NsrR Represses σE-Dependent Small RNAs and Interacts with RpoE via a Noncanonical Mechanism in Escherichia coli
Abstract
1. Introduction
2. Results
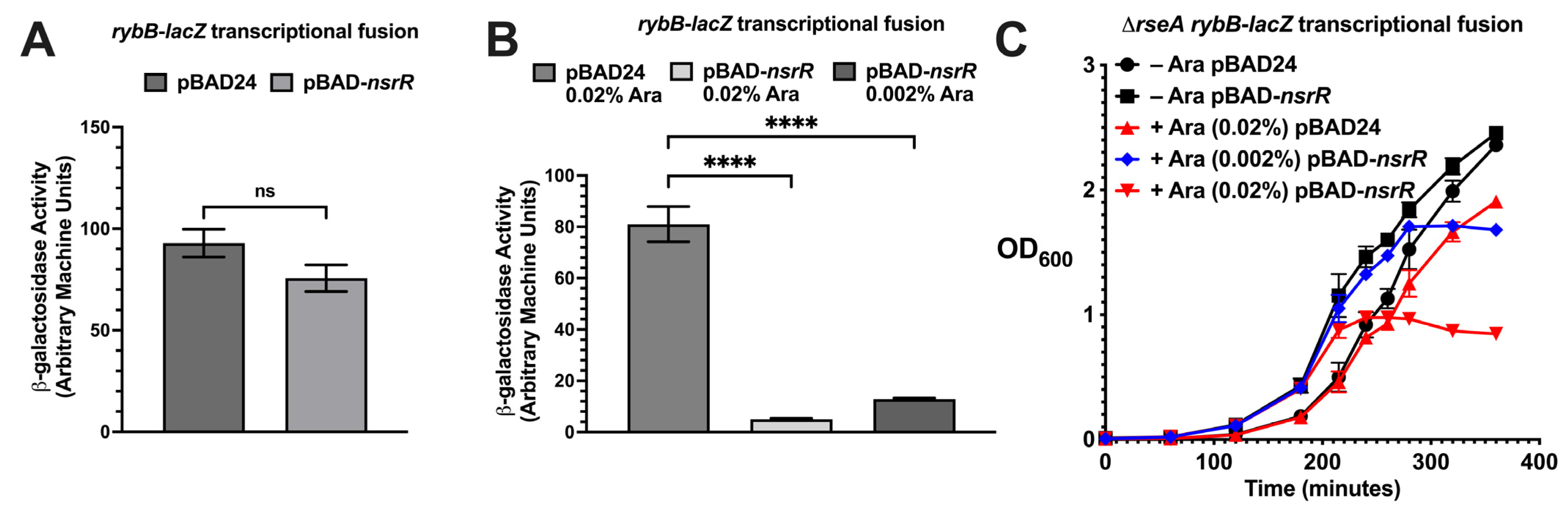
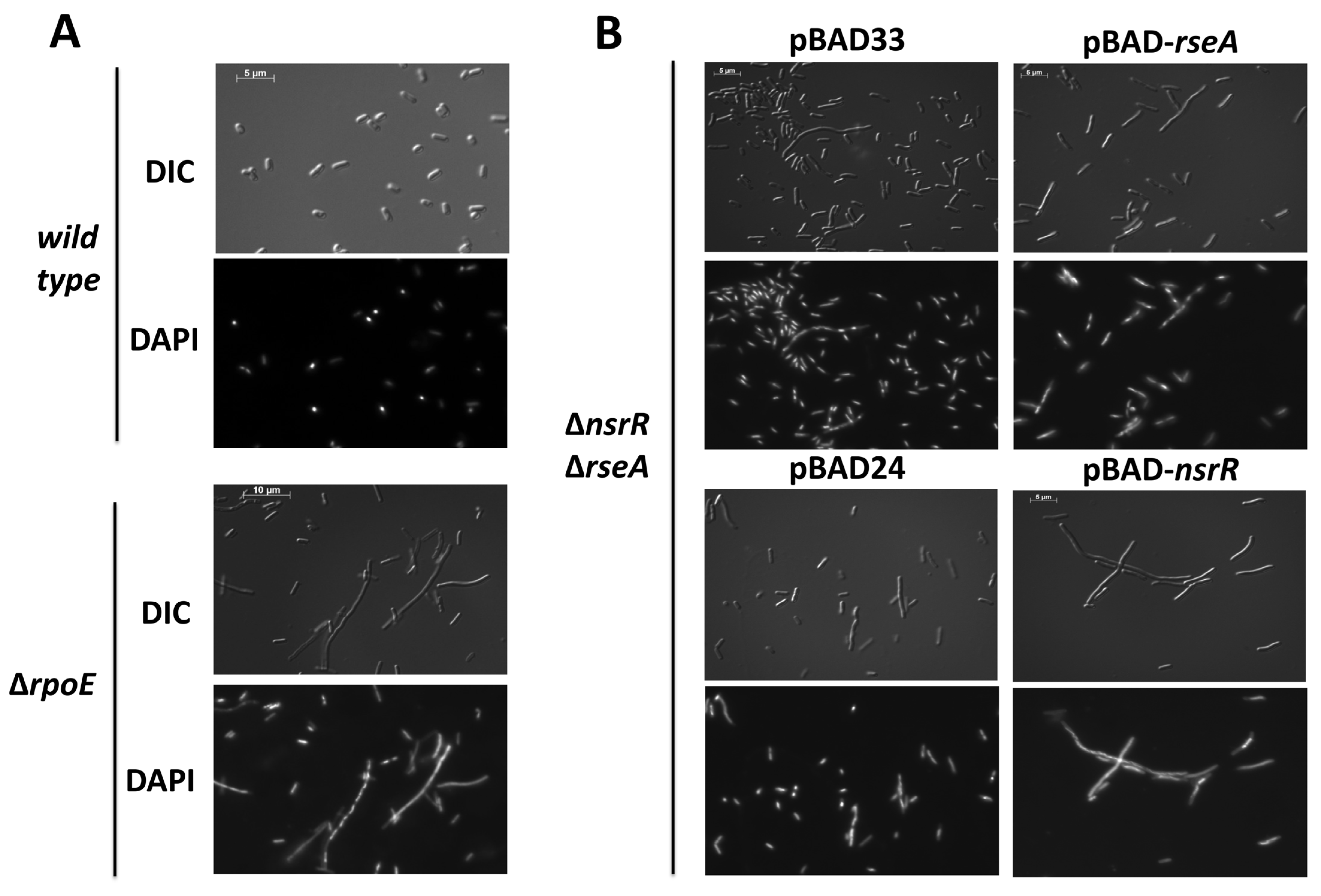
2.1. NsrR Overexpression Represses σE-Dependent Promoters and Disrupts Cell Division
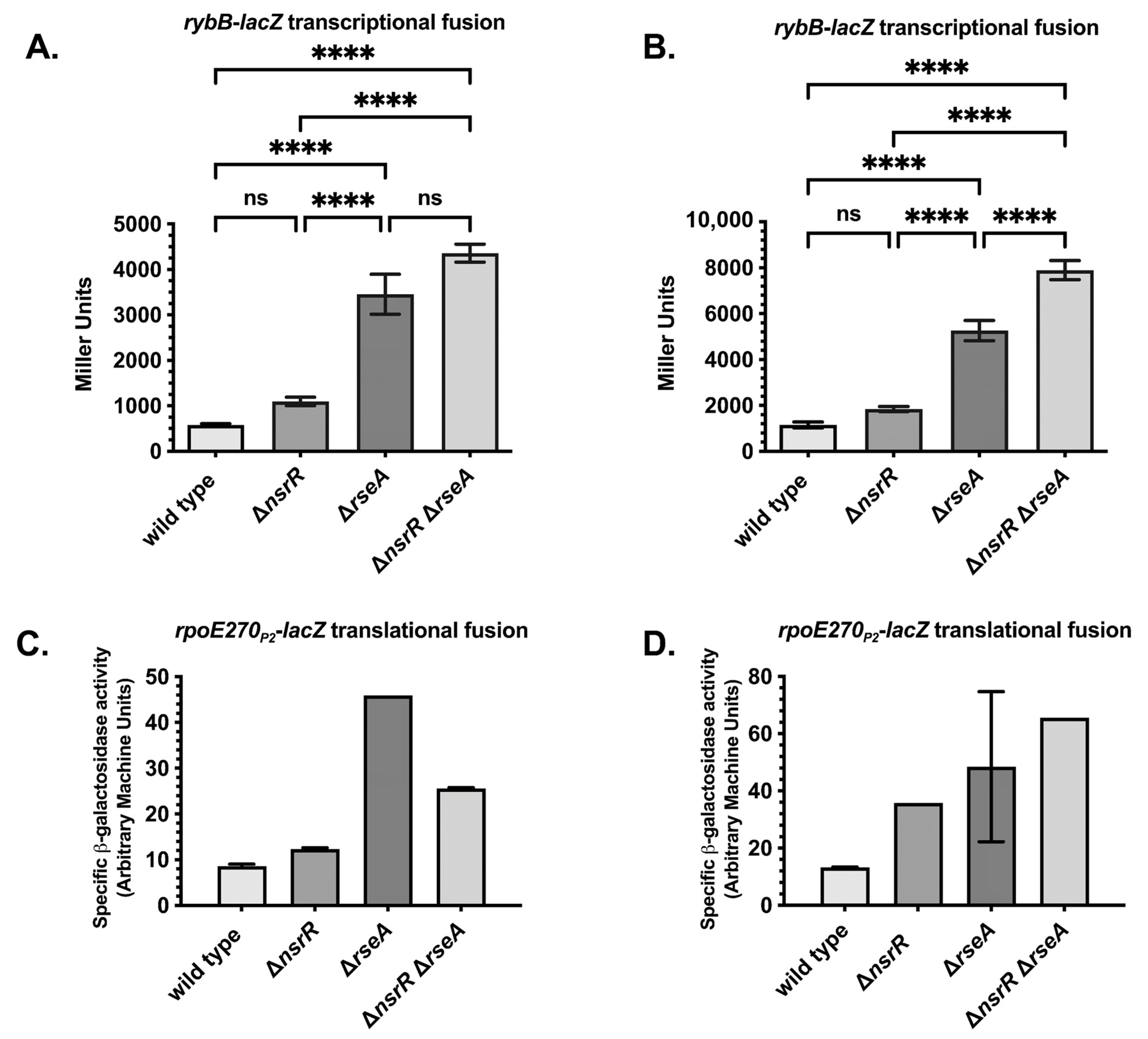
2.2. NsrR and RseA Independently Repress rybB and rpoE, Supporting a Parallel Regulatory Model
2.3. NsrR Interferes with RpoE-Dependent Promoter Activation in an Inducible Expression System
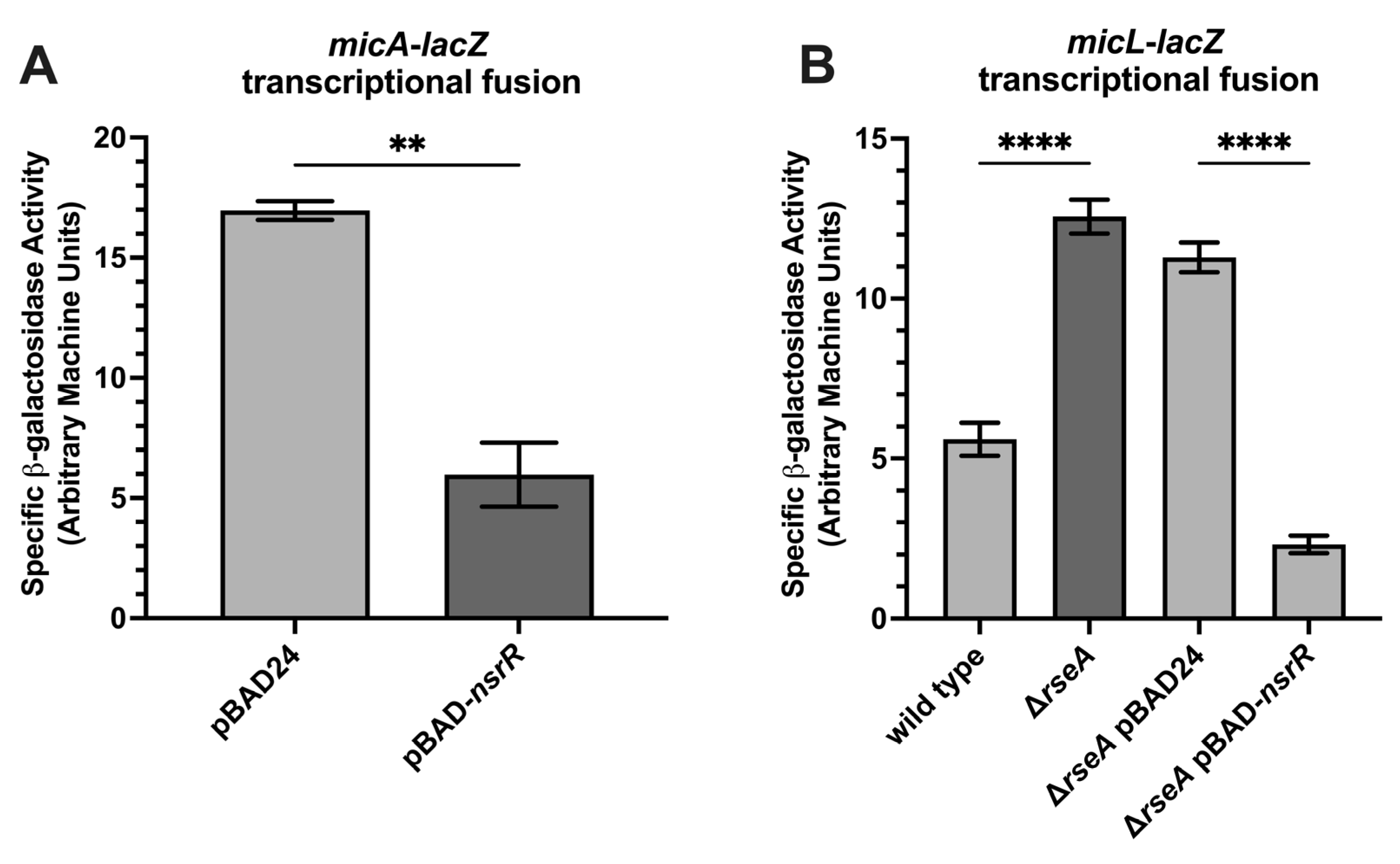
2.4. NsrR Represses Additional σE-Regulated sRNAs
2.5. NsrR Does Not Exhibit Robust Binding to σE Target Promoters In Vitro
2.6. NsrR Phenocopies RseA Repression Kinetics at the rybB Promoter
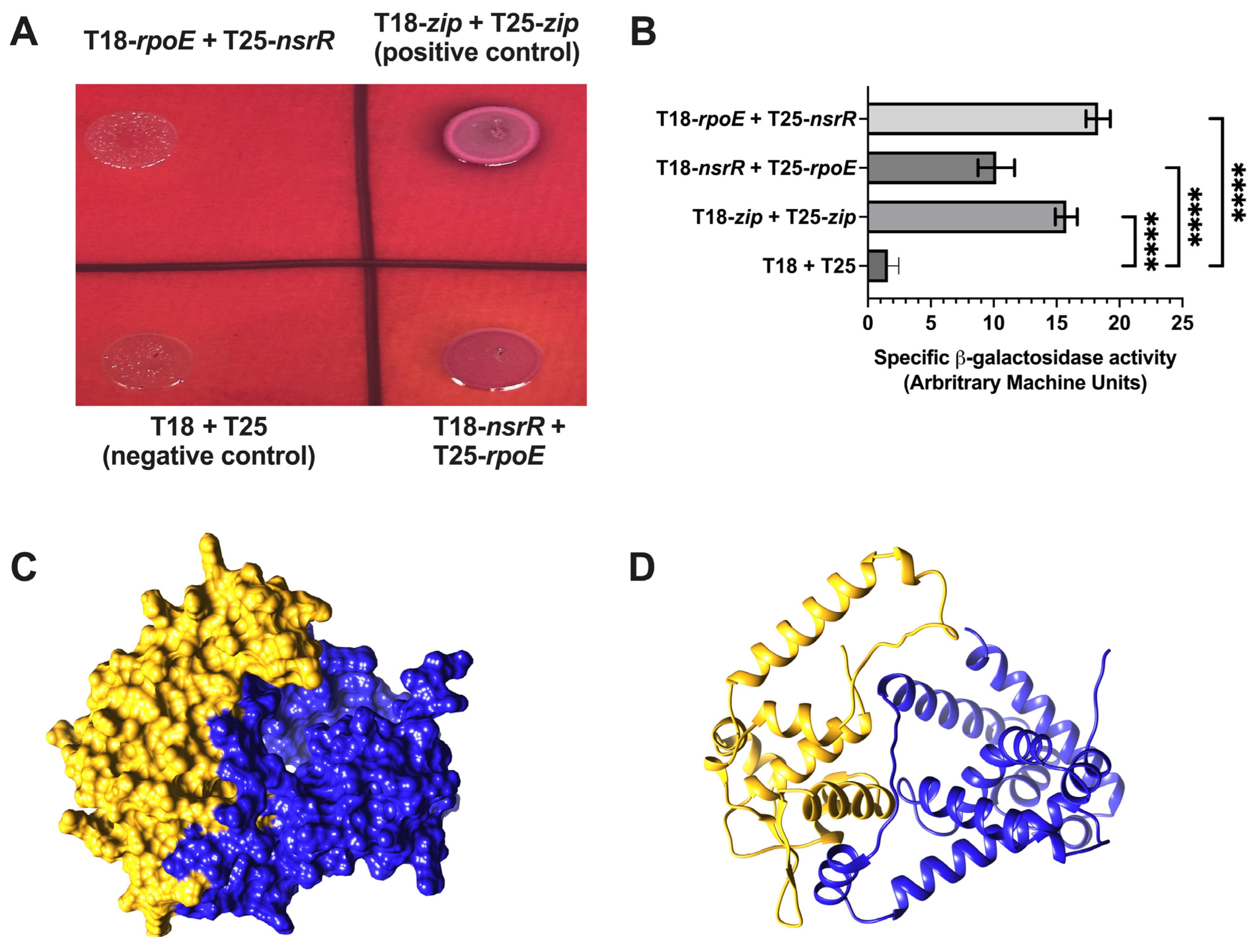
2.7. NsrR–RpoE Interaction Confirmed by Bacterial Adenylate Cyclase Two-Hybrid (BACTH) and Explored via AlphaFold3 Multimer Modeling
3. Discussion
3.1. NsrR Is a Dual Regulator of Envelope and Nitrosative Stress Responses
3.2. RybB Repression as a Model for the NsrR-Dependent Modulation of σE
3.3. NsrR May Act as a Noncanonical Anti-σE Factor
3.4. Physiological and Evolutionary Implications
3.5. Open Questions and Future Directions
4. Materials and Methods
4.1. Media and Growth Conditions
4.2. Strains, Plasmids, and Oligonucleotides
4.2.1. Chromosomal Deletion–Insertion Mutations of nsrR and rpoE
4.2.2. Construction of Plasmid-Based Arabinose-Inducible nsrR and rseA Alleles
4.2.3. Construction of Bacterial Adenylate Cyclase Two-Hybrid Assay (BACTH) Plasmids
4.3. Bacterial Two-Hybrid (BACTH) Assay
4.4. Quantitative β-Galactosidase Assays
4.5. Microscopy Experiments
4.6. Protein–DNA Interaction Assays
4.7. Structural Modeling Using AlphaFold3
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Raina, S.; Missiakas, D.; Georgopoulos, C. The rpoE gene encoding the sigma E (sigma 24) heat shock sigma factor of Escherichia coli. EMBO J. 1995, 14, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Mescas, J.; Rouviere, P.E.; Erickson, J.W.; Donohue, T.J.; Gross, C.A. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by experssion of out membrane proteins. Genes Dev. 1993, 7, 2618–2628. [Google Scholar]
- Erickson, J.W.; A Gross, C. Identification of the sigma E subunit of Escherichia coli RNA polymerase: A second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 1989, 3, 1462–1471. [Google Scholar] [CrossRef]
- Missiakas, D.; Mayer, M.P.; Lemaire, M.; Georgopoulos, C.; Raina, S. Modulation of the Escherichia coli sE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol. 1997, 24, 355–371. [Google Scholar] [CrossRef]
- De Las Penans, A.; Connolly, L.; Gross, C.A. The sE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of sE. Mol. Microbiol. 1997, 24, 373–385. [Google Scholar] [CrossRef]
- Alba, B.M.; Leeds, J.A.; Onufryk, C.; Lu, C.Z.; Gross, C.A. DegS and YaeL participate sequentially in the cleavage of RseA to activate the ςE-dependent extracytoplasmic stress response. Genes Dev. 2002, 16, 2156–2168. [Google Scholar] [CrossRef]
- Walsh, N.P.; Alba, B.M.; Bose, B.; A Gross, C.; Sauer, R.T. OMP Peptide Signals Initiate the Envelope-Stress Response by Activating DegS Protease via Relief of Inhibition Mediated by Its PDZ Domain. Cell 2003, 113, 61–71. [Google Scholar] [CrossRef]
- Snyder, W.B.; Davis, L.J.; Danese, P.N.; Cosma, C.L.; Silhavy, T.J. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J. Bacteriol. 1995, 177, 4216–4223. [Google Scholar] [CrossRef]
- Campbell, E.A.; Tupy, J.L.; Gruber, T.M.; Wang, S.; Sharp, M.M.; Gross, C.A.; Darst, S.A. Crystal Structure of Escherichia coli σE with the Cytoplasmic Domain of Its Anti-σ RseA. Mol. Cell 2003, 11, 1067–1078. [Google Scholar] [CrossRef]
- Kanehara, K.; Ito, K.; Akiyama, Y. YaeL (EcfE) activates the sE pathway of stress response through a site-2 cleavage of anti-sE, RseA. Genes Dev. 2002, 16, 2147–2155. [Google Scholar] [CrossRef]
- Ades, S.E.; Connolly, L.E.; Alba, B.M.; Gross, C.A. The Escherichia coli sigma E-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 1999, 13, 2449–2461. [Google Scholar] [CrossRef] [PubMed]
- A Rhodius, V.; Suh, W.C.; Nonaka, G.; West, J.; A Gross, C.; Eisen, J.A. Conserved and Variable Functions of the σE Stress Response in Related Genomes. PLoS Biol. 2005, 4, e2. [Google Scholar] [CrossRef] [PubMed]
- Johansen, J.; Eriksen, M.; Kallipolitis, B.; Valentin-Hansen, P. Down-regulation of Outer Membrane Proteins by Noncoding RNAs: Unraveling the cAMP–CRP- and σE-Dependent CyaR–ompX Regulatory Case. J. Mol. Biol. 2008, 383, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, G.; Blankschien, M.; Herman, C.; Gross, C.A.; Rhodius, V.A. Regulon and promoter analysis of the E. coli heat-shock factor, σ32, reveals a multifaceted cellular response to heat stress. Genes Dev. 2006, 20, 1776–1789. [Google Scholar] [CrossRef]
- Thompson, K.M.; Rhodius, V.A.; Gottesman, S. σ E Regulates and Is Regulated by a Small RNA in Escherichia coli. J. Bacteriol. 2007, 189, 4243–4256. [Google Scholar] [CrossRef]
- Gogol, E.B.; Rhodius, V.A.; Papenfort, K.; Vogel, J.; Gross, C.A. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc. Natl. Acad. Sci. USA 2011, 108, 12875–12880. [Google Scholar] [CrossRef]
- Udekwu, K.I.; Wagner, E.G. Sigma E controls biogenesis of the antisense RNA MicA. Nucleic Acids Res. 2007, 35, 1018–1037. [Google Scholar] [CrossRef]
- Coornaert, A.; Lu, A.; Mandin, P.; Springer, M.; Gottesman, S.; Guillier, M. MicA sRNA links the PhoP regulon to cell envelope stress. Mol. Microbiol. 2010, 76, 467–479. [Google Scholar] [CrossRef]
- Guo, M.S.; Updegrove, T.B.; Gogol, E.B.; Shabalina, S.A.; Gross, C.A.; Storz, G. MicL, a new σE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 2014, 28, 1620–1634. [Google Scholar] [CrossRef]
- Papenfort, K.; Pfeiffer, V.; Mika, F.; Lucchini, S.; Hinton, J.C.; Vogel, J. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 2006, 62, 1674–1688. [Google Scholar] [CrossRef]
- Udekwu, K.I.; Darfeuille, F.; Vogel, J.; Reimegård, J.; Holmqvist, E.; Wagner, E.G.H. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005, 19, 2355–2366. [Google Scholar] [CrossRef] [PubMed]
- Dartigalongue, C.; Missiakas, D.; Raina, S. Characterization of the Escherichia coli sigma E regulon. J. Biol. Chem. 2001, 276, 20866–20875. [Google Scholar] [CrossRef] [PubMed]
- Johansen, J.; Rasmussen, A.A.; Overgaard, M.; Valentin-Hansen, P. Conserved Small Non-coding RNAs that belong to the σE Regulon: Role in Down-regulation of Outer Membrane Proteins. J. Mol. Biol. 2006, 364, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Guillier, M.; Gottesman, S.; Storz, G. Modulating the outer membrane with small RNAs. Genes Dev. 2006, 20, 2338–2348. [Google Scholar] [CrossRef]
- Klein, G.; Raina, S. Regulated Control of the Assembly and Diversity of LPS by Noncoding sRNAs. BioMed Res. Int. 2015, 2015, 153561. [Google Scholar] [CrossRef]
- Klein, G.; Kobylak, N.; Lindner, B.; Stupak, A.; Raina, S. Assembly of Lipopolysaccharide in Escherichia coli Requires the Essential LapB Heat Shock Protein. J. Biol. Chem. 2014, 289, 14829–14853. [Google Scholar] [CrossRef]
- Klein, G.; Raina, S. Small regulatory bacterial RNAs regulating the envelope stress response. Biochem. Soc. Trans. 2017, 45, 417–425. [Google Scholar] [CrossRef]
- Wassarman, K.M.; Repoila, F.; Rosenow, C.; Storz, G.; Gottesman, S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001, 15, 1637–1651. [Google Scholar] [CrossRef]
- Gopalkrishnan, S.; Nicoloff, H.; Ades, S.E. Co-ordinated regulation of the extracytoplasmic stress factor, sigmaE, with other Escherichia coli sigma factors by (p)ppGpp and DksA may be achieved by specific regulation of individual holoenzymes. Mol. Microbiol. 2014, 93, 479–493. [Google Scholar] [CrossRef]
- Costanzo, A.; Nicoloff, H.; Barchinger, S.E.; Banta, A.B.; Gourse, R.L.; Ades, S.E. ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor σE in Escherichia coli by both direct and indirect mechanisms. Mol. Microbiol. 2007, 67, 619–632. [Google Scholar] [CrossRef]
- Vogel, J.; Papenfort, K. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 2006, 9, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Guillier, M.; Gottesman, S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 2005, 59, 231–247. [Google Scholar] [CrossRef]
- Filenko, N.; Spiro, S.; Browning, D.F.; Squire, D.; Overton, T.W.; Cole, J.; Constantinidou, C. The NsrR Regulon of Escherichia coli K-12 Includes Genes Encoding the Hybrid Cluster Protein and the Periplasmic, Respiratory Nitrite Reductase. J. Bacteriol. 2007, 189, 4410–4417. [Google Scholar] [CrossRef]
- Bodenmiller, D.M.; Spiro, S. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J. Bacteriol. 2006, 188, 874–881. [Google Scholar] [CrossRef]
- Efromovich, S.; Grainger, D.; Bodenmiller, D.; Spiro, S. Genome-wide identification of binding sites for the nitric oxide-sensitive transcriptional regulator NsrR. Methods Enzymol. 2008, 437, 211–233. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Hayden, J.D.; Ades, S.E.; Sandler, S. The Extracytoplasmic Stress Factor, σE, Is Required to Maintain Cell Envelope Integrity in Escherichia coli. PLoS ONE 2008, 3, e1573. [Google Scholar] [CrossRef]
- Partridge, J.D.; Bodenmiller, D.M.; Humphrys, M.S.; Spiro, S. NsrR targets in the Escherichia coli genome: New insights into DNA sequence requirements for binding and a role for NsrR in the regulation of motility. Mol. Microbiol. 2009, 73, 680–694. [Google Scholar] [CrossRef]
- Tucker, N.P.; Hicks, M.G.; Clarke, T.A.; Crack, J.C.; Chandra, G.; Le Brun, N.E.; Dixon, R.; Hutchings, M.I.; Yuan, A. The Transcriptional Repressor Protein NsrR Senses Nitric Oxide Directly via a [2Fe-2S] Cluster. PLoS ONE 2008, 3, e3623. [Google Scholar] [CrossRef]
- Fang, C.; Li, L.; Shen, L.; Shi, J.; Wang, S.; Feng, Y.; Zhang, Y. Structures and mechanism of transcription initiation by bacterial ECF factors. Nucleic Acids Res. 2019, 47, 7094–7104. [Google Scholar] [CrossRef]
- Panȳ-Farrȳ, J.; Jonas, B.; Hardwick, S.W.; Gronau, K.; Lewis, R.J.; Hecker, M.; Engelmann, S. Role of RsbU in Controlling SigB Activity in Staphylococcus aureus following Alkaline Stress. J. Bacteriol. 2009, 191, 2561–2573. [Google Scholar] [CrossRef] [PubMed]
- Hecker, M.; Pané-Farré, J.; Uwe, V. SigB-Dependent General Stress Response in Bacillus subtilis and Related Gram-Positive Bacteria. Annu. Rev. Microbiol. 2007, 61, 215–236. [Google Scholar] [CrossRef] [PubMed]
- Nicoloff, H.; Gopalkrishnan, S.; Ades, S.E.; de Boer, P.A.J. Appropriate Regulation of the σE-Dependent Envelope Stress Response Is Necessary To Maintain Cell Envelope Integrity and Stationary-Phase Survival in Escherichia coli. J. Bacteriol. 2017, 199, e00089-17. [Google Scholar] [CrossRef] [PubMed]
- Thomason, L.C.; Costantino, N.; Court, D.L. E. coli Genome Manipulation by P1 Transduction. Curr. Protoc. Mol. Biol. 2007, 79, 1.17.1–1.17.8. [Google Scholar] [CrossRef]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef]
- Sharan, S.K.; Thomason, L.C.; Kuznetsov, S.G.; Court, D.L. Recombineering: A homologous recombination-based method of genetic engineering. Nat. Protoc. 2009, 4, 206–223. [Google Scholar] [CrossRef]
- Court, D.L.; Swaminathan, S.; Yu, D.; Wilson, H.; Baker, T.; Bubunenko, M.; Sawitzke, J.; Sharan, S.K. Mini-λ: A tractable system for chromosome and BAC engineering. Gene 2003, 315, 63–69. [Google Scholar] [CrossRef]
- Yu, D.; Ellis, H.M.; Lee, E.-C.; Jenkins, N.A.; Copeland, N.G.; Court, D.L. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 2000, 97, 5978–5983. [Google Scholar] [CrossRef]
- Battesti, A.; Bouveret, E. The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 2012, 58, 325–334. [Google Scholar] [CrossRef]
- Battesti, A.; Bouveret, E. Improvement of bacterial two-hybrid vectors for detection of fusion proteins and transfer to pBAD-tandem affinity purification, calmodulin binding peptide, or 6-histidine tag vectors. Proteomics 2008, 8, 4768–4771. [Google Scholar] [CrossRef]
- Miller, J.H. A Short Course in Bacterial Genetics; Cold Spring Harbor Laboratory Press: Plainview, NY, USA, 1992. [Google Scholar]
- Thibodeau, S.A.; Fang, R.; Joung, J.K. High-throughput β-galactosidase assay for bacterial cell-based reporter systems. BioTechniques 2004, 36, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]


| σE Regulon | CpxR Regulon | Envelope Homeostasis |
|---|---|---|
| rpoE 1 | spy 2 | mipA (yeaF) 2,6 |
| rybB 1 | dsbA 2 | yfhB (pgpC) 2 |
| mica 1 | mepH (ydhO) 2 | |
| htrG (ygiM/ecfG) 2,3,4 | ygiH (inner membrane protein) 2 | |
| hcp 2,3,5 | cbrB 2 | |
| smpA 2,3 |
| Strain or Plasmid | Genotype | Source or Reference |
|---|---|---|
| Strains | ||
| MG1655 | Esherichia coli K12 | Thomspon et al. 2007 [15] |
| DJ480 | MG1655 ΔlacIZYA | Thomspon et al. 2007 [15] |
| JN100 | NM580 micL-lacZ WT | NM580 x micL-lacZ PCR gBlock by Electroporation |
| JN101 | NM580 micL-lacZ ΔrseA::kan | JN100 x P1 (ΔrseA::kan) |
| JN102 | NM580 micL-lacZ ΔrseA::kan pBAD24 | JN101 + pBAD24 (TSS Transformation) |
| JN103 | NM580 micL-lacZ ΔrseA::kan pBAD-nsrR | JN102 +pBAD-nsrR (TSS Transformation) |
| KMT243 | DJ480 mini-l::tet ΔrseA::kan | KMT295 (induced) + ΔrseA::kan (PCR w/oligos KT76 and KT77) cassette via electroporation |
| KMT414 | MG1655 ΔlacZYAfrtfrt lacIQ Δara714 ΔParaE::frtfrtPCP18araE | NM534 obtained from Nadim Majdalani as a recipient for frtkanfrt-PLlacO-rpoE |
| KMT422 | DJ480 ΔnsrR::tet | This work, x ΔnsrR::tet PCR |
| KMT456 | E. coli BTH101 pEB354 (T25 linker) KanR | T25 linker vector, EcoR I/Xho I, from Aureilia Battesti, Ph.D. |
| KMT457 | E. coli BTH101 pEB354 (T25-Zip) KanR | Positive control for pEB354 (T25 linker) Aureilia Battesti, Ph.D. |
| KMT458 | E. coli BTH101 pEB355 (T18 linker) AmpR | T18 linker vector, EcoR I/Xho I, from Aureilia Battesti, Ph.D. |
| KMT459 | E. coli BTH101 pEB355 (T18-Zip) AmpR | Positive control for pEB355 (T18 linker) Aureilia Battesti, Ph.D. |
| KMT470 | MG1655 ΔlacZYAfrtfrt lacIQ Δara714 ΔParaE::frtfrtPCP18araE ΔnsrR::tet | KMT414 x P1 (ΔnsrR::tet) |
| KMT471 | MG1655 ΔlacZYAfrtfrt lacIQ Δara714 ΔParaE::frtfrtPCP18araE ΔrpoE::cat | KMT414 x P1 (ΔrpoE::cat) |
| KMT473 | MG1655 ΔlacZYAfrtfrt lacIQ Δara714 ΔParaE::frtfrtPCP18araE ΔnsrR::tet ΔrseA::kan | KMT470 x P1 (ΔrseA::kan) |
| KMT10003 | MC1061 [ΦlrpoH P3::lacZ] rpoE+ with suppressor of rpoE::WCm | CAG41001-lab collection |
| KMT12000 | DJ480 rybB-lacZ long fusion (−69 to +22) | Thomspon et al. 2007 [15] |
| KMT12069 | KMT12000 Δara714 leu::Tn10(tet) | Thomspon et al. 2007 [15] |
| KMT12078 | KMT12069 ΔrseA::kan | Thomspon et al. 2007 [15] |
| KMT12090 | KMT12078 pBAD24 | Thomspon et al. 2007 [15] |
| KMT12091 | KMT12078 pBAD-nsrR | Thomspon et al. 2007 [15] |
| KMT12093 | KMT12000 Δlara714 leu+ | This work |
| KMT12094 | KMT12093 ΔrseA::kan | This work |
| KMT12099 | KMT12093 ΔnsrR::tet | This work |
| KMT12100 | KMT12094 ΔnsrR::tet | This work |
| KMT12121 | DJ480 rybB-lacZ [long fusion] Δara714 leu+ ΔrseA::kan ΔnsrR::tet pBAD24 | 12100 pBAD24 (TSS transformation) |
| KMT12122 | DJ480 rybB-lacZ [long fusion] Δara714 leu+ ΔrseA::kan ΔnsrR::tet pBAD-nsrR | 12100 pBAD-nsrR (TSS transformation) |
| KMT12123 | DJ480 rybB-lacZ [long fusion] Δara714 leu+ ΔrseA::kan ΔnsrR::tet pBAD33 | 12100 pBAD33 (TSS transformation) |
| KMT12124 | DJ480 rybB-lacZ [long fusion] Δara714 leu+ ΔrseA::kan ΔnsrR::tet pBAD33-rseA | 12100 pBAD33-rseA (TSS transformation) |
| KMT12131 | DJ480 rybB-lacZ [long fusion] Δara714 leu+ ΔnsrR::tet ΔrpoE::kan | KMT12130 x ΔrpoE::kan PCR kanR at 37 °C on LB-kan after ara induction |
| KMT12132 | DJ480 rybB-lacZ [long fusion] Δara714 leu+ ΔnsrR::tet ΔrpoE::kan pBAD33 pTrc99A | KMT12131 + pBAD33 + pTrc99A (Electroporation and AmpR, CmR) |
| KMT12133 | DJ480 rybB-lacZ [long fusion] Δara714 leu+ ΔnsrR::tet ΔrpoE::kan pBAD33 + pCL245 (pTrc99A-rpoE) | KMT12131 + pBAD33 + pCL245 (pTrc99A-rpoE) (Electroporation and AmpR, CmR) |
| KMT12134 | DJ480 rybB-lacZ [long fusion] Δara714 leu+ ΔnsrR::tet ΔrpoE::kan pBAD33-nsrR + pCL245 (pTrc99A-rpoE) | KMT12131 + pBAD33-nsrR + pCL245 (pTrc99A-rpoE) (Electroporation and AmpR, CmR) |
| KMT12218 | MG1655 lacIq micA-lacZ | NM580 x micA-lacZ PCR |
| KMT12294 | MG1655 lacIq micA-lacZ pBAD24 | KMT12218 + pBAD24 (TSS Transformation) |
| KMT12295 | MG1655 lacIq micA-lacZ pBAD24-nsrR | KMT12218 + pBAD24-nsrR (TSS Transformation) |
| KMT14000 | DJ480 Δara714 leu+ rpoE270P2-lacZ translational fusion | Thomspon et al. 2007 [15] |
| KMT14001 | KMT14000 ΔrseA::kan | Thomspon et al. 2007 [15] |
| KMT14021 | KMT14000 ΔnsrR::tet | This work |
| KMT14022 | KMT14021 ΔrseA::kan | This work |
| KMT14029 | DJ480 Δara leu+ ΔnsrR::tet ΔrpoE::kan rpoE270P2-lacZ translational fusion | KMT14021 x P1 (DJ480 ΔrpoE::kan) |
| KMT14031 | DJ480 Δara leu+ ΔnsrR::tet ΔrpoE::kan rpoE270P2-lacZ translational fusion pTrc99A pBAD33 | KMT14029 + pTrc99A & pBAD33 (double electroporation and selection on LB-Cm-Amp) |
| KMT14032 | DJ480 Δara leu+ ΔnsrR::tet ΔrpoE::kan rpoE270P2-lacZ translational fusion pCL245 (pTrc99A-rpoE), pBAD33 | KMT14029 + pCL245 (pTrc99A-rpoE) & pBAD33 (double electroporation and selection on LB-Cm-Amp) |
| KMT14033 | DJ480 Δara leu+ ΔnsrR::tet ΔrpoE::kan rpoE270P2-lacZ translational fusion pCL245 (pTrc99A-rpoE), pBAD33-nsrR | KMT14029 + pCL245 (pTrc99A-rpoE) & pBAD33-nsrR (double electroporation and selection on LB-Cm-Amp) |
| NM200 | DJ480 mini-l::cat | Nadim Majdalani, Ph.D. |
| NM300 | DJ480 mini-l::tet | “ ” |
| Plasmid | ||
| pBAD24 | araC+, ColE1 ori, bla, MCS, AmpR | Guzman et al., 1995 [45] |
| pBAD33 | araC+, p15A ori, cat, MCS, CmR | “ ” |
| pBAD24-nsrR | nsrR gene cloned into EcoR I/Pst I site of MCS in pBAD24, AmpR | Thompson et al., 2007 [15] |
| pBAD33-rseA | rseA gene cloned into Sac I/Xba I of MCS in pBAD33, CmR | This work |
| pCL245 | rpoE gene cloned into pTrc99A, AmpR | Rhodius et al. 2006 [14] |
| pEB354 (T25) | Bacterial Adenylate Cylase Two Hybrid Interaction Expression Vector, PLac, LacO site, CAP binding site, p15A ori, Adenylate Cyclase (cya) in T25 domain linker, (KanR) | |
| pEB354-zip (T25-zip) | zip domain cloned into the EcoR I/Xho I site of pEB354, (KanR) | This work |
| pEB354-nsrR (T25-nsrR) | nsrR gene cloned into the EcoR I/Xho I site of pEB354, (KanR) | This work |
| pEB354-zip (T25-rpoE) | rpoE gene domain cloned into the EcoR I/Xho I site of pEB354, (KanR) | This work |
| pEB355 (T18) | Bacterial Adenylate Cylase Two Hybrid Interaction Expression Vector, PLac, LacO site, CAP binding site, ColE1 ori, Adenylate Cyclase (cya) in T18 domain linker, bla (AmpR) | |
| pEB355-zip (T8-zip) | zip domain cloned into the EcoR I/Xho I site of pEB355, bla (AmpR) | This work |
| pEB355-zip (T8-nsrR) | nsrR gene cloned into the EcoR I/Xho I site of pEB355, bla (AmpR) | This work |
| pEB355-zip (T8-rpoE) | rpoE gene cloned into the EcoR I/Xho I site of pEB355, bla (AmpR) | This work |
| pTrc99A | lacIq ColE1 ori, bla MCS, IPTG inducible Ptrc | Thompson et al., 2007 [15] |
| Oligonucleotide ID | Sequence 5′-3′ | Purpose |
|---|---|---|
| KT75 | tttctagattactgcgattgcgttcctaaagtttg | reverse primer for the amplification and cloning of rseA into pBAD33 or pBAD24 Xba I site |
| KT76 | atgcagaaagaacaactttccgctttaatggatggcgaaacgctggatagaaagccacgttgtgtctcaa | ΔrseA::kan forward oligo for the amplification of recombineering substrate |
| KT77 | ttactgcgattgcgttcctaaagtttgaattcctggcacctgtacagcgggcgctgaggtctgcctcgtg | ΔrseA::kan reverse oligo for the amplification of recombineering substrate |
| KT95 | aagtccggagctcaggaggaattcaccatgcagaaagaacaactttccgctttaat | forward primer for the amplification and cloning of rseA into pBAD33 or pTrc99A Sac I site |
| KT237 | ttatcatcaatataaatgtattttttcccgatttcccttttgaggttgatctagacatcattaattccta | forward primer for the amplification of ΔnsrR::tet recombineering substrate |
| KT238 | tcttgtgacatctcggttcctccgttgtcatctctgatgaagattttcggaagctaaatcttctttatcg | reverse primer for the amplification of ΔnsrR::tet recombineering substrate |
| KT906 | gcgtgagctcgatttcccttttgaggttgatgtgcag | forward primer for the amplification and cloning of nsrR into pBAD33 (21 nucleotides upstream of the ATG) Sac I site |
| KT907 | gcgtctgcagtcactccaccagcaataatttataaag | reverse primer for the amplification and cloning of nsrR into pBAD33 Pst I site |
| KT926 | TGGCGTTTCGATAGCGCGTGGAAATTTGGTTTGGGGAGACTTTACCTCGGgcgctgaggtctgcctcgtg | forward primer for ΔrpoE::kan recombineering substrate |
| KT927 | AAGTTGTTCTTTCTGCATGCCTAATACCCTTATCCAGTATCCCGCTATCGaaagccacgttgtgtctcaa | reverse primer for the amplification of ΔrpoE::kan recombineering substrate |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aubee, J.I.; Nurse, J.; Lewis, D.; Tai, C.-H.; Thompson, K.M. NsrR Represses σE-Dependent Small RNAs and Interacts with RpoE via a Noncanonical Mechanism in Escherichia coli. Int. J. Mol. Sci. 2025, 26, 6318. https://doi.org/10.3390/ijms26136318
Aubee JI, Nurse J, Lewis D, Tai C-H, Thompson KM. NsrR Represses σE-Dependent Small RNAs and Interacts with RpoE via a Noncanonical Mechanism in Escherichia coli. International Journal of Molecular Sciences. 2025; 26(13):6318. https://doi.org/10.3390/ijms26136318
Chicago/Turabian StyleAubee, Joseph I., Jalisa Nurse, Dale Lewis, Chin-Hsien Tai, and Karl M. Thompson. 2025. "NsrR Represses σE-Dependent Small RNAs and Interacts with RpoE via a Noncanonical Mechanism in Escherichia coli" International Journal of Molecular Sciences 26, no. 13: 6318. https://doi.org/10.3390/ijms26136318
APA StyleAubee, J. I., Nurse, J., Lewis, D., Tai, C.-H., & Thompson, K. M. (2025). NsrR Represses σE-Dependent Small RNAs and Interacts with RpoE via a Noncanonical Mechanism in Escherichia coli. International Journal of Molecular Sciences, 26(13), 6318. https://doi.org/10.3390/ijms26136318








