Circulating Bacterial DNA as a Novel Blood-Based Biomarker in Type 2 Diabetes Mellitus (DM2): Results from the PROMOTERA Study
Abstract
1. Introduction
2. Results
2.1. Clinical Characteristics of the PROMOTERA Cohort
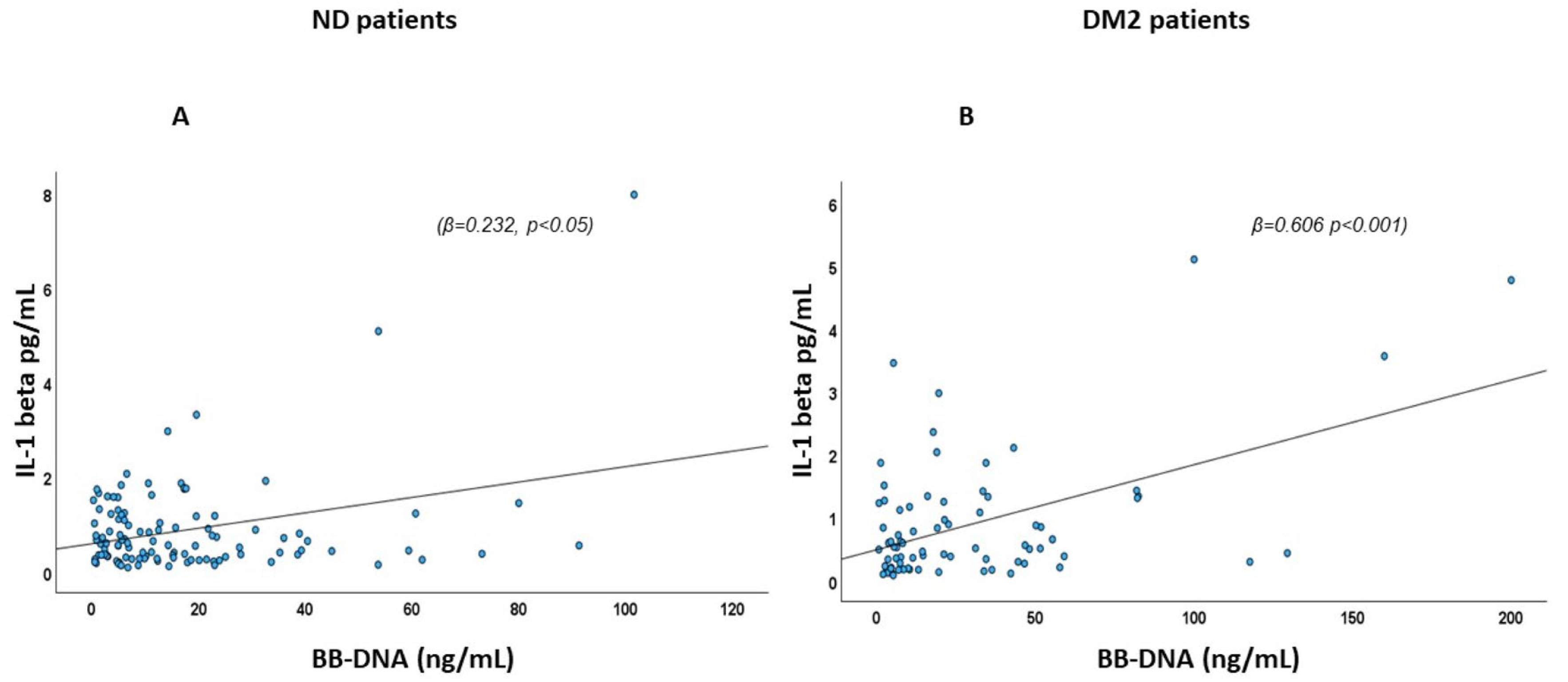
2.2. Association of BB-DNA with Plasma IL-1β Levels
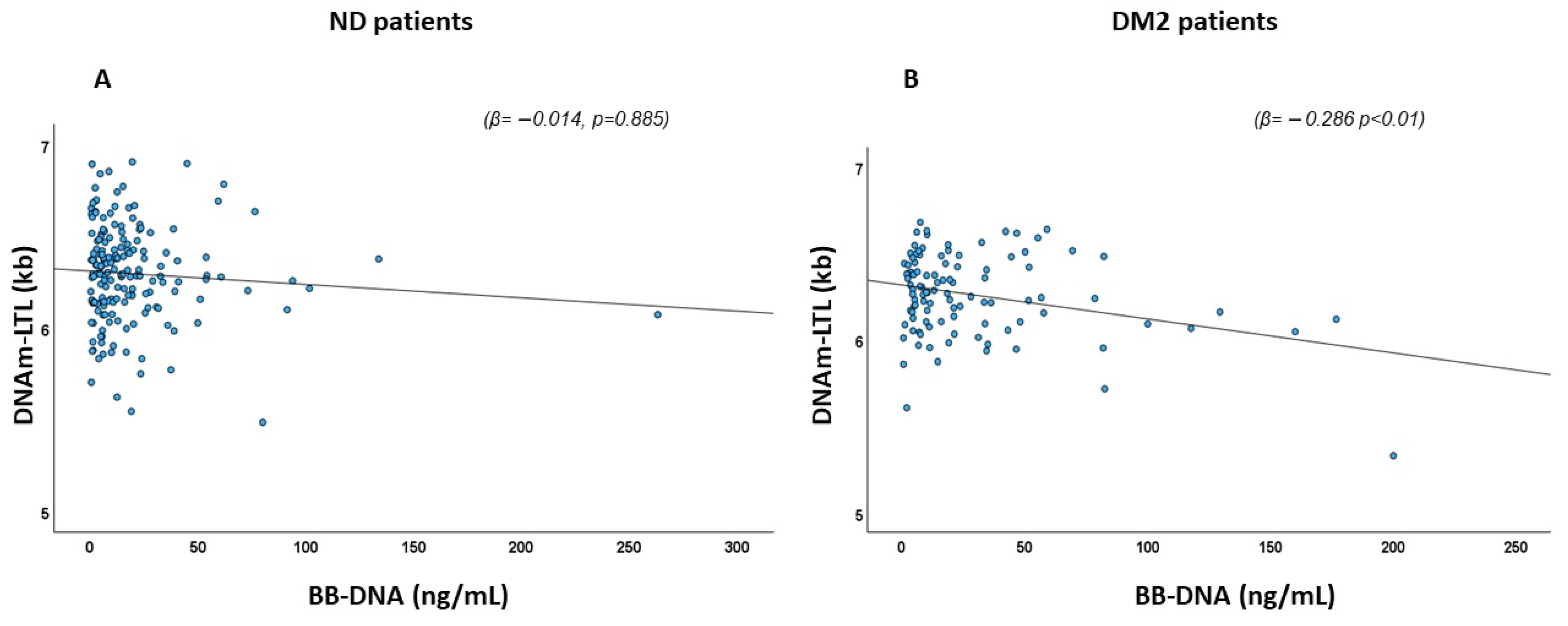
2.3. Association of BB-DNA and DNA Methylation-Based Estimator of Leukocyte Telomere Length (DNAm-LTL)
2.4. Analysis of the Association of BB-DNA and DNAm Biological Age Estimators
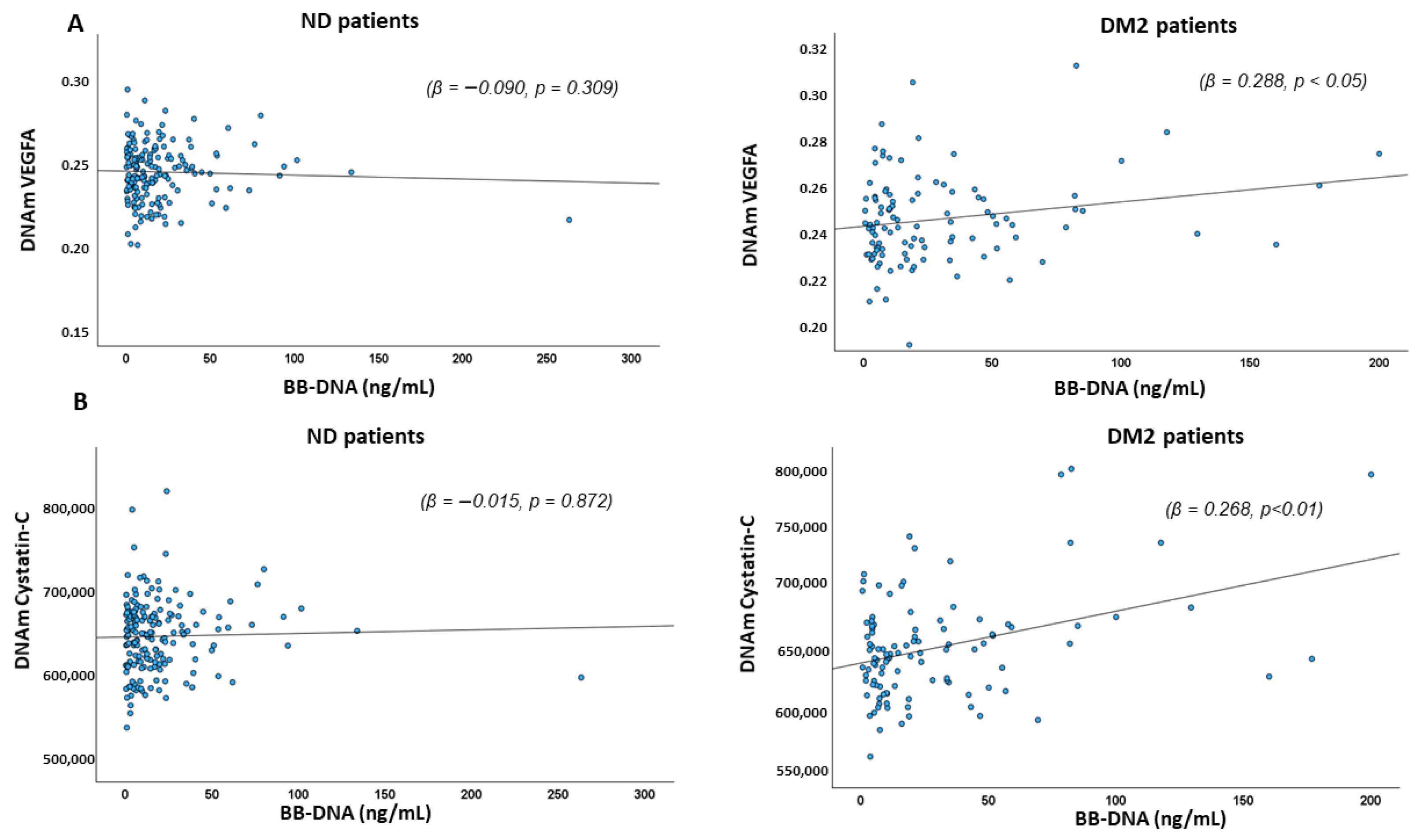
2.5. Association Between BB-DNA and DNAm-Based Estimators of Angiogenic, Pro-Inflammatory Factors, and DNAm-Based Estimators of Blood Cell Types
2.6. Association Between BB-DNA and Epigenetic Signatures of Type 2 Diabetes Mellitus
2.7. Associations Between BB-DNA and Epigenetic Markers in the Overall Cohort
3. Discussion
4. Materials and Methods
4.1. Study Population, Recruitment, Data, and Blood Collection
4.2. DNA Extraction and Methylation Assay
4.3. Blood Bacterial DNA (BB-DNA) Quantification
4.4. Estimation of Surrogate Biomarkers
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bellary, S.; Kyrou, I.; Brown, J.E.; Bailey, C.J. Type 2 diabetes mellitus in older adults: Clinical considerations and management. Nat. Rev. Endocrinol. 2021, 17, 534–548. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Z.; Ren, Y.; Wang, Y.; Fang, J.; Yue, H.; Ma, S.; Guan, F. Aging and age-related diseases: From mechanisms to therapeutic strategies. Biogerontology 2021, 22, 165–187. [Google Scholar] [CrossRef]
- Ikegami, H.; Hiromine, Y.; Noso, S. Insulin-dependent diabetes mellitus in older adults: Current status and future prospects. Geriatr. Gerontol. Int. 2022, 22, 549–553. [Google Scholar] [CrossRef]
- Halim, M.; Halim, A. The effects of inflammation, aging and oxidative stress on the pathogenesis of diabetes mellitus (type 2 diabetes). Diabetes Metab. Syndr. 2019, 13, 1165–1172. [Google Scholar] [CrossRef]
- Sanz-Cánovas, J.; López-Sampalo, A.; Cobos-Palacios, L.; Ricci, M.; Hernández-Negrín, H.; Mancebo-Sevilla, J.J.; Álvarez-Recio, E.; López-Carmona, M.D.; Pérez-Belmonte, L.M.; Gómez-Huelgas, R.; et al. Management of Type 2 Diabetes Mellitus in Elderly Patients with Frailty and/or Sarcopenia. Int. J. Environ. Res. Public Health 2022, 19, 8677. [Google Scholar] [CrossRef]
- Zatterale, F.; Longo, M.; Naderi, J.; Raciti, G.A.; Desiderio, A.; Miele, C.; Beguinot, F. Chronic Adipose Tissue Inflammation Linking Obesity to Insulin Resistance and Type 2 Diabetes. Front. Physiol. 2020, 10, 1607. [Google Scholar] [CrossRef]
- Kabacik, S.; Lowe, D.; Fransen, L.; Leonard, M.; Ang, S.L.; Whiteman, C.; Corsi, S.; Cohen, H.; Felton, S.; Bali, R.; et al. The relationship between epigenetic age and the hallmarks of aging in human cells. Nat. Aging 2022, 2, 484–493. [Google Scholar] [CrossRef]
- Johansson, A.; Enroth, S.; Gyllensten, U. Continuous Aging of the Human DNA Methylome Throughout the Human Lifespan. PLoS ONE 2013, 8, e67378. [Google Scholar] [CrossRef]
- Horvath, S.; Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev Genet. 2018, 19, 371–384. [Google Scholar] [CrossRef]
- Simpson, D.J.; Chandra, T. Epigenetic age prediction. Aging Cell 2021, 20, e13452. [Google Scholar] [CrossRef]
- Paparazzo, E.; Geracitano, S.; Lagani, V.; Bartolomeo, D.; Aceto, M.A.; D’Aquila, P.; Citrigno, L.; Bellizzi, D.; Passarino, G.; Montesanto, A. A Blood-Based Molecular Clock for Biological Age Estimation. Cells 2022, 12, 32. [Google Scholar] [CrossRef]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.B.; Gao, Y.; et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef]
- Oblak, L.; van der Zaag, J.; Higgins-Chen, A.T.; Levine, M.E.; Boks, M.P. A systematic review of biological, social and environmental factors associated with epigenetic clock acceleration. Ageing Res. Rev. 2021, 69, 101348. [Google Scholar] [CrossRef]
- Arpón, A.; Milagro, F.I.; Santos, J.L.; García-Granero, M.; Riezu-Boj, J.I.; Martínez, J.A. Interaction Among Sex, Aging, and Epigenetic Processes Concerning Visceral Fat, Insulin Resistance, and Dyslipidaemia. Front. Endocrinol. 2019, 10, 496. [Google Scholar] [CrossRef]
- Fraszczyk, E.; Thio, C.H.L.; Wackers, P.; Dollé, M.E.T.; Bloks, V.W.; Hodemaekers, H.; Picavet, H.S.; Stynenbosch, M.; Verschuren, W.M.M.; Snieder, H.; et al. DNA methylation trajectories and accelerated epigenetic aging in incident type 2 diabetes. Geroscience 2022, 44, 2671–2684. [Google Scholar] [CrossRef]
- Takahashi, K. Influence of bacteria on epigenetic gene control. Cell. Mol. Life Sci. 2014, 71, 1045–1054. [Google Scholar] [CrossRef]
- Pietrzak, B.; Kawacka, I.; Olejnik-Schmidt, A.; Schmidt, M. Circulating Microbial Cell-Free DNA in Health and Disease. Int. J. Mol. Sci. 2023, 24, 3051. [Google Scholar] [CrossRef]
- D’Aquila, P.; Giacconi, R.; Malavolta, M.; Piacenza, F.; Bürkle, A.; Villanueva, M.M.; Dollé, M.E.T.; Jansen, E.; Grune, T.; Gonos, E.S.; et al. Microbiome in Blood Samples from the General Population Recruited in the MARK-AGE Project: A Pilot Study. Front. Microbiol. 2021, 12, 707515. [Google Scholar] [CrossRef]
- Goraya, M.U.; Li, R.; Mannan, A.; Gu, L.; Deng, H.; Wang, G. Human circulating bacteria and dysbiosis in non-infectious diseases. Front. Cell. Infect. Microbiol. 2022, 12, 932702. [Google Scholar] [CrossRef]
- Giacconi, R.; D’Aquila, P.; Balietti, M.; Giuli, C.; Malavolta, M.; Piacenza, F.; Costarelli, L.; Postacchini, D.; Passarino, G.; Bellizzi, D.; et al. Bacterial DNAemia in Alzheimer’s Disease and Mild Cognitive Impairment: Association with Cognitive Decline, Plasma BDNF Levels, and Inflammatory Response. Int. J. Mol. Sci. 2022, 24, 78. [Google Scholar] [CrossRef]
- Giacconi, R.; D’Aquila, P.; Malavolta, M.; Piacenza, F.; Bürkle, A.; Villanueva, M.M.; Dollé, M.E.T.; Jansen, E.; Grune, T.; Gonos, E.S.; et al. Bacterial DNAemia in Older Participants and Nonagenarian Offspring and Association with Redox Biomarkers: Results From MARK-AGE Study. J. Gerontol. A Biol. Sci. Med. Sci. 2023, 78, 42–50. [Google Scholar] [CrossRef]
- Giacconi, R.; D’Aquila, P.; Cardelli, M.; Piacenza, F.; Pierpaoli, E.; Sena, G.; Di Rosa, M.; Bonfigli, A.R.; Galeazzi, R.; Cherubini, A.; et al. Blood circulating bacterial DNA in hospitalized old COVID-19 patients. Immun. Ageing 2023, 20, 76. [Google Scholar] [CrossRef]
- Lu, A.T.; Seeboth, A.; Tsai, P.C.; Sun, D.; Quach, A.; Reiner, A.P.; Kooperberg, C.; Ferrucci, L.; Hou, L.; Baccarelli, A.A.; et al. DNA methylation-based estimator of telomere length. Aging 2019, 11, 5895–5923. [Google Scholar] [CrossRef]
- Lu, A.T.; Quach, A.; Wilson, J.G.; Reiner, A.P.; Aviv, A.; Raj, K.; Hou, L.; Baccarelli, A.A.; Li, Y.; Stewart, J.D.; et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 2019, 11, 303–327. [Google Scholar] [CrossRef]
- Levine, M.E.; Lu, A.T.; Quach, A.; Chen, B.H.; Assimes, T.L.; Bandinelli, S.; Hou, L.; Baccarelli, A.A.; Stewart, J.D.; Li, Y.; et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging 2018, 10, 573–591. [Google Scholar] [CrossRef]
- Horvath, S.; Oshima, J.; Martin, G.M.; Lu, A.T.; Quach, A.; Cohen, H.; Felton, S.; Matsuyama, M.; Lowe, D.; Kabacik, S.; et al. Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies. Aging 2018, 10, 1758–1775. [Google Scholar] [CrossRef]
- Zhang, Q.; Vallerga, C.L.; Walker, R.M.; Lin, T.; Henders, A.K.; Montgomery, G.W.; He, J.; Fan, D.; Fowdar, J.; Kennedy, M.; et al. Improved precision of epigenetic clock estimates across tissues and its implication for biological ageing. Genome Med. 2019, 11, 54. [Google Scholar] [CrossRef]
- Lu, A.T.; Binder, A.M.; Zhang, J.; Yan, Q.; Reiner, A.P.; Cox, S.R.; Corley, J.; Harris, S.E.; Kuo, P.L.; Moore, A.Z.; et al. DNA methylation GrimAge version 2. Aging 2022, 14, 9484–9549. [Google Scholar] [CrossRef]
- Domingo-Relloso, A.; Gribble, M.O.; Riffo-Campos, A.L.; Haack, K.; Cole, S.A.; Tellez-Plaza, M.; Umans, J.G.; Fretts, A.M.; Zhang, Y.; Fallin, M.D.; et al. Epigenetics of type 2 diabetes and diabetes-related outcomes in the Strong Heart Study. Clin. Epigenetics 2022, 14, 177. [Google Scholar] [CrossRef]
- Seo, H.; Park, J.H.; Hwang, J.T.; Choi, H.K.; Park, S.H.; Lee, J. Epigenetic Profiling of Type 2 Diabetes Mellitus: An Epigenome-Wide Association Study of DNA Methylation in the Korean Genome and Epidemiology Study. Genes 2023, 14, 2207. [Google Scholar] [CrossRef]
- Sato, J.; Kanazawa, A.; Ikeda, F.; Yoshihara, T.; Goto, H.; Abe, H.; Komiya, K.; Kawaguchi, M.; Shimizu, T.; Ogihara, T.; et al. Gut dysbiosis and detection of "live gut bacteria" in blood of Japanese patients with type 2 diabetes. Diabetes Care 2014, 37, 2343–2350. [Google Scholar] [CrossRef]
- Amar, J.; Serino, M.; Lange, C.; Chabo, C.; Iacovoni, J.; Mondot, S.; Lepage, P.; Klopp, C.; Mariette, J.; Bouchez, O.; et al. Involvement of tissue bacteria in the onset of diabetes in humans: Evidence for a concept. Diabetologia 2011, 54, 3055–3061. [Google Scholar] [CrossRef]
- Qiu, J.; Zhou, H.; Jing, Y.; Dong, C. Association between blood microbiome and type 2 diabetes mellitus: A nested case-control study. J. Clin. Lab. Anal. 2019, 33, e22842. [Google Scholar] [CrossRef]
- Salameh, Y.; Bejaoui, Y.; El Hajj, N. DNA Methylation Biomarkers in Aging and Age-Related Diseases. Front. Genet. 2020, 11, 171. [Google Scholar] [CrossRef]
- Maghbooli, Z.; Hossein-Nezhad, A.; Larijani, B.; Amini, M.; Keshtkar, A. Global DNA methylation as a possible biomarker for diabetic retinopathy. Diabetes Metab. Res. Rev. 2015, 31, 183–189. [Google Scholar] [CrossRef]
- Sabbatinelli, J.; Giuliani, A.; Kwiatkowska, K.M.; Matacchione, G.; Belloni, A.; Ramini, D.; Prattichizzo, F.; Pellegrini, V.; Piacenza, F.; Tortato, E.; et al. DNA Methylation-derived biological age and long-term mortality risk in subjects with type 2 diabetes. Cardiovasc. Diabetol. 2024, 23, 250. [Google Scholar] [CrossRef]
- Sawicki, K.; Matysiak-Kucharek, M.; Gorczyca-Siudak, D.; Kruszewski, M.; Kurzepa, J.; Kapka-Skrzypczak, L.; Dziemidok, P. Leukocyte Telomere Length as a Marker of Chronic Complications in Type 2 Diabetes Patients: A Risk Assessment Study. Int. J. Mol. Sci. 2025, 26, 290. [Google Scholar] [CrossRef]
- Liang, X.; Aouizerat, B.E.; So-Armah, K.; Cohen, M.H.; Marconi, V.C.; Xu, K.; Justice, A.C. DNA methylation-based telomere length is associated with HIV infection, physical frailty, cancer, and all-cause mortality. Aging Cell 2024, 23, e14174. [Google Scholar] [CrossRef]
- Ricciardi, C.A.; Gnudi, L. Vascular growth factors as potential new treatment in cardiorenal syndrome in diabetes. Eur. J. Clin. Investig. 2021, 51, e13579. [Google Scholar] [CrossRef]
- Dong, H.; Guo, W.; Yue, R.; Sun, X.; Zhou, Z. Nuclear Nicotinamide Adenine Dinucleotide Deficiency by Nmnat1 Deletion Impaired Hepatic Insulin Signaling, Mitochondrial Function, and Hepatokine Expression in Mice Fed a High-Fat Diet. Lab. Investig. 2024, 104, 100329. [Google Scholar] [CrossRef]
- Linh, H.T.; Iwata, Y.; Senda, Y.; Sakai-Takemori, Y.; Nakade, Y.; Oshima, M.; Nakagawa-Yoneda, S.; Ogura, H.; Sato, K.; Minami, T.; et al. Intestinal Bacterial Translocation Contributes to Diabetic Kidney Disease. J. Am. Soc. Nephrol. 2022, 33, 1105–1119. [Google Scholar] [CrossRef]
- Hasegawa, K.; Tamaki, M.; Sakamaki, Y.; Wakino, S. Nmnat1 Deficiency Causes Mitoribosome Excess in Diabetic Nephropathy Mediated by Transcriptional Repressor HIC1. Int. J. Mol. Sci. 2024, 25, 6384. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.W.; Huang, W.C.; Wang, C.; Lin, C.Y. Associations of Serum Cystatin C, DNAm Cystatin C, Renal Function, and Mortality in U.S. Adults. Life 2024, 15, 13. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Levy, M.; Grosheva, I.; Zheng, D.; Soffer, E.; Blacher, E.; Braverman, S.; Tengeler, A.C.; Barak, O.; Elazar, M.; et al. Hyperglycemia drives intestinal barrier dysfunction and risk for enteric infection. Science 2018, 359, 1376–1383. [Google Scholar] [CrossRef] [PubMed]
- Al-Dabet, M.M.; Shahzad, K.; Elwakiel, A.; Sulaj, A.; Kopf, S.; Bock, F.; Gadi, I.; Zimmermann, S.; Rana, R.; Krishnan, S.; et al. Reversal of the renal hyperglycemic memory in diabetic kidney disease by targeting sustained tubular p21 expression. Nat. Commun. 2022, 13, 5062. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.X.; Liu, Z.J. Potential roles of neutrophils in regulating intestinal mucosal inflammation of inflammatory bowel disease. J. Dig. Dis. 2017, 18, 495–503. [Google Scholar] [CrossRef]
- Tian, Z.; Zhang, Y.; Zheng, Z.; Zhang, M.; Zhang, T.; Jin, J.; Zhang, X.; Yao, G.; Kong, D.; Zhang, C.; et al. Gut microbiome dysbiosis contributes to abdominal aortic aneurysm by promoting neutrophil extracellular trap formation. Cell Host Microbe. 2022, 30, 1450–1463.e8. [Google Scholar] [CrossRef]
- Dai, M.; Wu, J.; Ji, Z.; Chen, P.; Yang, C.; Luo, J.; Shan, P.; Xu, M. Construction of a metabolic-immune model for predicting the risk of diabetic nephropathy and study of gut microbiota. J Diabetes Investig. 2025, 16, 863–873. [Google Scholar] [CrossRef]
- Bustacchini, S.; Abbatecola, A.M.; Bonfigli, A.R.; Chiatti, C.; Corsonello, A.; Di Stefano, G.; Galeazzi, R.; Fabbietti, P.; Lisa, R.; Guffanti, E.E.; et al. The Report-AGE project: A permanent epidemiological observatory to identify clinical and biological markers of health outcomes in elderly hospitalized patients in Italy. Aging Clin. Exp. Res. 2015, 27, 893–901. [Google Scholar] [CrossRef]
- Di Rosa, M.; D’Alia, S.; Guarasci, F.; Soraci, L.; Pierpaoli, E.; Lenci, F.; Ricci, M.; Onder, G.; Volpato, S.; Ruggiero, C.; et al. Cognitive Impairment, Chronic Kidney Disease, and 1-Year Mortality in Older Patients Discharged from Acute Care Hospital. J. Clin. Med. 2020, 9, 2202. [Google Scholar] [CrossRef] [PubMed]
- Baldelli, L.; Pirazzini, C.; Sambati, L.; Ravaioli, F.; Gentilini, D.; Calandra-Buonaura, G.; Guaraldi, P.; Franceschi, C.; Cortelli, P.; Garagnani, P.; et al. Epigenetic clocks suggest accelerated aging in patients with isolated REM Sleep Behavior Disorder. NPJ Parkinsons Dis. 2023, 9, 48. [Google Scholar] [CrossRef] [PubMed]

| ND (178) | DM2 (107) | p Value | |
|---|---|---|---|
| Age (yrs) | 83.1± 0.6 | 83.4 ± 0.7 | NS |
| Sex, Male n (%) | 77 (43.3%) | 53 (49.5%) | NS |
| Hypertension n (%) | 137 (77.0%) | 90 (84.1%) | NS |
| AF n (%) | 0 (0%) | 55 (51.4%) | <0.001 |
| IHD n (%) | 18 (10.1%) | 20 (18.7%) | 0.039 |
| CHF n (%) | 20 (11.2%) | 24 (22.4%) | 0.011 |
| Stroke n (%) | 8 (5.6%) | 12 (14.5%) | 0.024 |
| CKD n (%) | 33 (18.5%) | 35 (32.7%) | 0.007 |
| COPD n (%) | 24 (13.5%) | 19 (17.8%) | NS |
| Anemia n (%) | 34 (19.1%) | 20 (18.7%) | NS |
| Infection n (%) | 54 (30.3%) | 29 (27.1%) | NS |
| Dementia n (%) | 25 (14.0%) | 23 (21.5%) | NS |
| NLR | 4.2 ± 0.3 | 5.2 ± 0.4 | 0.050 |
| Neutrophils (×103/μL) | 5.2 ± 0.2 | 6.5 ± 0.4 | <0.01 |
| Lymphocytes (×103/μL) | 1.54 ± 0.05 | 1.52 ± 0.07 | NS |
| SIRI index | 2.4 ± 0.9 | 3.9 ± 0.4 | 0.050 |
| Fasting Glucose (mg/dL) | 93.9 ± 1.6 | 128.5 ± 5.6 | <0.001 |
| Creatinine (mg/dL) | 1.14 ± 0.04 | 1.25 ± 0.06 | NS |
| eGFR, mL/min/1.73 m2 | 58.9 ± 1.7 | 52.5 ±1.9 | 0.014 |
| BB-DNA (ng/mL) | 19.8 ± 3.5 | 30.5 ± 4.1 | 0.012 |
| ND Patients | DM2 Patients | References | |||
|---|---|---|---|---|---|
| Beta | p Value | Beta | p Value | ||
| Age Acceleration Residual | −0.008 | 0.925 | 0.191 | 0.063 | [12] |
| Age Acceleration Grimage | −0.034 | 0.711 | 0.023 | 0.848 | [25] |
| Age Acceleration PhenoAge | −0.058 | 0.476 | 0.202 | 0.045 | [26] |
| DNAmAge | −0.057 | 0.582 | 0.157 | 0.191 | [13] |
| DNAmAge | −0.048 | 0.682 | 0.246 | 0.071 | [12] |
| IEAA | −0.063 | 0.430 | 0.087 | 0.391 | [13] |
| IEAA | −0.055 | 0.490 | 0.164 | 0.105 | [12] |
| DNAmPhenoAge | −0.074 | 0.468 | 0.224 | 0.045 | [26] |
| DNAmAge Skin Blood Clock | −0.017 | 0.882 | 0.455 | 0.004 | [27] |
| EpigeneticAge | −0.113 | 0.374 | 0.345 | 0.035 | [28] |
| DNAm GrimAge * | −0.116 | 0.394 | −0.031 | 0.487 | [25] |
| DNAm GrimAge2 * | −0.060 | 0.629 | −0.078 | 0.613 | [29] |
| CpG Sites | UCSC_RefGene_Group | Standardized Coefficients—Beta | p Value |
|---|---|---|---|
| IFNγ_cg09711238 | TSS200 | −0.528 | <0.001 |
| IFNγ_cg26227465 | TSS200 | 0.265 | 0.035 |
| IFNγ_cg12640631 | TSS1500 | −0.182 | 0.047 |
| TNFα_cg08553327 * | 1stExon | 0.408 | <0.001 |
| TNFα_cg12681001 * | 1stExon | −0.352 | 0.004 |
| IL6_cg05265849 | Body | −0.122 | <0.01 |
| IL6_cg21785978 | Body; TSS200 | 0.232 | 0.007 |
| IL6_cg01770232 | TSS1500 | −0.178 | 0.031 |
| IL10_cg17067005 | Body | −0.192 | 0.047 |
| IL1β_cg20983042 | TSS1500 | −0.237 | 0.010 |
| IL1β_cg07250315 | Body | 0.619 | <0.001 |
| IL1β_cg07935264 | TSS200 | −0.374 | 0.002 |
| IL1β_cg15218327 | Body | −0.269 | 0.005 |
| NFKB1_cg27333178 | 5′UTR | −0.11 | <0.001 |
| NFKB1_cg23462257 | TSS1500 | −0.205 | <0.001 |
| NFKB1_cg23898555 | Body | 0.129 | 0.011 |
| NFKB1_cg07955720 | Body | −0.111 | 0.028 |
| CRP_cg25257346 | Body | −0.477 | <0.001 |
| CRP_cg08474603 | TSS200 | 0.102 | 0.018 |
| CRP_cg24976805 | TSS1500 | −0.510 | <0.001 |
| CRP_cg09267046 | Body | 0.291 | <0.001 |
| CDKN1A/p21_cg13662121 | TSS1500 | −0.352 | <0.001 |
| CDKN1A/p21_cg09774179 | Body | 0.300 | <0.001 |
| CDKN1A/p21_cg17526952 * | - | −0.222 | 0.006 |
| CDKN1A/p21_cg06827361 | Body | −0.228 | 0.007 |
| CDKN2A/p16_cg27048359 | Body | −0.645 | <0.001 |
| CDKN2A/p16_cg13601799 * | 1stExon; Body | 0.217 | 0.003 |
| CDKN2A/p16_cg23426614 * | TSS200; TSS1500 | −0.187 | 0.011 |
| CDKN2A/p16_cg01694391 * | TSS1500; Body | −0.198 | 0.010 |
| TP53_cg27105645 | 5′UTR | −0.267 | 0.002 |
| TP53_cg10653997 | 5′UTR | −0.278 | <0.001 |
| TP53_cg08691422 | 5′UTR; TSS1500 | −0.239 | 0.006 |
| TP53_cg09168066 | Body; TSS1500; ExonBnd; 5′UTR | −0.303 | <0.001 |
| TP53_cg07343727 * | 5′UTR; TSS1500; TSS200 | −0.232 | 0.007 |
| TP53_cg02166782 * | TSS1500; 5′UTR; TSS200 | 0.225 | 0.008 |
| TP53_cg15206330 * | TSS1500; 5′UTR; TSS200 | −0.168 | 0.047 |
| CpG Sites | UCSC_RefGene_ Group | Gene Function | Standardized Coefficients | |
|---|---|---|---|---|
| Beta | p Value | |||
| IGF1_cg02823066 | Body | Metabolic regulation (glucose and lipid metabolism); growth and development | −0.270 | 0.004 |
| IGF1_cg25163611 * | TSS1500 | −0.319 | <0.001 | |
| IGF1_cg18504440 | 1stExon; 5′UTR; Body | −0.219 | 0.021 | |
| PDK4_cg22758834 | Body | Regulation of glucose and fatty acid metabolism | −0.215 | 0.025 |
| PDK4_cg17075888 | Body | −0.194 | 0.042 | |
| ABCG1_cg27641007 | Body | Regulation of efflux of phospholipids such as sphingomyelin and cholesterol | −0.253 | 0.006 |
| ABCG1_cg00222799 | Body | −0.494 | <0.001 | |
| ABCG1_cg00177237 | Body | −0.607 | <0.001 | |
| ABCG1_cg20727187 | Body | 0.260 | 0.001 | |
| ABCG1_cg18382690 | Body | −0.451 | 0.001 | |
| ABCG1_cg02494239 | 5′UTR; Body | 0.226 | 0.002 | |
| ABCG1_cg02370100 * | Body | 0.258 | 0.001 | |
| UFM1_cg07243519 * | 1stExon;5′UTR | Ufmylation (post-transcriptional modification) ER-associated degradation; regulation of transcription | −0.272 | 0.004 |
| UFM1_cg07350703 * | TSS1500 | 0.199 | 0.034 | |
| PFKFB2_cg15339972 * | Body; TSS1500 | Regulation of glycolysis; expressed in heart | −0.907 | <0.001 |
| PFKFB2_cg20198644 | Body | −0.346 | <0.001 | |
| PFKFB2_cg22944368 * | TSS200 | −0.245 | 0.005 | |
| PFKFB2_cg05398095 * | TSS1500 | 0.547 | 0.039 | |
| ARRDC4_cg01088608 | Body | Adapter recruiting ubiquitin-protein ligases; possible role in glucose uptake | −0.277 | 0.004 |
| ARRDC4_cg09442792 | 3′UTR | −0.196 | 0.041 | |
| SREBF1_cg23155675 | Body | Obesity, type 2 diabetes and insulin sensitivity | −0.792 | <0.001 |
| SREBF1_cg27407935 | Body | 0.197 | 0.004 | |
| SREBF1_cg04805065 | Body | −0.178 | 0.005 | |
| SREBF1_cg06619462 | Body | 0.164 | 0.014 | |
| SREBF1_cg14808739 * | TSS1500 | 0.157 | 0.014 | |
| SREBF1_cg01049850 | Body | −0.137 | 0.031 | |
| SREBF1_cg12244055 | 3′UTR | 0.155 | 0.042 | |
| PLAGL1_cg04895233 | TSS1500 | Transient neonatal diabetes mellitus | −0.502 | <0.001 |
| PLAGL1_cg18316621 | TSS1500; TSS200 5′UTR | −0.209 | 0.009 | |
| PLAGL1_cg21416120 | TSS1500; TSS200 5′UTR | −0.135 | 0.043 | |
| PLAGL1_cg15262884 | 5′UTR | −0.471 | <0.001 | |
| PLAGL1_cg01445838 | 5′UTR | 0.366 | <0.001 | |
| PLAGL1_cg10254692 | TSS1500 | 0.254 | 0.025 | |
| PLAGL1_cg04696964 | 5′UTR | 0.185 | 0.022 | |
| PLAGL1_cg01659632 | 3′UTR | 0.199 | 0.007 | |
| PLAGL1_cg03562868 | TSS1500; 5′UTR; TSS200 | −0.177 | 0.034 | |
| HEG1_cg20125761 | Body | Regulator of heart and vessel formation | −0.369 | <0.001 |
| HEG1_cg06477303 | Body | −0.342 | <0.001 | |
| HEG1_cg00213745 | Body | 0.280 | 0.002 | |
| HEG1_cg16044109 | Body | −0.270 | 0.002 | |
| HEG1_cg10294433 | Body | −0.229 | 0.005 | |
| OAZ2_cg13262282 | Body | Polyamine biosynthesis, type 2 diabetes | −0.459 | <0.001 |
| OAZ2_cg05353131 * | TSS200 | −0.295 | 0.005 | |
| OAZ2_cg23061600 | TSS1500 | 0.246 | 0.009 | |
| OAZ2_cg14909603 | Body | −0.216 | 0.023 | |
| OAZ2_cg24538975 | Body | 0.267 | 0.005 | |
| OAZ2_cg07031532 * | TSS1500 | 0.205 | <0.05 | |
| FAM3C_cg04873577 | Body | Type 2 diabetes and non-alcoholic fatty liver disease | −0.351 | <0.001 |
| POP7_cg05340629 * | TSS1500 | Ribosome biogenesis | 0.342 | <0.001 |
| POP7_cg04494750 | Body | −0.202 | 0.031 | |
| TCEB2_cg02026611 | Body | Transcription elongation and cellular senescence | −0.228 | 0.018 |
| COMMD7_cg23356674 | TSS1500 | NF-kappa-B complex activity | −0.279 | 0.004 |
| COMMD7_cg2339673 * | TSS1500 | −0.201 | 0.037 | |
| DECR2_cg04571183 | Body | Lipid metabolism | −0.561 | <0.001 |
| DECR2_cg27315249 | TSS1500; 3′UTR | 0.449 | <0.001 | |
| DECR2_cg00481259 | TSS1500 | −0.253 | 0.030 | |
| DECR2_cg10509880 | Body | −0.211 | 0.012 | |
| BSN_cg16885237 | Body | Spatial organization of synaptic vesicle cluster | −0.396 | <0.001 |
| BSN_cg04381190 | 3′UTR | −0.296 | 0.001 | |
| BSN_cg13465832 | Body | −0.353 | <0.001 | |
| BSN_cg11216396 | Body | 0.221 | 0.027 | |
| BSN_cg13444307 | Body | −0.228 | 0.005 | |
| BSN_cg19602139 | Body | 0.208 | 0.008 | |
| PRDX5_cg01708924 | 3′UTR | Cellular protection against oxidative stress | −0.385 | <0.001 |
| FBXO42_cg02207034 | 5′UTR | Protein–ubiquitin ligases | −0.449 | <0.001 |
| FBXO42_cg06216849 | Body | −0.219 | 0.013 | |
| FBXO42_cg22937685 | Body | −0.185 | 0.035 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giacconi, R.; D’Aquila, P.; Olivieri, F.; Gentilini, D.; Calzari, L.; Fortunato, C.; Badillo Pazmay, G.V.; Di Rosa, M.; Sena, G.; De Rose, E.; et al. Circulating Bacterial DNA as a Novel Blood-Based Biomarker in Type 2 Diabetes Mellitus (DM2): Results from the PROMOTERA Study. Int. J. Mol. Sci. 2025, 26, 6564. https://doi.org/10.3390/ijms26146564
Giacconi R, D’Aquila P, Olivieri F, Gentilini D, Calzari L, Fortunato C, Badillo Pazmay GV, Di Rosa M, Sena G, De Rose E, et al. Circulating Bacterial DNA as a Novel Blood-Based Biomarker in Type 2 Diabetes Mellitus (DM2): Results from the PROMOTERA Study. International Journal of Molecular Sciences. 2025; 26(14):6564. https://doi.org/10.3390/ijms26146564
Chicago/Turabian StyleGiacconi, Robertina, Patrizia D’Aquila, Fabiola Olivieri, Davide Gentilini, Luciano Calzari, Carlo Fortunato, Gretta Veronica Badillo Pazmay, Mirko Di Rosa, Giada Sena, Elisabetta De Rose, and et al. 2025. "Circulating Bacterial DNA as a Novel Blood-Based Biomarker in Type 2 Diabetes Mellitus (DM2): Results from the PROMOTERA Study" International Journal of Molecular Sciences 26, no. 14: 6564. https://doi.org/10.3390/ijms26146564
APA StyleGiacconi, R., D’Aquila, P., Olivieri, F., Gentilini, D., Calzari, L., Fortunato, C., Badillo Pazmay, G. V., Di Rosa, M., Sena, G., De Rose, E., Cherubini, A., Sarzani, R., Antonicelli, R., Pelliccioni, G., Bonfigli, A. R., Galeazzi, R., Lattanzio, F., Passarino, G., Bellizzi, D., & Piacenza, F. (2025). Circulating Bacterial DNA as a Novel Blood-Based Biomarker in Type 2 Diabetes Mellitus (DM2): Results from the PROMOTERA Study. International Journal of Molecular Sciences, 26(14), 6564. https://doi.org/10.3390/ijms26146564








