Abstract
Psoriasis involves complex epigenetic alterations, but detailed studies on histone methyltransferases and their role in disease progression are limited. We conducted a comprehensive analysis of nearly 300 transcriptomes, focusing mainly on differential expression of protein isoform-coding transcripts within the SET domain family of histone methyltransferases. Consistent with previous findings, EZH2 transcripts showed increased expression in lesional skin, indicating altered H3K27 methylation that may enhance gene silencing, promoting keratinocyte proliferation and inflammatory responses. In the SET2 family, ASH1L exhibited reversed expression patterns between non-lesional and lesional skin, while NSD1 and NSD2 were upregulated, and SETD2 downregulated in lesions, suggesting disrupted H3K36 methylation that may affect immune responses and keratinocyte proliferation. Among H3K9 methyltransferases, SUV39 members, SUV39H2 was upregulated in lesions, whereas EHMT1 transcripts increased in non-lesional skin, and SETDB2 decreased in lesions. Additionally, PRDM family members such as PRDM2, MECOM (PRDM3), PRDM6, and PRDM8 showed altered expression in lesional skin. The H4K20 methylating SUV4-20 subfamily member, a SUV420H1 transcript, and SETD8 belonging to the other SET domain-containing family of methyltransferases were significantly increased in non-lesional skin and in lesions, respectively. Overall, aberrant expression and isoform variability of histone methyltransferases likely contribute to psoriasis pathogenesis by dysregulating proliferation, differentiation, and immune responses.
1. Introduction
Psoriasis, affecting 2–3% of the population [1], is an immune-mediated skin disease characterized by inflammation and an exaggerated response to stressors, leading to abnormal keratinocyte proliferation and immune cell infiltration and response [2]. In psoriasis, numerous molecular and cellular alternations occur even in the symptom-free non-lesional skin, some of which stabilize the non-lesional skin state while others set the stage for lesion development [3]. Alterations of non-lesional psoriatic skin are not limited to the cells but also affect the extracellular matrix, including modified splicing [4], processing [5], and degradation [6] of extracellular molecules, some of which are known to have an impact on cell proliferation [7,8]. Considering these complex non-lesional skin abnormalities, epigenetic dysregulations were proposed to play a role [9], potentially affecting keratinocyte proliferation, differentiation, and immune responses.
Among epigenetic regulatory processes, histone methylation plays a crucial role in the regulation of proliferation. Histone methylation patterns of proliferating cells undergo specific modulation during different phases of the cell cycle [10]. Mono- and dimethylated ‘Lys-9’ of histone H3 (H3K9me1/2) remain unchanged throughout the cell cycle, while H3K9me3 shows a pronounced peak during the late G2 to mitosis (M) transition [11]. Monomethylated ‘Lys-20’ of histone H4 (H4K20me1) peaks during G2 to M transition but quickly converts to dimethylated ‘Lys-20’ of histone H4 (H4K20me2), which remains consistently high throughout the cell cycle, while H4K20me3 shows a slight increase in early G1 phase [12]. Disturbances in these regulatory processes can lead to an overturning of the cell division rate. During epidermal development, trimethylated ‘Lys-27’ of histone H3 (H3K27me3) and trimethylated ‘Lys-20’ of histone H4 (H4K20me3) methylation levels transition from low in basal cells to high in suprabasal cells, suggested to be important for the proper switch from proliferation to differentiation [13]. Indeed, abnormal keratinocyte differentiation in psoriasis has been shown to be accompanied by altered histone methylation patterns [14]. The higher levels of H3K27me3 described in psoriatic skin [15] suggest aberrant epigenetic regulation, which is likely to contribute to hyperproliferation and abnormal differentiation of keratinocytes.
Dynamic transitions of histone methylation patterns are also important regulators of both adaptive and innate immune responses. Dysregulation of immune cell functions plays a crucial role in the development and maintenance of psoriatic lesions, among which T cell-mediated immune responses are believed to play a central role [16]. The interplay of bivalent chromatin marks, such as trimethylated histone H3 (H3K4me3 and H3K27me3), appears to regulate important T cell functions, including differentiation, effector/memory T cell formation, T cell exhaustion processes, and differentiation of helper T cells into different subsets [17]. Dynamic regulation of H3K27me3 is also observed in natural killer cells and macrophages, and these processes balance pro-inflammatory and immunomodulatory activities to maintain immune homeostasis [18,19].
Despite the high impact of histone methylation-related alterations in psoriasis, a complete overview of this process in the context of the disease is still missing. Therefore, to fulfill this gap, we aimed to provide an overview of histone methyltransferases and their expression in psoriasis by analyzing a literature-based [20,21,22] psoriasis transcriptome database [23] of nearly 300 individuals. Finally, we also analyzed histone methyltransferases identified with altered expression for their possible associations with key disease-related processes, including cell proliferation/differentiation and immune regulation.
2. Results
To identify histone methyltransferases with altered expression in psoriasis, we analyzed the transcriptional profiles of all lysine and/or arginine methyltransferases belonging to the two known major families of SET domain (Figure 1a) and 7β-strand methyltransferases (Figure 1b). These molecules function in large complexes [24,25]; therefore, we have included components of these complexes in our analysis (Figure 1c). The resulting methyltransferases and methyltransferase complex members identified with differentially expressed transcripts in psoriasis are shown in Figure 1d (and Supplementary Table S1).
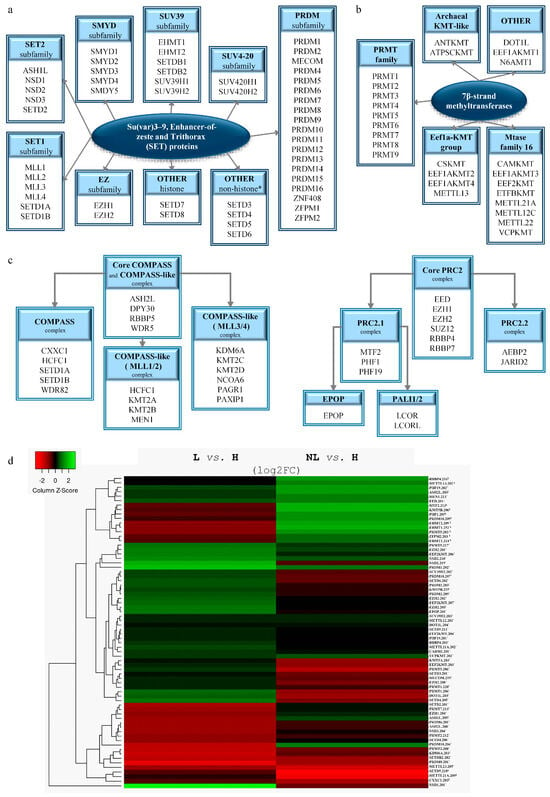
Figure 1.
Differentially expressed histone methylation-related molecules in psoriasis. Classification of SET domain (a) and 7β-strand (b) lysine and/or arginine methyltransferases and their complexes (COMPASS, COMPASS-like, and PRC2) required for their proper function (c) used for screening. The heatmap of differentially expressed transcripts of methyltransferases and associated complex members in psoriasis (d). (H: healthy, NL: non-lesional, L: lesional skin; ¥: transcript variants showing differential expression in NL vs. H; ‡: indicate transcript variants that are differentially expressed in both NL and L skin vs. to H; *: transcript variants showing disparate expression levels in L skin vs. H).
2.1. SET Domain-Containing Histone Lysine Methyltransferases with Altered Expression in Psoriasis
The most characteristic histone lysine methyltransferases belong to the SET domain family (Figure 1a). Among the family members, we identified EZH1/2, KMT5A, MECOM, NSD1-3, SETD2, SETDB2, SUV39H2, PRDM2, and PRDM8 with differential expression in lesional skin and EHMT1/2 in non-lesional skin (Figure 1d and Figure 2, Table 1 and Table S1). Meanwhile, ASH1L and SUV420H1 displayed abnormal expressions in both non-lesional and lesional skin (Figure 1d and Figure 2, Table 1 and Table S1). Our analysis also unveiled transcriptional disparities of family members with uncertain histone methyltransferase activity in lesional skin, including SETD3, SETD4, and SETD6, whereas SETD5 exhibited differences in both non-lesional and lesional skin compared to healthy controls (Figure 1d and Figure 2, and Supplementary Table S1).
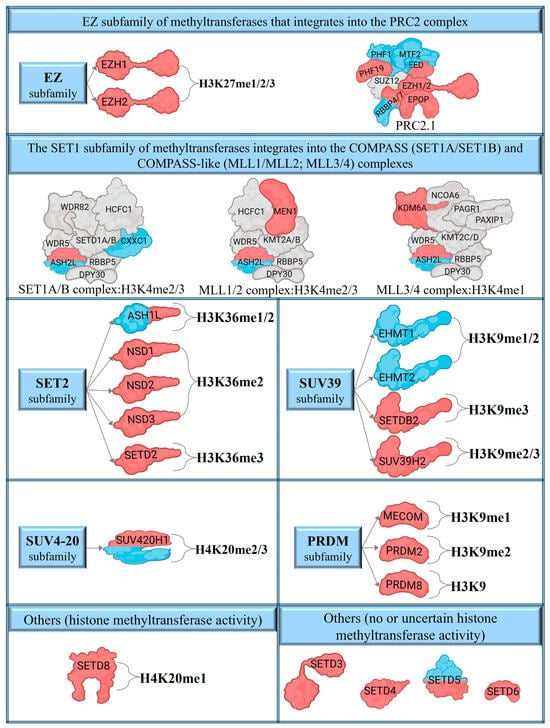
Figure 2.
The differentially expressed transcripts of SET domain catalytic methyltransferases and their complexes in psoriasis. The blue colors indicate differentially expressed transcripts in non-lesional skin, while red indicates lesional transcriptional alterations. Blue and red colors indicate the disparity in both non-lesional and lesional expression levels.

Table 1.
SET domain-containing histone methyltransferases and their constituents that target histone lysine residues for methylation. Methyltransferases identified with differentially expressed transcripts in non-lesional (NL) and/or lesional (L) skin are highlighted in bold.
2.2. Histone Lysine Methyltransferase Complex Members Affected by Altered Expression in Psoriasis
Among the members of the histone methyltransferase COMPASS complex (Figure 1c and Figure 2), CXXC1 shows changes in expression in non-lesional skin (Figure 1d and Figure 2, Table 2 and Table S1). The expression of ASH2L, which functions as a member of both the COMPASS and COMPASS-like complexes (Figure 1c and Figure 2), was found to be altered in both lesional and non-lesional skin (Figure 1d and Figure 2, Table 2 and Table S1), while KDM6A and MEN1, members of a COMPASS-like complex (Figure 1c and Figure 2), displays alteration only in lesional skin (Figure 1d, Table 2 and Table S1).

Table 2.
The molecular compositions of histone methyltransferase COMPASS/COMPASS-like and PRC2 complexes. (Transcripts exhibiting altered expression in psoriasis are shown in bold).
EZH1/2 are the catalytic methyltransferase subunits of the PRC2 complex [64,65] (Figure 1c and Figure 2), of which the non-catalytic members EED and RBBP4 show abnormal expression in non-lesional and lesional skin (Figure 1d, Table 2 and Table S1). The PRC2 complex has two modules, the PRC2.1 and the PRC2.2 subcomplexes [66] (Figure 1c). In psoriatic skin, we have observed expression changes of the PRC2.1 components PHF1 and MTF2 in non-lesional samples, and PHF19 and EPOP in skin lesions (Figure 1d and Figure 2, Table 2 and Table S1). In contrast, members of the PRC2.2 subcomplex were found to be unaffected in both psoriatic non-lesional and lesional skin.
2.3. Alterations in the Expression of Seven-β-Strand Lysine Methyltransferases in Psoriasis
While seven-β-strand methyltransferases are predominantly recognized as non-histone-specific enzymes, several members have been identified as histone methyltransferases [67,68] (Figure 1b and Table 3). Among the members known for their histone methyltransferase activity, only DOT1L was identified with altered expression in lesional (but not in non-lesional) psoriatic skin, compared to the healthy controls (Figure 1d, Table 3 and Table S1).

Table 3.
Lysine methyltransferases characterized by seven-β-strand structures, together with their targets on histone or non-histone proteins and the type of modifications [67,68]. Methyltransferases identified with differentially expressed transcripts in non-lesional (NL) and/or lesional (L) skin are highlighted in bold.
We observed abnormal transcriptional expression of the known non-histone-modifying lysine methyltransferases EEF2KMT, METTL12, and VCPKMT in psoriatic lesional skin, while METTL13 showed alterations in non-lesional skin (Figure 1d, Table 3 and Table S1). In addition, both non-lesional and lesional skin exhibited abnormalities in METTL21A expression (Figure 1d, Table 3 and Table S1). Table 3 summarizes the differentially expressed lysine seven-β-strand methyltransferases, including their targets, types, and modification sites.
2.4. Modifications in the Expression of Seven-β-Strand Arginine Methyltransferases in Psoriasis
Protein arginine methyltransferases belonging to the seven-β-strand group possess histone- and non-histone-specific methyltransferase activity (Figure 1b). Among the histone-arginine methyltransferases (PRMTs), four members (CARM1 also known as PRMT4 and PRMT1/2/7) exhibited altered expression only in lesional skin (Figure 1d and Figure 3, Table 4 and Table S1), whereas PRMT5 displayed expression changes in both lesional and non-lesional skin (Figure 1d and Figure 3, Table 4 and Table S1). In addition, the METTL23 arginine methyltransferase, which shares only distant homology with PRMTs, was identified with expression changes in lesional skin (Figure 1d and Supplementary Table S1). Differentially expressed PRMTs, along with their target histones, modification types, and sites, are summarized in Table 4 and Figure 3.
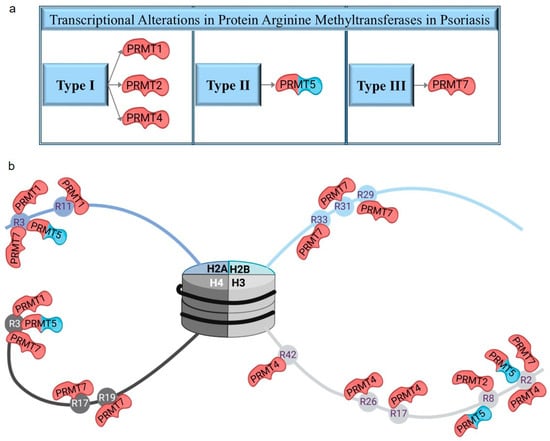
Figure 3.
Protein arginine methyltransferases with altered expression in psoriasis (a) and their target histones with arginine methylation sites (b). The red colors highlight differentially expressed transcripts in lesions, while the blue and red colors reflect differences in expression levels in both lesional and non-lesional skin areas compared to healthy controls.

Table 4.
The classification of PRMT histone methyltransferases and their targets on histones and the type of modifications [69]. Methyltransferases identified with differentially expressed transcripts in non-lesional (NL) and/or lesional (L) skin are highlighted in bold.
2.5. Diversity of Methyltransferase Transcript Variants and Encoded Isoforms in Psoriasis
Alternative splicing generates a diverse pool of transcript variants, including non-protein-coding transcripts and those that encode protein isoforms with altered or novel functionalities compared to the canonical form [70]. These alternative isoforms can exhibit distinct or even antagonistic biological roles relative to their canonical counterparts [71,72]. To investigate this phenomenon in psoriasis, we analyzed the composition of potential protein isoforms coded by differentially expressed transcripts (DETs) in both non-lesional and lesional skin samples, compared to healthy controls.
Within the EZ family, we identified a non-protein-coding transcript of EZH1 containing a retained intron, which was significantly decreased in lesional skin (Table 5). Conversely, four DETs of EZH2, with increased expression in lesional skin, encoded the canonical isoform (Q15910-1) along with three additional transcript variant-coded isoforms (Q15910-2-4) that retained all essential functional domains; however, differences from the canonical isoform affect their DNMT binding site, suggesting potential functional diversification (Table 5 and Table S2 and Figure 4A). In case of Q15910-4 isoform, a glycosylation and two phosphorylation post-translational modification sites are missing that may affect its localization or catalytic activity.

Table 5.
Differential expression of transcripts from the SET domain methyltransferase family observed in psoriasis, and the protein isoforms they encode.
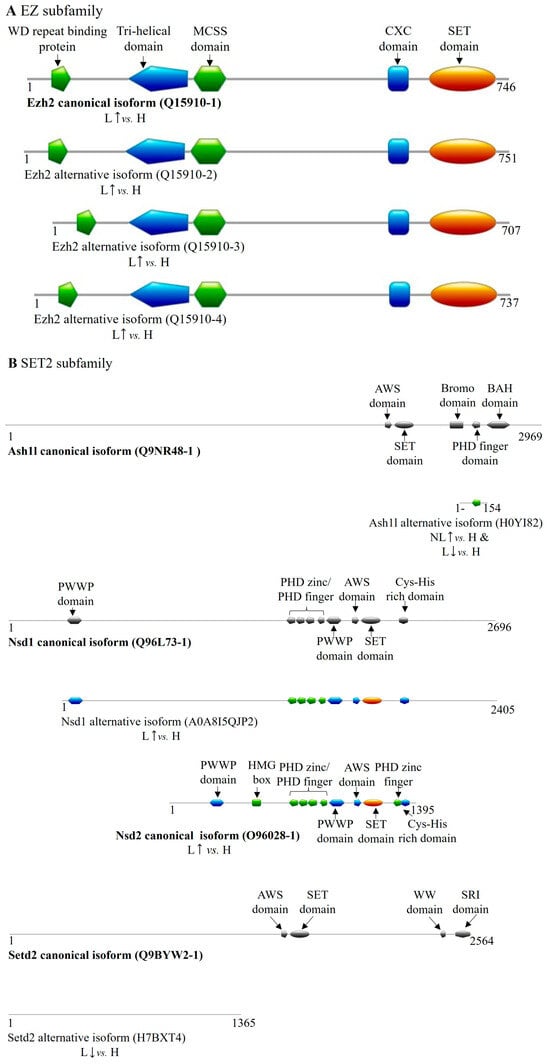
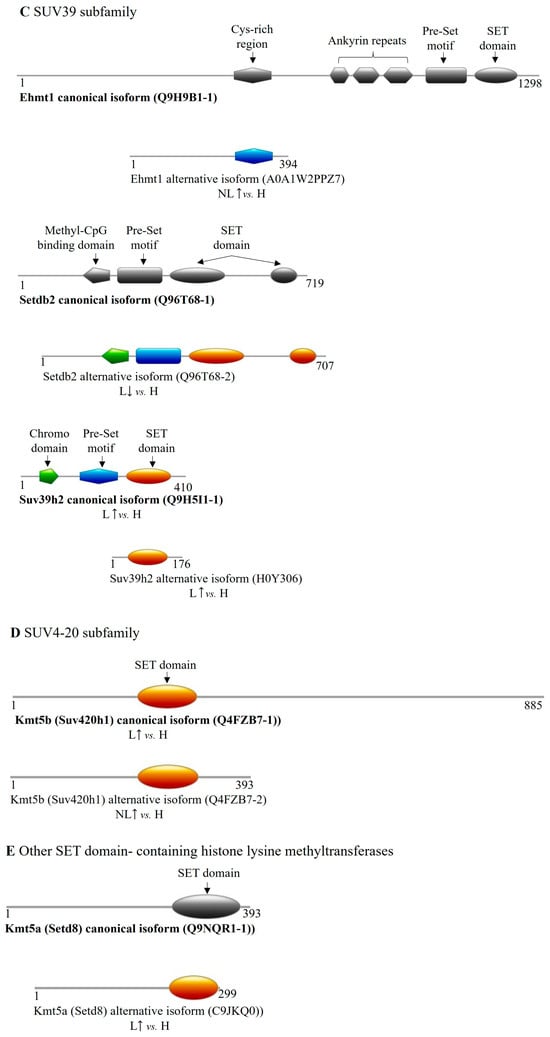
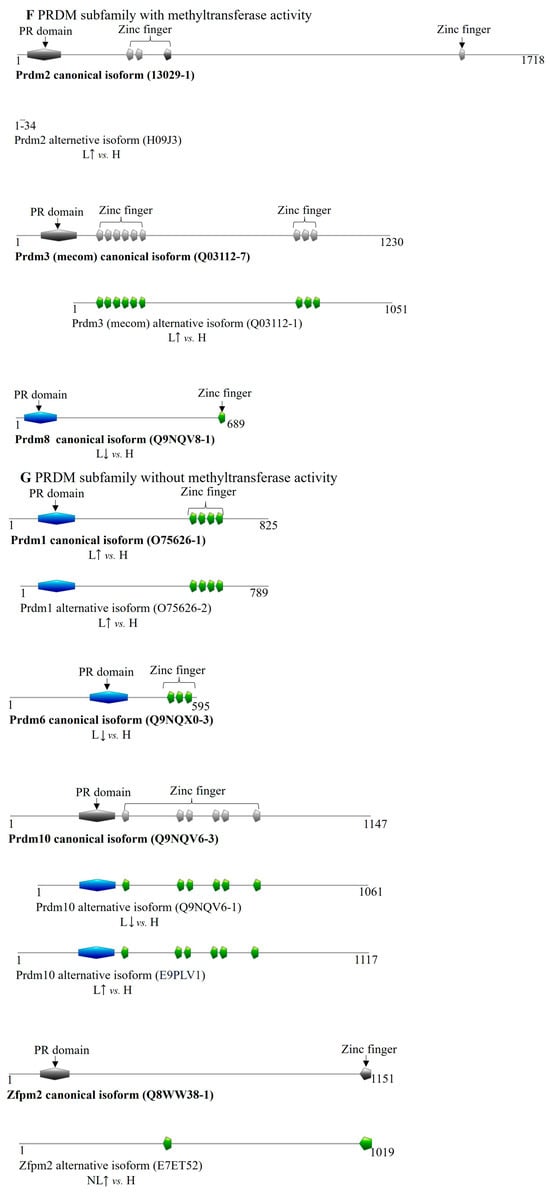
Figure 4.
Differentially expressed transcript variant-encoded protein isoforms of SET domain family methyltransferases in psoriasis. DET-encoded isoforms of (A) EZ, (B) SET2, (C) SUV39, (D) SUV4-20, (E) other SET domain-containing methyltransferases; the (F,G) PRDM (PR/SET domain) subfamily members are illustrated in color. Canonical isoforms, if expressed normally, are depicted in gray for isoform comparison. (H: healthy, NL: non-lesional, L: lesional, : increased expression of coding transcript, decreased expression of coding transcript).
In the SET2 family, we identified DETs of ASH1L, NSD1, NSD2, and SETD2 in our database. The same ASH1L transcript variant was upregulated in non-lesional skin and downregulated in lesional tissue, coding for a 154-amino-acid-long isoform (H0YI82) that partially overlaps with the Bromo domain and fully with the PHD finger domain of the canonical protein (Table 5 and Table S2, Figure 4B). The NSD1 DET observed in lesional skin encodes a truncated isoform (A0A8I5QJP2) that lacks the first 291 amino acids. For NSD2, increased expression was observed for both the canonical protein isoform (O96028-1)-coding transcript and for a non-protein-coding DET in lesional samples. Additionally, a non-protein-coding processed transcript of NSD3 was found in lesions with decreased expression (Table 5). The SETD2 DET found in lesions encodes an isoform (H7BXT4) that overlaps with the canonical protein sequence from amino acids 130 to 1487 but lacks any known functional domains, suggesting it may be an inactive and, potentially, a regulatory isoform (Figure 4B and Supplementary Table S2).
Analyzing the SUV39 family, we identified DETs of EHMT1, EHMT2, SETDB2, and SUV39H2. Specifically, EHMT1 transcripts showed increased expression of both a non-protein-coding and a protein-coding transcript in non-lesional skin. The latter variant encodes a truncated isoform (A0A1W2PPZ7) that lacks nearly all functional domains except for a Cys-rich region overlapping with the canonical Ehmt1 isoform (Table 5 and Table S2, Figure 4C). A non-protein-coding processed transcript of EHMT2 was found to be upregulated in non-lesional skin samples. A SETDB2-derived transcript exhibited decreased expression in lesional samples, coding for an 11-amino-acid shorter isoform (Q96T68-2) (Table 5 and Table S2, Figure 4C). SUV39H2 presented two isoform-coding transcripts with elevated expression in lesional samples, one encoding the canonical protein (Q9H5I1-1) and the other a shorter isoform (H0Y306) containing a SET domain (Table 5 and Table S2, Figure 4C).
Within the SUV4-20 family, the canonical SUV420H1 (KMT5B) isoform-coding transcript was upregulated in lesional skin, while a transcript encoding a C-terminal truncated isoform containing a SET domain (Q4FZB7-2) was increased in non-lesional tissue (Table 5 and Figure 4D).
Among other SET domain-containing histone methyltransferases, only a SETD8 (KMT5A) transcript variant showed elevated expression in lesional skin, encoding a shorter isoform (C9JKQ0) with a reduced-sized SET domain (Table 5 and Table S2, Figure 4E).
Regarding the PRDM family members with methyltransferase activity, we identified DETs of PRDM2, MECOM (PRDM3), and PRDM8 exclusively in lesional skin (Table 5). Notably, the PRDM2 transcript variant upregulated in lesions encodes a 44-amino-acid-sized micropeptide (H09J3) that overlaps the N-terminal region of the canonical isoform by 35 amino acids (10–44). In lesions, MECOM’s overexpressed transcript variant encodes an isoform (Q03112-1) that lacks the PR domain at the N-terminus (Table 5 and Table S2, Figure 4F). Conversely, transcripts coding for the canonical isoforms of PRDM8 (Q9NQV8-1) showed decreased expression in lesional tissue.
Among PRDM family members lacking known methyltransferase activity, two PRDM1 transcripts were increased in lesions, encoding the canonical isoform (O75626-1) and a shorter isoform (O75626-2) missing 36 amino acids from the N-terminus. The canonical isoform of PRDM6 (Q9NQX0-3) displayed reduced expression in lesional samples. Additionally, two transcripts of PRDM10, both coding for shorter isoforms that retain all functional domains, exhibited altered expression in lesions. In non-lesional skin, an increase of a ZFPM2 transcript was detected, coding for a shorter isoform of Zfpm2 (E7ET52) with a partial PR domain and an additional zinc finger domain not present in the canonical protein (Table 5 and Table S2, Figure 4G).
3. Discussion
Several studies have investigated epigenetic modifications and aberrant methylation patterns in psoriasis [73,74,75]. However, based on the available knowledge to date, a comprehensive investigation of the histone methyltransferases responsible for shaping histone methylation patterns has not yet been conducted, and only a limited amount of information on how they may regulate proliferation and the immune system dysfunction in psoriasis. Therefore, we performed a detailed analysis of a literature-based psoriasis transcriptome database of nearly 300 individuals to identify differential expression of histone methyltransferases. To provide a complete overview, we discuss the observed expressional alterations and their potential implications in psoriasis of each methyltransferase family (Table 6).

Table 6.
The function of differentially expressed SET domain-containing histone methyltransferases and their known or potential roles in the pathogenesis of psoriasis.
3.1. Histone Methyltransferase-Related Alterations in Psoriasis
3.1.1. SET Domain Methyltransferases
The SET domain MTase family is recognized to encompass all known lysine methyltransferases involved in the methylation of flexible histone tails [112,113]. Within the SET domain family, several subfamilies are distinguished by structural differences, including EZ, SET1, SET2, SMYD, SUV39, SUV420, and RIZ (PRDM). Some members are not classified into these subfamilies, such as SET7/9 and SET8 [113]. We refer to these as “other SET domain-containing histone methyltransferases” in our discussion.
EZ Subfamily of Methyltransferases
EZH1/2 in the EZ subfamily of methyltransferases is initially inactive [24,26,114] but activates within the PRC2 complex to methylate H3K27 [26], crucial for PRC2-mediated gene silencing to maintain stem cell functions [115,116]. Our analysis showed differential expression of EED, EZH1/2, and RBBP4 in the PRC2 complex, likely to affect stem cell self-renewal [117] and possibly contributing to keratinocyte hyperproliferation in psoriasis [118]. EED modulates T cell immune responses, impacting thymocyte maturation and CD4+ T cells [119,120]. EZH2 was previously shown to relate to keratinocyte proliferation and inflammatory responses in psoriasis [15,76], and it may also affect CD4+ and CD8+ T cell differentiation [121,122] and epidermal stratification [123,124], potentially contributing to psoriatic hyperkeratosis [125,126]. We found increased expression of the canonical and three functional EZH2 isoform-coding transcripts in lesional skin. Sequential differences at the DNMT binding sites of the three non-canonical isoforms may suggest potential functional diversification and may influence DNA methylation. Two DET-coded isoforms (Q15910-2 and Q15910-3) were previously characterized as EZH2α and β, which participate in similar biological processes, but form separate repressive complexes capable of cell-specific gene regulation [127].
PRC2 has two subcomplexes: PRC2.1 and PRC2.2 [128]. In our study, PRC2.1 subcomplex (EPOP, MTF2, PHF1, PHF19) showed transcriptional abnormalities. EPOP influences the chromatin environment and gene expression during the cell cycle G1 phase and aids in the induction of cell differentiation [129]. In addition, MTF2 and PHF19 promote, whereas PHF1 suppresses, cell proliferation and may impact keratinocyte proliferation [130].
SET1 Subfamily of Methyltransferases
The SET1 family influences euchromatin-like H3K4 methylation associated with transcriptional activation [131]. SET1 proteins, with low intrinsic activity, assemble into COMPASS and COMPASS-like complexes for enhanced catalytic function [25]. COMPASS di- and trimethylates H3K4 globally [132], while COMPASS-like complexes mono- and dimethylate development-specific genes [28]. These complexes include SET1A/B and four COMPASS-like multiprotein complexes: MLL1-4 [28,133]. Although we found no differences in catalytic subunit expressions, other components of the complex, including ASH2L, CXXC1, KDM6A, and MEN1, exhibited differential expression.
ASH2L regulates pluripotency and cellular reprogramming genes [134]. CXXC1 is crucial for thymocyte development [135], balancing Th1/Th2 [136] and Th17/Treg dynamics [137] relevant to psoriasis [90,111,138,139,140]. KDM6A, a member of the COMPASS-like complex, possesses demethylase activity that counteracts the PRC2 complex by demethylating H3K27me3 and facilitating H3K4me, thereby enhancing IFN responses and tumor-suppressive gene expression [141]. Additionally, KDM6A is vital for lineage-specific differentiation and hematopoietic balance [142,143,144] and contributes to age-related keratinocyte proliferation/differentiation imbalances [88,98,99] that may be important in the late-onset of the disease [145]. Its role in H3K27me3 demethylation affects T cell development [146] and migration [147] and may influence psoriasis pathology through IFN-γ-induced chemokines and T cell recruitment [148,149].
SET2 Subfamily of Methyltransferases
The SET2 subfamily, including ASH1L, NSD1-3, and SETD2, orchestrates H3K36 methylation [150], critical for transcriptional activation by SETD2 and H3K36me3 [151]. Our analysis revealed altered expression of ASH1L, NSD1-3, and SETD2. ASH1L maintains epidermal homeostasis, regulates keratinocyte proliferation and differentiation activity [77], and suppresses TLR-mediated inflammatory responses [78]. The ASH1L transcript variant was upregulated in non-lesional skin and downregulated in lesions. Although this ASH1L transcript encodes a non-functional isoform, its PHD finger domain may interfere with the recognition of histones and chromatin modifications, in a contrary manner in non-lesional and lesional skin. The NSD1 DET overexpressed in lesional skin encodes a truncated but functional isoform that may influence chemokine expression and immune cell infiltration via the NF-κB pathway [81,152,153]. Reduced expression of Wnt10b has been detected in psoriatic skin compared to healthy skin [83], and plays a pivotal role in cell proliferation and migration through the NSD1/H3/Wnt10b pathway [82]. In the case of NSD2, we detected the increased expression of the canonical protein isoform-coding transcript in lesions. NSD2 also modulates cell proliferation through the Wnt signaling pathway by regulating cyclin D1 transcription [84], known to be increased in psoriatic lesions [85]. A decreased expression of the SETD2 transcript codes for a non-functional isoform that may interfere with the interaction of the functional isoform in lesions. SETD2 deficiency was previously shown to trigger enhanced keratinocyte proliferation [87], and it influences Th17/Treg balance [86]. Therefore, dysregulated SETD2 may contribute to psoriasis symptoms and immune dysregulation [89,90,139].
SMYD Subfamily of Methyltransferases
The SMYD subfamily, comprising SET and MYND domain-containing proteins, plays a dual role, controlling both transcriptional activation and repression of genes [154]. Based on our analysis, none of the members (SMYD1-5) show significant alterations in either non-lesional or involved skin.
SUV39 Subfamily of Methyltransferases
The SUV39 subfamily deposits methyl groups onto histone H3 at lysine 9, forming H3K9me2 and H3K9me3 marks [155]. These marks are linked to transcriptional repression and heterochromatin formation [156,157] and are inherited following cell division [158].
We found altered expression of SUV39 subfamily members, including EHMT1, SETDB2, and SUV39H2, in our analysis. H3K9 methylation regulates IL-23 expression through the TNF/N-WASP/EHMT1-2 pathway [73].
An increased expression of EHMT1 transcript variant was observed in non-lesional skin, coding for a shorter isoform containing a Cys-rich region. Cys-rich regions of methyltransferases are known to play a role in maintaining their activity and specificity. Therefore, the shorter isoform may interfere with these properties of EHMT1. This might be relevant in psoriasis since EHMT1 negatively regulates gene induction pathways mediated by NF-κB and type I interferon [91], and is involved in Treg cell differentiation [92]. EHMT1 via CDKN1A modulation regulates the cell cycle [159].
SETDB2 is involved in proliferation-associated chromosome condensation and segregation [43] and inhibits inflammatory cytokine gene transcription via NF-κB [95]. Therefore, the decreased expression of a shorter but likely functional isoform-coding SETDB2 transcript found with reduced expression in lesions may influence these processes. Meanwhile, SUV39H2, found to be elevated in lesions, may modulate the suppression of key genes for epidermal differentiation [97]. Therefore, SUV39 subfamily-related alterations will likely impact immune responses and keratinocyte proliferation and differentiation in psoriasis [160].
SUV4-20 Subfamily Methyltransferases
The SUV4-20 subfamily members, SUV420H1 and -H2, serve as methyltransferases primarily responsible for the di- and trimethylation of histone H4K20 for heterochromatin formation and gene silencing [47,48]. In our RNA sequencing dataset, SUV420H1 showed increased expression of the canonical isoform-coding transcript in lesional samples. While in non-lesional skin samples, a shorter isoform-coding transcript expression is elevated, missing the C-terminal region following the SET domain implicated in protein–protein interactions. While the isoform differentially expressed in non-lesional skin increases H4K20me3 levels globally in the nucleus, the canonical isoform-mediated methylation is mainly restricted to pericentric regions [161]. These alterations may impact psoriasis since SUV4-20 members are crucial for DNA replication [100], developmental DNA rearrangements [101], and telomeric chromatin formation [102].
PRDM Subfamily of Methyltransferases
The PRDMs are part of the SET domain family of histone methyltransferases, comprising 19 distinct transcription factors [162,163]. Although classified as methyltransferases, only some members exhibit this activity, including PRDM2 [53,54,164], MECOM (PRDM3) [55], PRDM7 [56], PRDM8 [57], PRDM9 [58,59,60,61,62], and PRDM16 [55,63]. These proteins are critical in regulating cell proliferation, differentiation, and gene expression through various signaling pathways [165,166]. In lesional skin, altered expression levels of PRDM2, MECOM, and PRDM8 were observed. PRDM2 is vital for stem cell self-renewal and cellular quiescence [108], and it regulates T cell-specific transcription factor GATA3 activity [109], whose levels are reduced in psoriatic lesional skin compared to non-lesional samples. Tape-stripping non-lesional areas also decreases GATA3 expression, indicating its role in inflammation and epidermal regeneration [167]. Interestingly, the increased expression of a PRDM2 transcript variant coding a 44-amino-acid-sized micropeptide with unknown regulatory function was detected in lesions. MECOM’s altered expression in psoriasis was previously shown to correlate with excessive keratinocyte proliferation [105]. In addition, MECOM is essential for hematopoiesis [168], inhibiting monocyte differentiation into macrophages [106]. However, the transcript variant of MECOM with elevated expression in lesions codes for an inactive isoform where the PR domain is missing, based on our database. Such isoforms typically act as inhibitors in a competitive manner. Therefore, further studies are required to elucidate the precise function of MECOM. PRDM8 is a key player in inducing trained immunity in response to damage-associated molecular patterns, relevant to chronic inflammatory diseases [169]. We found decreased expression of the canonical PRDM8 isoform-coding transcript.
Other SET Domain-Containing Histone Lysine Methyltransferases: SETD7 and SETD8
SETD7 is expressed normally in non-lesional and lesional psoriatic skin, while SETD8 shows altered expression in lesions. In particular, the expression of a shorter isoform-coding transcript is elevated, containing a reduced-size SET domain with unknown activity. SETD8 specifically catalyzes H4K20me1 methylation [50], while SUV4-20H1/H2 (discussed above) converts H4K20me1 to H4K20me2/3, which is essential for pre-replication complex formation and cell cycle progression [170]. SETD8 also supports the survival and differentiation of epidermal stem cells [104], suggesting that its altered expression may contribute to psoriatic changes in cell proliferation and differentiation.
Further discussion on SET domain methyltransferases with no or uncertain histone methyltransferase activity is provided as Supplementary Information [171,172,173,174,175,176,177,178,179,180].
3.1.2. Seven-β-Strand (7BS) Methyltransferases
The seven-β-strand methyltransferases are primarily known as non-histone-specific methyltransferases; nevertheless, several members may also function as lysine or arginine histone methyltransferases [68,69,181,182]. Therefore, further discussion on seven-β-strand methyltransferases is provided as Supplementary Information [67,69,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206,207,208,209,210,211,212,213,214,215,216,217].
4. Materials and Methods
4.1. Guidelines for Establishing a Combined Psoriasis Transcriptome Sequencing Dataset Based on Literature Sources
Our comprehensive transcriptome sequencing database was assembled as described previously [9,23]. In brief, data from three psoriatic transcriptome studies [20,21,22] of randomly recruited patients with chronic plaque psoriasis and healthy individuals were merged. In these studies, RNA sequencing data were obtained from 6 mm skin punch biopsies collected from various regions of the body, without any age (>18) or gender criteria (non-lesional psoriatic: n = 27, lesional psoriatic: n = 99, and healthy individuals: n = 172). PASI constitutes at least 1% of the total body surface area. To ensure that our data accurately reflect the general alterations associated with chronic plaque psoriasis, defined washout periods were implemented before biopsy collection for both topical (1 week) and systemic (2 weeks) treatments.
4.2. Processing and Differential Expression Analysis of RNA Sequencing Data
Data processing was performed as described previously [9,23]. Differential expression analysis was performed on the previously published dataset [23]. In brief, the RNA sequencing data were sourced from the Sequence Read Archive under accession numbers SRP035988, SRP050971, and SRP055813 utilizing SRA-tools (version 2.9.2). All samples of the datasets were uniformly reprocessed to ensure consistent analysis. Transcript levels were assessed employing Kallisto [218] (version 0.43.0) and the GENCODE [219] v27 transcriptome annotation, with Kallisto (defined parameters: --bias --single -l 120 -s 20 -b 100). Subsequently, transcript-level length-scaled TPM (Transcripts Per Million) expression estimates computed by Kallisto were transferred into the R statistical environment (version 3.4.3.) using the tximport [220] package (version 1.6.0). Following TMM normalization (using edgeR [221] v3.20.9) and voom transformation (limma [222,223] v3.34.9), the voomWithQualityWeights() function was employed, integrating sample-specific weights with transcript-level weights to accommodate lower-quality samples while mitigating their influence. Expression differences across sample groups were assessed using Limma. A linear model was applied via the lmFit function, and moderated t-statistics were computed using eBayes. Transcripts with an FDR-corrected [222,224] p-value of <0.05 were considered as differentially expressed.
4.3. Screening for Histone Methylation-Related DETs in Psoriasis
Datasets downloaded from https://amigo.geneontology.org/amigo (accessed on 24 April 2024) were employed to analyze differentially expressed transcripts (DETs) from the non-lesional/uninvolved (NL) vs. healthy (H) and lesional (L) vs. healthy (H) comparisons. The downloaded methyltransferase dataset (GO:0042054 and Supplementary Table S3) was augmented and verified with relevant information extracted from the literature that also includes methyltransferase-specific complexes. The literature references used are listed in Table 1. Detailed information about the dataset used for the screening is presented in Figure 1a–c; where data from both the GO database and the literature [26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,67,69,225,226,227] are presented. Intersection analysis was used to filter and identify matches between non-lesional and healthy, as well as lesional and healthy samples, and the downloaded methyltransferase datasets in Python (Python 3.13.0).
4.4. Analysis of Protein Isoforms Derived from Differentially Expressed Transcripts
Differentially expressed transcripts in our psoriasis database were matched with Transcript IDs from the Ensembl database (https://www.ensembl.org/ accessed on 20 May 2025). Protein-coding transcripts were assigned their corresponding UniProt identifiers. In our analysis, the “canonical” protein isoform listed in UniProt served as the reference for comparing alternative isoforms. In our comparative analysis of DET-encoded protein isoforms, we considered amino acid deletions and sequence variations documented in the UniProt database for all alternative isoforms where these differences were explicitly noted. For isoforms lacking detailed sequence information in UniProt, we performed sequence comparisons against the canonical sequence using the Protein BLAST tool (https://blast.ncbi.nlm.nih.gov accessed on 21 May 2025), with particular focus on identifying gaps and mismatches.
Using the protein-coding differentially expressed transcripts (DETs), the domains of various isoforms were identified based on UniProt protein sequences, utilizing the Pfam databases of InterPro (https://www.ebi.ac.uk/interpro/ accessed on 22 May 2025). To ensure accurate proportional representation in the figures, the Prosite MyDomains Image Creator tool (https://prosite.expasy.org/ accessed on 24 May 2025) was employed for visualization.
5. Conclusions
In summary, various subfamilies of histone methyltransferases, including the EZ, SET1, SET2, SMYD, SUV39, SUV4-20, and PRDM subfamilies, play crucial roles in regulating gene expression, cell proliferation, and differentiation. Dysregulation of key proteins, such as SETD8 and EZH2, may contribute to hyperproliferation of keratinocytes and inflammatory responses related to the disease. Abnormal expressions of proteins like MECOM and PRDM2 further indicate their significance in immune modulation and stem cell functions, likely to influence the pathogenesis of psoriasis. Our analysis not only confirmed previously reported expressional alteration of EHMT1/2 in non-lesional skin but also revealed abnormal transcription of two SET domain family members and a β7 histone MTase in non-lesional skin. Transcriptional changes of these MTases highlight their potential involvement in early dysregulation of keratinocyte and T cell proliferation and differentiation. Additionally, the interactions of these methyltransferases with other signaling pathways highlight their potential as therapeutic targets for managing psoriasis symptoms. It is important to note that since our study is based on mRNA expression data analysis, further research is required to determine how these transcriptional changes manifest at the protein level and whether they affect enzymatic activity, function, and downstream cellular processes. However, if translated, the differentially expressed transcripts identified in this study may give rise to histone methyltransferase isoforms that influence the modification of key histone lysine residues, including H3K27 (EZH2, EZH1), H3K36 (ASH1L, NSD1, NSD2, SETD2), H3K9 (EHMT1, EHMT2, SUV39H2, SETDB2), and H4K20 (SUV420H1, SETD8). Understanding these complex regulatory mechanisms will be essential for developing new strategies to treat and combat psoriasis effectively.
Supplementary Materials
The following Supporting Information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms26136329/s1.
Author Contributions
Conceptualization, G.G.; methodology, G.G. and D.R.; software, G.G. and D.R.; validation, G.G. and D.R.; formal analysis, G.G. and D.R.; investigation, G.G. and D.R.; resources, G.G. and D.R.; data curation, G.G. and D.R.; writing—original draft preparation, D.R., Á.B. and G.G.; writing—review and editing, D.R., Á.B., K.S., L.K., R.G. and G.G.; visualization, D.R. and Á.B.; supervision, G.G.; project administration, G.G. and D.R.; funding acquisition, G.G. and D.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Research, Development, and Innovation Office (NKFIH, OTKA K143576 research grant). This research also received funding from the EU’s Horizon 2020 research and innovation program under grant agreement No. 739593. This project also received funding from the HUN-REN Hungarian Research Network, and the National Research Development and Innovation Fund (project no. TKP2021-EGA-28), and was also supported by the Géza Hetényi Research Grant of the Albert Szent-Györgyi Medical School, University of Szeged.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Only publicly available data were used in the study (Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accessed on 15 November 2021); study ID: SRP035988, SRP050971, and SRP055813).
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| 7BS | seven-β-strand |
| AEBP2 | AE (Adipocyte Enhancer)-Binding Protein 2 |
| ANTKMT | Adenine Nucleotide Translocase Lysine Methyltransferase |
| ASH1L | ASH1 Like Histone Lysine Methyltransferase |
| ASH2L | ASH2 Like Histone Lysine Methyltransferase Complex Subunit |
| ATPSCKMT | ATP Synthase C Subunit Lysine N-Methyltransferase |
| CAMKMT | Calmodulin-Lysine N-Methyltransferase |
| CD4+ T cells | Cluster of Differentiation 4 Positive T cells |
| CD8+ T cells | Cluster of Differentiation 8 Positive T cells |
| CDK2 | Cyclin Dependent Kinase 2 |
| CDKN1A | Cyclin Dependent Kinase Inhibitor 1A |
| CSKMT | Citrate Synthase Lysine Methyltransferase |
| COMPASS | Complex Proteins Associated with SET1 |
| c-MYC | MYC Proto-Oncogene, BHLH transcription Factor |
| CXCL10 | C-X-C Motif Chemokine Ligand 10 |
| CXXC1 | CXXC Finger Protein 1 |
| DET | Differentially Expressed Transcript |
| DNA | Deoxyribonucleic acid |
| DOT1L | DOT1 Like Histone Lysine Methyltransferase |
| DPY30 | Dpy-30 Histone Methyltransferase Complex Regulatory Subunit |
| E2F | E2F Transcription Factor |
| EED | Embryonic Ectoderm Development |
| EEF1AKMT1 | EEF1A Lysine Methyltransferase 1 |
| EEF1AKMT2 | EEF1A Lysine Methyltransferase 2 |
| EEF1AKMT3 | EEF1A Lysine Methyltransferase 3 |
| EEF1AKMT4 | EEF1A Lysine Methyltransferase 4 |
| EEF2KMT | Eukaryotic Elongation Factor 2 Lysine Methyltransferase |
| EHMT1 | Euchromatic Histone Lysine Methyltransferase 1 |
| EHMT2 | Euchromatic Histone Lysine Methyltransferase 2 |
| EPOP | Elongin BC And Polycomb Repressive Complex 2 Associated Protein |
| ETFBKMT | Electron Transfer Flavoprotein Beta Subunit Kinase Methyltransferase |
| EZH1 | Enhancer of Zeste 1 Polycomb Repressive Complex 2 Subunit |
| EZH2 | Enhancer of Zeste 2 Polycomb Repressive Complex 2 Subunit |
| GATA3 | GATA Binding Protein 3 |
| GO | Gene Ontology |
| H2AR11me2 | Histone 2A Arginine 11 Dimethylation |
| H2AR29me2 | Histone 2A Arginine 29 Dimethylation |
| H2AR3me1 | Histone 2A Arginine 3 Monomethylation |
| H2AR3me2 | Histone 2A Arginine 3 Dimethylation |
| H2ARme1/2 | Histone 2A Arginine Monomethylation/Dimethylation |
| H2AZ | Histone variantH2A.Z |
| H2AZK7 | Histone variantH2A.Z Lysine 7 Methylation |
| H2B | Histone 2B |
| H2BR29me1 | Histone 2B Arginine 29 Monomethylation |
| H2BR31me1 | Histone 2B Arginine 31 Monomethylation |
| H2BR33me1 | Histone 2B Arginine 33 Monomethylation |
| H3 | Histone 3 |
| H3K18me1 | Histone 3 Lysine 18 Monomethylation |
| H3K27 | Histone 3 Lysine 27 |
| H3K27me1/2/3 | Histone 3 Lysine 27 Mono-/Di-/Trimethylation |
| H3K27me3 | Histone 3 Lysine 27 Trimethylation |
| H3K36 | Histone 3 Lysine 36 |
| H3K36me1/2 | Histone 3 Lysine 36 Mono-/Dimethylation |
| H3K36me2 | Histone 3 Lysine 36 Dimethylation |
| H3K36me3 | Histone 3 Lysine 36 Trimethylation |
| H3K4 | Histone 3 Lysine 4 |
| H3K4me1 | Histone 3 Lysine 4 Monomethylation |
| H3K4me1/2/3 | Histone 3 Lysine 4 Mono-/Di-/Trimethylation |
| H3K4me2/3 | Histone 3 Lysine 4 Di-/Trimethylation |
| H3K4me3 | Histone 3 Lysine 4 Trimethylation |
| H3K9ac | Histone 3 Lysine 9 Acetylation |
| H3K9 | Histone 3 Lysine 9 |
| H3K9me1 | Histone 3 Lysine 9 Monomethylation |
| H3K9me1/2 | Histone 3 Lysine 9 Mono-/Dimethylation |
| H3K9me1/2/3 | Histone 3 Lysine 9 Mono-/Di-/Trimethylation |
| H3K9me1/3 | Histone 3 Lysine 9 Mono-/Trimethylation |
| H3K9me2 | Histone 3 Lysine 9 Dimethylation |
| H3K9me2/3 | Histone 3 Lysine 9 Dimethylation/Trimethylation |
| H3K9me3 | Histone 3 Lysine 9 Trimethylation |
| H3K9ac | Histone 3 Lysine 9 Acetylation |
| H3R17me2 | Histone 3 Arginine 17 Dimethylation |
| H3R26me2 | Histone 3 Arginine 26 Dimethylation |
| H3R2me1/2 | Histone 3 Arginine 2 Monomethylation/Dimethylation |
| H3R2me1/2 | Histone 3 Arginine 2 Monomethylation/Dimethylation |
| H3R2me2 | Histone 3 Arginine 2 Dimethylation |
| H3R42me2 | Histone 3 Arginine 42 Dimethylation |
| H3R8me2 | Histone 3 Arginine 8 Dimethylation |
| H4K20 | Histone 4 Lysine 20 |
| H4K20me1 | Histone 4 Lysine 20 Monomethylation |
| H4K20me1/2 | Histone 4 Lysine 20 Monomethylation/Dimethylation |
| H4K20me2/3 | Histone 4 Lysine 20 Dimethylation/Trimethylation |
| H4K20me3 | Histone 4 Lysine 20 Trimethylation |
| H4K5 | Histone 4 Lysine 5 Methylation |
| H4R17me1 | Histone 4 Arginine 17 Monomethylation |
| H4R19me1 | Histone 4 Arginine 19 Monomethylation |
| H4R3me1 | Histone 4 Arginine 3 Monomethylation |
| H4R3me2 | Histone 4 Arginine 3 Dimethylation |
| H4 | Histone 4 |
| HCFC1 | Host Cell Factor C1 |
| H | Healthy |
| IFN-γ | Interferon Gamma |
| IL-17A | Interleukin 17A |
| IL-23 | Interleukin 23 |
| IRF3 | Interferon Regulatory Factor 3 |
| JARID2 | Jumonji And AT-Rich Interaction Domain Containing 2 |
| K6 | K6 Keratin |
| K16 | K16 Keratin |
| KDM6A | Lysine-specific Demethylase 6A |
| KMT2A | Lysine Methyltransferase 2A |
| KMT2B | Lysine Methyltransferase 2B |
| KMT2C | Lysine Methyltransferase 2C |
| KMT2D | Lysine Methyltransferase 2D |
| L | Lesional |
| LCOR | Ligand-Dependent Nuclear Receptor Corepressor |
| LCORL | Ligand-Dependent Nuclear Receptor Corepressor Like |
| MAPK | Mitogen-activated protein kinase |
| MECOM | MDS1 and EVI1 Complex Locus |
| MEN1 | Menin 1 |
| MEP50 | WD Repeat Domain 77 |
| METTL13 | Methyltransferase 13, EEF1A N-Terminus And K55 |
| METTL21A | Methyltransferase 21A, HSPA Lysine |
| METTL21C | Methyltransferase 21C, AARS1 Lysine |
| METTL22 | Methyltransferase 22, Kin17 Lysine |
| MHC-II | Major Histocompatibility Complex Class II |
| MLL1 | Lysine Methyltransferase 2A |
| MLL2 | Lysine Methyltransferase 2B |
| MLL3 | Lysine Methyltransferase 2C |
| MLL4 | Lysine Methyltransferase 2D |
| mRNA | Messenger Ribonucleic Acid |
| Mtase | Methyltransferases |
| MTF2 | Metal Response Element Binding Transcription Factor 2 |
| N6AMT1 | N-6 Adenine-Specific DNA Methyltransferase 1 |
| NCOA6 | Nuclear Receptor Coactivator 6 |
| NF-κB | Nuclear Factor Kappa B |
| NL | Non-leional/uninvolved |
| NSD1 | Nuclear Receptor Binding SET Domain Protein 1 |
| NSD2 | Nuclear Receptor Binding SET Domain Protein 2 |
| NSD3 | Nuclear Receptor Binding SET Domain Protein 3 |
| N-WASP | Neural Wiskott-Aldrich Syndrome Protein (WASP Like Actin Nucleation Promoting Factor) |
| PAGR1 | PAXIP1 Associated Glutamate Rich Protein 1 |
| PASI | Psoriasis Area and Severity Index |
| PAXIP1 | PAX Interacting Protein 1 |
| PHF1 | PHD Finger Protein 1 |
| PHF19 | PHD Finger Protein 19 |
| PRC2 | Polycomb Repressive Complex 2 |
| PRDM1 | PR/SET Domain 1 |
| PRDM10 | PR/SET Domain 10 |
| PRDM11 | PR/SET Domain 11 |
| PRDM12 | PR/SET Domain 12 |
| PRDM13 | PR/SET Domain 13 |
| PRDM14 | PR/SET Domain 14 |
| PRDM15 | PR/SET Domain 15 |
| PRDM16 | PR/SET Domain 16 |
| PRDM2 | PR/SET Domain 2 |
| PRDM4 | PR/SET Domain 4 |
| PRDM5 | PR/SET Domain 5 |
| PRDM6 | PR/SET Domain 6 |
| PRDM7 | PR/SET Domain 7 |
| PRDM8 | PR/SET Domain 8 |
| PRDM9 | PR/SET Domain 9 |
| PRMT1 | Protein Arginine Methyltransferase 1 |
| PRMT2 | Protein Arginine Methyltransferase 2 |
| PRMT3 | Protein Arginine Methyltransferase 3 |
| PRMT4 | Protein Arginine Methyltransferase 4 |
| PRMT5 | Protein Arginine Methyltransferase 5 |
| PRMT6 | Protein Arginine Methyltransferase 6 |
| PRMT7 | Protein Arginine Methyltransferase 7 |
| PRMT8 | Protein Arginine Methyltransferase 8 |
| PRMT9 | Protein Arginine Methyltransferase 9 |
| RBBP4 | Retinoblastoma Binding Protein 4, Chromatin Remodeling Factor |
| RBBP5 | Retinoblastoma Binding Protein 5, Histone Lysine Methyltransferase Complex Subunit |
| RBBP7 | Retinoblastoma Binding Protein 7, Chromatin Remodeling Factor |
| RelA | RELA Proto-Oncogene, NF-KB Subunit |
| RNA | Ribonucleic Acid |
| SET-domain | Suppressor of variegation 3–9, Enhancer of zeste, and Trithorax |
| SETD3 | SET Domain Containing 3, Actin N3(Tau)-Histidine Methyltransferase |
| SETD4 | SET Domain Containing 4 |
| SETD5 | SET Domain Containing 5 |
| SETD6 | SET Domain Containing 6, Protein Lysine Methyltransferase |
| SETD1A | SET Domain Containing 1A, Histone Lysine Methyltransferase |
| SETD1B | SET Domain Containing 1B, Histone Lysine Methyltransferase |
| SETD2 | SET Domain Containing 2, Histone Lysine Methyltransferase |
| SETD7 | SET Domain Containing Lysine Methyltransferase 7 |
| SETD8 | Lysine Methyltransferase 5A (SET Domain Containing Lysine Methyltransferase 8) |
| SETDB1 | SET Domain Bifurcated Histone Lysine Methyltransferase 1 |
| SETDB2 | SET Domain Bifurcated Histone Lysine Methyltransferase 2 |
| SMYD1 | SET and MYND Domain Containing 1 |
| SMYD2 | SET and MYND Domain Containing 2 |
| SMYD3 | SET and MYND Domain Containing 3 |
| SMYD4 | SET and MYND Domain Containing 4 |
| SMYD5 | SMYD Family Member 5 |
| SRA | Sequence Read Archive |
| SUV39H1 | SUV39H1 Histone Lysine Methyltransferase (Suppressor of Variegation 3-9 Homolog 1) |
| SUV39H2 | SUV39H2 Histone Lysine Methyltransferase (Suppressor of Variegation 3-9 Homolog 2) |
| SUV420H1 | Lysine Methyltransferase 5B (Suppressor of Variegation 4-20 Homolog 1) |
| SUV420H2 | Lysine Methyltransferase 5C (Suppressor of Variegation 4-20 Homolog 2) |
| SUZ12 | SUZ12 Polycomb Repressive Complex 2 Subunit |
| Th1 | T Helper Type 1 |
| Th17 | T Helper Type 17 |
| Th2 | T Helper Type 2 |
| TLR | Toll-like Receptor |
| TLR4 | Toll-like Receptor 4 |
| TMM | trimmed mean of M-values |
| TNF | Tumor Necrosis Factor |
| TNFα | Tumor Necrosis Factor Alpha |
| TPM | Transcripts Per Million |
| Treg | Regulatory T cell |
| VCPKMT | Valosin Containing Protein Lysine Methyltransferase |
| WDR5 | WD Repeat Domain 5 |
| WDR82 | WD Repeat Domain 82 |
| WNT10B | Wnt Family Member 10B (Wingless-type MMTV Integration Site Family, Member 10B) |
| ZFPM1 | Zinc Finger Protein, FOG Family Member 1 |
| ZFPM2 | Zinc Finger Protein, FOG Family Member 2 |
| ZNF408 | Zinc Finger Protein 408 |
References
- Sewerin, P.; Brinks, R.; Schneider, M.; Haase, I.; Vordenbäumen, S. Prevalence and incidence of psoriasis and psoriatic arthritis. Ann. Rheum. Dis. 2019, 78, 286–287. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, S.; Dogra, S.; Sharma, K.; Raychaudhuri, S.K.; Raychaudhuri, S.P. Recent Update on Immunopathogenesis of Psoriasis. Indian J. Dermatol. 2022, 67, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Kelemen, E.; Bozó, R.; Groma, G.; Bata-Csörgő, Z.; Kemény, L.; Danis, J.; Széll, M. The Psoriatic Nonlesional Skin: A Battlefield between Susceptibility and Protective Factors. J. Investig. Dermatol. 2021, 141, 2785–2790. [Google Scholar] [CrossRef]
- Szlavicz, E.; Szabo, K.; Groma, G.; Bata-Csorgo, Z.; Pagani, F.; Kemeny, L.; Szell, M. Splicing factors differentially expressed in psoriasis alter mRNA maturation of disease-associated EDA+ fibronectin. Mol. Cell. Biochem. 2017, 436, 189–199. [Google Scholar] [CrossRef]
- Groma, G.; Grskovic, I.; Schael, S.; Ehlen, H.W.A.; Wagener, R.; Fosang, A.; Aszodi, A.; Paulsson, M.; Brachvogel, B.; Zaucke, F. Matrilin-4 is processed by ADAMTS-5 in late Golgi vesicles present in growth plate chondrocytes of defined differentiation state. Matrix Biol. J. Int. Soc. Matrix Biol. 2011, 30, 275–280. [Google Scholar] [CrossRef]
- Fleischmajer, R.; Kuroda, K.; Hazan, R.; Gordon, R.E.; Lebwohl, M.G.; Sapadin, A.N.; Unda, F.; Iehara, N.; Yamada, Y. Basement membrane alterations in psoriasis are accompanied by epidermal overexpression of MMP-2 and its inhibitor TIMP-2. J. Investig. Dermatol. 2000, 115, 771–777. [Google Scholar] [CrossRef]
- Gubán, B.; Vas, K.; Balog, Z.; Manczinger, M.; Bebes, A.; Groma, G.; Széll, M.; Kemény, L.; Bata-Csörgő, Z. Abnormal regulation of fibronectin production by fibroblasts in psoriasis. Br. J. Dermatol. 2016, 174, 533–541. [Google Scholar] [CrossRef]
- Bozó, R.; Szél, E.; Danis, J.; Gubán, B.; Bata-Csörgő, Z.; Szabó, K.; Kemény, L.; Groma, G. Cartilage Oligomeric Matrix Protein Negatively Influences Keratinocyte Proliferation via α5β1-Integrin: Potential Relevance of Altered Cartilage Oligomeric Matrix Protein Expression in Psoriasis. J. Investig. Dermatol. 2020, 140, 1733–1742.e7. [Google Scholar] [CrossRef]
- Romhányi, D.; Szabó, K.; Kemény, L.; Groma, G. Histone and Histone Acetylation-Related Alterations of Gene Expression in Uninvolved Psoriatic Skin and Their Effects on Cell Proliferation, Differentiation, and Immune Responses. Int. J. Mol. Sci. 2023, 24, 14551. [Google Scholar] [CrossRef] [PubMed]
- Hublitz, P.; Albert, M.; Peters, A.H.F.M. Mechanisms of transcriptional repression by histone lysine methylation. Int. J. Dev. Biol. 2009, 53, 335–354. [Google Scholar] [CrossRef]
- McManus, K.J.; Biron, V.L.; Heit, R.; Underhill, D.A.; Hendzel, M.J. Dynamic Changes in Histone H3 Lysine 9 Methylations: Identification of a Mitosis-Specific Function for Dynamic Methylation in Chromosome Congression and Segregation. J. Biol. Chem. 2006, 281, 8888–8897. [Google Scholar] [CrossRef] [PubMed]
- Evertts, A.G.; Manning, A.L.; Wang, X.; Dyson, N.J.; Garcia, B.A.; Coller, H.A. H4K20 methylation regulates quiescence and chromatin compaction. Mol. Biol. Cell 2013, 24, 3025–3037. [Google Scholar] [CrossRef] [PubMed]
- Shue, Y.T.; Lee, K.T.; Walters, B.W.; Ong, H.B.; Silvaraju, S.; Lam, W.J.; Lim, C.Y. Dynamic shifts in chromatin states differentially mark the proliferative basal cells and terminally differentiated cells of the developing epidermis. Epigenetics 2020, 15, 932–948. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Wang, Y.; Huang, B.; Chen, X.; Jiang, R.; Yin, M. Depletion of G9A attenuates imiquimod-induced psoriatic dermatitis via targeting EDAR-NF-κB signaling in keratinocyte. Cell Death Dis. 2023, 14, 627. [Google Scholar] [CrossRef]
- Zhang, T.; Yang, L.; Ke, Y.; Lei, J.; Shen, S.; Shao, S.; Zhang, C.; Zhu, Z.; Dang, E.; Wang, G. EZH2-dependent epigenetic modulation of histone H3 lysine-27 contributes to psoriasis by promoting keratinocyte proliferation. Cell Death Dis. 2020, 11, 826. [Google Scholar] [CrossRef]
- Zhang, P.; Su, Y.; Li, S.; Chen, H.; Wu, R.; Wu, H. The roles of T cells in psoriasis. Front. Immunol. 2023, 14, 1081256. [Google Scholar] [CrossRef]
- Xiao, C.; Fan, T.; Zheng, Y.; Tian, H.; Deng, Z.; Liu, J.; Li, C.; He, J. H3K4 trimethylation regulates cancer immunity: A promising therapeutic target in combination with immunotherapy. J. Immunother. Cancer 2023, 11, e005693. [Google Scholar] [CrossRef]
- Cribbs, A.; Hookway, E.S.; Wells, G.; Lindow, M.; Obad, S.; Oerum, H.; Prinjha, R.K.; Athanasou, N.; Sowman, A.; Philpott, M.; et al. Inhibition of histone H3K27 demethylases selectively modulates inflammatory phenotypes of natural killer cells. J. Biol. Chem. 2018, 293, 2422–2437. [Google Scholar] [CrossRef]
- Rondeaux, J.; Groussard, D.; Renet, S.; Tardif, V.; Dumesnil, A.; Chu, A.; Di Maria, L.; Lemarcis, T.; Valet, M.; Henry, J.-P.; et al. Ezh2 emerges as an epigenetic checkpoint regulator during monocyte differentiation limiting cardiac dysfunction post-MI. Nat. Commun. 2023, 14, 4461. [Google Scholar] [CrossRef]
- Li, B.; Tsoi, L.C.; Swindell, W.R.; Gudjonsson, J.E.; Tejasvi, T.; Johnston, A.; Ding, J.; Stuart, P.E.; Xing, X.; Kochkodan, J.J.; et al. Transcriptome Analysis of Psoriasis in a Large Case–Control Sample: RNA-Seq Provides Insights into Disease Mechanisms. J. Investig. Dermatol. 2014, 134, 1828–1838. [Google Scholar] [CrossRef]
- Tsoi, L.C.; Iyer, M.K.; Stuart, P.E.; Swindell, W.R.; Gudjonsson, J.E.; Tejasvi, T.; Sarkar, M.K.; Li, B.; Ding, J.; Voorhees, J.J.; et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015, 16, 24. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Tsoi, L.C.; Xing, X.; Beamer, M.A.; Swindell, W.R.; Sarkar, M.K.; Berthier, C.C.; Stuart, P.E.; Harms, P.W.; Nair, R.P.; et al. A gene network regulated by the transcription factor VGLL3 as a promoter of sex-biased autoimmune diseases. Nat. Immunol. 2017, 18, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Romhányi, D.; Szabó, K.; Kemény, L.; Sebestyén, E.; Groma, G. Transcriptional Analysis-Based Alterations Affecting Neuritogenesis of the Peripheral Nervous System in Psoriasis. Life 2022, 12, 111. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Zhang, Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 2004, 15, 57–67. [Google Scholar] [CrossRef]
- Takahashi, Y.; Westfield, G.H.; Oleskie, A.N.; Trievel, R.C.; Shilatifard, A.; Skiniotis, G. Structural analysis of the core COMPASS family of histone H3K4 methylases from yeast to human. Proc. Natl. Acad. Sci. USA 2011, 108, 20526–20531. [Google Scholar] [CrossRef]
- Margueron, R.; Li, G.; Sarma, K.; Blais, A.; Zavadil, J.; Woodcock, C.L.; Dynlacht, B.D.; Reinberg, D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 2008, 32, 503–518. [Google Scholar] [CrossRef]
- Yu, J.-R.; Lee, C.-H.; Oksuz, O.; Stafford, J.M.; Reinberg, D. PRC2 is high maintenance. Genes Dev. 2019, 33, 903–935. [Google Scholar] [CrossRef]
- Ford, D.J.; Dingwall, A.K. The cancer COMPASS: Navigating the functions of MLL complexes in cancer. Cancer Genet. 2015, 208, 178–191. [Google Scholar] [CrossRef]
- Tanaka, Y.; Katagiri, Z.; Kawahashi, K.; Kioussis, D.; Kitajima, S. Trithorax-group protein ASH1 methylates histone H3 lysine 36. Gene 2007, 397, 161–168. [Google Scholar] [CrossRef]
- Li, Y.; Trojer, P.; Xu, C.-F.; Cheung, P.; Kuo, A.; Drury, W.J.; Qiao, Q.; Neubert, T.A.; Xu, R.-M.; Gozani, O.; et al. The Target of the NSD Family of Histone Lysine Methyltransferases Depends on the Nature of the Substrate. J. Biol. Chem. 2009, 284, 34283–34295. [Google Scholar] [CrossRef]
- Kuo, A.J.; Cheung, P.; Chen, K.; Zee, B.M.; Kioi, M.; Lauring, J.; Xi, Y.; Park, B.H.; Shi, X.; Garcia, B.A.; et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol. Cell 2011, 44, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Edmunds, J.W.; Mahadevan, L.C.; Clayton, A.L. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008, 27, 406–420. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.A.; Sims, R.J.; Gottlieb, P.D.; Tucker, P.W. Identification and characterization of Smyd2: A split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol. Cancer 2006, 5, 26. [Google Scholar] [CrossRef]
- Van Aller, G.S.; Reynoird, N.; Barbash, O.; Huddleston, M.; Liu, S.; Zmoos, A.-F.; McDevitt, P.; Sinnamon, R.; Le, B.; Mas, G.; et al. Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics 2012, 7, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Foreman, K.W.; Brown, M.; Park, F.; Emtage, S.; Harriss, J.; Das, C.; Zhu, L.; Crew, A.; Arnold, L.; Shaaban, S.; et al. Structural and Functional Profiling of the Human Histone Methyltransferase SMYD3. PLoS ONE 2011, 6, e22290. [Google Scholar] [CrossRef]
- Stender, J.D.; Pascual, G.; Liu, W.; Kaikkonen, M.U.; Do, K.; Spann, N.J.; Boutros, M.; Perrimon, N.; Rosenfeld, M.G.; Glass, C.K. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol. Cell 2012, 48, 28–38. [Google Scholar] [CrossRef]
- Aljazi, M.B.; Gao, Y.; Wu, Y.; He, J. SMYD5 is a histone H3-specific methyltransferase mediating mono-methylation of histone H3 lysine 36 and 37. Biochem. Biophys. Res. Commun. 2022, 599, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, M.; Ueda, J.; Fukuda, M.; Takeda, N.; Ohta, T.; Iwanari, H.; Sakihama, T.; Kodama, T.; Hamakubo, T.; Shinkai, Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005, 19, 815–826. [Google Scholar] [CrossRef]
- Tachibana, M.; Sugimoto, K.; Nozaki, M.; Ueda, J.; Ohta, T.; Ohki, M.; Fukuda, M.; Takeda, N.; Niida, H.; Kato, H.; et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002, 16, 1779–1791. [Google Scholar] [CrossRef]
- Schultz, D.C.; Ayyanathan, K.; Negorev, D.; Maul, G.G.; Rauscher, F.J. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002, 16, 919–932. [Google Scholar] [CrossRef]
- Wang, H.; An, W.; Cao, R.; Xia, L.; Erdjument-Bromage, H.; Chatton, B.; Tempst, P.; Roeder, R.G.; Zhang, Y. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell 2003, 12, 475–487. [Google Scholar] [CrossRef]
- Basavapathruni, A.; Gureasko, J.; Porter Scott, M.; Hermans, W.; Godbole, A.; Leland, P.A.; Boriack-Sjodin, P.A.; Wigle, T.J.; Copeland, R.A.; Riera, T.V. Characterization of the Enzymatic Activity of SETDB1 and Its 1:1 Complex with ATF7IP. Biochemistry 2016, 55, 1645–1651. [Google Scholar] [CrossRef]
- Falandry, C.; Fourel, G.; Galy, V.; Ristriani, T.; Horard, B.; Bensimon, E.; Salles, G.; Gilson, E.; Magdinier, F. CLLD8/KMT1F Is a Lysine Methyltransferase That Is Important for Chromosome Segregation. J. Biol. Chem. 2010, 285, 20234–20241. [Google Scholar] [CrossRef]
- Lehnertz, B.; Ueda, Y.; Derijck, A.A.H.A.; Braunschweig, U.; Perez-Burgos, L.; Kubicek, S.; Chen, T.; Li, E.; Jenuwein, T.; Peters, A.H.F.M. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. CB 2003, 13, 1192–1200. [Google Scholar] [CrossRef]
- Peters, A.H.; O’Carroll, D.; Scherthan, H.; Mechtler, K.; Sauer, S.; Schöfer, C.; Weipoltshammer, K.; Pagani, M.; Lachner, M.; Kohlmaier, A.; et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 2001, 107, 323–337. [Google Scholar] [CrossRef]
- García-Cao, M.; O’Sullivan, R.; Peters, A.H.F.M.; Jenuwein, T.; Blasco, M.A. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 2004, 36, 94–99. [Google Scholar] [CrossRef]
- Schotta, G.; Lachner, M.; Sarma, K.; Ebert, A.; Sengupta, R.; Reuter, G.; Reinberg, D.; Jenuwein, T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004, 18, 1251–1262. [Google Scholar] [CrossRef]
- Yang, H.; Pesavento, J.J.; Starnes, T.W.; Cryderman, D.E.; Wallrath, L.L.; Kelleher, N.L.; Mizzen, C.A. Preferential dimethylation of histone H4 lysine 20 by Suv4-20. J. Biol. Chem. 2008, 283, 12085–12092. [Google Scholar] [CrossRef]
- Wang, H.; Cao, R.; Xia, L.; Erdjument-Bromage, H.; Borchers, C.; Tempst, P.; Zhang, Y. Purification and Functional Characterization of a Histone H3-Lysine 4-Specific Methyltransferase. Mol. Cell 2001, 8, 1207–1217. [Google Scholar] [CrossRef]
- Nishioka, K.; Rice, J.C.; Sarma, K.; Erdjument-Bromage, H.; Werner, J.; Wang, Y.; Chuikov, S.; Valenzuela, P.; Tempst, P.; Steward, R.; et al. PR-Set7 Is a Nucleosome-Specific Methyltransferase that Modifies Lysine 20 of Histone H4 and Is Associated with Silent Chromatin. Mol. Cell 2002, 9, 1201–1213. [Google Scholar] [CrossRef]
- Yin, Y.; Liu, C.; Tsai, S.N.; Zhou, B.; Ngai, S.M.; Zhu, G. SET8 Recognizes the Sequence RHRK20VLRDN within the N Terminus of Histone H4 and Mono-methylates Lysine 20. J. Biol. Chem. 2005, 280, 30025–30031. [Google Scholar] [CrossRef]
- Di Tullio, F.; Schwarz, M.; Zorgati, H.; Mzoughi, S.; Guccione, E. The duality of PRDM proteins: Epigenetic and structural perspectives. FEBS J. 2022, 289, 1256–1275. [Google Scholar] [CrossRef]
- Kim, K.-C.; Geng, L.; Huang, S. Inactivation of a histone methyltransferase by mutations in human cancers. Cancer Res. 2003, 63, 7619–7623. [Google Scholar]
- Pandzic, T.; Rendo, V.; Lim, J.; Larsson, C.; Larsson, J.; Stoimenov, I.; Kundu, S.; Ali, M.A.; Hellström, M.; He, L.; et al. Somatic PRDM2 c.4467delA mutations in colorectal cancers control histone methylation and tumor growth. Oncotarget 2017, 8, 98646–98659. [Google Scholar] [CrossRef]
- Pinheiro, I.; Margueron, R.; Shukeir, N.; Eisold, M.; Fritzsch, C.; Richter, F.M.; Mittler, G.; Genoud, C.; Goyama, S.; Kurokawa, M.; et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 2012, 150, 948–960. [Google Scholar] [CrossRef]
- Blazer, L.L.; Lima-Fernandes, E.; Gibson, E.; Eram, M.S.; Loppnau, P.; Arrowsmith, C.H.; Schapira, M.; Vedadi, M. PR Domain-containing Protein 7 (PRDM7) Is a Histone 3 Lysine 4 Trimethyltransferase. J. Biol. Chem. 2016, 291, 13509–13519. [Google Scholar] [CrossRef]
- Eom, G.H.; Kim, K.; Kim, S.-M.; Kee, H.J.; Kim, J.-Y.; Jin, H.M.; Kim, J.-R.; Kim, J.H.; Choe, N.; Kim, K.-B.; et al. Histone methyltransferase PRDM8 regulates mouse testis steroidogenesis. Biochem. Biophys. Res. Commun. 2009, 388, 131–136. [Google Scholar] [CrossRef]
- Hayashi, K.; Yoshida, K.; Matsui, Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 2005, 438, 374–378. [Google Scholar] [CrossRef]
- Koh-Stenta, X.; Joy, J.; Poulsen, A.; Li, R.; Tan, Y.; Shim, Y.; Min, J.-H.; Wu, L.; Ngo, A.; Peng, J.; et al. Characterization of the histone methyltransferase PRDM9 using biochemical, biophysical and chemical biology techniques. Biochem. J. 2014, 461, 323–334. [Google Scholar] [CrossRef]
- Eram, M.S.; Bustos, S.P.; Lima-Fernandes, E.; Siarheyeva, A.; Senisterra, G.; Hajian, T.; Chau, I.; Duan, S.; Wu, H.; Dombrovski, L.; et al. Trimethylation of Histone H3 Lysine 36 by Human Methyltransferase PRDM9 Protein. J. Biol. Chem. 2014, 289, 12177–12188. [Google Scholar] [CrossRef]
- Powers, N.R.; Parvanov, E.D.; Baker, C.L.; Walker, M.; Petkov, P.M.; Paigen, K. The Meiotic Recombination Activator PRDM9 Trimethylates Both H3K36 and H3K4 at Recombination Hotspots In Vivo. PLoS Genet. 2016, 12, e1006146. [Google Scholar] [CrossRef]
- Wu, H.; Mathioudakis, N.; Diagouraga, B.; Dong, A.; Dombrovski, L.; Baudat, F.; Cusack, S.; de Massy, B.; Kadlec, J. Molecular basis for the regulation of the H3K4 methyltransferase activity of PRDM9. Cell Rep. 2013, 5, 13–20. [Google Scholar] [CrossRef]
- Zhou, B.; Wang, J.; Lee, S.Y.; Xiong, J.; Bhanu, N.; Guo, Q.; Ma, P.; Sun, Y.; Rao, R.C.; Garcia, B.A.; et al. PRDM16 suppresses MLL leukemia via intrinsic histone methyltransferase activity. Mol. Cell 2016, 62, 222–236. [Google Scholar] [CrossRef]
- Müller, J.; Hart, C.M.; Francis, N.J.; Vargas, M.L.; Sengupta, A.; Wild, B.; Miller, E.L.; O’Connor, M.B.; Kingston, R.E.; Simon, J.A. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 2002, 111, 197–208. [Google Scholar] [CrossRef]
- Völkel, P.; Bary, A.; Raby, L.; Chapart, A.; Dupret, B.; Le Bourhis, X.; Angrand, P.-O. Ezh1 arises from Ezh2 gene duplication but its function is not required for zebrafish development. Sci. Rep. 2019, 9, 4319. [Google Scholar] [CrossRef]
- Flora, P.; Dalal, G.; Cohen, I.; Ezhkova, E. Polycomb Repressive Complex(es) and Their Role in Adult Stem Cells. Genes 2021, 12, 1485. [Google Scholar] [CrossRef]
- Falnes, P.Ø.; Małecki, J.M.; Herrera, M.C.; Bengtsen, M.; Davydova, E. Human seven-β-strand (METTL) methyltransferases—Conquering the universe of protein lysine methylation. J. Biol. Chem. 2023, 299, 104661. [Google Scholar] [CrossRef]
- Lukinović, V.; Casanova, A.G.; Roth, G.S.; Chuffart, F.; Reynoird, N. Lysine Methyltransferases Signaling: Histones are Just the Tip of the Iceberg. Curr. Protein Pept. Sci. 2020, 21, 655–674. [Google Scholar] [CrossRef]
- Zheng, K.; Chen, S.; Ren, Z.; Wang, Y. Protein arginine methylation in viral infection and antiviral immunity. Int. J. Biol. Sci. 2023, 19, 5292–5318. [Google Scholar] [CrossRef]
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463, 457–463. [Google Scholar] [CrossRef]
- Jacob, A.G.; Smith, C.W.J. Intron retention as a component of regulated gene expression programs. Hum. Genet. 2017, 136, 1043–1057. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.J.-L.; Au, A.Y.M.; Ritchie, W.; Rasko, J.E.J. Intron retention in mRNA: No longer nonsense. BioEssays 2016, 38, 41–49. [Google Scholar] [CrossRef]
- Li, H.; Yao, Q.; Mariscal, A.G.; Wu, X.; Hülse, J.; Pedersen, E.; Helin, K.; Waisman, A.; Vinkel, C.; Thomsen, S.F.; et al. Epigenetic control of IL-23 expression in keratinocytes is important for chronic skin inflammation. Nat. Commun. 2018, 9, 1420. [Google Scholar] [CrossRef]
- Zhang, P.; Su, Y.; Zhao, M.; Huang, W.; Lu, Q. Abnormal histone modifications in PBMCs from patients with psoriasis vulgaris. Eur. J. Dermatol. 2011, 21, 552–557. [Google Scholar] [CrossRef]
- Ovejero-Benito, M.C.; Reolid, A.; Sánchez-Jiménez, P.; Saiz-Rodríguez, M.; Muñoz-Aceituno, E.; Llamas-Velasco, M.; Martín-Vilchez, S.; Cabaleiro, T.; Román, M.; Ochoa, D.; et al. Histone modifications associated with biological drug response in moderate-to-severe psoriasis. Exp. Dermatol. 2018, 27, 1361–1371. [Google Scholar] [CrossRef]
- Qu, S.; Liu, Z.; Wang, B. EZH2 is involved in psoriasis progression by impairing miR-125a-5p inhibition of SFMBT1 and leading to inhibition of the TGFβ/SMAD pathway. Ther. Adv. Chronic Dis. 2021, 12, 2040622320987348. [Google Scholar] [CrossRef]
- Li, G.; Ye, Z.; Shi, C.; Sun, L.; Han, M.; Zhuang, Y.; Xu, T.; Zhao, S.; Wu, X. The Histone Methyltransferase Ash1l is Required for Epidermal Homeostasis in Mice. Sci. Rep. 2017, 7, 45401. [Google Scholar] [CrossRef]
- Xia, M.; Liu, J.; Wu, X.; Liu, S.; Li, G.; Han, C.; Song, L.; Li, Z.; Wang, Q.; Wang, J.; et al. Histone Methyltransferase Ash1l Suppresses Interleukin-6 Production and Inflammatory Autoimmune Diseases by Inducing the Ubiquitin-Editing Enzyme A20. Immunity 2013, 39, 470–481. [Google Scholar] [CrossRef]
- Matsumoto, R.; Dainichi, T.; Tsuchiya, S.; Nomura, T.; Kitoh, A.; Hayden, M.S.; Ishii, K.J.; Tanaka, M.; Honda, T.; Egawa, G.; et al. Epithelial TRAF6 drives IL-17-mediated psoriatic inflammation. JCI Insight 2018, 3, e121175. [Google Scholar] [CrossRef]
- Goldminz, A.M.; Au, S.C.; Kim, N.; Gottlieb, A.B.; Lizzul, P.F. NF-κB: An essential transcription factor in psoriasis. J. Dermatol. Sci. 2013, 69, 89–94. [Google Scholar] [CrossRef]
- Lu, T.; Jackson, M.W.; Wang, B.; Yang, M.; Chance, M.R.; Miyagi, M.; Gudkov, A.V.; Stark, G.R. Regulation of NF-κB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc. Natl. Acad. Sci. USA 2010, 107, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, F.; Chen, Q.; Wan, C.; Xiong, J.; Xu, J. CRISPR/Cas9-mediated knockout of NSD1 suppresses the hepatocellular carcinoma development via the NSD1/H3/Wnt10b signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 467. [Google Scholar] [CrossRef] [PubMed]
- Assarsson, M.; Söderman, J.; Duvetorp, A.; Mrowietz, U.; Skarstedt, M.; Seifert, O. Narrowband UVB treatment induces expression of WNT7B, WNT10B and TCF7L2 in psoriasis skin. Arch. Dermatol. Res. 2019, 311, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Toyokawa, G.; Cho, H.-S.; Masuda, K.; Yamane, Y.; Yoshimatsu, M.; Hayami, S.; Takawa, M.; Iwai, Y.; Daigo, Y.; Tsuchiya, E.; et al. Histone lysine methyltransferase Wolf-Hirschhorn syndrome candidate 1 is involved in human carcinogenesis through regulation of the Wnt pathway. Neoplasia 2011, 13, 887–898. [Google Scholar] [CrossRef]
- Kim, S.A.; Ryu, Y.W.; Kwon, J.I.; Choe, M.S.; Jung, J.W.; Cho, J.W. Differential expression of cyclin D1, Ki-67, pRb, and p53 in psoriatic skin lesions and normal skin. Mol. Med. Rep. 2018, 17, 735–742. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, K.; Zhu, H.; Qin, H.; Liu, J.; Cao, X. Methyltransferase Setd2 prevents T cell–mediated autoimmune diseases via phospholipid remodeling. Proc. Natl. Acad. Sci. USA 2024, 121, e2314561121. [Google Scholar] [CrossRef]
- Li, X.; Liu, C.; Zhu, Y.; Rao, H.; Liu, M.; Gui, L.; Feng, W.; Tang, H.; Xu, J.; Gao, W.-Q.; et al. SETD2 epidermal deficiency promotes cutaneous wound healing via activation of AKT/mTOR Signalling. Cell Prolif. 2021, 54, e13045. [Google Scholar] [CrossRef]
- Buerger, C.; Shirsath, N.; Lang, V.; Berard, A.; Diehl, S.; Kaufmann, R.; Boehncke, W.-H.; Wolf, P. Inflammation dependent mTORC1 signaling interferes with the switch from keratinocyte proliferation to differentiation. PLoS ONE 2017, 12, e0180853. [Google Scholar] [CrossRef]
- Morhenn, V.B.; Nelson, T.E.; Gruol, D.L. The rate of wound healing is increased in psoriasis. J. Dermatol. Sci. 2013, 72, 87–92. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, X.-Q.; Cheng, J.; Hui, R.-S.; Gao, T.-W. Increased Th17 cells are accompanied by FoxP3+ Treg cell accumulation and correlated with psoriasis disease severity. Clin. Immunol. 2010, 135, 108–117. [Google Scholar] [CrossRef]
- Ea, C.-K.; Hao, S.; Yeo, K.S.; Baltimore, D. EHMT1 Protein Binds to Nuclear Factor-κB p50 and Represses Gene Expression. J. Biol. Chem. 2012, 287, 31207–31217. [Google Scholar] [CrossRef] [PubMed]
- Karl, M.; Sommer, C.; Gabriel, C.H.; Hecklau, K.; Venzke, M.; Hennig, A.F.; Radbruch, A.; Selbach, M.; Baumgrass, R. Recruitment of Histone Methyltransferase Ehmt1 to Foxp3 TSDR Counteracts Differentiation of Induced Regulatory T Cells. J. Mol. Biol. 2019, 431, 3606–3625. [Google Scholar] [CrossRef]
- Chen, L.; Shen, Z.; Wang, G.; Fan, P.; Liu, Y. Dynamic frequency of CD4+CD25+Foxp3+ Treg cells in Psoriasis vulgaris. J. Dermatol. Sci. 2008, 51, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Richetta, A.G.; Mattozzi, C.; Salvi, M.; Giancristoforo, S.; D’epiro, S.; Milana, B.; Carboni, V.; Zampetti, M.; Calvieri, S.; Morrone, S. CD4+ CD25+ T-regulatory cells in psoriasis. Correlation between their numbers and biologics-induced clinical improvement. Eur. J. Dermatol. 2011, 21, 344–348. [Google Scholar] [CrossRef]
- Kimball, A.S.; Davis, F.M.; den Dekker, A.; Joshi, A.D.; Schaller, M.A.; Bermick, J.; Xing, X.; Burant, C.F.; Obi, A.T.; Nysz, D.; et al. The Histone Methyltransferase Setdb2 Modulates Macrophage Phenotype and Uric Acid Production in Diabetic Wound Repair. Immunity 2019, 51, 258–271.e5. [Google Scholar] [CrossRef]
- Lin, S.-H.; Chuang, H.-Y.; Ho, J.-C.; Lee, C.-H.; Hsiao, C.-C. Treatment with TNF-α inhibitor rectifies M1 macrophage polarization from blood CD14+ monocytes in patients with psoriasis independent of STAT1 and IRF-1 activation. J. Dermatol. Sci. 2018, 91, 276–284. [Google Scholar] [CrossRef]
- Balmer, P.; Hariton, W.V.J.; Sayar, B.S.; Jagannathan, V.; Galichet, A.; Leeb, T.; Roosje, P.; Müller, E.J. SUV39H2 epigenetic silencing controls fate conversion of epidermal stem and progenitor cells. J. Cell Biol. 2021, 220, e201908178. [Google Scholar] [CrossRef]
- Leuner, K.; Kraus, M.; Woelfle, U.; Beschmann, H.; Harteneck, C.; Boehncke, W.-H.; Schempp, C.M.; Müller, W.E. Reduced TRPC channel expression in psoriatic keratinocytes is associated with impaired differentiation and enhanced proliferation. PLoS ONE 2011, 6, e14716. [Google Scholar] [CrossRef] [PubMed]
- Mercurio, L.; Morelli, M.; Scarponi, C.; Scaglione, G.L.; Pallotta, S.; Albanesi, C.; Madonna, S. PI3Kδ Sustains Keratinocyte Hyperproliferation and Epithelial Inflammation: Implications for a Topically Druggable Target in Psoriasis. Cells 2021, 10, 2636. [Google Scholar] [CrossRef]
- Brustel, J.; Kirstein, N.; Izard, F.; Grimaud, C.; Prorok, P.; Cayrou, C.; Schotta, G.; Abdelsamie, A.F.; Déjardin, J.; Méchali, M.; et al. Histone H4K20 tri-methylation at late-firing origins ensures timely heterochromatin replication. EMBO J. 2017, 36, 2726–2741. [Google Scholar] [CrossRef]
- Schotta, G.; Sengupta, R.; Kubicek, S.; Malin, S.; Kauer, M.; Callén, E.; Celeste, A.; Pagani, M.; Opravil, S.; De La Rosa-Velazquez, I.A.; et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008, 22, 2048–2061. [Google Scholar] [CrossRef]
- Benetti, R.; Gonzalo, S.; Jaco, I.; Schotta, G.; Klatt, P.; Jenuwein, T.; Blasco, M.A. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J. Cell Biol. 2007, 178, 925–936. [Google Scholar] [CrossRef]
- Mehmetbeyoglu, E.; Kianmehr, L.; Borlu, M.; Yilmaz, Z.; Basar Kılıc, S.; Rajabi-Maham, H.; Taheri, S.; Rassoulzadegan, M. Decrease in RNase HII and Accumulation of lncRNAs/DNA Hybrids: A Causal Implication in Psoriasis? Biomolecules 2022, 12, 368. [Google Scholar] [CrossRef]
- Driskell, I.; Oda, H.; Blanco, S.; Nascimento, E.; Humphreys, P.; Frye, M. The histone methyltransferase Setd8 acts in concert with c-Myc and is required to maintain skin. EMBO J. 2012, 31, 616–629. [Google Scholar] [CrossRef]
- Manczinger, M.; Kemény, L. Novel Factors in the Pathogenesis of Psoriasis and Potential Drug Candidates Are Found with Systems Biology Approach. PLoS ONE 2013, 8, e80751. [Google Scholar] [CrossRef]
- Shen, C.; Chen, M.-T.; Zhang, X.-H.; Yin, X.-L.; Ning, H.-M.; Su, R.; Lin, H.-S.; Song, L.; Wang, F.; Ma, Y.-N.; et al. The PU.1-Modulated MicroRNA-22 Is a Regulator of Monocyte/Macrophage Differentiation and Acute Myeloid Leukemia. PLoS Genet. 2016, 12, e1006259. [Google Scholar] [CrossRef]
- Wang, Y.; Edelmayer, R.; Wetter, J.; Salte, K.; Gauvin, D.; Leys, L.; Paulsboe, S.; Su, Z.; Weinberg, I.; Namovic, M.; et al. Monocytes/Macrophages play a pathogenic role in IL-23 mediated psoriasis-like skin inflammation. Sci. Rep. 2019, 9, 5310. [Google Scholar] [CrossRef]
- Cheedipudi, S.; Puri, D.; Saleh, A.; Gala, H.P.; Rumman, M.; Pillai, M.S.; Sreenivas, P.; Arora, R.; Sellathurai, J.; Schrøder, H.D.; et al. A fine balance: Epigenetic control of cellular quiescence by the tumor suppressor PRDM2/RIZ at a bivalent domain in the cyclin a gene. Nucleic Acids Res. 2015, 43, 6236–6256. [Google Scholar] [CrossRef]
- Shapiro, V.S.; Lee, P.; Winoto, A. Identification and cloning of the G3B cDNA encoding a 3′ segment of a protein binding to GATA-3. Gene 1995, 163, 329–330. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Wu, R.; Zhang, S.; Zhao, M.; Wu, H.; Lu, Q.; Fu, S.; Su, Y. Wilms’ tumor 1-associating protein contributes to psoriasis by promoting keratinocytes proliferation via regulating cyclinA2 and CDK2. Int. Immunopharmacol. 2020, 88, 106918. [Google Scholar] [CrossRef] [PubMed]
- Vanaki, E.; Ataei, M.; Sanati, M.; Mansouri, P.; Mahmoudi, M.; Zarei, F.; Jadali, Z. Expression patterns of Th1/Th2 transcription factors in patients with guttate psoriasis. Acta Microbiol. Immunol. Hung. 2013, 60, 163–174. [Google Scholar] [CrossRef]
- Xiao, B.; Wilson, J.R.; Gamblin, S.J. SET domains and histone methylation. Curr. Opin. Struct. Biol. 2003, 13, 699–705. [Google Scholar] [CrossRef]
- Dillon, S.C.; Zhang, X.; Trievel, R.C.; Cheng, X. The SET-domain protein superfamily: Protein lysine methyltransferases. Genome Biol. 2005, 6, 227. [Google Scholar] [CrossRef]
- Pasini, D.; Bracken, A.P.; Jensen, M.R.; Denchi, E.L.; Helin, K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004, 23, 4061–4071. [Google Scholar] [CrossRef]
- Di Croce, L.; Helin, K. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 2013, 20, 1147–1155. [Google Scholar] [CrossRef]
- Margueron, R.; Reinberg, D. The Polycomb Complex PRC2 and its Mark in Life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef]
- Ura, H.; Murakami, K.; Akagi, T.; Kinoshita, K.; Yamaguchi, S.; Masui, S.; Niwa, H.; Koide, H.; Yokota, T. Eed/Sox2 regulatory loop controls ES cell self-renewal through histone methylation and acetylation. EMBO J. 2011, 30, 2190–2204. [Google Scholar] [CrossRef]
- Li, J.; Xing, J.; Lu, F.; Chang, W.; Liang, N.; Li, J.; Wang, Y.; Li, X.; Zhao, X.; Hou, R.; et al. Psoriatic Dermal-derived Mesenchymal Stem Cells Reduce Keratinocyte Junctions, and Increase Glycolysis. Acta Derm. Venereol. 2020, 100, adv00122. [Google Scholar] [CrossRef]
- Naito, T.; Muroi, S.; Taniuchi, I.; Kondo, M. Loss of Eed leads to lineage instability and increased CD8 expression of mouse CD4+ T cells upon TGFβ signaling. Mol. Immunol. 2018, 94, 140–152. [Google Scholar] [CrossRef]
- Richie, E.R.; Schumacher, A.; Angel, J.M.; Holloway, M.; Rinchik, E.M.; Magnuson, T. The Polycomb-group gene eed regulates thymocyte differentiation and suppresses the development of carcinogen-induced T-cell lymphomas. Oncogene 2002, 21, 299–306. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, J.; Guo, Z.; Li, C.; Tan, Z.; Wang, J.; Yang, J.; Xue, L. Easy or Not—The Advances of EZH2 in Regulating T Cell Development, Differentiation, and Activation in Antitumor Immunity. Front. Immunol. 2021, 12, 741302. [Google Scholar] [CrossRef]
- DuPage, M.; Chopra, G.; Quiros, J.; Rosenthal, W.L.; Morar, M.M.; Holohan, D.; Zhang, R.; Turka, L.; Marson, A.; Bluestone, J.A. The Chromatin-Modifying Enzyme Ezh2 Is Critical for the Maintenance of Regulatory T Cell Identity after Activation. Immunity 2015, 42, 227–238. [Google Scholar] [CrossRef]
- Cohen, I.; Zhao, D.; Menon, G.; Nakayama, M.; Koseki, H.; Zheng, D.; Ezhkova, E. PRC1 preserves epidermal tissue integrity independently of PRC2. Genes Dev. 2019, 33, 55–60. [Google Scholar] [CrossRef]
- Ezhkova, E.; Pasolli, H.A.; Parker, J.S.; Stokes, N.; Su, I.; Hannon, G.; Tarakhovsky, A.; Fuchs, E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 2009, 136, 1122–1135. [Google Scholar] [CrossRef]
- Sagdeo, A.; Wanat, K.; Seykora, J. Inflammatory Reaction Patterns and Diseases of Skin. In Pathobiology of Human Disease; McManus, L.M., Mitchell, R.N., Eds.; Academic Press: San Diego, CA, USA, 2014; pp. 1160–1167. [Google Scholar] [CrossRef]
- Ayala, F. Clinical Presentation of Psoriasis. Reumatismo 2007, 59, 40–45. [Google Scholar] [CrossRef]
- Grzenda, A.; Lomberk, G.; Svingen, P.; Mathison, A.; Calvo, E.; Iovanna, J.; Xiong, Y.; Faubion, W.; Urrutia, R. Functional characterization of EZH2β reveals the increased complexity of EZH2 isoforms involved in the regulation of mammalian gene expression. Epigenetics Chromatin 2013, 6, 3. [Google Scholar] [CrossRef]
- Petracovici, A.; Bonasio, R. Distinct PRC2 subunits regulate maintenance and establishment of Polycomb repression during differentiation. Mol. Cell 2021, 81, 2625–2639.e5. [Google Scholar] [CrossRef]
- Asenjo, H.G.; Gallardo, A.; López-Onieva, L.; Tejada, I.; Martorell-Marugán, J.; Carmona-Sáez, P.; Landeira, D. Polycomb regulation is coupled to cell cycle transition in pluripotent stem cells. Sci. Adv. 2020, 6, eaay4768. [Google Scholar] [CrossRef]
- Brien, G.L.; Healy, E.; Jerman, E.; Conway, E.; Fadda, E.; O’Donovan, D.; Krivtsov, A.V.; Rice, A.M.; Kearney, C.J.; Flaus, A.; et al. A chromatin-independent role of Polycomb-like 1 to stabilize p53 and promote cellular quiescence. Genes Dev. 2015, 29, 2231–2243. [Google Scholar] [CrossRef]
- Ruthenburg, A.J.; Allis, C.D.; Wysocka, J. Methylation of Lysine 4 on Histone H3: Intricacy of Writing and Reading a Single Epigenetic Mark. Mol. Cell 2007, 25, 15–30. [Google Scholar] [CrossRef]
- Wu, M.; Wang, P.F.; Lee, J.S.; Martin-Brown, S.; Florens, L.; Washburn, M.; Shilatifard, A. Molecular Regulation of H3K4 Trimethylation by Wdr82, a Component of Human Set1/COMPASS. Mol. Cell. Biol. 2008, 28, 7337–7344. [Google Scholar] [CrossRef]
- Shilatifard, A. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 2012, 81, 65–95. [Google Scholar] [CrossRef]
- Tsai, P.-H.; Chien, Y.; Wang, M.-L.; Hsu, C.-H.; Laurent, B.; Chou, S.-J.; Chang, W.-C.; Chien, C.-S.; Li, H.-Y.; Lee, H.-C.; et al. Ash2l interacts with Oct4-stemness circuitry to promote super-enhancer-driven pluripotency network. Nucleic Acids Res. 2019, 47, 10115–10133. [Google Scholar] [CrossRef]
- Cao, W.; Guo, J.; Wen, X.; Miao, L.; Lin, F.; Xu, G.; Ma, R.; Yin, S.; Hui, Z.; Chen, T.; et al. CXXC finger protein 1 is critical for T-cell intrathymic development through regulating H3K4 trimethylation. Nat. Commun. 2016, 7, 11687. [Google Scholar] [CrossRef]
- Kiuchi, M.; Onodera, A.; Kokubo, K.; Ichikawa, T.; Morimoto, Y.; Kawakami, E.; Takayama, N.; Eto, K.; Koseki, H.; Hirahara, K.; et al. The Cxxc1 subunit of the Trithorax complex directs epigenetic licensing of CD4+ T cell differentiation. J. Exp. Med. 2021, 218, e20201690. [Google Scholar] [CrossRef]
- Lin, F.; Meng, X.; Guo, Y.; Cao, W.; Liu, W.; Xia, Q.; Hui, Z.; Chen, J.; Hong, S.; Zhang, X.; et al. Epigenetic initiation of the TH17 differentiation program is promoted by Cxxc finger protein 1. Sci. Adv. 2019, 5, eaax1608. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Y.; Yang, X.; Wei, J.; Zhou, S.; Zhao, Z.; Cheng, J.; Duan, H.; Jia, T.; Lei, Q.; et al. Characterization of Th17 and FoxP3(+) Treg Cells in Paediatric Psoriasis Patients. Scand. J. Immunol. 2016, 83, 174–180. [Google Scholar] [CrossRef]
- Qiao, Z.; Zhao, W.; Liu, Y.; Feng, W.; Ma, Y.; Jin, H. Low-dose Interleukin-2 For Psoriasis Therapy Based on the Regulation of Th17/Treg Cell Balance in Peripheral Blood. Inflammation 2023, 46, 2359–2373. [Google Scholar] [CrossRef]
- Szegedi, A.; Aleksza, M.; Gonda, A.; Irinyi, B.; Sipka, S.; Hunyadi, J.; Antal-Szalmás, P. Elevated rate of Thelper1 (TH1) lymphocytes and serum IFN-γ levels in psoriatic patients. Immunol. Lett. 2003, 86, 277–280. [Google Scholar] [CrossRef]
- Punnia-Moorthy, G.; Hersey, P.; Emran, A.A.; Tiffen, J. Lysine Demethylases: Promising Drug Targets in Melanoma and Other Cancers. Front. Genet. 2021, 12, 680633. [Google Scholar] [CrossRef]
- Thieme, S.; Gyárfás, T.; Richter, C.; Özhan, G.; Fu, J.; Alexopoulou, D.; Muders, M.H.; Michalk, I.; Jakob, C.; Dahl, A.; et al. The histone demethylase UTX regulates stem cell migration and hematopoiesis. Blood 2013, 121, 2462–2473. [Google Scholar] [CrossRef]
- Liu, J.; Mercher, T.; Scholl, C.; Brumme, K.; Gilliland, D.G.; Zhu, N. A functional role for the histone demethylase UTX in normal and malignant hematopoietic cells. Exp. Hematol. 2012, 40, 487–498.e3. [Google Scholar] [CrossRef]
- Sera, Y.; Nakata, Y.; Ueda, T.; Yamasaki, N.; Koide, S.; Kobayashi, H.; Ikeda, K.; Kobatake, K.; Iwasaki, M.; Oda, H.; et al. UTX maintains the functional integrity of the murine hematopoietic system by globally regulating aging-associated genes. Blood 2021, 137, 908–922. [Google Scholar] [CrossRef]
- Fatema, F.; Ghoshal, L.; Saha, A.; Agarwal, S.; Bandyopadhyay, D. Early-Onset Versus Late-Onset Psoriasis: A Comparative Study of Clinical Variables, Comorbidities, and Association with HLA CW6 in a Tertiary Care Center. Indian J. Dermatol. 2021, 66, 705. [Google Scholar] [CrossRef]
- Manna, S.; Kim, J.K.; Baugé, C.; Cam, M.; Zhao, Y.; Shetty, J.; Vacchio, M.S.; Castro, E.; Tran, B.; Tessarollo, L.; et al. Histone H3 Lysine 27 demethylases Jmjd3 and Utx are required for T-cell differentiation. Nat. Commun. 2015, 6, 8152. [Google Scholar] [CrossRef]
- Yi, J.; Shi, X.; Xuan, Z.; Wu, J. Histone demethylase UTX/KDM6A enhances tumor immune cell recruitment, promotes differentiation and suppresses medulloblastoma. Cancer Lett. 2021, 499, 188–200. [Google Scholar] [CrossRef]
- Goebeler, M.; Toksoy, A.; Spandau, U.; Engelhardt, E.; Bröcker, E.B.; Gillitzer, R. The C-X-C chemokine Mig is highly expressed in the papillae of psoriatic lesions. J. Pathol. 1998, 184, 89–95. [Google Scholar] [CrossRef]
- Giustizieri, M.L.; Mascia, F.; Frezzolini, A.; De Pità, O.; Chinni, L.M.; Giannetti, A.; Girolomoni, G.; Pastore, S. Keratinocytes from patients with atopic dermatitis and psoriasis show a distinct chemokine production profile in response to T cell–derived cytokines. J. Allergy Clin. Immunol. 2001, 107, 871–877. [Google Scholar] [CrossRef]
- Husmann, D.; Gozani, O. Histone lysine methyltransferases in biology and disease. Nat. Struct. Mol. Biol. 2019, 26, 880–889. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Zhu, B. Roles of H3K36-specific histone methyltransferases in transcription: Antagonizing silencing and safeguarding transcription fidelity. Biophys. Rep. 2018, 4, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Brennan, K.; Shin, J.H.; Tay, J.K.; Prunello, M.; Gentles, A.J.; Sunwoo, J.B.; Gevaert, O. NSD1 inactivation defines an immune cold, DNA hypomethylated subtype in squamous cell carcinoma. Sci. Rep. 2017, 7, 17064. [Google Scholar] [CrossRef]
- Chen, C.; Shin, J.H.; Fang, Z.; Brennan, K.; Horowitz, N.B.; Pfaff, K.L.; Welsh, E.L.; Rodig, S.J.; Gevaert, O.; Gozani, O.; et al. Targeting KDM2A Enhances T-cell Infiltration in NSD1-Deficient Head and Neck Squamous Cell Carcinoma. Cancer Res. 2023, 83, 2645–2655. [Google Scholar] [CrossRef] [PubMed]
- Tracy, C.M.; Warren, J.S.; Szulik, M.; Wang, L.; Garcia, J.; Makaju, A.; Russell, K.; Miller, M.; Franklin, S. The Smyd Family of Methyltransferases: Role in Cardiac and Skeletal Muscle Physiology and Pathology. Curr. Opin. Physiol. 2017, 1, 140–152. [Google Scholar] [CrossRef] [PubMed]
- Saha, N.; Muntean, A.G. Insight into the multi-faceted role of the SUV family of H3K9 methyltransferases in carcinogenesis and cancer progression. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188498. [Google Scholar] [CrossRef]
- Ninova, M.; Fejes Tóth, K.; Aravin, A.A. The control of gene expression and cell identity by H3K9 trimethylation. Dev. Camb. Engl. 2019, 146, dev181180. [Google Scholar] [CrossRef] [PubMed]
- Eissenberg, J.C.; Elgin, S.C.R. HP1a: A structural chromosomal protein regulating transcription. Trends Genet. 2014, 30, 103–110. [Google Scholar] [CrossRef]
- Cheedipudi, S.; Genolet, O.; Dobreva, G.D. Epigenetic inheritance of cell fates during embryonic development. Front. Genet. 2014, 5, 19. [Google Scholar] [CrossRef]
- Lee, J.; Kim, K.; Ryu, T.Y.; Jung, C.-R.; Lee, M.-S.; Lim, J.H.; Park, K.; Kim, D.-S.; Son, M.-Y.; Hamamoto, R.; et al. EHMT1 knockdown induces apoptosis and cell cycle arrest in lung cancer cells by increasing CDKN1A expression. Mol. Oncol. 2021, 15, 2989–3002. [Google Scholar] [CrossRef]
- Ekman, A.-K.; Bivik Eding, C.; Rundquist, I.; Enerbäck, C. IL-17 and IL-22 Promote Keratinocyte Stemness in the Germinative Compartment in Psoriasis. J. Invest. Dermatol. 2019, 139, 1564–1573.e8. [Google Scholar] [CrossRef]
- Tsang, L.W.K.; Hu, N.; Underhill, D.A. Comparative Analyses of SUV420H1 Isoforms and SUV420H2 Reveal Differences in Their Cellular Localization and Effects on Myogenic Differentiation. PLoS ONE 2010, 5, e14447. [Google Scholar] [CrossRef]
- Fog-Tonnesen, C.K.; Galli, G.K.; Lund, A.H. PRDM proteins: Important players in differentiation and disease. BioEssays 2012, 34, 50–60. [Google Scholar] [CrossRef]
- Sorrentino, A.; Federico, A.; Rienzo, M.; Gazzerro, P.; Bifulco, M.; Ciccodicola, A.; Casamassimi, A.; Abbondanza, C. PR/SET Domain Family and Cancer: Novel Insights from The Cancer Genome Atlas. Int. J. Mol. Sci. 2018, 19, 3250. [Google Scholar] [CrossRef] [PubMed]
- Derunes, C.; Briknarová, K.; Geng, L.; Li, S.; Gessner, C.R.; Hewitt, K.; Wu, S.; Huang, S.; Woods, V.I.; Ely, K.R. Characterization of the PR domain of RIZ1 histone methyltransferase. Biochem. Biophys. Res. Commun. 2005, 333, 925–934. [Google Scholar] [CrossRef]
- Casamassimi, A.; Rienzo, M.; Di Zazzo, E.; Sorrentino, A.; Fiore, D.; Proto, M.C.; Moncharmont, B.; Gazzerro, P.; Bifulco, M.; Abbondanza, C. Multifaceted Role of PRDM Proteins in Human Cancer. Int. J. Mol. Sci. 2020, 21, 2648. [Google Scholar] [CrossRef] [PubMed]
- Di Zazzo, E.; De Rosa, C.; Abbondanza, C.; Moncharmont, B. PRDM Proteins: Molecular Mechanisms in Signal Transduction and Transcriptional Regulation. Biology 2013, 2, 107–141. [Google Scholar] [CrossRef]
- Rácz, E.; Kurek, D.; Kant, M.; Baerveldt, E.M.; Florencia, E.; Mourits, S.; de Ridder, D.; Laman, J.D.; van der Fits, L.; Prens, E.P. GATA3 expression is decreased in psoriasis and during epidermal regeneration; induction by narrow-band UVB and IL-4. PLoS ONE 2011, 6, e19806. [Google Scholar] [CrossRef]
- Yuasa, H.; Oike, Y.; Iwama, A.; Nishikata, I.; Sugiyama, D.; Perkins, A.; Mucenski, M.L.; Suda, T.; Morishita, K. Oncogenic transcription factor Evi1 regulates hematopoietic stem cell proliferation through GATA-2 expression. EMBO J. 2005, 24, 1976–1987. [Google Scholar] [CrossRef]
- Neidhart, M.; Pajak, A.; Laskari, K.; Riksen, N.P.; Joosten, L.A.B.; Netea, M.G.; Lutgens, E.; Stroes, E.S.G.; Ciurea, A.; Distler, O.; et al. Oligomeric S100A4 Is Associated With Monocyte Innate Immune Memory and Bypass of Tolerance to Subsequent Stimulation With Lipopolysaccharides. Front. Immunol. 2019, 10, 791. [Google Scholar] [CrossRef]
- van Nuland, R.; Gozani, O. Histone H4 Lysine 20 (H4K20) Methylation, Expanding the Signaling Potential of the Proteome One Methyl Moiety at a Time. Mol. Cell. Proteomics 2016, 15, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Eom, G.H.; Kim, K.-B.; Kim, J.H.; Kim, J.-Y.; Kim, J.-R.; Kee, H.J.; Kim, D.-W.; Choe, N.; Park, H.-J.; Son, H.-J.; et al. Histone methyltransferase SETD3 regulates muscle differentiation. J. Biol. Chem. 2011, 286, 34733–34742. [Google Scholar] [CrossRef]
- Kwiatkowski, S.; Seliga, A.K.; Vertommen, D.; Terreri, M.; Ishikawa, T.; Grabowska, I.; Tiebe, M.; Teleman, A.A.; Jagielski, A.K.; Veiga-Da-Cunha, M.; et al. SETD3 protein is the actin-specific histidine N-methyltransferase. eLife 2018, 7, e37921. [Google Scholar] [CrossRef]
- Wu, C.; Wu, L.; Ha, Y.; Zou, Y.; Shi, K.; Xing, J.; Zhao, Y.; Guo, J.; Shen, Z.; Jie, W. Methyltransferase SETD4 mediates macrophages proliferation through EGFR signaling. Res. Sq. 2023, preprint. [Google Scholar] [CrossRef]
- Wang, Y.; Shen, Z. Unmasking the mammalian SET domain-containing protein 4. NAR Cancer 2022, 4, zcac021. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Hou, Y.; Zhang, Z.; Zhang, B.; Huang, T.; Sun, A.; Shao, G.; Lin, Q. Structure, activity and function of the lysine methyltransferase SETD5. Front. Endocrinol. 2023, 14, 1089527. [Google Scholar] [CrossRef] [PubMed]
- Binda, O.; Sevilla, A.; LeRoy, G.; Lemischka, I.R.; Garcia, B.A.; Richard, S. SETD6 monomethylates H2AZ on lysine 7 and is required for the maintenance of embryonic stem cell self-renewal. Epigenetics 2013, 8, 177–183. [Google Scholar] [CrossRef]
- Chang, Y.; Levy, D.; Horton, J.R.; Peng, J.; Zhang, X.; Gozani, O.; Cheng, X. Structural basis of SETD6-mediated regulation of the NF-kB network via methyl-lysine signaling. Nucleic Acids Res. 2011, 39, 6380–6389. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.; Kuo, A.J.; Chang, Y.; Schaefer, U.; Kitson, C.; Cheung, P.; Espejo, A.; Zee, B.M.; Liu, C.L.; Tangsombatvisit, S.; et al. SETD6 lysine methylation of RelA couples GLP activity at chromatin to tonic repression of NF-κB signaling. Nat. Immunol. 2011, 12, 29–36. [Google Scholar] [CrossRef]
- Lizzul, P.F.; Aphale, A.; Malaviya, R.; Sun, Y.; Masud, S.; Dombrovskiy, V.; Gottlieb, A.B. Differential Expression of Phosphorylated NF-κB/RelA in Normal and Psoriatic Epidermis and Downregulation of NF-κB in Response to Treatment with Etanercept. J. Investig. Dermatol. 2005, 124, 1275–1283. [Google Scholar] [CrossRef]
- Nguyen, T.U.; Koo, J. Etanercept in the treatment of plaque psoriasis. Clin. Cosmet. Investig. Dermatol. CCID 2009, 2, 77–84. [Google Scholar][Green Version]
- Min, J.; Feng, Q.; Li, Z.; Zhang, Y.; Xu, R.-M. Structure of the Catalytic Domain of Human DOT1L, a Non-SET Domain Nucleosomal Histone Methyltransferase. Cell 2003, 112, 711–723. [Google Scholar] [CrossRef]
- Metzger, E.; Wang, S.; Urban, S.; Willmann, D.; Schmidt, A.; Offermann, A.; Allen, A.; Sum, M.; Obier, N.; Cottard, F.; et al. KMT9 monomethylates histone H4 lysine 12 and controls proliferation of prostate cancer cells. Nat. Struct. Mol. Biol. 2019, 26, 361–371. [Google Scholar] [CrossRef] [PubMed]
- van Leeuwen, F.; Gafken, P.R.; Gottschling, D.E. Dot1p Modulates Silencing in Yeast by Methylation of the Nucleosome Core. Cell 2002, 109, 745–756. [Google Scholar] [CrossRef]
- Kim, W.; Kim, R.; Park, G.; Park, J.-W.; Kim, J.-E. Deficiency of H3K79 histone methyltransferase Dot1-like protein (DOT1L) inhibits cell proliferation. J. Biol. Chem. 2012, 287, 5588–5599. [Google Scholar] [CrossRef] [PubMed]
- Wille, C.K.; Sridharan, R. DOT1L inhibition enhances pluripotency beyond acquisition of epithelial identity and without immediate suppression of the somatic transcriptome. Stem Cell Rep. 2022, 17, 384–396. [Google Scholar] [CrossRef]
- Tian, Y.; Meng, L.; Yu, H.; Hexner, E.O.; Zheng, L.; Hu, S.; Zhang, Y.; Zhang, Y. Graft-Versus-Host Disease Impairs the Histone Methyltransferase Dot1l-Regulated Reconstitution of Plasmacytoid Dendritic Cells in Mice Undergoing Allo-HSCT. Blood 2018, 132, 477. [Google Scholar] [CrossRef]
- Scheer, S.; Runting, J.; Bramhall, M.; Russ, B.; Zaini, A.; Ellemor, J.; Rodrigues, G.; Ng, J.; Zaph, C. The Methyltransferase DOT1L Controls Activation and Lineage Integrity in CD4+ T Cells during Infection and Inflammation. Cell Rep. 2020, 33, 108505. [Google Scholar] [CrossRef]
- Kwesi-Maliepaard, E.M.; Aslam, M.A.; Alemdehy, M.F.; Brand, T.v.D.; McLean, C.; Vlaming, H.; van Welsem, T.; Korthout, T.; Lancini, C.; Hendriks, S.; et al. The histone methyltransferase DOT1L prevents antigen-independent differentiation and safeguards epigenetic identity of CD8+ T cells. Proc. Natl. Acad. Sci. USA 2020, 117, 20706–20716. [Google Scholar] [CrossRef] [PubMed]
- Willemsen, L.; Prange, K.H.; Neele, A.E.; van Roomen, C.P.; Gijbels, M.; Griffith, G.R.; Toom, M.D.; Beckers, L.; Siebeler, R.; Spann, N.J.; et al. DOT1L regulates lipid biosynthesis and inflammatory responses in macrophages and promotes atherosclerotic plaque stability. Cell Rep. 2022, 41, 111703. [Google Scholar] [CrossRef]
- Yang, Y.; Bedford, M.T. Protein arginine methyltransferases and cancer. Nat Rev Cancer Nat. Rev. Cancer 2013, 13, 37–50. [Google Scholar] [CrossRef]
- Al-Hamashi, A.A.; Diaz, K.; Huang, R. Non-Histone Arginine Methylation by Protein Arginine Methyltransferases. Curr. Protein Pept. Sci. 2020, 21, 699–712. [Google Scholar] [CrossRef]
- Wei, H.-H.; Fan, X.-J.; Hu, Y.; Tian, X.-X.; Guo, M.; Mao, M.-W.; Fang, Z.-Y.; Wu, P.; Gao, S.-X.; Peng, C.; et al. A systematic survey of PRMT interactomes reveals the key roles of arginine methylation in the global control of RNA splicing and translation. Sci. Bull. 2021, 66, 1342–1357. [Google Scholar] [CrossRef] [PubMed]
- Bedford, M.T.; Richard, S. Arginine methylation an emerging regulator of protein function. Mol. Cell 2005, 18, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Geoghegan, V.; Guo, A.; Trudgian, D.; Thomas, B.; Acuto, O. Comprehensive identification of arginine methylation in primary T cells reveals regulatory roles in cell signalling. Nat. Commun. 2015, 6, 6758. [Google Scholar] [CrossRef]
- Stojadinovic, O.; Brem, H.; Vouthounis, C.; Lee, B.; Fallon, J.; Stallcup, M.; Merchant, A.; Galiano, R.D.; Tomic-Canic, M. Molecular Pathogenesis of Chronic Wounds: The Role of β-Catenin and c-myc in the Inhibition of Epithelialization and Wound Healing. Am. J. Pathol. 2005, 167, 59–69. [Google Scholar] [CrossRef]
- Stojadinovic, O.; Vouthounis, C.; Lee, B.; Stallcup, M.; Tomic-Canic, M. 142 β-Catenin and Carm-1 as Co-Repressors of Glucocorticoid Receptor Lead to Inhibition of Keratinocyte Migration. Wound Repair Regen. 2004, 12, A37. [Google Scholar] [CrossRef]
- Zika, E.; Fauquier, L.; Vandel, L.; Ting, J.P.-Y. Interplay among coactivator-associated arginine methyltransferase 1, CBP, and CIITA in IFN-gamma-inducible MHC-II gene expression. Proc. Natl. Acad. Sci. USA 2005, 102, 16321–16326. [Google Scholar] [CrossRef]
- Li, J.; Zhao, Z.; Carter, C.; Ehrlich, L.I.; Bedford, M.T.; Richie, E.R. Coactivator-associated arginine methyltransferase 1 regulates fetal hematopoiesis and thymocyte development. J. Immunol. 2013, 190, 597–604. [Google Scholar] [CrossRef]
- Bao, X.; Siprashvili, Z.; Zarnegar, B.J.; Shenoy, R.M.; Rios, E.J.; Nady, N.; Qu, K.; Mah, A.; Webster, D.E.; Rubin, A.J.; et al. CSNK1a1 Regulates PRMT1 to Maintain the Progenitor State in Self-Renewing Somatic Tissue. Dev. Cell 2017, 43, 227–239.e5. [Google Scholar] [CrossRef]
- Albrecht, L.V.; Zhang, L.; Shabanowitz, J.; Purevjav, E.; Towbin, J.A.; Hunt, D.F.; Green, K.J. GSK3- and PRMT-1–dependent modifications of desmoplakin control desmoplakin–cytoskeleton dynamics. J. Cell Biol. 2015, 208, 597–612. [Google Scholar] [CrossRef]
- Mowen, K.A.; Schurter, B.T.; Fathman, J.W.; David, M.; Glimcher, L.H. Arginine Methylation of NIP45 Modulates Cytokine Gene Expression in Effector T Lymphocytes. Mol. Cell 2004, 15, 559–571. [Google Scholar] [CrossRef]
- Sen, S.; He, Z.; Ghosh, S.; Dery, K.J.; Yang, L.; Zhang, J.; Sun, Z. PRMT1 Plays a Critical Role in Th17 Differentiation by Regulating Reciprocal Recruitment of STAT3 and STAT5. J. Immunol. 2018, 201, 440–450. [Google Scholar] [CrossRef]
- Fan, Z.; Li, J.; Li, P.; Ye, Q.; Xu, H.; Wu, X.; Xu, Y. Protein arginine methyltransferase 1 (PRMT1) represses MHC II transcription in macrophages by methylating CIITA. Sci. Rep. 2017, 7, 40531. [Google Scholar] [CrossRef] [PubMed]
- Reintjes, A.; Fuchs, J.E.; Kremser, L.; Lindner, H.H.; Liedl, K.R.; Huber, L.A.; Valovka, T. Asymmetric arginine dimethylation of RelA provides a repressive mark to modulate TNFα/NF-κB response. Proc. Natl. Acad. Sci. USA 2016, 113, 4326–4331. [Google Scholar] [CrossRef] [PubMed]
- Covic, M.; Hassa, P.O.; Saccani, S.; Buerki, C.; Meier, N.I.; Lombardi, C.; Imhof, R.; Bedford, M.T.; Natoli, G.; Hottiger, M.O. Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-κB-dependent gene expression. EMBO J. 2004, 24, 85–96. [Google Scholar] [CrossRef]
- Wang, J.; Hua, H.; Wang, F.; Yang, S.; Zhou, Q.; Wu, X.; Feng, D.; Peng, H. Arginine methylation by PRMT2 promotes IFN-β production through TLR4/IRF3 signaling pathway. Mol. Immunol. 2021, 139, 202–210. [Google Scholar] [CrossRef]
- Oliveira, A.L.d.B.; Monteiro, V.V.S.; Navegantes-Lima, K.C.; Reis, J.F.; Gomes, R.D.S.; Rodrigues, D.V.S.; Gaspar, S.L.d.F.; Monteiro, M.C. Resveratrol Role in Autoimmune Disease—A Mini-Review. Nutrients 2017, 9, 1306. [Google Scholar] [CrossRef]
- Tong, W.; Chen, X.; Song, X.; Chen, Y.; Jia, R.; Zou, Y.; Li, L.; Yin, L.; He, C.; Liang, X.; et al. Resveratrol inhibits LPS-induced inflammation through suppressing the signaling cascades of TLR4-NF-κB/MAPKs/IRF3. Exp. Ther. Med. 2020, 19, 1824–1834. [Google Scholar] [CrossRef]
- Saha, K.; Eckert, R.L. Methylosome Protein 50 and PKCδ/p38δ Protein Signaling Control Keratinocyte Proliferation via Opposing Effects on p21Cip1 Gene Expression. J. Biol. Chem. 2015, 290, 13521–13530. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Kong, X.; Xia, J.; Wu, X.; Li, H.; Xu, H.; Fang, M.; Xu, Y. The arginine methyltransferase PRMT5 regulates CIITA-dependent MHC II transcription. Biochim. Biophys. Acta (BBA)—Gene Regul. Mech. 2016, 1859, 687–696. [Google Scholar] [CrossRef]
- Sengupta, S.; Kennemer, A.; Patrick, K.; Tsichlis, P.; Guerau-De-Arellano, M. Protein Arginine Methyltransferase 5 in T Lymphocyte Biology. Trends Immunol. 2020, 41, 918–931. [Google Scholar] [CrossRef]
- Lee, S.-H.; Chen, T.-Y.; Dhar, S.S.; Gu, B.; Chen, K.; Kim, Y.Z.; Li, W.; Lee, M.G. A feedback loop comprising PRMT7 and miR-24-2 interplays with Oct4, Nanog, Klf4 and c-Myc to regulate stemness. Nucleic Acids Res. 2016, 44, 10603–10618. [Google Scholar] [CrossRef] [PubMed]
- Vuong, T.A.; Jeong, H.-J.; Lee, H.-J.; Kim, B.-G.; Leem, Y.-E.; Cho, H.; Kang, J.-S. PRMT7 methylates and suppresses GLI2 binding to SUFU thereby promoting its activation. Cell Death Differ. 2020, 27, 15–28. [Google Scholar] [CrossRef]
- Blanc, R.S.; Vogel, G.; Chen, T.; Crist, C.; Richard, S. PRMT7 Preserves Satellite Cell Regenerative Capacity. Cell Rep. 2016, 14, 1528–1539. [Google Scholar] [CrossRef]
- Jeridi, A.; Conlon, T.M.; Günsel, G.G.; Lang, N.J.; Burgstaller, G.; Van Eeckhoutte, H.P.; Verleden, S.E.; Stöger, T.; Königshoff, M.; Eickelberg, O.; et al. Monocyte migration and COPD pathogenesis are epigenetically regulated by PRMT7. ERJ Open Res. 2022, 8 (Suppl. S8), 178. [Google Scholar] [CrossRef]
- Günsel, G.G.; Conlon, T.M.; Jeridi, A.; Kim, R.; Ertüz, Z.; Lang, N.J.; Ansari, M.; Novikova, M.; Jiang, D.; Strunz, M.; et al. The arginine methyltransferase PRMT7 promotes extravasation of monocytes resulting in tissue injury in COPD. Nat. Commun. 2022, 13, 1303. [Google Scholar] [CrossRef] [PubMed]
- Golden, J.B.; Groft, S.G.; Squeri, M.V.; Debanne, S.M.; Ward, N.L.; McCormick, T.S.; Cooper, K.D. Chronic psoriatic skin inflammation leads to increased monocyte adhesion and aggregation. J. Immunol. 2015, 195, 2006–2018. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S.; et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012, 22, 1760–1774. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2016, 4, 1521. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Schwämmle, V.; León, I.R.; Jensen, O.N. Assessment and Improvement of Statistical Tools for Comparative Proteomics Analysis of Sparse Data Sets with Few Experimental Replicates. J. Proteome Res. 2013, 12, 3874–3883. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Holik, A.Z.; Su, S.; Jansz, N.; Chen, K.; Leong, H.S.; Blewitt, M.E.; Asselin-Labat, M.-L.; Smyth, G.K.; Ritchie, M.E. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 2015, 43, e97. [Google Scholar] [CrossRef] [PubMed]
- Cyrus, S.; Burkardt, D.; Weaver, D.D.; Gibson, W.T. PRC2-complex related dysfunction in overgrowth syndromes: A review of EZH2, EED, and SUZ12 and their syndromic phenotypes. Am. J. Med. Genet. C Semin. Med. Genet. 2019, 181, 519–531. [Google Scholar] [CrossRef]
- Conway, E.; Jerman, E.; Healy, E.; Ito, S.; Holoch, D.; Oliviero, G.; Deevy, O.; Glancy, E.; Fitzpatrick, D.J.; Mucha, M.; et al. A Family of Vertebrate-Specific Polycombs Encoded by the LCOR/LCORL Genes Balance PRC2 Subtype Activities. Mol. Cell 2018, 70, 408–421.e8. [Google Scholar] [CrossRef]
- Luo, Z.; Lin, C.; Shilatifard, A. The super elongation complex (SEC) family in transcriptional control. Nat. Rev. Mol. Cell Biol. 2012, 13, 543–547. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).