Regulation of the Microbiome in Soil Contaminated with Diesel Oil and Gasoline
Abstract
1. Introduction
2. Results
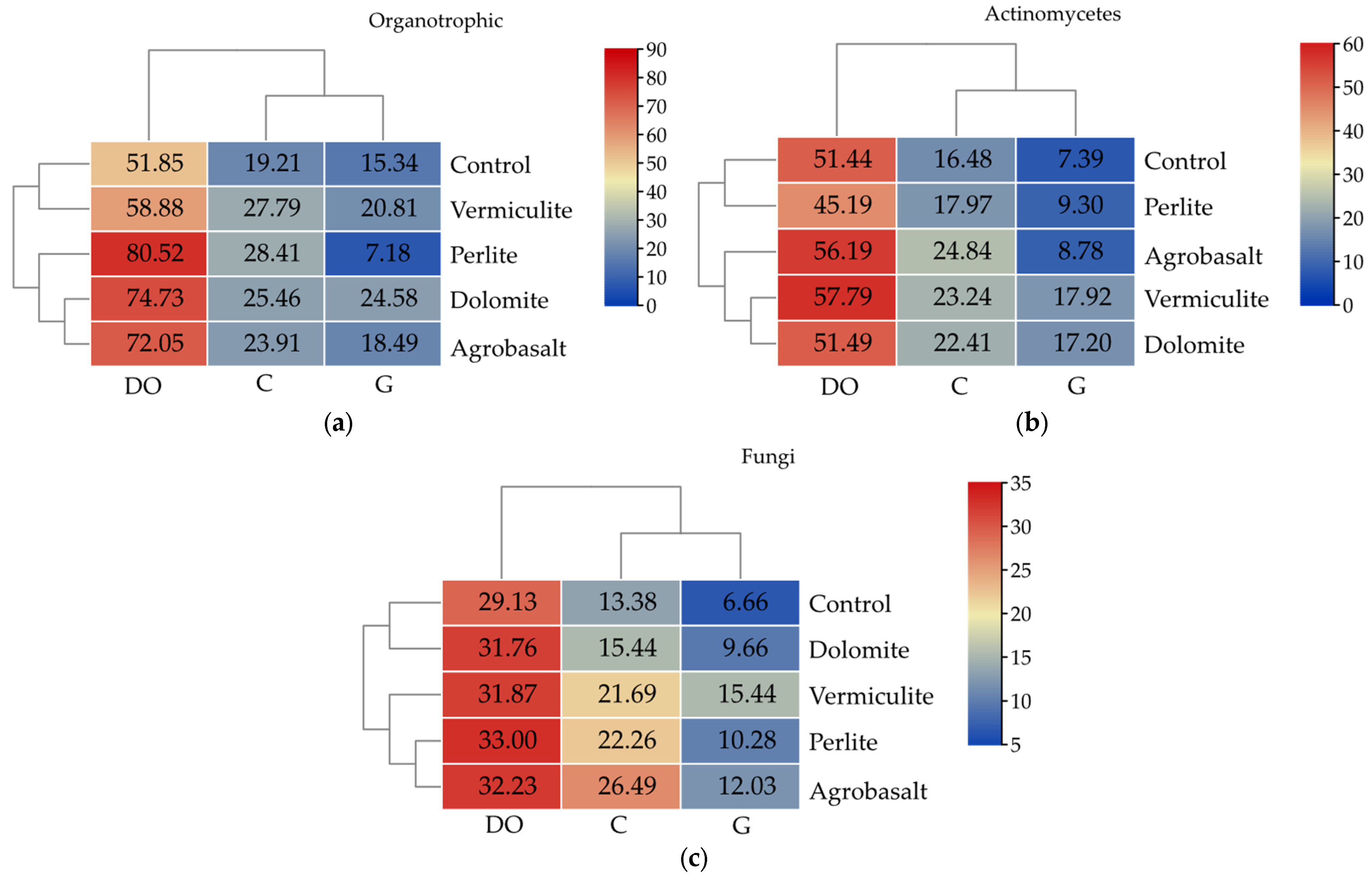
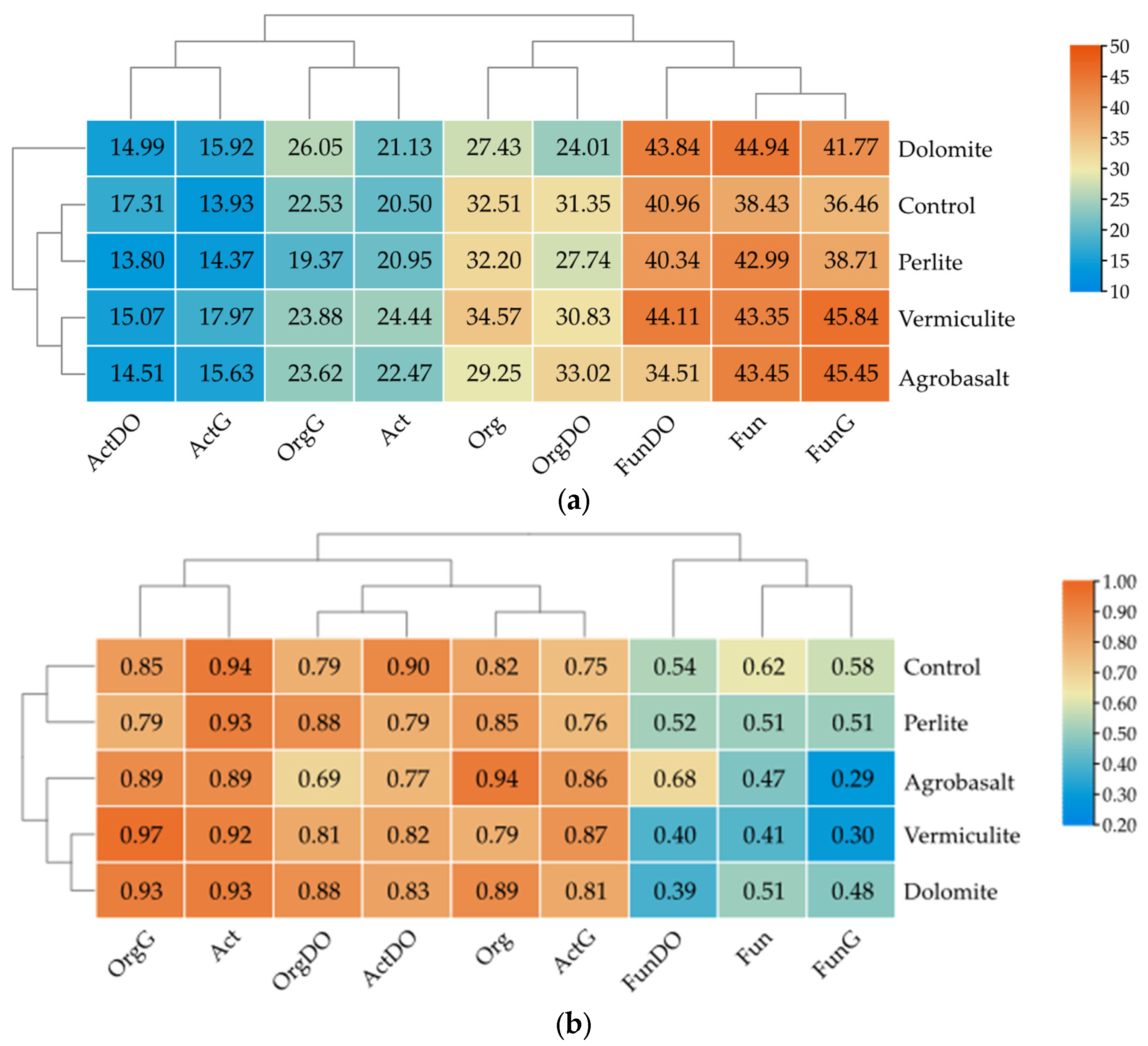
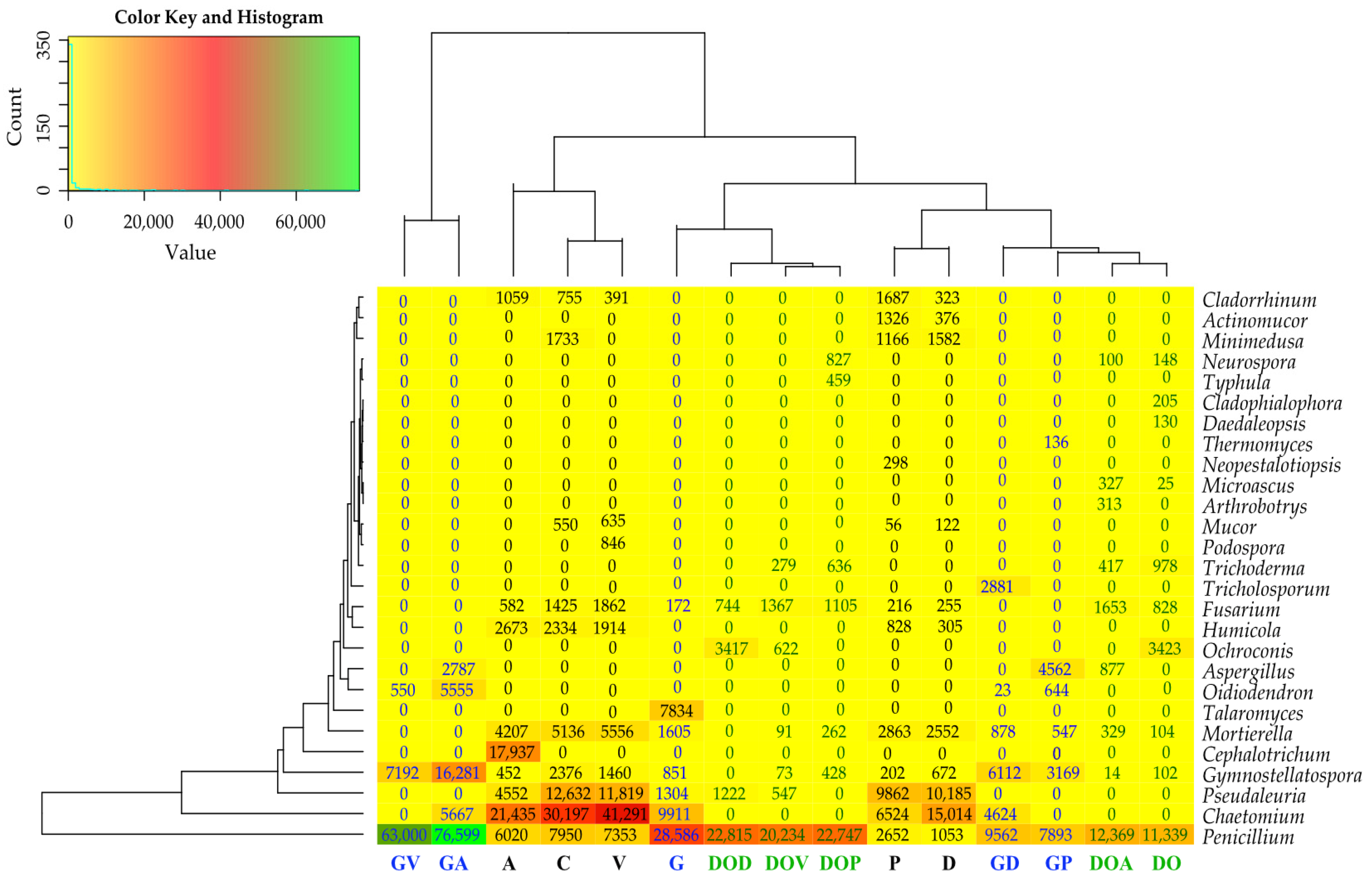
2.1. Culturable Microorganisms
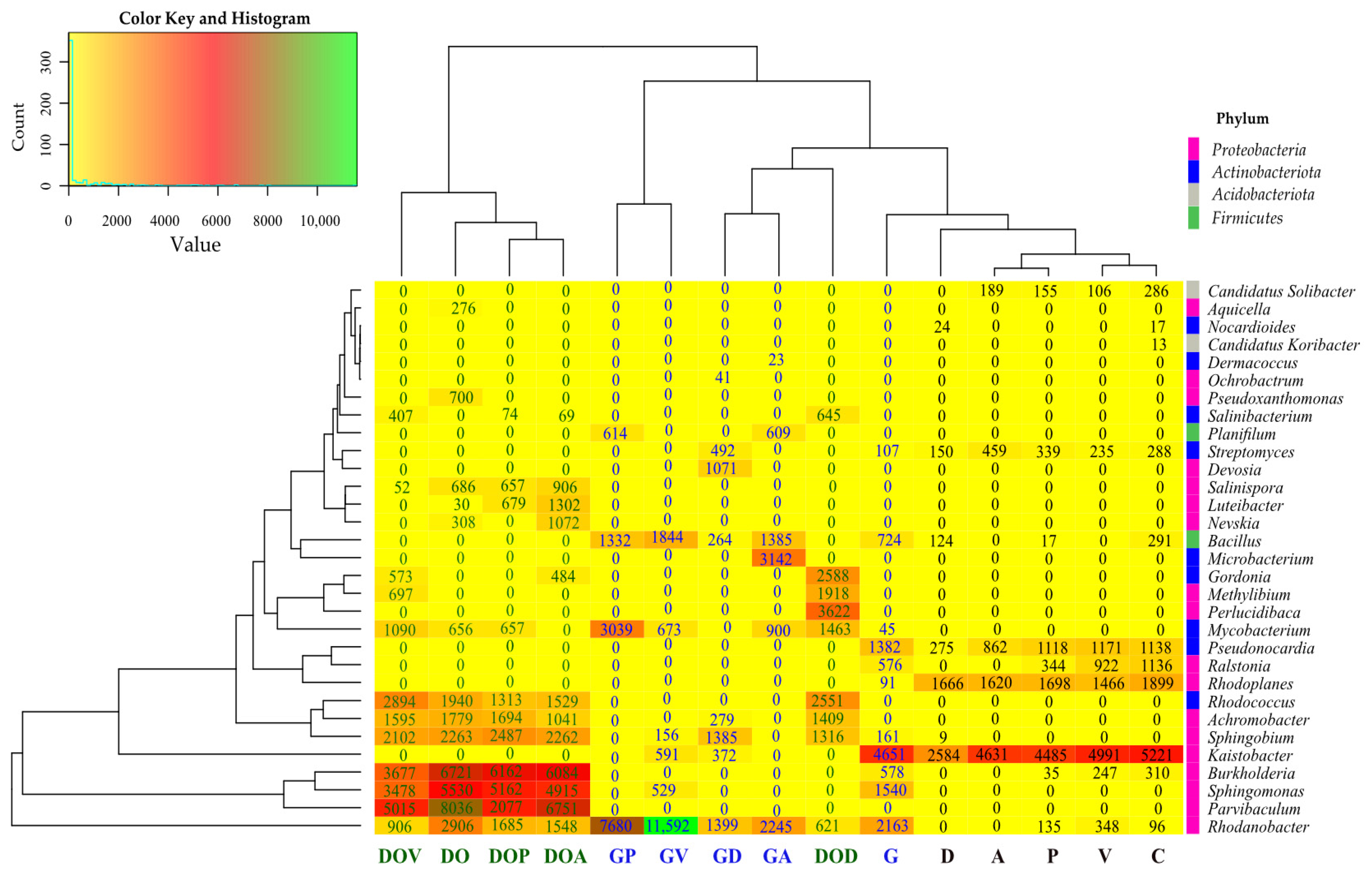
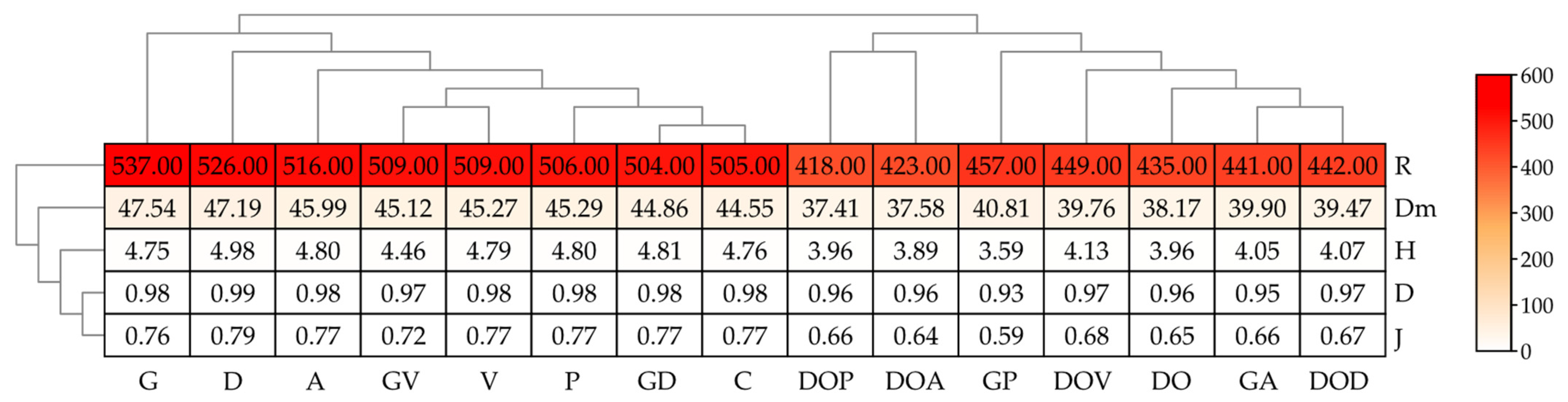
2.2. The Abundance of Non-Cultivable Microorganisms
2.3. Prediction of Protein Properties Based on Nucleotide Sequences
3. Discussion
3.1. The Influence of Petroleum Products on the Cultivable Microbiome
3.2. The Influence of Petroleum Products on the Non-Cultivable Microbiome
3.2.1. Bacteria
3.2.2. Fungi
3.3. The Impact of Sorbents on Soil Microbiome
3.4. Implications of Protein Property Predictions Derived from Nucleotide Sequence Data
4. Materials and Methods
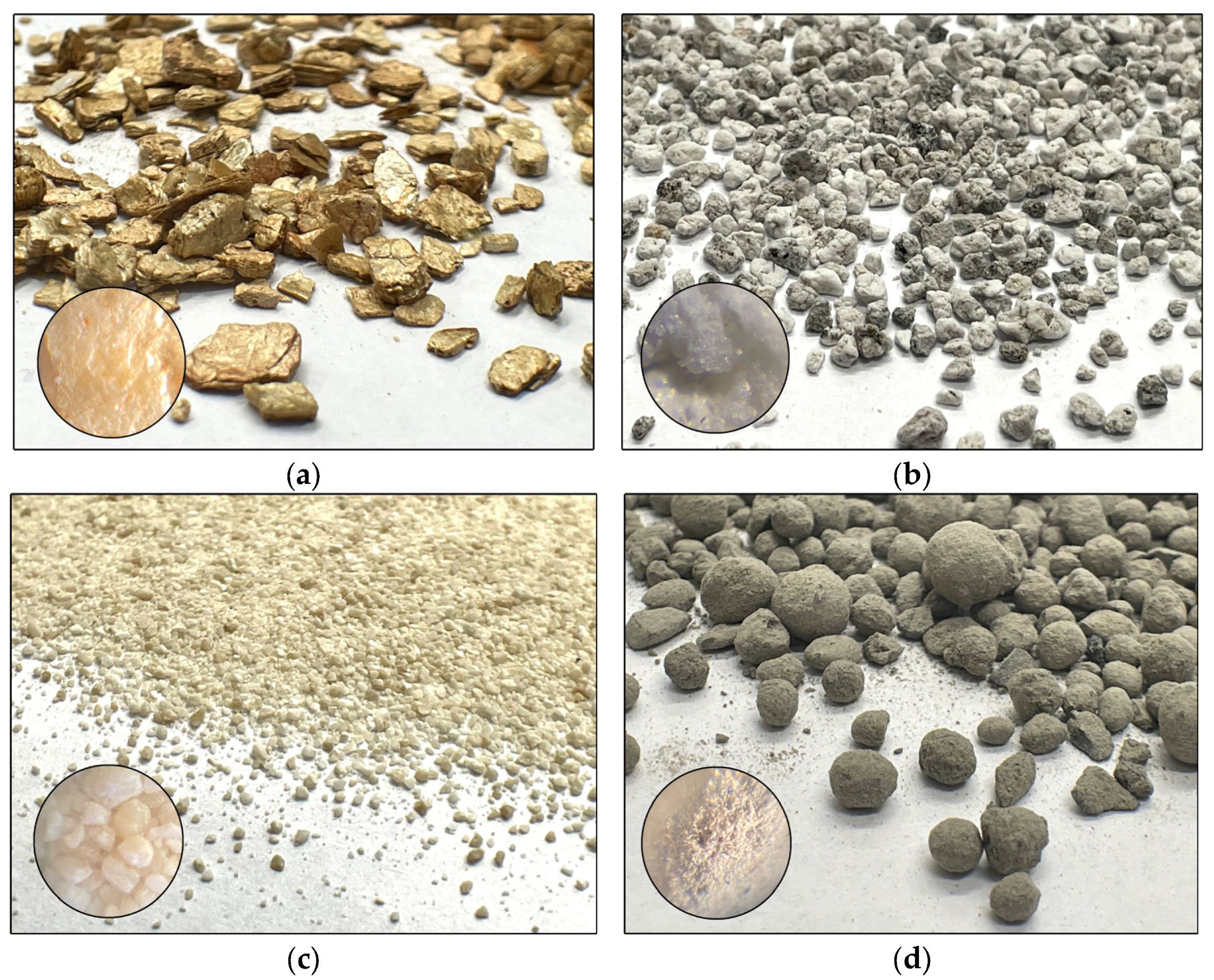
4.1. Characterization of Soil Material, Sorbents, and Petroleum-Derived Substances
4.2. Experimental Design
4.3. Cultivable Microorganisms
4.4. Non-Cultivable Microorganisms
4.5. Data Analysis and Statistical Processing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ossai, I.C.; Ahmed, A.; Hassan, A.; Hamid, F.S. Remediation of Soil and Water Contaminated with Petroleum Hydrocarbon: A Review. Environ. Technol. Innov. 2020, 17, 100526. [Google Scholar] [CrossRef]
- BP. Peak Oil Demand and Long-Run Oil Prices; BP: London, UK, 2023; Available online: https://www.bp.com/content/dam/bp/business-sites/en/global/corporate/pdfs/energy-economics/bp-peak-oil-demand-and-long-run-oil-prices.pdf (accessed on 25 June 2025).
- Global Carbon Project JV. Fossil Fuel CO2 Emissions Increase Again in 2024. Available online: https://globalcarbonbudget.org (accessed on 27 June 2025).
- Kostianoy, A.G.; Lavrova, O.Y. Introduction. In Oil Pollution in the Baltic Sea; Springer: Berlin/Heidelberg, Germany, 2014; pp. 1–13. ISBN 978-3-642-38476-9. Available online: https://link.springer.com/chapter/10.1007/698_2013_233 (accessed on 25 June 2025).[Green Version]
- George, I.I.; Nawawi, M.G.M.; Mohd, Z.J.; Farah, B.S. Environmental Effects from Petroleum Product Transportation Spillage in Nigeria: A Critical Review. Environ. Sci. Pollut. Res. 2024, 31, 1719–1747. [Google Scholar] [CrossRef] [PubMed]
- Falih, K.T.; Mohd Razali, S.F.; Abdul Maulud, K.N.; Abd Rahman, N.; Abba, S.I.; Yaseen, Z.M. Assessment of Petroleum Contamination in Soil, Water, and Atmosphere: A Comprehensive Review. Int. J. Environ. Sci. Technol. 2024, 21, 8803–8832. [Google Scholar] [CrossRef]
- Pal, D.; Sen, S. In-Depth Coverage of Petroleum Waste Sources, Characteristics, Environmental Impact, and Sustainable Remediation Process. In Impact of Petroleum Waste on Environmental Pollution and Its Sustainable Management Through Circular Economy; Behera, I.D., Das, A.P., Eds.; Springer Nature: Cham, Switzerland, 2023; pp. 1–38. ISBN 978-3-031-48220-5. [Google Scholar][Green Version]
- Hidalgo-Martinez, K.; Giachini, A.J.; Schneider, M.; Soriano, A.; Baessa, M.P.; Martins, L.F.; de Oliveira, V.M. Shifts in Structure and Dynamics of the Soil Microbiome in Biofuel/Fuel Blend–Affected Areas Triggered by Different Bioremediation Treatments. Environ. Sci. Pollut. Res. 2024, 31, 33663–33684. [Google Scholar] [CrossRef]
- Daza, B.X.D.; Mendoza, A.J.D.; Zambrano, J.J.Z.; Aransiola, S.A.; Maddela, N.R. Effects of Soil Contaminants on Soil Microbiome. In Soil Microbiome in Green Technology Sustainability; Aransiola, S.A., Atta, H.I., Maddela, N.R., Eds.; Springer Nature: Cham, Switzerland, 2024; pp. 183–199. ISBN 978-3-031-71844-1. [Google Scholar][Green Version]
- Parmar, P.; Dhurandhar, R.; Naik, S. Environmental Fate and Microbial Reactions to Petroleum Hydrocarbon Contamination in Terrestrial Ecosystems. In Impact of Petroleum Waste on Environmental Pollution and Its Sustainable Management Through Circular Economy; Behera, I.D., Das, A.P., Eds.; Springer Nature: Cham, Switzerland, 2023; pp. 139–158. ISBN 978-3-031-48220-5. [Google Scholar][Green Version]
- Marschmann, G.L.; Tang, J.; Zhalnina, K.; Karaoz, U.; Cho, H.; Le, B.; Pett-Ridge, J.; Brodie, E.L. Predictions of Rhizosphere Microbiome Dynamics with a Genome-Informed and Trait-Based Energy Budget Model. Nat. Microbiol. 2024, 9, 421–433. [Google Scholar] [CrossRef]
- Kornienko, I.V.; Aramova, O.Y.; Tishchenko, A.A.; Rudoy, D.V.; Chikindas, M.L. RNA Stability: A Review of the Role of Structural Features and Environmental Conditions. Molecules 2024, 29, 5978. [Google Scholar] [CrossRef]
- Saon, S.; Boehm, K.; Fu, G.; Hou, I.; Yu, J.; Znosko, B.; Hou, J. Exploring the Efficiency of Deep Graph Neural Networks for RNA Secondary Structure Prediction. bioRxiv 2024. [Google Scholar] [CrossRef]
- Vasilyeva, G.; Mikhedova, E.; Zinnatshina, L.; Strijakova, E.; Akhmetov, L.; Sushkova, S.; Ortega-Calvo, J.-J. Use of Natural Sorbents for Accelerated Bioremediation of Grey Forest Soil Contaminated with Crude Oil. Sci. Total. Environ. 2022, 850, 157952. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J.; Kucharski, M.; Kucharski, J. Implications of Soil Pollution with Diesel Oil and BP Petroleum with ACTIVE Technology for Soil Health. Int. J. Environ. Res. Public Health 2019, 16, 2474. [Google Scholar] [CrossRef]
- Wyszkowska, J.; Borowik, A.; Zaborowska, M.; Kucharski, J. The Usability of Sorbents in Restoring Enzymatic Activity in Soils Polluted with Petroleum-Derived Products. Materials 2023, 16, 3738. [Google Scholar] [CrossRef]
- Gertsen, M.M.; Arlyapov, V.A.; Perelomov, L.V.; Kharkova, A.S.; Golysheva, A.N.; Atroshchenko, Y.M.; Cardinale, A.M.; Reverberi, A.P. Environmental Implications of Energy Sources: A Review on Technologies for Cleaning Oil-Contaminated Ecosystems. Energies 2024, 17, 3561. [Google Scholar] [CrossRef]
- Sawicka, B.; Vambol, V.; Krochmal-Marczak, B.; Messaoudi, M.; Skiba, D.; Pszczółkowski, P.; Barbaś, P.; Farhan, A.K. Green Technology as a Way of Cleaning the Environment from Petroleum Substances in South-Eastern Poland. Front. Biosci-Elite 2022, 14, 28. [Google Scholar] [CrossRef] [PubMed]
- Mathur, J.; Panwar, R. Synergistic Effect of Pyrene and Heavy Metals (Zn, Pb, and Cd) on Phytoremediation Potential of Medicago sativa L. (Alfalfa) in Multi-Contaminated Soil. Environ. Sci. Pollut. Res. 2024, 31, 21012–21027. [Google Scholar] [CrossRef] [PubMed]
- Dąbrowski, P.; Małuszyńska, I.; Małuszyński, M.J.; Pawluśkiewicz, B.; Gnatowski, T.; Baczewska-Dąbrowska, A.H.; Kalaji, H.M. Photosynthetic Efficiency of Plants as an Indicator of Tolerance to Petroleum-Contaminated Soils. Sustainability 2024, 16, 10811. [Google Scholar] [CrossRef]
- Das, N.; Kumar, V.; Chaure, K.; Pandey, P. Environmental Restoration of Polyaromatic Hydrocarbon-Contaminated Soil through Sustainable Rhizoremediation: Insights into Bioeconomy and High-Throughput Systematic Analysis. Environ. Sci. Adv. 2025, 4, 842–883. [Google Scholar] [CrossRef]
- Ahmad, M.S.; Ghazala; Rasul, F.; Ibrahim, M.; Mahmood, A. Organic Amendments Enhanced Phytoremediation, Physio-Chemical Functions, Growth and Productivity of Zea mays L. in Diesel-Contaminated Soil. J. Soil Sci. Plant Nutr. 2025, 25, 550–561. [Google Scholar] [CrossRef]
- Ionata, E.; Caputo, E.; Mandrich, L.; Marcolongo, L. Moving towards Biofuels and High-Value Products through Phytoremediation and Biocatalytic Processes. Catalysts 2024, 14, 118. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J.; Zaborowska, M.; Kucharski, J. Soil Enzyme Response and Calorific Value of Zea Mays Used for the Phytoremediation of Soils Contaminated with Diesel Oil. Energies 2024, 17, 2552. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J.; Zaborowska, M.; Kucharski, J. Energy Quality of Corn Biomass from Gasoline-Contaminated Soils Remediated with Sorbents. Energies 2024, 17, 5322. [Google Scholar] [CrossRef]
- Wyszkowska, J.; Borowik, A.; Zaborowska, M.; Kucharski, J. The Potential for Restoring the Activity of Oxidoreductases and Hydrolases in Soil Contaminated with Petroleum Products Using Perlite and Dolomite. Appl. Sci. 2024, 14, 3591. [Google Scholar] [CrossRef]
- Garcia-Valles, M.; Alfonso, P.; Martínez, S.; Roca, N. Mineralogical and Thermal Characterization of Kaolinitic Clays from Terra Alta (Catalonia, Spain). Minerals 2020, 10, 142. [Google Scholar] [CrossRef]
- Kovaleva, E.I.; Perebasova, P.M.; Avdulov, D.A.; Ladonin, D.V.; Trofimov, S.Y. The Effectiveness of Remediation Agents for Detoxification of Heavy-Metal-Contaminated Soils According to Experimental Results. Mosc. Univ. Soil Sci. Bull. 2024, 79, 177–189. [Google Scholar] [CrossRef]
- Imam, A.; Kanaujia, P.K.; Ray, A.; Suman, S.K. Removal of Petroleum Contaminants Through Bioremediation with Integrated Concepts of Resource Recovery: A Review. Indian J. Microbiol. 2021, 61, 250–261. [Google Scholar] [CrossRef]
- Malandrino, M.; Abollino, O.; Giacomino, A.; Aceto, M.; Mentasti, E. Adsorption of Heavy Metals on Vermiculite: Influence of pH and Organic Ligands. J. Colloid Interface Sci. 2006, 299, 537–546. [Google Scholar] [CrossRef]
- Topka, P.; Soukup, K.; Hejtmánek, V.; Hlásenský, I.; Kaštánek, F.; Šolcová, O. Remediation of Brownfields Contaminated by Organic Compounds and Heavy Metals: A Bench-Scale Test of a Sulfur/Vermiculite Sorbent for Mercury Vapor Removal. Environ. Sci. Pollut. Res. Int. 2020, 27, 42182–42188. [Google Scholar] [CrossRef]
- Brião, G.d.V.; da Silva, M.G.C.; Vieira, M.G.A. Efficient and Selective Adsorption of Neodymium on Expanded Vermiculite. Ind. Eng. Chem. Res. 2021, 60, 4962–4974. [Google Scholar] [CrossRef]
- Szerement, J.; Kowalski, A.; Mokrzycki, J.; Marcińska-Mazur, L.; Mierzwa-Hersztek, M. Restoration of Soils Contaminated with PAHs by the Mixture of Zeolite Composites Mixed with Exogenous Organic Matter and Mineral Salts. Sci. Rep. 2023, 13, 14227. [Google Scholar] [CrossRef]
- Raza, A.; Glatz, G.; Gholami, R.; Mahmoud, M.; Alafnan, S. Carbon Mineralization and Geological Storage of CO2 in Basalt: Mechanisms and Technical Challenges. Earth-Sci. Rev. 2022, 229, 104036. [Google Scholar] [CrossRef]
- Sundaramoorthi, A.; Thangaraju, P. Optimising Cement-Based Electrolytes: Ionic Strength Analysis and Electrical Performance in Cement-Based Battery Applications. Eur. J. Environ. Civ. Eng. 2025, 1–24. [Google Scholar] [CrossRef]
- Szadkowski, B.; Marzec, A.; Rybiński, P.; Żukowski, W.; Zaborski, M. Characterization of Ethylene–Propylene Composites Filled with Perlite and Vermiculite Minerals: Mechanical, Barrier, and Flammability Properties. Materials 2020, 13, 585. [Google Scholar] [CrossRef]
- Faleh, S.T.; Fahmi, A.H.; Abood, M.A. The Role of Biochar and Perlite in Improving Some Chemical Properties of Clay Loam Soil and Sandy Loam Soil. IOP Conf. Ser. Earth Environ. Sci. 2023, 1262, 082011. [Google Scholar] [CrossRef]
- Cedeño, J.M.; Magán, J.-J.; Thompson, R.B.; Fernández, M.-D.; Gallardo, M. Comparison of Methods to Determine Nutrient Uptake of Tomato Grown in Free-Draining Perlite Substrate—Key Information for Optimal Fertigation Management. Horticulturae 2024, 10, 232. [Google Scholar] [CrossRef]
- Wyszkowska, J.; Borowik, A.; Zaborowska, M.; Kucharski, J. Revitalization of Soil Contaminated by Petroleum Products Using Materials That Improve the Physicochemical and Biochemical Properties of the Soil. Molecules 2024, 29, 5838. [Google Scholar] [CrossRef]
- Duarte, H.; Aliaño-González, M.J.; Romano, A.; Medronho, B. Advancements in Detection and Mitigation Strategies for Petroleum-Derived Contaminants in Aquatic Environments: A Comprehensive Review. Sensors 2024, 24, 3284. [Google Scholar] [CrossRef]
- Yi, P.; Zuo, X.; Liang, N.; Wu, M.; Chen, Q.; Zhang, L.; Pan, B. Molecular clusters played an important role in the adsorption of polycyclic aromatic hydrocarbons (PAHs) on carbonaceous materials. Chemosphere 2022, 302, 134772. [Google Scholar] [CrossRef]
- Denison, S.B.; Jin, P.; Zygourakis, K.; Senftle, T.P.; Alvarez, P.J.J. Mechanistic Implications of the Varying Susceptibility of PAHs to Pyro-Catalytic Treatment as a Function of Their Ionization Potential and Hydrophobicity. Environ. Sci. Technol. 2024, 58, 13521–13528. [Google Scholar] [CrossRef]
- Hua, F.; Wang, H.Q. Uptake and Trans-Membrane Transport of Petroleum Hydrocarbons by Microorganisms. Biotechnol. Biotechnol. Equip. 2014, 28, 165–175. [Google Scholar] [CrossRef]
- Nourbakhsh, F.; Lotfalizadeh, M.; Badpeyma, M.; Shakeri, A.; Soheili, V. From Plants to Antimicrobials: Natural Products against Bacterial Membranes. Phytother. Res. 2022, 36, 33–52. [Google Scholar] [CrossRef]
- Devi, A.; Ferreira, L.F.R.; Saratale, G.D.; Mulla, S.I.; More, N.; Bharagava, R.N. Chapter1—Microbe-Assisted Phytoremediation of Environmental Contaminants. In Advances in Microbe-Assisted Phytoremediation of Polluted Sites; Bauddh, K., Ma, Y., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 3–26. ISBN 978-0-12-823443-3. [Google Scholar]
- Tarigholizadeh, S.; Sushkova, S.; Rajput, V.D.; Ranjan, A.; Arora, J.; Dudnikova, T.; Barbashev, A.; Mandzhieva, S.; Minkina, T.; Wong, M.H. Transfer and Degradation of PAHs in the Soil–Plant System: A Review. J. Agric. Food Chem. 2024, 72, 46–64. [Google Scholar] [CrossRef]
- Chen, B.-H.; Inbaraj, B.S.; Hsu, K.-C. Recent Advances in the Analysis of Polycyclic Aromatic Hydrocarbons in Food and Water. J. Food Drug Anal. 2022, 30, 494–522. [Google Scholar] [CrossRef]
- de Melo, A.P.Z.; Hoff, R.B.; Molognoni, L.; de Oliveira, T.; Daguer, H.; Barreto, P.L.M. Disasters with Oil Spills in the Oceans: Impacts on Food Safety and Analytical Control Methods. Food Res. Int. 2022, 157, 111366. [Google Scholar] [CrossRef] [PubMed]
- Patton, J.S.; Stone, B.; Papa, C.; Abramowitz, R.; Yalkowsky, S.H. Solubility of Fatty Acids and Other Hydrophobic Molecules in Liquid Trioleoylglycerol. J. Lipid Res. 1984, 25, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C. Lipophilicity: Understanding the Role of Lipid Affinity in Drug Design and Absorption. J. Pharmacokinet. Exp. Ther. 2024, 8, 1000278. [Google Scholar]
- Del Río-Arrillaga, R.; García-Figueroa, A.A.; López-Cervantes, J.L.; Albijanic, B.; Gracia-Fadrique, J. Hydrophobicity of Benzene-Based Surfactants and Its Effect on Bubble Coalescence Inhibition. Molecules 2024, 29, 5042. [Google Scholar] [CrossRef]
- Zhu, Q.; Wang, S.; Fu, G.; Guo, F.; Huang, W.; Zhang, T.; Dong, H.; Jin, Z.; Zhang, D. Highly Flexible Cell Membranes Are the Key to Efficient Production of Lipophilic Compounds. J. Lipid Res. 2024, 65, 100597. [Google Scholar] [CrossRef]
- Schloter, M.; Nannipieri, P.; Sørensen, S.J.; van Elsas, J.D. Microbial Indicators for Soil Quality. Biol. Fertil. Soils 2018, 54, 1–10. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J.; Kucharski, J. Microbiological Study in Petrol-Spiked Soil. Molecules 2021, 26, 2664. [Google Scholar] [CrossRef]
- van Dillewijn, P.; Hurtado-Fernández, I.; Garrido-Peláez, P.; Molina, L.; Udaondo, Z.; Velando, F.; Segura, A. Effect of Diesel Contamination on Bacterial Populations in a Pristine Soil during Rhizoremediation. bioRxiv 2025. [Google Scholar] [CrossRef]
- Song, Q.; Zhou, B.; Song, Y.; Du, X.; Chen, H.; Zuo, R.; Zheng, J.; Yang, T.; Sang, Y.; Li, J. Microbial Community Dynamics and Bioremediation Strategies for Petroleum Contamination in an In-Service Oil Depot, Middle-Lower Yellow River Basin. Front. Microbiol. 2025, 16, 1544233. [Google Scholar] [CrossRef]
- Xue, J.; Dun, Y.; Wang, S.; Cheng, D.; Qiao, Y.; Liu, Y.; Wang, F.; Zhang, X.; Li, Y. Enhancement of Bioremediation Efficacy in Petroleum-Contaminated Marine Environments via Quorum Sensing: Mechanistic Insights and Efficacy. J. Environ. Chem. Eng. 2025, 13, 115281. [Google Scholar] [CrossRef]
- Galitskaya, P.; Biktasheva, L.; Blagodatsky, S.; Selivanovskaya, S. Response of Bacterial and Fungal Communities to High Petroleum Pollution in Different Soils. Sci. Rep. 2021, 11, 164. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, D.K.; Bajagain, R.; Jeong, S.-W.; Kim, J. Insights into the Biodegradation of Diesel Oil and Changes in Bacterial Communities in Diesel-Contaminated Soil as a Consequence of Various Soil Amendments. Chemosphere 2021, 285, 131416. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, D.K.; Li, C.; Jiang, C.; Chakraborty, A.; Grasby, S.E.; Hubert, C.R.J. Natural Attenuation of Spilled Crude Oil by Cold-Adapted Soil Bacterial Communities at a Decommissioned High Arctic Oil Well Site. Sci. Total Environ. 2020, 722, 137258. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.S.; Lamare, E.; Chopade, B.A.; Chanda, R.; Devi, N.R.; Upadhyay, K.K.; Devi, K.V.; Tripathi, S.K. The Hidden Players: Soil Microbes in Ecosystems Sustainability. In Climate Change and Soil Microorganisms for Environmental Sustainability; Saraf, M., Goswami, D., Maheshwari, D.K., Eds.; Springer Nature: Singapore, 2025; pp. 295–320. ISBN 978-981-96-3425-5. [Google Scholar]
- Mori, J.F.; Kanaly, R.A. Multispecies Diesel Fuel Biodegradation and Niche Formation Are Ignited by Pioneer Hydrocarbon-Utilizing Proteobacteria in a Soil Bacterial Consortium. Appl. Environ. Microbiol. 2020, 87, e02268-20. [Google Scholar] [CrossRef]
- Maeda, A.H.; Nishi, S.; Hatada, Y.; Ohta, Y.; Misaka, K.; Kunihiro, M.; Mori, J.F.; Kanaly, R.A. Chemical and Genomic Analyses of Polycyclic Aromatic Hydrocarbon Biodegradation in Sphingobium barthaii KK22 Reveals Divergent Pathways in Soil Sphingomonads. Int. Biodeterior. Biodegrad. 2020, 151, 104993. [Google Scholar] [CrossRef]
- Liu, Q.; Tang, J.; Gao, K.; Gurav, R.; Giesy, J.P. Aerobic Degradation of Crude Oil by Microorganisms in Soils from Four Geographic Regions of China. Sci. Rep. 2017, 7, 14856. [Google Scholar] [CrossRef]
- Costa, G.d.S.; Martinez-Burgos, W.J.; dos Reis, G.A.; Puche, Y.P.; Vega, F.R.; Rodrigues, C.; Serra, J.L.; Campos, S.d.M.; Soccol, C.R. Advances in Biomass and Microbial Lipids Production: Trends and Prospects. Processes 2024, 12, 2903. [Google Scholar] [CrossRef]
- Varjani, S.J.; Upasani, V.N. A New Look on Factors Affecting Microbial Degradation of Petroleum Hydrocarbon Pollutants. Int. Biodeterior. Biodegrad. 2017, 120, 71–83. [Google Scholar] [CrossRef]
- Kato, H.; Miura, D.; Kato, M.; Shimizu, M. Metabolic Mechanism of Lignin-Derived Aromatics in White-Rot Fungi. Appl. Microbiol. Biotechnol. 2024, 108, 532. [Google Scholar] [CrossRef]
- Rani, M.H.S.; Nandana, R.K.; Khatun, A.; Brindha, V.; Midhun, D.; Gowtham, P.; Mani, S.S.D.; Kumar, S.R.; Aswini, A.; Muthukumar, S. Three Strategy Rules of Filamentous Fungi in Hydrocarbon Remediation: An Overview. Biodegradation 2024, 35, 833–861. [Google Scholar] [CrossRef]
- Vo, P.H.N.; Le, G.K.; Huy, L.N.; Zheng, L.; Chaiwong, C.; Nguyen, N.N.; Nguyen, H.T.M.; Ralph, P.J.; Kuzhiumparambil, U.; Danaee, S.; et al. Occurrence, Spatiotemporal Trends, Fate, and Treatment Technologies for Microplastics and Organic Contaminants in Biosolids: A Review. J. Hazard. Mater. 2024, 466, 133471. [Google Scholar] [CrossRef] [PubMed]
- Aranda, E.; Godoy, P.; Reina, R.; Badia-Fabregat, M.; Rosell, M.; Marco-Urrea, E.; García-Romera, I. Isolation of Ascomycota Fungi with Capability to Transform PAHs: Insights into the Biodegradation Mechanisms of Penicillium oxalicum. Int. Biodeterior. Biodegrad. 2017, 122, 141–150. [Google Scholar] [CrossRef]
- Huarte-Bonnet, C.; Kumar, S.; Saparrat, M.C.N.; Girotti, J.R.; Santana, M.; Hallsworth, J.E.; Pedrini, N. Insights into Hydrocarbon Assimilation by Eurotialean and Hypocrealean Fungi: Roles for CYP52 and CYP53 Clans of Cytochrome P450 Genes. Appl. Biochem. Biotechnol. 2018, 184, 1047–1060. [Google Scholar] [CrossRef]
- Ostrem Loss, E.M.; Lee, M.-K.; Wu, M.-Y.; Martien, J.; Chen, W.; Amador-Noguez, D.; Jefcoate, C.; Remucal, C.; Jung, S.; Kim, S.-C.; et al. Cytochrome P450 Monooxygenase-Mediated Metabolic Utilization of Benzo[a]Pyrene by Aspergillus Species. mBio 2019, 10, e00558-19. [Google Scholar] [CrossRef]
- Gul, Z.; Bora, S. Exploiting Pre-Trained Convolutional Neural Networks for the Detection of Nutrient Deficiencies in Hydroponic Basil. Sensors 2023, 23, 5407. [Google Scholar] [CrossRef]
- Soleimani, L.; Salehi, H.; Pasternak, T. Optimizing In Vitro Propagation of Haworthia truncata Schönland Using Leaf, Root, and Inflorescence. Plants 2025, 14, 212. [Google Scholar] [CrossRef]
- Tarigholizadeh, S.; Motafakkerazad, R.; Salehi-Lisar, S.Y.; Mohajel Kazemi, E.; Sushkova, S.; Minkina, T. Phenanthrene Uptake and Translocation by Panicum miliaceum L. Tissues: An Experimental Study in an Artificial Environment. Environ. Geochem. Health 2023, 45, 9281–9292. [Google Scholar] [CrossRef]
- Wu, H.; Hu, J.; Shaaban, M.; Xu, P.; Zhao, J.; Hu, R. The Effect of Dolomite Amendment on Soil Organic Carbon Mineralization Is Determined by the Dolomite Size. Ecol. Process. 2021, 10, 8. [Google Scholar] [CrossRef]
- Lewis, A.L.; Sarkar, B.; Wade, P.; Kemp, S.J.; Hodson, M.E.; Taylor, L.L.; Yeong, K.L.; Davies, K.; Nelson, P.N.; Bird, M.I.; et al. Effects of Mineralogy, Chemistry and Physical Properties of Basalts on Carbon Capture Potential and Plant-Nutrient Element Release via Enhanced Weathering. Appl. Geochem. 2021, 132, 105023. [Google Scholar] [CrossRef]
- Vienne, A.; Poblador, S.; Portillo-Estrada, M.; Hartmann, J.; Ijiehon, S.; Wade, P.; Vicca, S. Enhanced Weathering Using Basalt Rock Powder: Carbon Sequestration, Co-Benefits and Risks in a Mesocosm Study with Solanum tuberosum. Front. Clim. 2022, 4, 869456. [Google Scholar] [CrossRef]
- Wu, M.; Feng, S.; Liu, Z.; Tang, S. Bioremediation of Petroleum-Contaminated Soil Based on Both Toxicity Risk Control and Hydrocarbon Removal—Progress and Prospect. Environ. Sci. Pollut. Res. 2024, 31, 59795–59818. [Google Scholar] [CrossRef] [PubMed]
- Erofeevskya, L.A.; Aleksandrov, A.R. Vermiculite of the Inagli Field as a Promising Material for Environmental Use at Reclamation Sites. IOP Conf. Ser. Earth Environ. Sci. 2021, 666, 032077. [Google Scholar] [CrossRef]
- Giagnoni, L.; dos Anjos Borges, L.G.; Giongo, A.; de Oliveira Silveira, A.; Ardissone, A.N.; Triplett, E.W.; Mench, M.; Renella, G. Dolomite and Compost Amendments Enhance Cu Phytostabilization and Increase Microbiota of the Leachates from a Cu-Contaminated Soil. Agronomy 2020, 10, 719. [Google Scholar] [CrossRef]
- Bucheli, T.D.; Blum, F.; Desaules, A.; Gustafsson, Ö. Polycyclic Aromatic Hydrocarbons, Black Carbon, and Molecular Markers in Soils of Switzerland. Chemosphere 2004, 56, 1061–1076. [Google Scholar] [CrossRef]
- Mohanta, S.; Pradhan, B.; Behera, I.D. Impact and Remediation of Petroleum Hydrocarbon Pollutants on Agricultural Land: A Review. Geomicrobiol. J. 2023, 44, 345–359. [Google Scholar] [CrossRef]
- Wu, X.; Liu, Y.; Shang, Y.; Liu, D.; Liesack, W.; Cui, Z.; Peng, J.; Zhang, F. Peat-Vermiculite Alters Microbiota Composition towards Increased Soil Fertility and Crop Productivity. Plant Soil 2022, 470, 21–34. [Google Scholar] [CrossRef]
- Moscoso, M.A.P.; Yzquierdo, G.A.R.; Vásquez, M.B. Identificación de microorganismos asociados al suelo, sustrato y agua de un sistema productivo de Cannabis sativa L. Temas Agrar. 2022, 27, 272–286. [Google Scholar] [CrossRef]
- Azmi, N.S.A.; Hisham, A.A. Isolation and morphological identification of soil fungi from agricultural soil in Kuantan. J. Clean WAS 2021, 5, 31–34. [Google Scholar] [CrossRef]
- Fang, W.; Zhu, Y.; Liang, C.; Shao, S.; Chen, J.; Qing, H.; Xu, Q. Deciphering Differences in Microbial Community Characteristics and Main Factors Between Healthy and Root Rot-Infected Carya cathayensis Rhizosphere Soils. Front. Microbiol. 2024, 15, 1448675. [Google Scholar] [CrossRef]
- Rice, A.V.; Currah, R.S. Full Article: Two New Species of Pseudogymnoascus with Geomyces Anamorphs and Their Phylogenetic Relationship with Gymnostellatospora. Mycologia 2006, 98, 307–318. [Google Scholar] [CrossRef]
- Sharma, K.; Shah, G.; Singh, H.; Bhatt, U.; Singhal, K.; Soni, V. Advancements in Natural Remediation Management Techniques for Oil Spills: Challenges, Innovations, and Future Directions. Environ. Pollut. Manag. 2024, 1, 128–146. [Google Scholar] [CrossRef]
- Vimal, S.R.; Singh, J.S.; Kumar, A.; Prasad, S.M. Plant Genotype-Microbiome Engineering as Nature-Based Solution (NbS) for Regeneration of Stressed Agriculture: A Review. Sci. Hortic. 2023, 321, 112258. [Google Scholar] [CrossRef]
- Kabir, A.H.; Baki, M.Z.I.; Ahmed, B.; Mostofa, M.G. Current, Faltering, and Future Strategies for Advancing Microbiome-Assisted Sustainable Agriculture and Environmental Resilience. New Crop. 2024, 1, 100013. [Google Scholar] [CrossRef]
- Liu, P.; Xu, H.; Zhang, X. Metabolic Engineering of Microorganisms for L-Alanine Production. J. Ind. Microbiol. Biotechnol. 2022, 49, kuab057. [Google Scholar] [CrossRef]
- Ugwuodo, C.J.; Colosimo, F.; Adhikari, J.; Purvine, S.O.; Eder, E.K.; Hoyt, D.W.; Wright, S.A.; Lipton, M.S.; Mouser, P.J. Aromatic Amino Acid Metabolism and Active Transport Regulation Are Implicated in Microbial Persistence in Fractured Shale Reservoirs. ISME Commun. 2024, 4, ycae149. [Google Scholar] [CrossRef]
- Giustarini, D.; Milzani, A.; Dalle-Donne, I.; Rossi, R. How to Increase Cellular Glutathione. Antioxidants 2023, 12, 1094. [Google Scholar] [CrossRef]
- Idrees, M.; Mohammad, A.R.; Karodia, N.; Rahman, A. Multimodal Role of Amino Acids in Microbial Control and Drug Development. Antibiotics 2020, 9, 330. [Google Scholar] [CrossRef]
- Wangsanut, T.; Pongpom, M. The Role of the Glutathione System in Stress Adaptation, Morphogenesis and Virulence of Pathogenic Fungi. Int. J. Mol. Sci. 2022, 23, 10645. [Google Scholar] [CrossRef]
- Aarti, C.; Khusro, A.; Agastian, P.; Darwish, N.M.; Al Farraj, D.A. Molecular Diversity and Hydrolytic Enzymes Production Abilities of Soil Bacteria. Saudi J. Biol. Sci. 2020, 27, 3235–3248. [Google Scholar] [CrossRef]
- Skinnider, M.A.; Johnston, C.W.; Gunabalasingam, M.; Merwin, N.J.; Kieliszek, A.M.; MacLellan, R.J.; Li, H.; Ranieri, M.R.M.; Webster, A.L.H.; Cao, M.P.T.; et al. Comprehensive Prediction of Secondary Metabolite Structure and Biological Activity from Microbial Genome Sequences. Nat. Commun. 2020, 11, 6058. [Google Scholar] [CrossRef]
- Solar Venero, E.C.; Matera, G.; Vogel, J.; López, N.I.; Tribelli, P.M. Small RNAs in the Antarctic Bacterium Pseudomonas extremaustralis Responsive to Oxygen Availability and Oxidative Stress. Environ. Microbiol. Rep. 2022, 14, 604–615. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Ji, Y.; Stormo, G.D. Discovering cis-Regulatory RNAs in Shewanella Genomes by Support Vector Machines. PLoS Comput. Biol. 2009, 5, e1000338. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Nayarisseri, A.; Singh, S.K. Characterization and Optimization of Azo Dyes Degrading Microbes Isolated from Textile Effluent. Sci. Rep. 2025, 15, 11241. [Google Scholar] [CrossRef] [PubMed]
- Chittoor, S.S.; Giunta, S. Comparative Analysis of Predicted DNA Secondary Structures Infers Complex Human Centromere Topology. Am. J. Hum. Genet. 2024, 111, 2707–2719. [Google Scholar] [CrossRef]
- Karaiyan, P.; Chang, C.C.H.; Chan, E.-S.; Tey, B.T.; Ramanan, R.N.; Ooi, C.W. In Silico Screening and Heterologous Expression of Soluble Dimethyl Sulfide Monooxygenases of Microbial Origin in Escherichia coli. Appl. Microbiol. Biotechnol. 2022, 106, 4523–4537. [Google Scholar] [CrossRef]
- Bunt, J.S.; Rovira, A.D. Microbiological Studies of Some Subantarctic Soils. J. Soil Sci. 1955, 6, 119–128. [Google Scholar] [CrossRef]
- Küster, E.; Williams, S.T. Selection of Media for Isolation of Streptomycetes. Nature 1964, 202, 928–929. [Google Scholar] [CrossRef]
- Martin, J.P. Use of acid, rose bengal, and streptomycin in the plate method for estimating soil fungi. Soil Sci. 1950, 69, 215–232. [Google Scholar] [CrossRef]
- Estensmo, E.L.F.; Maurice, N.; Morgado, L.; Martin-Sanchez, P.M.; Skrede, I.; Kauserud, H. The influence of intraspecific sequence variation during DNA metabarcoding: A case study of eleven fungal species. Mol. Ecol. Resour 2021, 21, 1141–1148. [Google Scholar] [CrossRef]
- Rzehak, T.; Praeg, N.; Galla, G.; Seeber, J.; Hauffe, H.C.; Illmer, P. Comparison of commonly used software pipelines for analyzing fungal metabarcoding data. BMC Genom. 2024, 25, 1085. [Google Scholar] [CrossRef]
- Tibco Software Inc. Statistica; Version 13; Data Analysis Software System; Tibco Software Inc.: Palo Alto, CA, USA, 2021; Available online: https://www.tibco.com/ (accessed on 8 July 2024).
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Sarathchandra, S.U.; Burch, G.; Cox, N.R. Growth Patterns of Bacterial Communities in the Rhizoplane and Rhizosphere of White Clover (Trifolium repens L.) and Perennial Ryegrass (Lolium perenne L.) in Long-Term Pasture. Appl. Soil Ecol. 1997, 6, 293–299. [Google Scholar] [CrossRef]
- De Leij, F.A.A.M.; Whipps, J.M.; Lynch, J.M. The Use of Colony Development for the Characterization of Bacterial Communities in Soil and on Roots. Microb. Ecol. 1994, 27, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. gplots: Various R Programming Tools for Plotting Data. 2024. Available online: https://cran.r-project.org/web/packages/gplots/gplots.pdf (accessed on 27 May 2025).
- Jiang, X.; Qu, Y.; Zeng, H.; Yang, J.; Liu, L.; Deng, D.; Ma, Y.; Chen, D.; Jian, B.; Guan, L.; et al. Long-Term Ecological Restoration Increased Plant Diversity and Soil Total Phosphorus Content of the Alpine Flowing Sand Land in Northwest Sichuan, China. Heliyon 2024, 10, e24035. [Google Scholar] [CrossRef]
- Yang, X.; Turmuhan, R.-G.; Wang, L.; Li, J.; Wan, L. Five Years of Natural Vegetation Recovery in Three Forests of Karst Graben Area and Its Effects on Plant Diversity and Soil Properties. Forests 2025, 16, 91. [Google Scholar] [CrossRef]
- Microsoft. MS Excel® for Microsoft 365 MSO; Microsoft Corporation: Albuquerque, NM, USA, 2021. [Google Scholar]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the Expasy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- RNAfold Web Server. Available online: http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi (accessed on 27 June 2025).

| Treatment | Amino Acid Composition, % | Molecular Weight (kDa) | Instability Index | Aliphatic Index | GRAVY | |||
|---|---|---|---|---|---|---|---|---|
| Ala | Cys | Gly | Thr | |||||
| C | 24.86 | 21.27 | 34.76 | 19.11 | 32.52 | 40.89 | 24.85 | 0.71 |
| DO | 26.11 | 20.69 | 33.58 | 19.62 | 32.96 | 40.38 | 26.11 | 0.72 |
| G | 27.05 | 20.05 | 32.70 | 20.30 | 32.76 | 40.66 | 27.02 | 0.71 |
| G, DO | 25.83 | 20.73 | 33.90 | 19.53 | 32.31 | 42.18 | 25.81 | 0.71 |
| G, C | 24.83 | 21.28 | 35.10 | 18.78 | 32.83 | 40.99 | 24.83 | 0.71 |
| G, DO, C | 26.20 | 20.10 | 34.20 | 19.40 | 33.57 | 38.62 | 26.23 | 0.70 |
| average | 25.81 | 20.69 | 34.04 | 19.46 | 32.83 | 40.62 | 25.81 | 0.71 |
| Treatment | MFE | Diversity (pz) | The Secondary Structure | MFE | Diversity (pz) | The Secondary Structure |
|---|---|---|---|---|---|---|
| Temperature −1 °C | Temperature 17 °C | |||||
| C | −150.77 | 64.14 | −138.14 | −217.27 | 53.74 | −204.75 |
| DO | −146.10 | 75.98 | −126.84 | −211.97 | 53.72 | −197.58 |
| G | −136.85 | 94.73 | −99.10 | −200.95 | 59.44 | −194.02 |
| G, DO | −144.55 | 79.26 | −124.95 | −209.78 | 60.55 | −188.39 |
| G, C | −151.30 | 82.57 | −125.78 | −218.26 | 63.03 | −199.13 |
| G, DO, C | −135.70 | 89.09 | −106.70 | −199.55 | 69.20 | −198.02 |
| average | −144.21 | 80.96 | −120.25 | −209.63 | 59.95 | −196.98 |
| Treatment | Amino Acid Composition, % | Molecular Weight (kDa) | Instability Index | Aliphatic Index | GRAVY | |||
|---|---|---|---|---|---|---|---|---|
| Ala | Cys | Gly | Thr | |||||
| C | 28.51 | 23.91 | 20.48 | 26.96 | 20.29 | 55.15 | 28.52 | 0.84 |
| DO | 21.80 | 29.45 | 26.40 | 22.30 | 21.56 | 55.54 | 21.82 | 0.87 |
| G | 23.13 | 27.67 | 26.10 | 23.10 | 22.75 | 51.00 | 23.14 | 0.84 |
| G, DO | 20.90 | 30.70 | 27.00 | 21.30 | 20.40 | 50.29 | 30.90 | 0.89 |
| G, C | 25.33 | 26.98 | 23.13 | 24.52 | 20.92 | 50.29 | 30.90 | 0.89 |
| G, DO, C | 21.33 | 31.63 | 26.47 | 20.53 | 20.55 | 47.19 | 21.34 | 0.93 |
| average | 23.50 | 28.39 | 24.93 | 23.12 | 21.08 | 51.58 | 26.10 | 0.88 |
| Treatment | MFE | Diversity (pz) | The Secondary Structure | MFE | Diversity (pz) | The Secondary Structure |
|---|---|---|---|---|---|---|
| Temperature −1 °C | Temperature 17 °C | |||||
| C | −60.63 | 43.84 | −54.40 | −93.46 | 39.34 | −81.14 |
| DO | −84.30 | 50.55 | −75.40 | −121.44 | 45.61 | −109.17 |
| G | −87.57 | 57.37 | −80.07 | −129.76 | 35.18 | −122.79 |
| G, DO | −88.30 | 59.64 | −64.70 | −125.27 | 49.78 | −105.72 |
| G, C | −72.67 | 54.97 | −56.35 | −108.11 | 47.69 | −95.90 |
| G, DO, C | −85.13 | 52.49 | −67.97 | −120.76 | 48.75 | −104.82 |
| average | −79.77 | 53.14 | −66.48 | −116.47 | 44.39 | −103.26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borowik, A.; Wyszkowska, J.; Zaborowska, M.; Kucharski, J. Regulation of the Microbiome in Soil Contaminated with Diesel Oil and Gasoline. Int. J. Mol. Sci. 2025, 26, 6491. https://doi.org/10.3390/ijms26136491
Borowik A, Wyszkowska J, Zaborowska M, Kucharski J. Regulation of the Microbiome in Soil Contaminated with Diesel Oil and Gasoline. International Journal of Molecular Sciences. 2025; 26(13):6491. https://doi.org/10.3390/ijms26136491
Chicago/Turabian StyleBorowik, Agata, Jadwiga Wyszkowska, Magdalena Zaborowska, and Jan Kucharski. 2025. "Regulation of the Microbiome in Soil Contaminated with Diesel Oil and Gasoline" International Journal of Molecular Sciences 26, no. 13: 6491. https://doi.org/10.3390/ijms26136491
APA StyleBorowik, A., Wyszkowska, J., Zaborowska, M., & Kucharski, J. (2025). Regulation of the Microbiome in Soil Contaminated with Diesel Oil and Gasoline. International Journal of Molecular Sciences, 26(13), 6491. https://doi.org/10.3390/ijms26136491









