (%) | 94.0 (100.0) |
| Redundancy | 11.4 (11.8) |
| Rmerge (%) | 0.092 (2.788) |
| Average I/σ (I) | 13.2 (0.9) |
| CC1/2 | 0.998 (0.405) |
| Anom. completeness (%) | 93.7 (100.0) |
| Anom. redundancy | 6.4 (6.3) |
| Refinement | |
| Resolution range (Å) | 56.87–2.12 |
| N. of reflections | 6183 |
| N. of reflections (working set) | 382 |
| R-factor/R-free (%) | 19.8/26.1 |
| N. of atoms | 1043 |
| Average B-factors (Å2) | |
| All atoms | 61.2 |
| Rh occupancy | 0.50/1.00/1.00/0.60/0.50 |
| Rh atoms | 100.5/125.1/159.9/103.2/115.7/134.7/139.1 |
| R.m.s.d. from ideality | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.57 |
| Ramachandran values (%) | |
| Most favored/Additionally allowed | 91.9/7.3 |
| Outliers | 0.8 |
| Rh/HEWL Adduct | Crystal at 310 K | Data from Literature | Data from Literature | Data from Literature | Data from Literature | Data from Literature | Data from Literature |
|---|---|---|---|---|---|---|---|
| Crystallization condition | 0.01 M HEPES pH 7.5 and 2.0 M sodium formate | 0.1 M sodium acetate, 20% ethylene glicol, 0.1 M sodium nitrate | |||||
| PDB code | 9RUV | 7BE2 | 7BDZ | 7BE0 | 7BE1 | 7BEB | 7BEC |
| Binding sites | Metal fragments at binding sites | ||||||
| C-ter site | Rh (0.50) Rh * (0.50) Leu129 (1) Leu129 * (1) Lys13 (1) Lys13 * (1) | Rh (0.45) Rh * (0.45) Leu129 (1) Leu129 * (1) Lys13 (1) Lys13 * (1) H2O (0.5) H2O * (0.5) | Rh (0.65) Rh * (0.65) Leu129 (1) Leu129 * (1) Lys13 (1) Lys13 * (1) Act (0.60) Act (0.60) H2O (0.65) H2O * (0.65) | Rh (0.65) Rh * (0.65) Leu129 (1) Leu129 * (1) Lys13 (1) Lys13 * (1) Act (0.65) Act (0.65) H2O (0.65) H2O* (0.65) | Rh (0.65) Rh* (0.65) Leu129 (1) Leu129* (1) Lys13 (1) Lys13* (1) Act (0.65) Act (0.65) H2O (0.65) H2O * (0.65) | Rh (0.7) Rh * (0.7) Leu129 (1) Leu129 * (1) Lys13 (1) Lys13 * (1) Act (0.7) Act (0.7) H2O (0.7) H2O * (0.7) | Rh (0.4) Rh * (0.4) Leu129 (1) Leu129 * (1) Lys13 (1) Lys13 * (1) |
| Lys33 site | Rh (1) Lys33 (1) H2O (1) H2O (1) H2O (1) H2O (1) | Rh (0.20) Lys33 (1) Act (0.33) | - | - | - | - | - |
| His15 site | Rh (1) His15 (1) Arg14 (1) H2O (1) H2O (1) | Rh (0.35) His15 (1) Arg14 (1) H2O (0.35) H2O (0.35) | Rh (0.35) His15 (1) Arg14 (1) Act (1) H2O (0.35) H2O (0.65) | Rh (0.5) His15 (1) Arg14 (1) Act (1) H2O (0.5) | Rh (0.3) His15 (1) Arg14 (0.5) Act (1) | Rh (0.3) His15 (1) Arg14 (0.5) Act (1) H2O (0.3) H2O (0.3) | Rh (0.25) His15 (1) Arg14 (0.5) Act (1) H2O (1) |
| Asp101 site | Rh (0.60) Rh (0.60) Asp101 (1) | Rh (0.40) Rh (0.40) Asp101 (1) H2O (0.40) | - | - | - | - | - |
| Asn93 site | Rh (0.50) Rh (0.50) Asn93 (1) Lys96 (1) | Rh (0.30) Rh (0.30) Asn93 (1) Lys96 (1) H2O (1) | - | - | - | - | - |
| Asp18 site | - | - | Rh (0.40) Rh (0.40) Asp18 (1) | Rh (0.20) Rh (0.20) Asp18 (1) H2O (0.20) | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tito, G.; Ferraro, G.; Merlino, A. Dirhodium Tetraacetate Binding to Lysozyme at Body Temperature. Int. J. Mol. Sci. 2025, 26, 6582. https://doi.org/10.3390/ijms26146582
Tito G, Ferraro G, Merlino A. Dirhodium Tetraacetate Binding to Lysozyme at Body Temperature. International Journal of Molecular Sciences. 2025; 26(14):6582. https://doi.org/10.3390/ijms26146582
Chicago/Turabian StyleTito, Gabriella, Giarita Ferraro, and Antonello Merlino. 2025. "Dirhodium Tetraacetate Binding to Lysozyme at Body Temperature" International Journal of Molecular Sciences 26, no. 14: 6582. https://doi.org/10.3390/ijms26146582
APA StyleTito, G., Ferraro, G., & Merlino, A. (2025). Dirhodium Tetraacetate Binding to Lysozyme at Body Temperature. International Journal of Molecular Sciences, 26(14), 6582. https://doi.org/10.3390/ijms26146582








