Identification and Expression Pattern Analysis of AsSWEET Gene Family in Achnatherum splendens
Abstract
1. Introduction
2. Results
2.1. Identification and Physicochemical Characterization of the AsSWEET Gene Family
2.2. Secondary Structure Analysis of the AsSWEET Gene Family
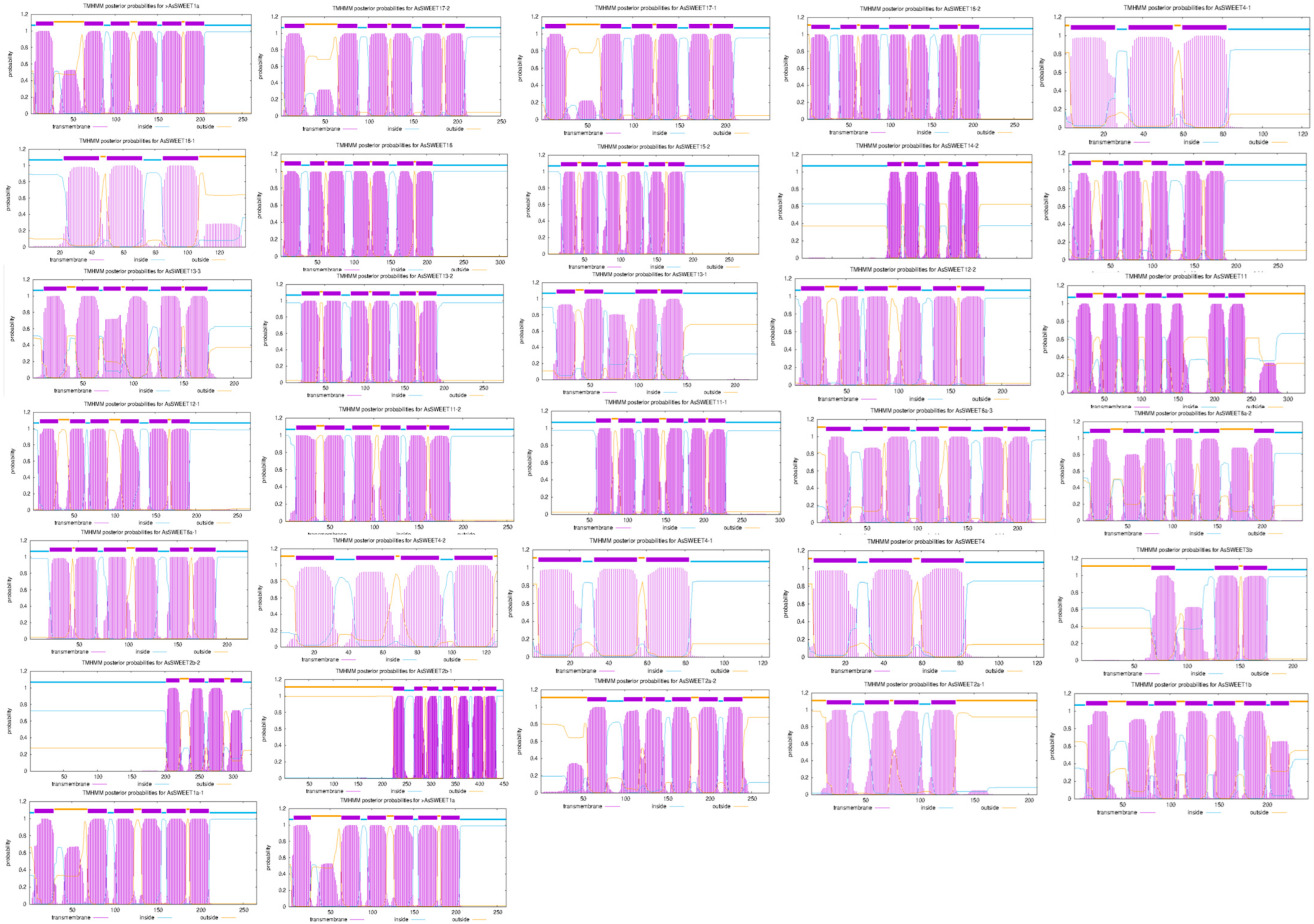
2.3. Analysis of Transmembrane Domains in the AsSWEET Family Proteins
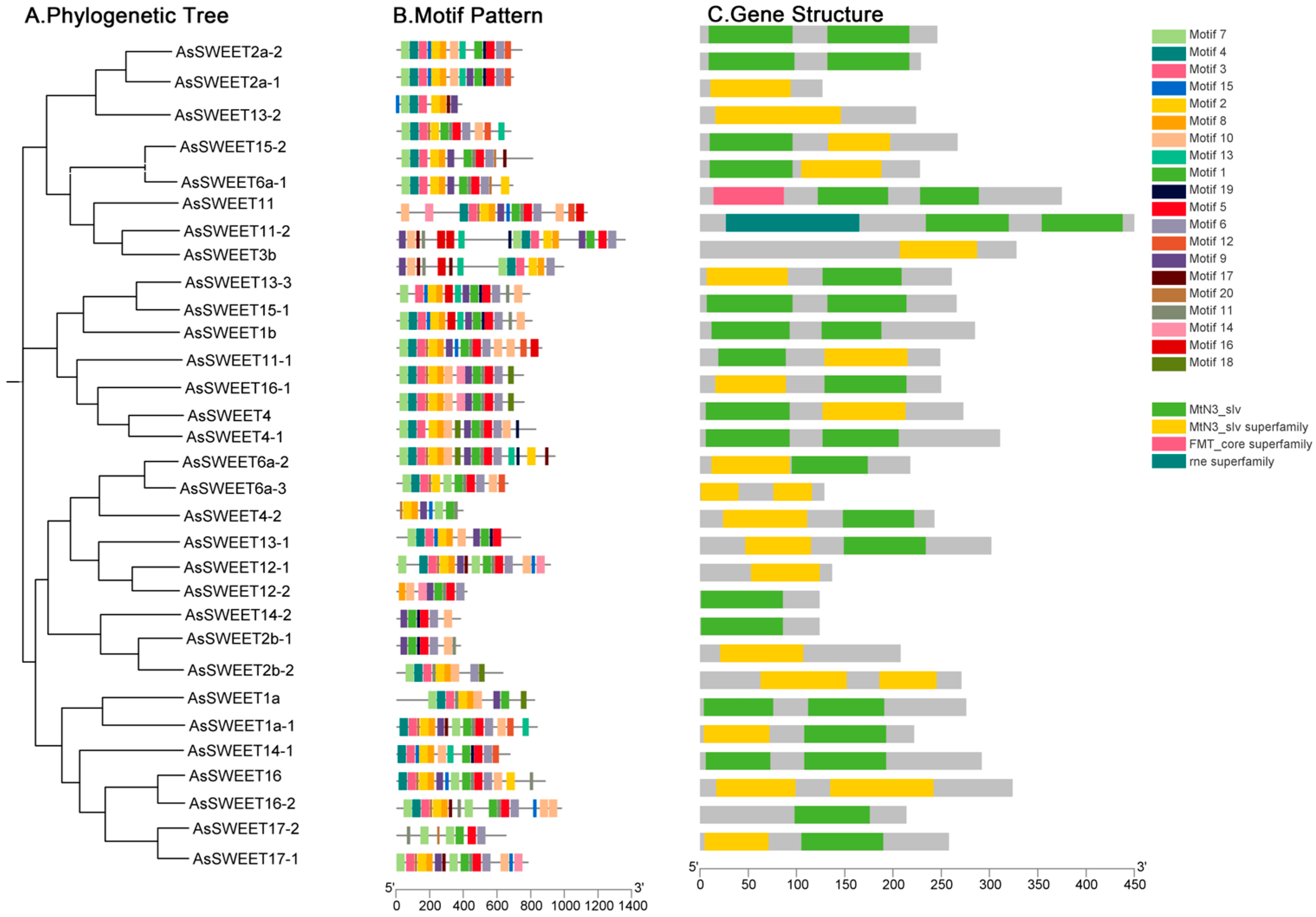
2.4. Analysis of AsSWEET Gene Structure and Protein Conserved Motifs
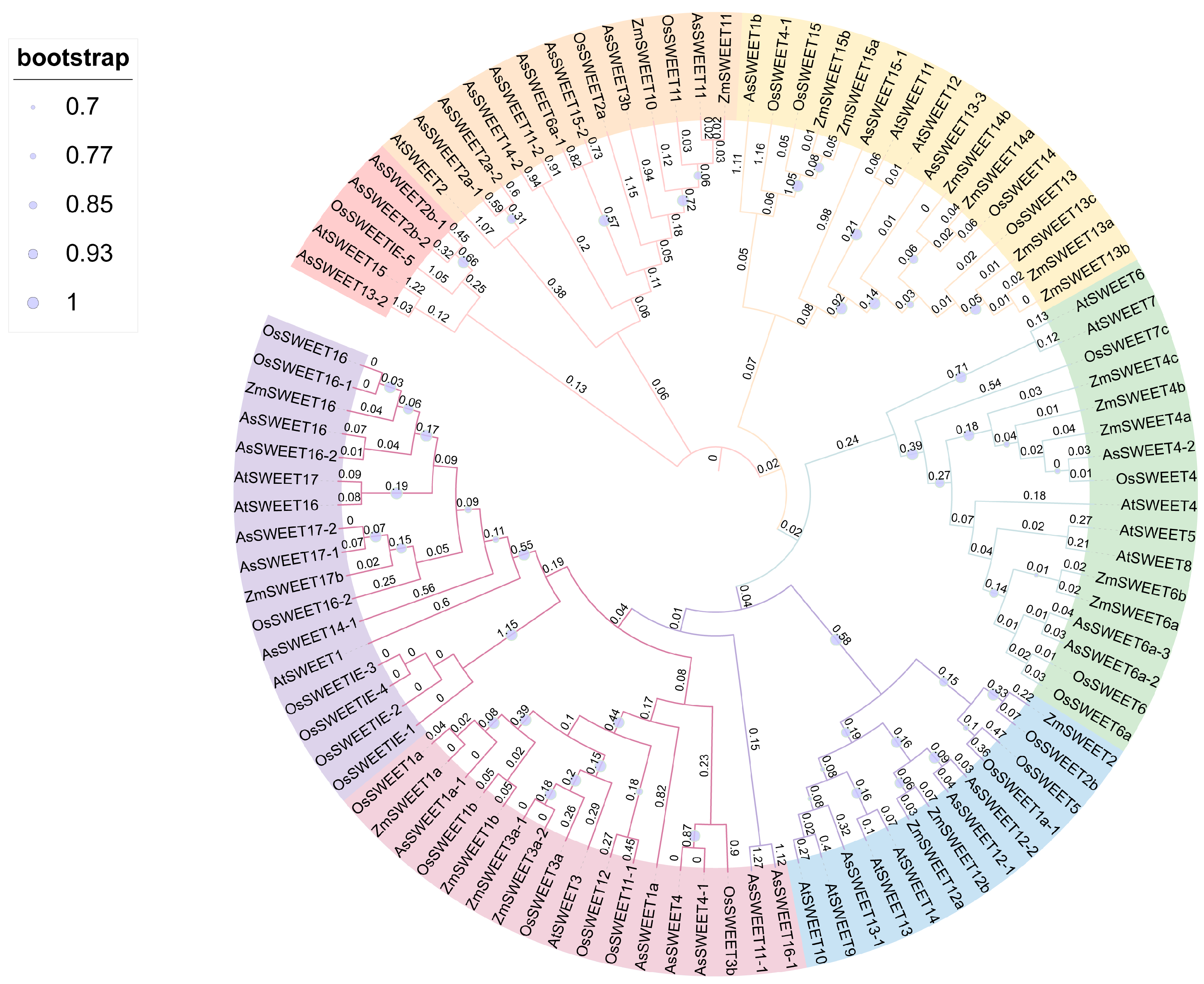
2.5. Phylogenetic Analysis of the AsSWEET Gene Family
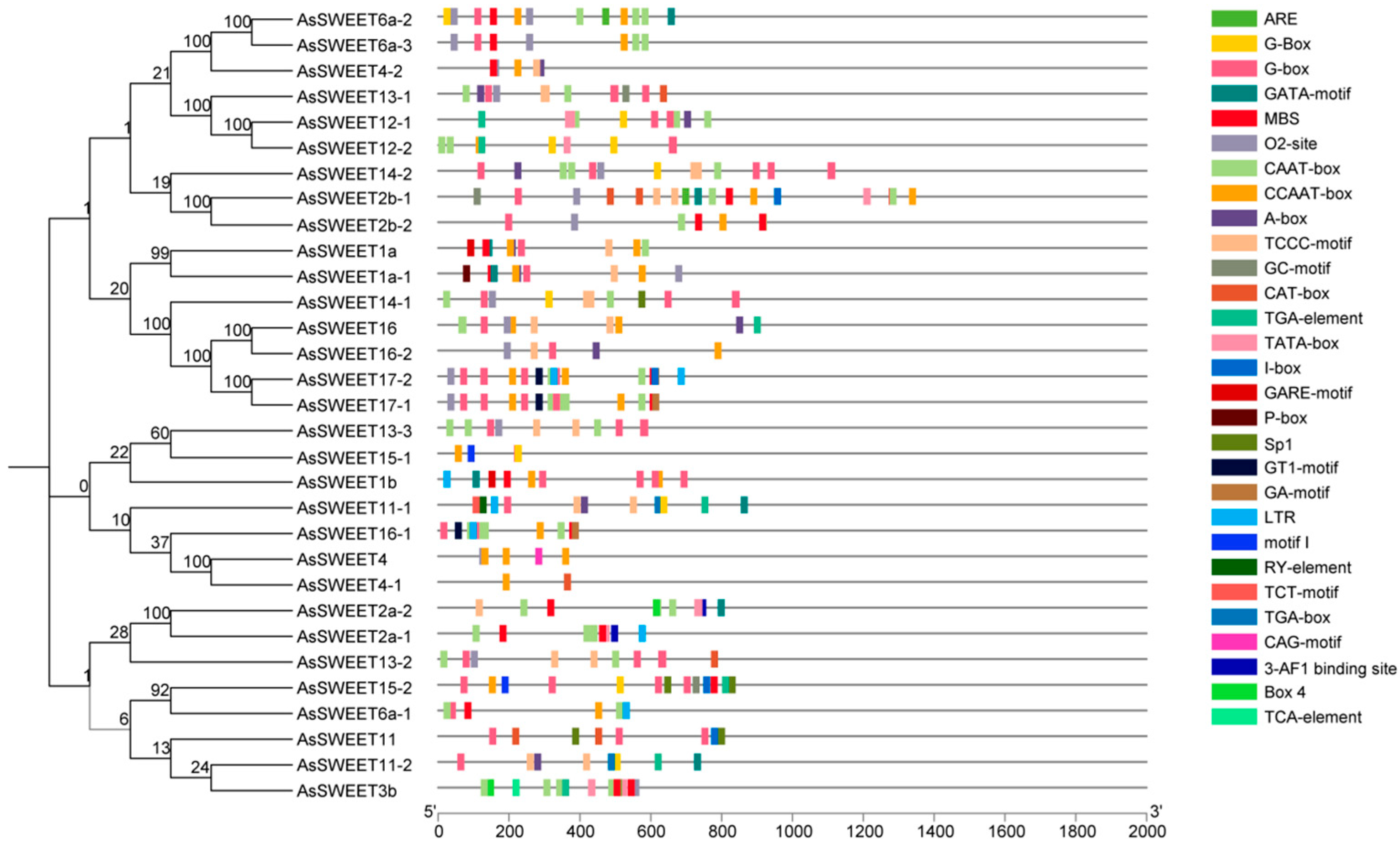
2.6. Analysis of Cis-Acting Elements in the Promoter Region of the AsSWEETs Gene Family
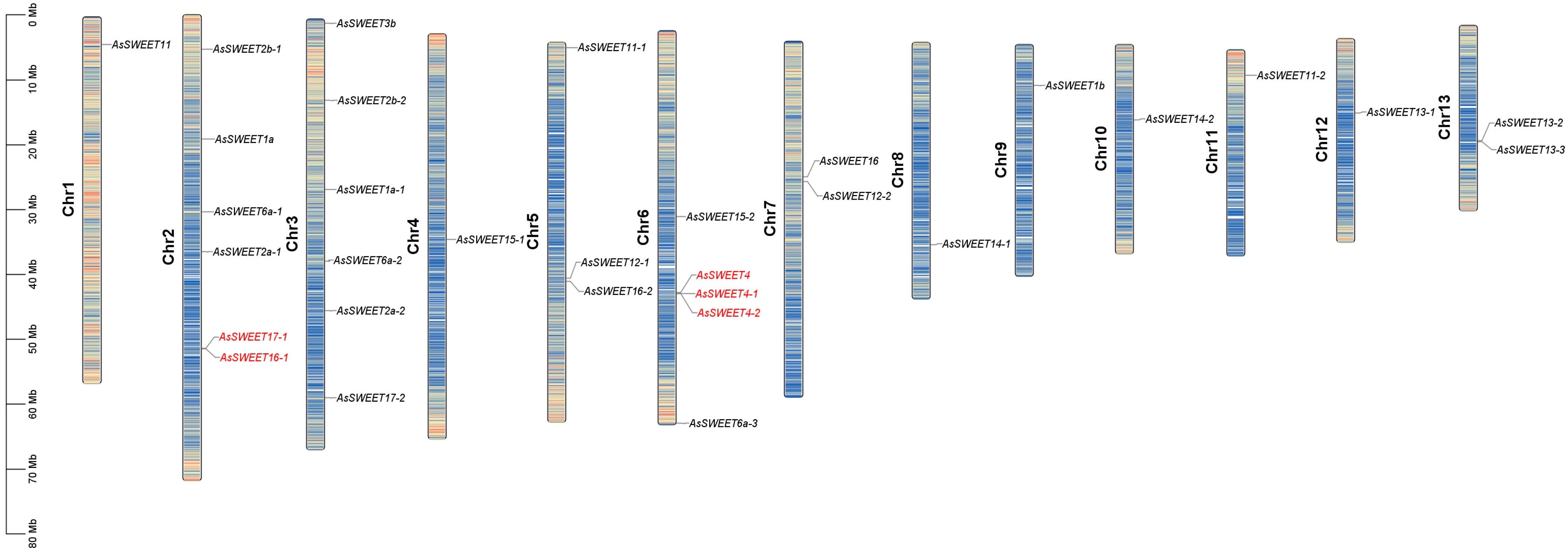
2.7. Chromosomal Localization Analysis of the AsSWEETs Gene Family
2.8. Subcellular Localization Prediction Analysis of the AsSWEETs Gene Family
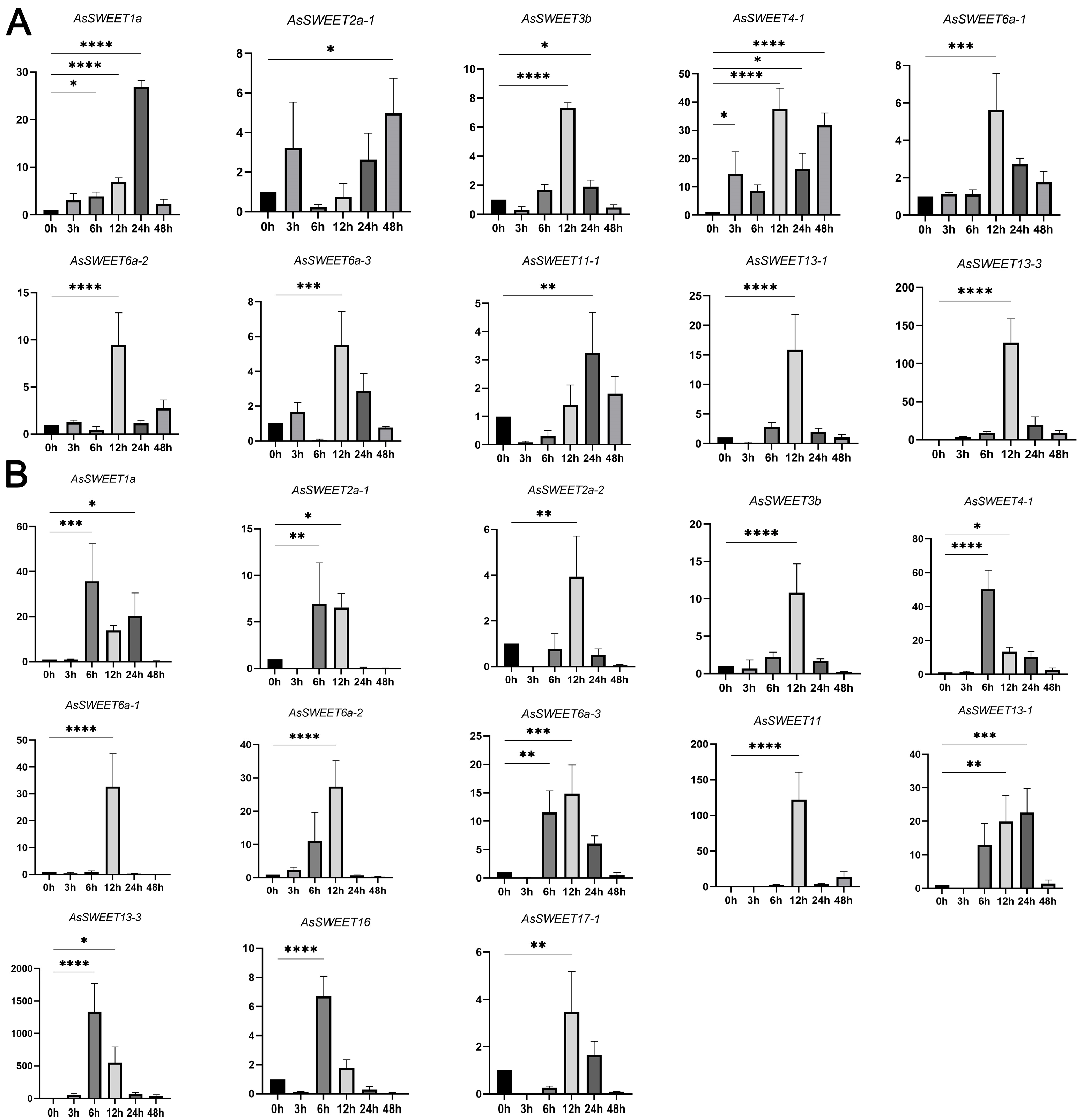
2.9. Expression Analysis of AsSWEETs Gene Family Members Under Abiotic Stress
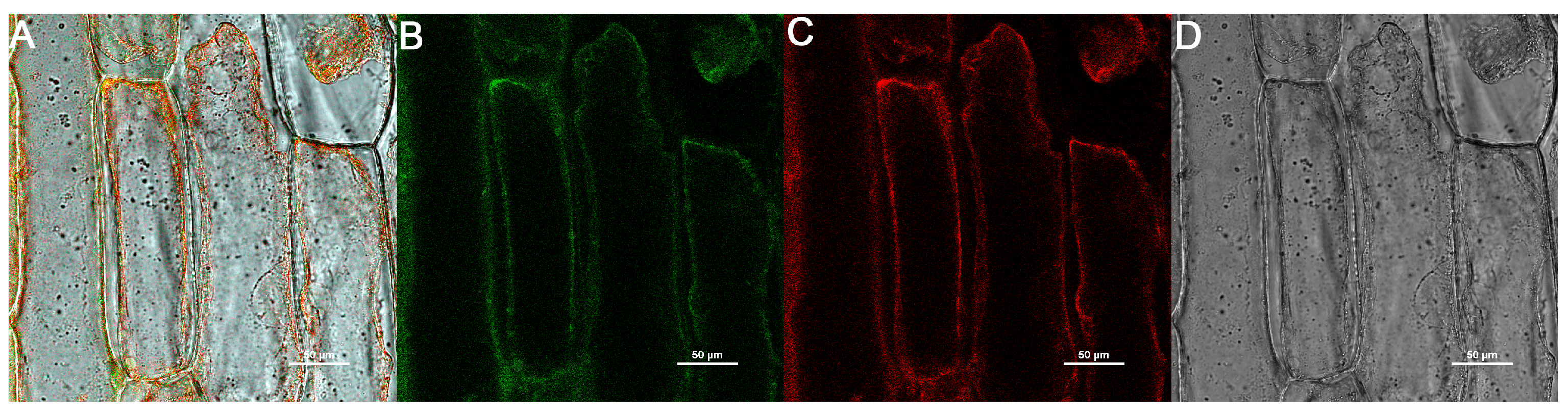
2.10. Subcellular Localization
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Experimental Methods
4.2.1. Identification and Physicochemical Characterization of the SWEET Gene Family in A. splendens
4.2.2. Secondary Structure Analysis of the SWEET Gene Family in A. splendens
4.2.3. Transmembrane Domain Analysis of A. splendens SWEET Proteins
4.2.4. Gene Structure and Protein Motif Analysis of A. splendens SWEET Family
4.2.5. Phylogenetic Analysis of A. splendens SWEET Gene Family
4.2.6. Promoter cis-Acting Element Analysis of A. splendens SWEET Genes
4.2.7. Chromosomal Localization of A. splendens SWEET Genes
4.2.8. Subcellular Localization Prediction of A. splendens SWEET Gene Family Members
4.2.9. Abiotic Stress Expression Analysis of SWEET Gene Family Members in A. splendens
4.2.10. Construction of Plant Expression Vector
4.2.11. Subcellular Localization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, J.H.; Peng, T.; Dai, W. Critical cis-acting elements and interacting transcription factors: Key players associated with abiotic stress responses in plants. Plant Mol. Biol. Rep. 2013, 32, 303–317. [Google Scholar] [CrossRef]
- Chen, L.Q.; Cheung, L.S.; Feng, L.; Tanner, W.; Frommer, W.B. Transport of sugars. Annu Rev Biochem. 2015, 84, 865–894. [Google Scholar] [CrossRef]
- Zhu, J.; Zhou, L.; Li, T.; Ruan, Y.; Zhang, A.; Dong, X.; Zhu, Y.; Li, C.; Fan, J. Genome-Wide Investigation and Characterization of SWEET Gene Family with Focus on Their Evolution and Expression during Hormone and Abiotic Stress Response in Maize. Genes 2022, 13, 1682. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Beena, R.; Laksmi, G.; Soni, K.B.; Alex, S.; Viji, M.M. Changes in Sucrose Metabolic Enzymes to Water Stress in Contrasting Rice Genotypes. Plant Stress 2022, 5, 100088. [Google Scholar] [CrossRef]
- Sambe, M.A.; He, X.; Tu, Q.; Guo, Z. A cold-induced myo-inositol transporter-like gene confers tolerance to multiple abiotic stresses in transgenic tobacco plants. Physiol. Plant. 2015, 153, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef]
- Wang, J.-G.; Zhao, T.-T.; Zhang, S.-Z.; Yang, B.-P.; Huang, C.-X. Plant Monosaccharide Transporters. Plant Physiol. J. 2007, 8, 1195–1201. [Google Scholar]
- Eom, J.S.; Chen, L.Q.; Sosso, D.; Julius, B.T.; Lin, I.W.; Qu, X.Q.; Braun, D.M.; Frommer, W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015, 25, 53–62. [Google Scholar] [CrossRef]
- Li, C.; Liu, Y.; Tian, J.; Zhu, Y.S.; Fan, J.J. Changes in sucrose metabolism in maize varieties with different cadmium sensitivities under cadmium stress. PLoS ONE 2020, 15, e0243835. [Google Scholar] [CrossRef]
- Sosso, D.; Luo, D.; Li, Q.B.; Sasse, J.; Yang, J.; Gendrot, G.; Suzuki, M.; Koch, K.E.; McCarty, D.R.; Chourey, P.S.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef]
- Zhang, Z.; Zou, L.; Ren, C.; Ren, F.; Wang, Y.; Fan, P.; Li, S.; Liang, Z. VvSWEET10 Mediates Sugar Accumulation in Grapes. Genes 2019, 10, 255. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.-Y.; Ho, L.-H.; Neuhaus, H.E.; Guo, W.-J. Transporter SlSWEET15 unloads sucrose from phloem and seed coat for fruit and seed development in tomato. Plant Physiol. 2021, 187, 2230–2245. [Google Scholar] [CrossRef]
- Li, Y.; Liu, H.; Yao, X.; Wang, J.; Feng, S.; Sun, L.; Ma, S.; Xu, K.; Chen, L.-Q.; Sui, X. Hexose transporter CsSWEET7a in cucumber mediates phloem unloading in companion cells for fruit development. Plant Physiol. 2021, 186, 640–654. [Google Scholar] [CrossRef]
- Zhou, A.; Ma, H.; Feng, S.; Gong, S.; Wang, J. DsSWEET17, a Tonoplast-Localized Sugar Transporter from Dianthus spiculifolius, Affects Sugar Metabolism and Confers Multiple Stress Tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 1564. [Google Scholar] [CrossRef]
- Zhou, A.; Ma, H.; Feng, S.; Gong, S.; Wang, J. A Novel Sugar Transporter from Dianthus spiculifolius, DsSWEET12, Affects Sugar Metabolism and Confers Osmotic and Oxidative Stress Tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 497. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Lyu, T.; Lyu, Y. LoSWEET14, a Sugar Transporter in Lily, Is Regulated by Transcription Factor LoABF2 to Participate in the ABA Signaling Pathway and Enhance Tolerance to Multiple Abiotic Stresses in Tobacco. Int. J. Mol. Sci. 2022, 23, 15093. [Google Scholar] [CrossRef] [PubMed]
- Fakher, B.; Jakada, B.H.; Greaves, J.G.; Wang, L.; Niu, X.; Cheng, Y.; Zheng, P.; Aslam, M.; Qin, Y.; Wang, X. Identification and expression analysis of pineapple sugar transporters reveal their role in the development and environmental response. Front. Plant Sci. 2022, 13, 964897. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Hou, B.-H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.-Q.; Guo, W.-J.; Kim, J.-G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Qu, X.-Q.; Hou, B.-H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Li, X.; Si, W.; Qin, Q.; Wu, H.; Jiang, H. Deciphering evolutionary dynamics of SWEET genes in diverse plant lineages. Sci. Rep. 2018, 8, 13440. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, C.; Wang, M.; Li, T.; Liu, X.; Jiang, J. Plasma Membrane-Localized SlSWEET7a and SlSWEET14 Regulate Sugar Transport and Storage in Tomato Fruits. Hortic. Res. 2021, 8, 186. [Google Scholar] [CrossRef] [PubMed]
- Kanika, K.; Puja, O.; Renu, B. Decoding Sugar Regulation and Homeostasis in Plants: Cracking Functional Roles Under Stresses. J. Plant Growth Regul. 2022, 42, 4797–4817. [Google Scholar]
- Yuan, M.; Wang, S. Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms. Mol. Plant 2013, 6, 665–674. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, H.; Zhao, K.; Wei, X.; Li, L.; Tang, Y.; Xiong, Y.; Xu, J. Expression Profiling Analysis of the SWEET Gene Family in In Vitro Pitaya Under Low-Temperature Stress and Study of Its Cold Resistance Mechanism. Plants 2024, 13, 3092. [Google Scholar] [CrossRef]
- Patil, G.; Valliyodan, B.; Deshmukh, R.; Prince, S.; Nicander, B.; Zhao, M.; Sonah, H.; Song, L.; Lin, L.; Chaudhary, J.; et al. Soybean (Glycine max) SWEET gene family: Insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genom. 2015, 16, 520. [Google Scholar] [CrossRef]
- Manck-Götzenberger, J.; Requena, N. Arbuscular mycorrhiza Symbiosis Induces a Major Transcriptional Reprogramming of the Potato SWEET Sugar Transporter Family. Front. Plant Sci. 2016, 7, 487. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Frommer, W.B. Structure and function of SemiSWEET and SWEET sugar transporters. Trends Biochem. Sci. 2015, 40, 480–486. [Google Scholar] [CrossRef]
- Mizuno, H.; Kasuga, S.; Kawahigashi, H. The sorghum SWEET gene family: Stem sucrose accumulation as revealed through transcriptome profiling. Biotechnol. Biofuels 2016, 9, 127. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, L.; Huang, W.; Yuan, M.; Zhou, F.; Li, X.; Lin, Y.; Zhang, J. Overexpression of OsSWEET5 in rice causes growth retardation and precocious senescence. PLoS ONE 2014, 9, e94210. [Google Scholar] [CrossRef]
- Chong, J.; Piron, M.-C.; Meyer, S.; Merdinoglu, D.; Bertsch, C.; Mestre, P. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J. Exp. Bot. 2014, 65, 6589–6601. [Google Scholar] [CrossRef]
- Irfan, M.; Chen, Q.; Yue, Y.; Pang, R.; Lin, Q.; Zhao, X.; Chen, H. Co-Production of Biochar, Bio-Oil and Syngas from Halophyte Grass (Achnatherum splendens L.) under Three Different Pyrolysis Temperatures. Bioresour. Technol. 2016, 211, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Ju, Y.; Xia, C.; Zhong, R.; Christensen, M.J.; Zhang, X.; Nan, Z. The Effect of Epichloë Endophyte on Phyllosphere Microbes and Leaf Metabolites in Achnatherum Inebrians. iScience 2022, 25, 104144. [Google Scholar] [CrossRef]
- Yang, J.; Xie, J.; Chen, H.; Zhu, S.; Hou, X.; Zhang, Z. Diversity and Biological Characteristics of Seed-Borne Bacteria of Achnatherum splendens. Microorganisms 2024, 12, 339. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L.; Bai, X.-M.; Zhang, J.-L.; Kang, R.-Q.; Zheng, W.-J.; Li, P.; Yan, Y.B.; Chen, H. Effects of Different Salt Solutions on Seed Germination Characteristics of Three Wild Achnatherum Species. Grassl. Turf 2024, 44, 183–189. [Google Scholar]
- Yuan, X.-H.; Sun, N.; Teng, W.-J.; Yang, X.-J.; Wu, J.-Y. Evaluation of Salt Tolerance at Seedling Stage in Nine Ornamental Grass Species and Effects of NaCl Stress on Achnatherum splendens Growth. J. Plant Resour. Environ. 2011, 20, 69–75. [Google Scholar]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of plant responses to salt stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Fujita, M. Plant Responses and Tolerance to Salt Stress: Physiological and Molecular Interventions. Int. J. Mol. Sci. 2022, 23, 4810. [Google Scholar] [CrossRef]
- Ozturk, M.; Unal, B.T.; García-Caparrós, P.; Khursheed, A.; Gul, A.; Hasanuzzaman, M. Osmoregulation and its actions during the drought stress in plants. Physiol. Plant. 2021, 172, 1321–1335. [Google Scholar] [CrossRef]
- Jiang, Y.-L. Effects of Saline-Alkali Stress on Seed Germination and Seedling Growth of Achnatherum splendens. Master’s Thesis, Lanzhou Jiaotong University, Lanzhou, China, 2014. [Google Scholar]
- Ma, X.-L. Responses of Gas Exchange Characteristics to Drought Stress in Different Plant Life Forms. Master’s Thesis, School of Life Sciences, Lanzhou University, Lanzhou, China, 2023. [Google Scholar]
- Pan, Y.; Niu, K.; Miao, P.; Zhao, G.; Zhang, Y.; Ju, Z.; Chai, J.; Yang, J.; Cui, X.; Zhang, R. Genome-wide analysis of the SWEET gene family and its response to powdery mildew and leaf spot infection in the common oat (Avena sativa L.). BMC Genom. 2024, 25, 995. [Google Scholar] [CrossRef]
- Han, X.; Han, S.; Zhu, Y.; Liu, Y.; Gao, S.; Yin, J.; Wang, F.; Yao, M. Genome-Wide Identification and Expression Analysis of the SWEET Gene Family in Capsicum annuum L. Int. J. Mol. Sci. 2023, 24, 17408. [Google Scholar] [CrossRef]
- Du, B.-S.; Zou, X.-H.; Wang, Z.-H.; Zhang, X.-Y.; Cao, Y.-B.; Zhang, L.-Y. Genome-Wide Identification and Expression Analysis of SWEET Gene Family in Camellia oleifera. Biotechnol. Bull. 2024, 40, 179–190. [Google Scholar] [CrossRef]
- Yin, Y.; Qin, X.-Y.; Zhao, J.-H.; Yang, P.; He, J.; An, W. Genome-Wide Identification of Lycium SWEET Gene Family and Its Correlation with Soluble Sugar Content. Acta Bot. Boreali-Occident. Sin. 2024, 44, 396–407. [Google Scholar]
- Xia, X.-Y.; Su, L.-H.; Ye, W.-X.; Duan, J.; Shi, H.-F.; Hu, H.-Y.; Zhang, N.; Yang, Z.; Ji, W. Genome-wide identification and expression analysis of SWEETs gene family in Vitis vinifera. Plant Physiol. J. 2023, 59, 2333–2343. [Google Scholar] [CrossRef]
- Li, G.; Niu, K.-J. Genome-wide identification and expression pattern analysis of SWEET gene family in triticale under various abiotic stresses. Acta Agrestia Sin. 2023, 31, 3310–3321. [Google Scholar]
- Li, M.; Zheng, D.-Z.; Hao, L.-D.; Guo, H.; Sun, Y. Genome-Wide Identification and Analysis of SWEET Gene Family in Maize. Molecular Plant Breeding [Online First], 1–11. Available online: http://kns.cnki.net/kcms/detail/46.1068.S.20221031.1009.008.html (accessed on 1 May 2025).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

| Gene Name | Protein/ (aa) | MW(Da) | PI | Instability Index | Aliphatic Index | GRAVY |
|---|---|---|---|---|---|---|
| AsSWEET1a | 785 | 65,294.44 | 5.05 | 50.48 | 17.43 | 0.736 |
| AsSWEET1a-1 | 801 | 66,709.30 | 5.04 | 54.29 | 16.73 | 0.728 |
| AsSWEET1b | 732 | 61,692.02 | 5.08 | 48.68 | 15.85 | 0.829 |
| AsSWEET2a-1 | 627 | 52,648.77 | 5.11 | 48.12 | 20.26 | 0.963 |
| AsSWEET2a-2 | 816 | 69,883.04 | 5.07 | 43.95 | 18.38 | 0.915 |
| AsSWEET2b-1 | 1353 | 109,286.91 | 4.99 | 41.24 | 18.99 | 0.839 |
| AsSWEET2b-2 | 987 | 79,015.14 | 5.06 | 41.72 | 20.67 | 0.980 |
| AsSWEET3b | 645 | 53,983.53 | 5.14 | 35.81 | 24.19 | 0.949 |
| AsSWEET4 | 375 | 31,499.38 | 5.18 | 57.86 | 21.07 | 0.933 |
| AsSWEET4-1 | 375 | 31,521.36 | 5.18 | 56.29 | 18.93 | 0.939 |
| AsSWEET4-2 | 384 | 33,170.84 | 5.14 | 59.34 | 14.06 | 0.949 |
| AsSWEET6a-1 | 669 | 56,383.84 | 5.08 | 54.43 | 19.73 | 0.817 |
| AsSWEET6a-2 | 741 | 63,173.95 | 5.05 | 53.99 | 18.35 | 0.786 |
| AsSWEET6a-3 | 690 | 58,671.54 | 5.07 | 53.71 | 19.28 | 0.79 |
| AsSWEET11 | 975 | 83,415.84 | 4.98 | 52.94 | 16.10 | 0.849 |
| AsSWEET11-1 | 909 | 77,606.83 | 5.00 | 55.62 | 16.28 | 0.837 |
| AsSWEET11-2 | 777 | 66,672.56 | 5.02 | 56.25 | 14.67 | 0.871 |
| AsSWEET12-1 | 804 | 68,695.44 | 5.03 | 58.07 | 17.91 | 0.799 |
| AsSWEET12-2 | 687 | 57,957.26 | 5.08 | 52.75 | 17.61 | 0.860 |
| AsSWEET13-1 | 675 | 55,438.35 | 5.08 | 51.67 | 20.59 | 1.004 |
| AsSWEET13-2 | 831 | 69,651.03 | 5.03 | 52.59 | 16.49 | 0.814 |
| AsSWEET13-3 | 657 | 55,396.52 | 5.08 | 52.79 | 16.89 | 0.892 |
| AsSWEET14-1 | 858 | 71,819.04 | 5.01 | 55.83 | 18.65 | 0.853 |
| AsSWEET14-2 | 1128 | 92,667.56 | 4.99 | 48.41 | 19.95 | 0.909 |
| AsSWEET15-1 | 390 | 33,558.76 | 5.16 | 53.69 | 13.59 | 0.927 |
| AsSWEET15-2 | 879 | 72,421.47 | 5.03 | 47.57 | 17.41 | 0.731 |
| AsSWEET16 | 936 | 76,825.64 | 5.02 | 53.29 | 17.74 | 0.663 |
| AsSWEET16-1 | 414 | 34,697.08 | 5.21 | 50.47 | 20.53 | 0.923 |
| AsSWEET16-2 | 844 | 70,557.10 | 4.78 | 50.61 | 18.79 | 0.696 |
| AsSWEET17-1 | 750 | 62,989.42 | 5.08 | 52.43 | 20.27 | 0.714 |
| AsSWEET17-2 | 753 | 62,918.27 | 5.08 | 51.34 | 20.85 | 0.728 |
| Gene Name | Alpha Helix (Hh) | Beta Turn (Tt) | Random Coil (Ee) | Extended Strand (Cc) |
|---|---|---|---|---|
| AsSWEET1a | 48.66 | 3.83 | 31.03 | 16.47 |
| AsSWEET1a-1 | 55.26 | 3.01 | 26.69 | 16.36 |
| AsSWEET1b | 47.33 | 4.12 | 29.22 | 19.05 |
| AsSWEET2a-1 | 36.54 | 8.65 | 35.10 | 25.81 |
| AsSWEET2a-2 | 41.61 | 10.22 | 20.44 | 21.83 |
| AsSWEET2b-1 | 45.78 | 8.67 | 26.00 | 19.34 |
| AsSWEET2b-2 | 36.59 | 6.71 | 34.45 | 27.74 |
| AsSWEET3b | 41.59 | 3.27 | 38.79 | 23.98 |
| AsSWEET4 | 42.74 | 3.23 | 30.65 | 22.26 |
| AsSWEET4-1 | 43.55 | 4.03 | 26.61 | 23.18 |
| AsSWEET4-2 | 51.18 | 7.87 | 21.26 | 23.39 |
| AsSWEET6a-1 | 39.19 | 4.05 | 34.68 | 18.48 |
| AsSWEET6a-2 | 39.84 | 4.47 | 31.71 | 16.48 |
| AsSWEET6a-3 | 43.23 | 4.37 | 30.57 | 17.83 |
| AsSWEET11 | 40.74 | 3.70 | 33.64 | 19.56 |
| AsSWEET11-1 | 37.75 | 4.64 | 34.44 | 19.19 |
| AsSWEET11-2 | 48.84 | 1.55 | 31.78 | 19.72 |
| AsSWEET12-1 | 37.45 | 0.75 | 43.82 | 17.98 |
| AsSWEET12-2 | 55.26 | 2.63 | 26.75 | 19.71 |
| AsSWEET13-1 | 41.07 | 5.36 | 35.27 | 37.98 |
| AsSWEET13-2 | 47.10 | 3.26 | 31.16 | 18.30 |
| AsSWEET13-3 | 40.37 | 1.83 | 38.07 | 20.02 |
| AsSWEET14-1 | 45.96 | 3.86 | 34.39 | 19.69 |
| AsSWEET14-2 | 40.96 | 5.17 | 34.69 | 20.40 |
| AsSWEET15-1 | 26.23 | 7.07 | 46.68 | 22.07 |
| AsSWEET15-2 | 42.47 | 3.42 | 37.67 | 16.44 |
| AsSWEET16 | 44.63 | 3.68 | 33.44 | 15.04 |
| AsSWEET16-1 | 30.23 | 2.33 | 29.46 | 21.91 |
| AsSWEET16-2 | 42.49 | 3.66 | 34.80 | 15.35 |
| AsSWEET17-1 | 49.80 | 4.02 | 29.72 | 15.79 |
| AsSWEET17-2 | 38.40 | 3.60 | 37.60 | 16.40 |
| Gene Name | Subcellular Localization |
|---|---|
| AsSWEET1a | Plasma membrane |
| AsSWEET1a-1 | Plasma membrane |
| AsSWEET1b | Tonoplast |
| AsSWEET2a-1 | Plasma membrane |
| AsSWEET2a-2 | Tonoplast |
| AsSWEET2b-1 | Plasma membrane |
| AsSWEET2b-2 | Nucleus |
| AsSWEET3b | Chloroplast |
| AsSWEET4 | Extracellular matrix |
| AsSWEET4-1 | Extracellular matrix |
| AsSWEET4-2 | Tonoplast |
| AsSWEET6a-1 | Plasma membrane |
| AsSWEET6a-2 | Plasma membrane |
| AsSWEET6a-3 | Plasma membrane |
| AsSWEET11 | Plasma membrane |
| AsSWEET11-1 | Plasma membrane |
| AsSWEET11-2 | Plasma membrane |
| AsSWEET12-1 | Plasma membrane |
| AsSWEET12-2 | Chloroplast |
| AsSWEET13-1 | Plasma membrane |
| AsSWEET13-2 | Plasma membrane |
| AsSWEET13-3 | Plasma membrane |
| AsSWEET14-1 | Plasma membrane |
| AsSWEET14-2 | Plasma membrane |
| AsSWEET15-1 | Tonoplast |
| AsSWEET15-2 | Plasma membrane |
| AsSWEET16 | Plasma membrane |
| AsSWEET16-1 | Tonoplast |
| AsSWEET16-2 | Tonoplast |
| AsSWEET17-1 | Tonoplast |
| AsSWEET17-2 | Tonoplast |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, M.; Kou, W.; Chen, M.; Li, X.; Wang, J.; Niu, J.; Wang, F.; Li, H.; Li, R. Identification and Expression Pattern Analysis of AsSWEET Gene Family in Achnatherum splendens. Int. J. Mol. Sci. 2025, 26, 6438. https://doi.org/10.3390/ijms26136438
Hu M, Kou W, Chen M, Li X, Wang J, Niu J, Wang F, Li H, Li R. Identification and Expression Pattern Analysis of AsSWEET Gene Family in Achnatherum splendens. International Journal of Molecular Sciences. 2025; 26(13):6438. https://doi.org/10.3390/ijms26136438
Chicago/Turabian StyleHu, Ming, Wei Kou, Mingsu Chen, Xiaoying Li, Jingru Wang, Jiahuan Niu, Fei Wang, Hongbin Li, and Rong Li. 2025. "Identification and Expression Pattern Analysis of AsSWEET Gene Family in Achnatherum splendens" International Journal of Molecular Sciences 26, no. 13: 6438. https://doi.org/10.3390/ijms26136438
APA StyleHu, M., Kou, W., Chen, M., Li, X., Wang, J., Niu, J., Wang, F., Li, H., & Li, R. (2025). Identification and Expression Pattern Analysis of AsSWEET Gene Family in Achnatherum splendens. International Journal of Molecular Sciences, 26(13), 6438. https://doi.org/10.3390/ijms26136438







