The Whole Blood Transcriptomic Analysis in Sickle Cell Disease Reveals RUNX3 as a Potential Marker for Vaso-Occlusive Crises
Abstract
1. Introduction
2. Results
2.1. Characteristics of Participants
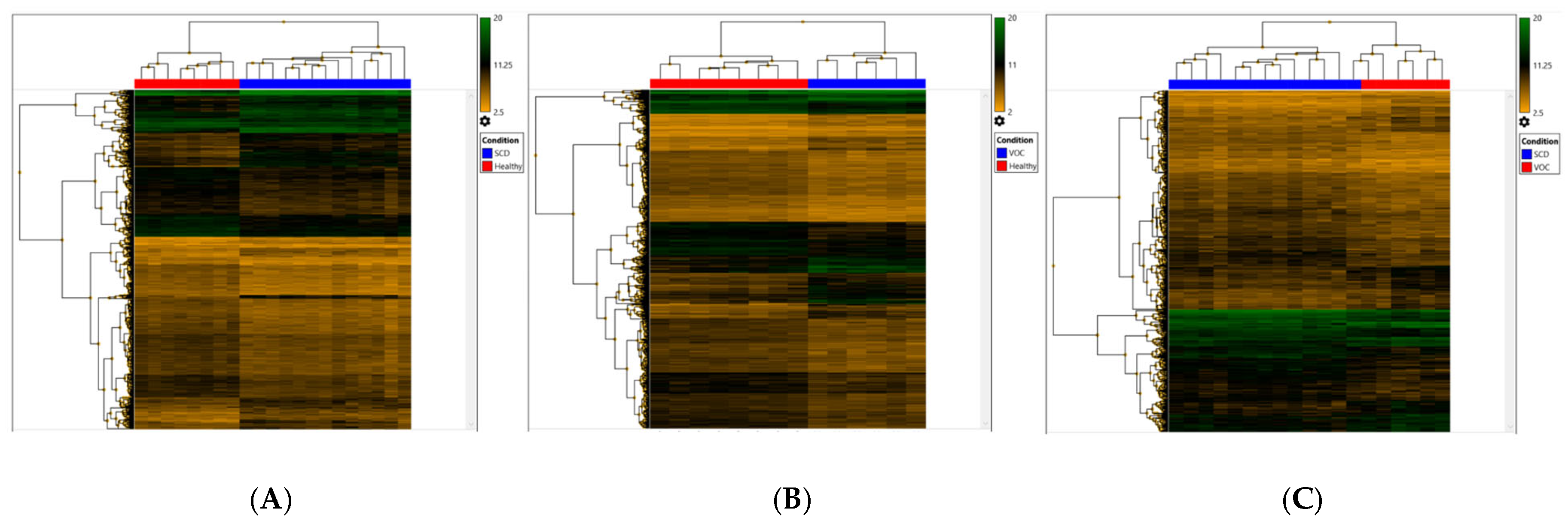
2.2. Determination of the Differentially-Expressed Genes
2.3. Potential Genetic Marker for Vaso-Occlusive Crisis
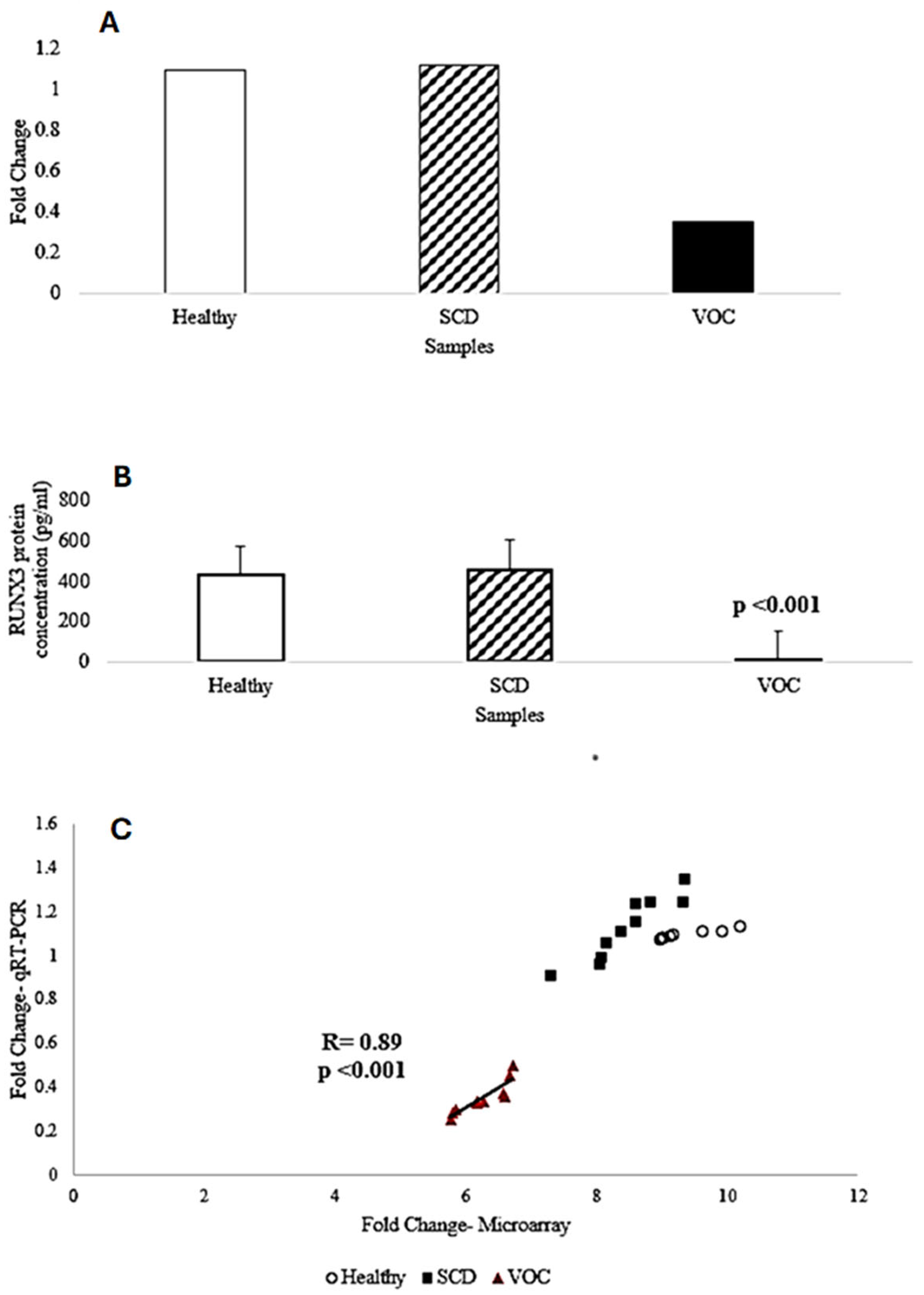
2.4. Validation of RUNX3 Through qRT-PCR and ELISA
3. Discussion
3.1. Molecular Mechanisms of RUNX3 in VOC Pathophysiology
- (1)
- Immune System Regulation: Based on our GO term enrichment analysis (Figure 2), RUNX3 down-regulation significantly impacts immune system processes, particularly through: (a) T-Cell Development and Function: RUNX3′s role in CD8+ T cell development aligns with our finding of down-regulated T cell receptor binding pathways.This is supported by previous studies showing RUNX3′s critical function in T-cell lineage specification [30]. The significant down-regulation of IL7R (12.76-fold) and CD3E (11.28-fold) in our study further supports this immune dysregulation pathway
- (2)
- Inflammatory Response: Our findings showed significant alterations in inflammatory pathways, specifically: (a) Cytokine Regulation: RUNX3 down-regulation correlates with altered inflammatory mediator expression. This is evidenced by the substantial down-regulation of CCL5 (9.24-fold) in our VOC samples. Previous studies have shown RUNX3′s protective role against inflammatory response through regulation of the JAK2/STAT3 pathway [33].
- (3)
- Cellular Response Mechanisms: The GO term enrichment analysis revealed: (a) Signal Transduction: Up-regulation of cellular localization and signal transduction regulation pathways. These changes suggest adaptive responses to the inflammatory and hypoxic environment during VOC. The diagram below illustrates the implications of RUNX3 gene downregulation on various molecular pathways and biological processes, as suggested by our g:Profiler results.
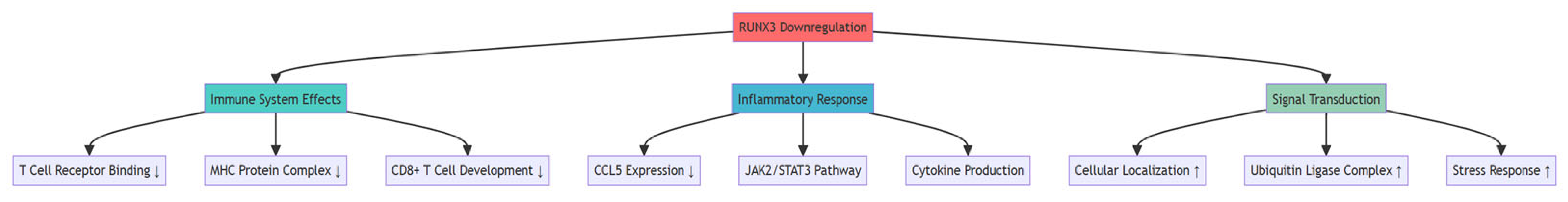
3.2. Integration with VOC Pathophysiology
- (a)
- Immune System Disruption: Impaired T-cell development and function, evidenced by down-regulation of key T-cell markers (IL7R, CD3E). Altered MHC protein complex binding, affecting immune response regulation
- (b)
- Inflammatory Cascade: Disrupted cytokine production and signaling. Altered inflammatory mediator expression (CCL5 down-regulation).
- (c)
- Cellular Stress Response: Enhanced cellular localization and signal transduction
3.3. Molecular Mechanisms Based on GO Term Enrichment Analysis
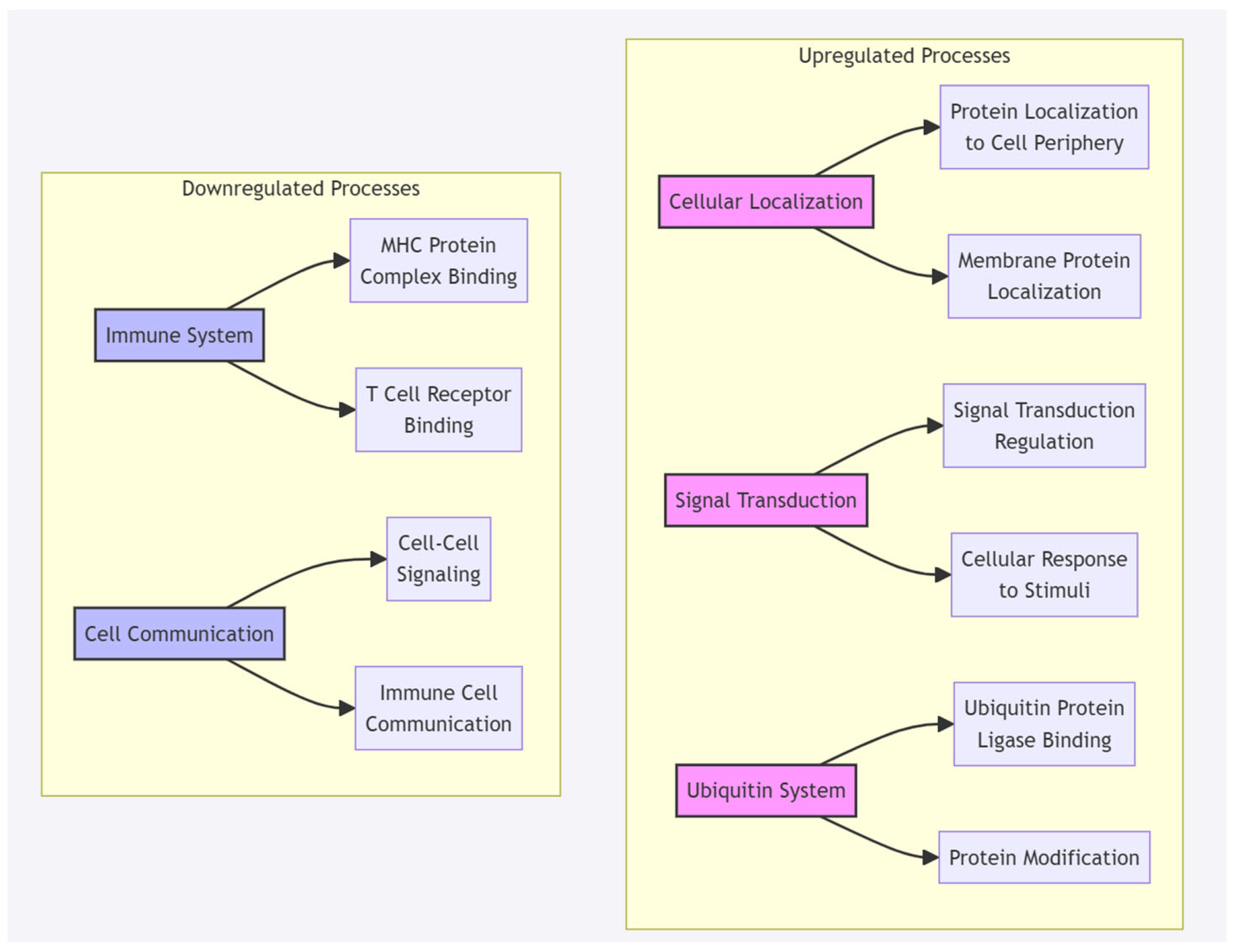
3.4. Timeline Limitations
3.5. Functional Confirmation Needs
4. Materials and Methods
4.1. Study Population
4.2. Sample Collection
4.3. RNA Extraction and Gene Expression Analysis
4.4. Quantitative Real-Time Polymerase Chain Reaction
4.5. Enzyme-Linked Immunosorbent Assay
4.6. Statistical Analysis
4.7. Gene Ontology (GO) Term Enrichment Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mason, V.R. Sickle cell anemia. JAMA 1922, 79, 1318. [Google Scholar] [CrossRef]
- Rees, D.C.; Williams, T.N.; Gladwin, M.T. Sickle-cell disease. Lancet 2010, 376, 2018–2031. [Google Scholar] [CrossRef] [PubMed]
- Manwani, D.; Frenette, P.S. Vaso-occlusion in sickle cell disease: Pathophysiology and novel targeted therapies. Blood 2013, 122, 3892–3898. [Google Scholar] [CrossRef]
- Pinto, V.M.; Balocco, M.; Quintino, S.; Forni, G.L. Sickle cell disease: A review for the internist. Intern. Emerg. Med. 2019, 14, 1051–1064. [Google Scholar] [CrossRef]
- Toledo, S.L.d.O.; Guedes, J.V.M.; Alpoim, P.N.; Rios, D.R.A.; de Pinheiro, B.M. Sickle cell disease: Hemostatic and inflammatory changes, and their interrelation. Clin. Chim. Acta 2019, 493, 129–137. [Google Scholar] [CrossRef]
- Allali, S.; Maciel, T.; Hermine, O.; de Montalembert, M. Innate immune cells, major protagonists of sickle cell disease pathophysiology. Haematologica 2020, 105, 273. [Google Scholar] [CrossRef]
- Benenson, I.; Porter, S. Sickle Cell Disease: Bone, Joint, Muscle, and Motor Complications. Orthop. Nurs. 2018, 37, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Renoux, C.; Joly, P.; Faes, C.; Mury, P.; Eglenen, B.; Turkay, M.; Yavas, G.; Yalcin, O.; Bertrand, Y.; Garnier, N.; et al. Association between Oxidative Stress, Genetic Factors, and Clinical Severity in Children with Sickle Cell Anemia. J. Pediatr. 2018, 195, 228–235. [Google Scholar] [CrossRef]
- Coelho, A.; Dias, A.; Morais, A.; Nunes, B.; Faustino, P.; Lavinha, J. Sickle cell disease severity scoring: A yet unsolved problem. Eur. J. Haematol. 2012, 89, 501–502. [Google Scholar] [CrossRef]
- Kalpatthi, R.; Novelli, E.M. Measuring success: Utility of biomarkers in sickle cell disease clinical trials and care. Hematol. Am. Soc. Hematol. Educ. Program. 2018, 2018, 482–492. [Google Scholar] [CrossRef]
- Rees, D.C.; Gibson, J.S. Biomarkers in sickle cell disease. Br. J. Haematol. 2012, 156, 433–445. [Google Scholar] [CrossRef]
- Damanhouri, G.A.; Jarullah, J.; Marouf, S.; Hindawi, S.I.; Mushtaq, G.; Kamal, M.A. Clinical biomarkers in sickle cell disease. Saudi J. Biol. Sci. 2015, 22, 24–31. [Google Scholar] [CrossRef]
- Hounkpe, B.W.; Fiusa, M.M.L.; Colella, M.P.; da Costa, L.N.G.; de Oliveira Benatti, R.; Saad, S.T.O.; Costa, F.F.; dos Santos, M.N.N.; De Paula, E.V. Role of innate immunity-triggered pathways in the pathogenesis of Sickle Cell Disease: A meta-analysis of gene expression studies. Sci. Rep. 2015, 5, 17822. [Google Scholar] [CrossRef] [PubMed]
- Hamda, C.B.; Sangeda, R.; Mwita, L.; Meintjes, A.; Nkya, S.; Panji, S.; Mulder, N.; Guizani-Tabbane, L.; Benkahla, A.; Makani, J.; et al. A common molecular signature of patients with sickle cell disease revealed by microarray meta-analysis and a genome-wide association study. PLoS ONE 2018, 13, e0199461. [Google Scholar]
- Schlissel, M.S.; Durum, S.D.; Muegge, K. The Interleukin 7 Receptor Is Required for T Cell Receptor γ Locus Accessibility to the V(D)j Recombinase. J. Exp. Med. 2000, 191, 1045–1050. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.E.; Spasova, D.S.; Frimpong-Boateng, K.; Kim, H.O.; Lee, M.; Kim, K.S.; Surh, C.D. Interleukin-7 Availability Is Maintained by a Hematopoietic Cytokine Sink Comprising Innate Lymphoid Cells and T Cells. Immunity 2017, 47, 171–182.e4. [Google Scholar] [CrossRef]
- Leung, G.A.; Cool, T.; Valencia, C.H.; Worthington, A.; Beaudin, A.E.; Forsberg, E.C. The lymphoid-associated interleukin 7 receptor (IL7R) regulates tissue-resident macrophage development. Development 2019, 146, dev176180. [Google Scholar] [CrossRef]
- Wang, M.; Windgassen, D.; Papoutsakis, E.T. Comparative analysis of transcriptional profiling of CD3+, CD4+ and CD+ T cells identifies novel immune response players in T-Cell activation. BMC Genom. 2008, 9, 225. [Google Scholar] [CrossRef]
- de la Hera, A.; Müller, U.; Olsson, C.; Isaaz, S.; Tunnacliffe, A. Structure of the T cell antigen receptor (TCR): Two CD3 epsilon subunits in a functional TCR/CD3 complex. J. Exp. Med. 1991, 173, 7–17. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- Maglott, D.; Ostell, J.; Pruitt, K.; Tatusova, T. Entrez Gene: Gene-centered information at NCBI. Nucleic Acids Res. 2005, 33, D54–D58. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Mabuchi, M.; Kataoka, H.; Miura, Y.; Kim, T.; Kawaguchi, M.; Ebi, M.; Tanaka, M.; Mori, Y.; Kubota, E.; Mizushima, T.; et al. Tumor suppressor, AT motif binding factor 1 (ATBF1), translocates to the nucleus with runt domain transcription factor 3 (RUNX3) in response to TGF-β signal transduction. Biochem. Biophys. Res. Commun. 2010, 398, 321–325. [Google Scholar] [CrossRef]
- Xue, M.; Chen, L.; Wang, W.; Su, T.; Shi, L.; Wang, L.; Zhang, W.; Si, J.-M.; Wang, L.-J.; Chen, S.-J. HOTAIR induces the ubiquitination of Runx3 by interacting with Mex3b and enhances the invasion of gastric cancer cells. Gastric Cancer 2018, 21, 756–764. [Google Scholar] [CrossRef]
- Liang, Y.; He, L.; Yuan, H.; Jin, Y.; Yao, Y. Association between RUNX3 promoter methylation and non-small cell lung cancer: A meta-analysis. J. Thorac. Dis. 2014, 6, 694–705. [Google Scholar]
- Wang, C.Q.; Krishnan, V.; Tay, L.S.; Chin, D.W.L.; Koh, C.P.; Chooi, J.Y.; Nah, G.S.S.; Du, L.; Jacob, B.; Yamashita, N.; et al. Disruption of Runx1 and Runx3 Leads to Bone Marrow Failure and Leukemia Predisposition due to Transcriptional and DNA Repair Defects. Cell Rep. 2014, 8, 767–782. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, L.; Gu, G. The correlation between Runx3 and bronchial asthma. Clin. Chim. Acta 2018, 487, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Fu, J.; Yao, L.; Hou, A.; Xue, X. Runx3 is a key modulator during the epithelial-mesenchymal transition of alveolar type II cells in animal models of BPD. Int. J. Mol. Med. 2017, 40, 1466–1476. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhu, J.; Bian, T.; Wang, R.; Gao, F. Mislocalization of Runt-related transcription factor 3 results in airway inflammation and airway hyper-responsiveness in a murine asthma model. Exp. Ther. Med. 2017, 14, 2695–2701. [Google Scholar] [CrossRef][Green Version]
- Serroukh, Y.; Gu-Trantien, C.; Kashani, B.H.; Defrance, M.; Manh, T.P.V.; Azouz, A.; Detavernier, A.; Hoyois, A.; Das, J.; Bizet, M.; et al. The transcription factors Runx3 and ThPOK cross-regulate acquisition of cytotoxic function by human Th1 lymphocytes. eLife 2018, 7, e30496. [Google Scholar] [CrossRef]
- Ebihara, T.; Song, C.; Ryu, S.H.; Plougastel-Douglas, B.; Yang, L.; Levanon, D.; Groner, Y.; Bern, M.D.; Stappenbeck, T.S.; Colonna, M.; et al. Runx3 specifies lineage commitment of innate lymphoid cells. Nat. Immunol. 2015, 16, 1124–1133. [Google Scholar] [CrossRef]
- Tanriver, Y.; Diefenbach, A. Transcription factors controlling development and function of innate lymphoid cells. Int. Immunol. 2014, 26, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Cui, H.Z.; Xu, C.M.; Sun, Z.W.; Tang, Z.K.; Chen, H.L. RUNX3 protects against acute lung injury by inhibiting the JAK2/STAT3 pathway in rats with severe acute pancreatitis. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5382–5391. [Google Scholar] [PubMed]
- Das, S.S.; Sinha, R.; Chakravorty, N. Integrative microRNA and gene expression analysis identifies new drug repurposing candidates for fetal hemoglobin induction in β-hemoglobinopathies. Gene 2019, 706, 77–83. [Google Scholar] [CrossRef]
- Kang, B.; Tan, F.; Samit, G.; Archer, D.; Sutliff, R.; Ofori-Acquah, S.; Hart, C.M. microRNA-301b reduces PPARγ expression in transgenic sickle mice and in hemin-treated human pulmonary artery endothelial cells (1089.22). FASEB J. 2014, 28, 1089–1111. [Google Scholar] [CrossRef]
- Guo, Y.J.; Liu, J.X.; Guan, Y.W. Hypoxia induced upregulation of miR-301a/b contributes to increased cell autophagy and viability of prostate cancer cells by targeting NDRG2. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 101–108. [Google Scholar]
- World Health Organization. WHO Guidelines on Pharmacological Treatment of Persisting Pain in Children with Medical Illnesses; World Health Organization: Geneva, Switzerland, 2012. [Google Scholar]
- Karcioglu, O.; Topacoglu, H.; Dikme, O.; Dikme, O. A systematic review of the pain scales in adults: Which to use? Am. J. Emerg. Med. 2018, 36, 707–714. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. g:Profiler—Interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]


| Parameters | Steady-State | VOC | p-Values | |
|---|---|---|---|---|
| Gender [n (%)] | Male | 9 (90) | 9 (90) | 1 |
| Female | 1 (10) | 1 (10) | ||
| Age Mean in years ± SD | 33 ± 10.82 | 34.9 ± 9.3 | 0.68 | |
| No. of VOC per year ± SD | 9.7 ± 6.16 | 8.3 ± 5.38 | 0.59 | |
| No. of Hospital admissions per year Mean ± SD | 3.3 ± 1.83 | 3.7 ± 1.64 | 0.61 | |
| White Blood Cell counts Mean in × 109/L ± SD | 5.4 ± 2.95 | 6.05 ± 3.91 | 0.68 | |
| Red Blood Cell counts Mean in × 1012/L ± SD | 4.89 ± 0.94 | 4.07 ± 0.95 | 0.07 | |
| Hemoglobin Mean in g/dL ± SD | 11.17 ± 1.14 | 10.48 ± 1.51 | 0.27 | |
| Platelet Mean in × 109/L ± SD | 309.19 ± 205.84 | 201.2 ± 118.04 | 0.17 | |
| Hemoglobin F Mean in % ± SD | 13.88 ± 8.3 | 18.26 ± 6.02 | 0.2 | |
| Hemoglobin S Mean in % ± SD | 79.81± 7.97 | 76.25± 5.41 | 0.26 | |
| ID | Gene Symbol | Chromosome | Group | p-Values | Fold Change |
|---|---|---|---|---|---|
| SCD Patients in Steady-State Compared to Healthy Controls | |||||
| TC1100010092.hg.1 | EIF4G2; SNORD97 | chr11 | Multiple_Complex | 1.31 × 10−10 | −16.33 |
| TC0200007835.hg.1 | ACTR2 | chr2 | Multiple_Complex | 3.00 × 10−11 | −8.22 |
| TC1500009865.hg.1 | ANP32A | chr15 | Multiple_Complex | 1.79 × 10−9 | −6.67 |
| TC1700007383.hg.1 | RPL23A; SNORD4B; SNORD42B; SNORD42A | chr17 | Multiple_Complex | 1.93 × 10−11 | −6.6 |
| TSUnmapped00000264.hg.1 | RPL7A | Coding | 2.88 × 10−12 | −6.47 | |
| TC0700010538.hg.1 | HNRNPA2B1 | chr7 | Multiple_Complex | 3.42 × 10−12 | −6.07 |
| TC0600007378.hg.1 | HIST1H4J | chr6 | Coding | 1.34 × 10−8 | −5.81 |
| TC1500008312.hg.1 | IQGAP1 | chr15 | Multiple_Complex | 1.18 × 10−5 | −5.64 |
| TC0800010667.hg.1 | PDE7A | chr8 | Multiple_Complex | 2.26 × 10−8 | −5.64 |
| TC0600011227.hg.1 | HIST1H4K | chr6 | Coding | 1.66 × 19−8 | −5.6 |
| SCD Patients in VOC Compared to Healthy Controls | |||||
| TC0500007138.hg.1 | IL7R | chr5 | Multiple_Complex | 2.75 × 10−10 | −12.76 |
| TC1100009200.hg.1 | CD3E | chr11 | Multiple_Complex | 2.61 × 10−12 | −11.28 |
| TC1700012052.hg.1 | ACTG1 | chr17 | Multiple_Complex | 2.01 × 10−7 | −10.4 |
| TC1200007758.hg.1 | HNRNPA1 | chr12 | Multiple_Complex | 6.70 × 10−10 | −9.75 |
| TC0600011173.hg.1 | GUSBP2 | chr6 | Multiple_Complex | 2.47 × 10−7 | −9.28 |
| TC1700010447.hg.1 | CCL5 | chr17 | Coding | 4.51 × 10−8 | −9.24 |
| TC0500013430.hg.1 | GNB2L1; SNORD95; SNORD96A | chr5 | Multiple_Complex | 6.41 × 10−9 | −8.96 |
| TC0100007676.hg.1 | LCK | chr1 | Multiple_Complex | 8.11 × 10−10 | −8.92 |
| TC0400011548.hg.1 | LEF1 | chr4 | Multiple_Complex | 7.22 × 10−9 | −8.88 |
| TC0600011517.hg.1 | HLA-DPA1 | chr6 | Multiple_Complex | 6.94 × 10−7 | −8.79 |
| ID | Gene Symbol | Chromo-Some | p-Values | Fold Change | ||
|---|---|---|---|---|---|---|
| VOC vs. Steady-State | VOC vs. Healthy | Steady-State vs Healthy | ||||
| TC0100017110.hg.1 | FCMR | chr1 | 2.75 × 10−7 | −10.98 | −9.24 | −1.98 |
| TC0200008268.hg.1 | GNLY | chr2 | 1.76 × 10−5 | −7.37 | −8.92 | −1.48 |
| TC1200010850.hg.1 | TESPA1 | chr12 | 2.22 × 10−5 | −6.3 | −8.72 | −1.48 |
| TC1700010447.hg.1 | CCL5 | chr17 | 7.39 × 10−7 | −6.24 | −7.92 | −1.46 |
| TC0600007657.hg.1 | HLA-DQA1 | chr6 | 3.97 × 10−5 | −6.1 | −6.5 | −1.41 |
| TC1900011774.hg.1 | EMP3 | chr19 | 2.17 × 10−7 | −5.99 | −6 | −1.34 |
| TC0500012470.hg.1 | CD74 | chr5 | 1.06 × 10−6 | −5.89 | −5.9 | −1.32 |
| TC1200006738.hg.1 | KLRG1 | chr12 | 0.0001 | −5.72 | −5.83 | −1.19 |
| TC1200012571.hg.1 | ITFG2 | chr12 | 8.44 × 10−9 | −5.52 | −5.82 | −1.15 |
| TC2200008641.hg.1 | RAC2 | chr22 | 9.48 × 10−7 | −5.43 | −5.8 | −1.07 |
| TC0200011075.hg.1 | PTMA | chr2 | 3.16 × 10−8 | −5.41 | −5.76 | −1.07 |
| TC1000011904.hg.1 | ABLIM1 | chr10 | 5.35 × 10−7 | −5.22 | −5.69 | −1.06 |
| TC1600006888.hg.1 | CIITA | chr16 | 1.76 × 10−7 | −5.21 | −5.69 | 1 |
| TC1200012801.hg.1 | CS | chr12 | 2.63 × 10−6 | −5.19 | −5.34 | 1.01 |
| TC1200012583.hg.1 | CD27 | chr12 | 2.89 × 10−5 | −5.14 | −4.99 | 1.03 |
| TC1100013190.hg.1 | CFL1 | chr11 | 6.80 × 10−7 | −4.97 | −4.59 | 1.05 |
| TC0600007650.hg.1 | HLA-DRA | chr6 | 7.56 × 10−6 | −4.96 | −4.53 | 1.1 |
| TC1900007839.hg.1 | FXYD5 | chr19 | 2.52 × 10−8 | −4.95 | −4.53 | 1.12 |
| TC1200010616.hg.1 | TUBA1B | chr12 | 1.20 × 10−7 | −4.8 | −4.36 | 1.14 |
| TC0100007676.hg.1 | LCK | chr1 | 6.31 × 10−7 | −4.52 | −4.31 | 1.14 |
| TC0300006993.hg.1 | CRTAP | chr3 | 2.83 × 10−6 | −4.4 | −4.27 | 1.15 |
| TC1100007790.hg.1 | CD5 | chr11 | 8.94 × 10−8 | −4.39 | −4.18 | 1.16 |
| TC1400006656.hg.1 | OXA1L | chr14 | 1.78 × 10−8 | −4.36 | −4.16 | 1.18 |
| TC0100013339.hg.1 | RUNX3 | chr1 | 2.33 × 10−8 | −4.12 | −4.08 | 1.22 |
| TC0100018246.hg.1 | LRRC8C | chr1 | 1.94 × 10−6 | −4.09 | −4.04 | 1.3 |
| TC0800009819.hg.1 | DOK2 | chr8 | 3.24 × 10−6 | −4.07 | −4.03 | 1.32 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taha, S.; Abdulwahab, H.; Aljishi, M.; Sultan, A.; Bakhiet, M.; Spicuglia, S.; Belhocine, M. The Whole Blood Transcriptomic Analysis in Sickle Cell Disease Reveals RUNX3 as a Potential Marker for Vaso-Occlusive Crises. Int. J. Mol. Sci. 2025, 26, 6338. https://doi.org/10.3390/ijms26136338
Taha S, Abdulwahab H, Aljishi M, Sultan A, Bakhiet M, Spicuglia S, Belhocine M. The Whole Blood Transcriptomic Analysis in Sickle Cell Disease Reveals RUNX3 as a Potential Marker for Vaso-Occlusive Crises. International Journal of Molecular Sciences. 2025; 26(13):6338. https://doi.org/10.3390/ijms26136338
Chicago/Turabian StyleTaha, Safa, Hawra Abdulwahab, Muna Aljishi, Ameera Sultan, Moiz Bakhiet, Salvatore Spicuglia, and Mohamed Belhocine. 2025. "The Whole Blood Transcriptomic Analysis in Sickle Cell Disease Reveals RUNX3 as a Potential Marker for Vaso-Occlusive Crises" International Journal of Molecular Sciences 26, no. 13: 6338. https://doi.org/10.3390/ijms26136338
APA StyleTaha, S., Abdulwahab, H., Aljishi, M., Sultan, A., Bakhiet, M., Spicuglia, S., & Belhocine, M. (2025). The Whole Blood Transcriptomic Analysis in Sickle Cell Disease Reveals RUNX3 as a Potential Marker for Vaso-Occlusive Crises. International Journal of Molecular Sciences, 26(13), 6338. https://doi.org/10.3390/ijms26136338







