Nuclear Fraction Proteome Analyses During rAAV Production of AAV2-Plasmid-Transfected HEK-293 Cells
Abstract
1. Introduction
2. Results
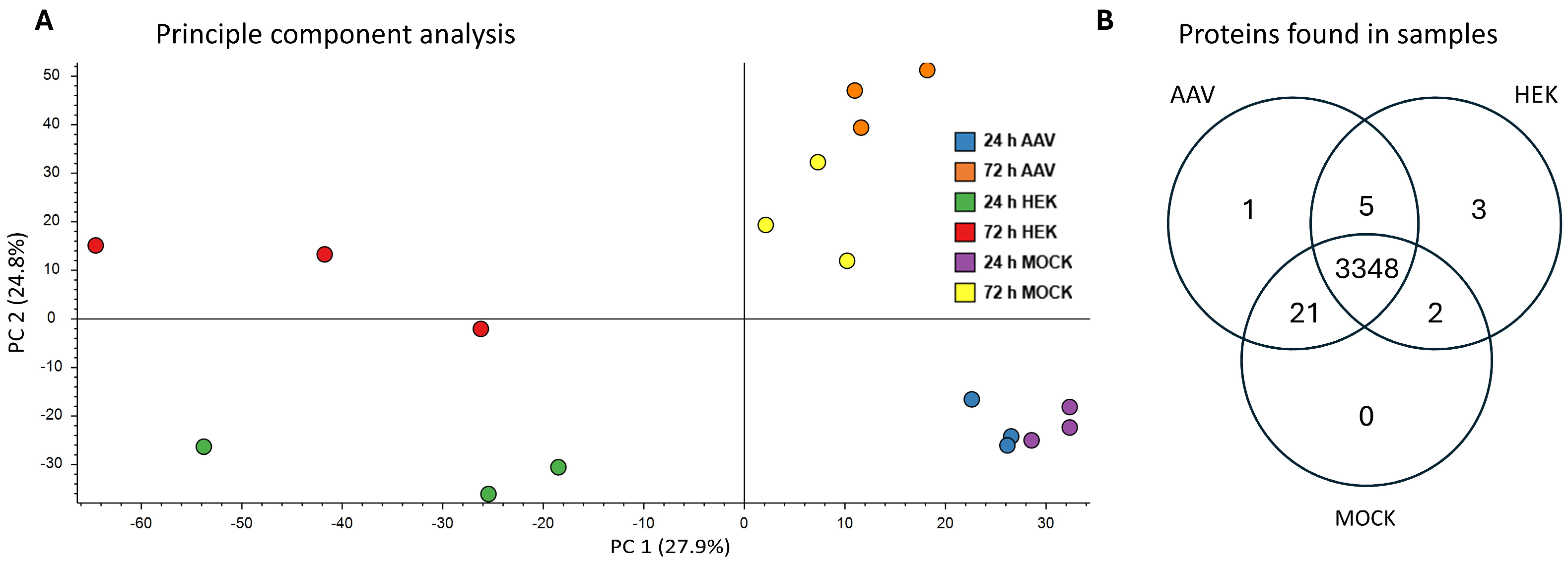
2.1. Protein Identification and Sample Variability
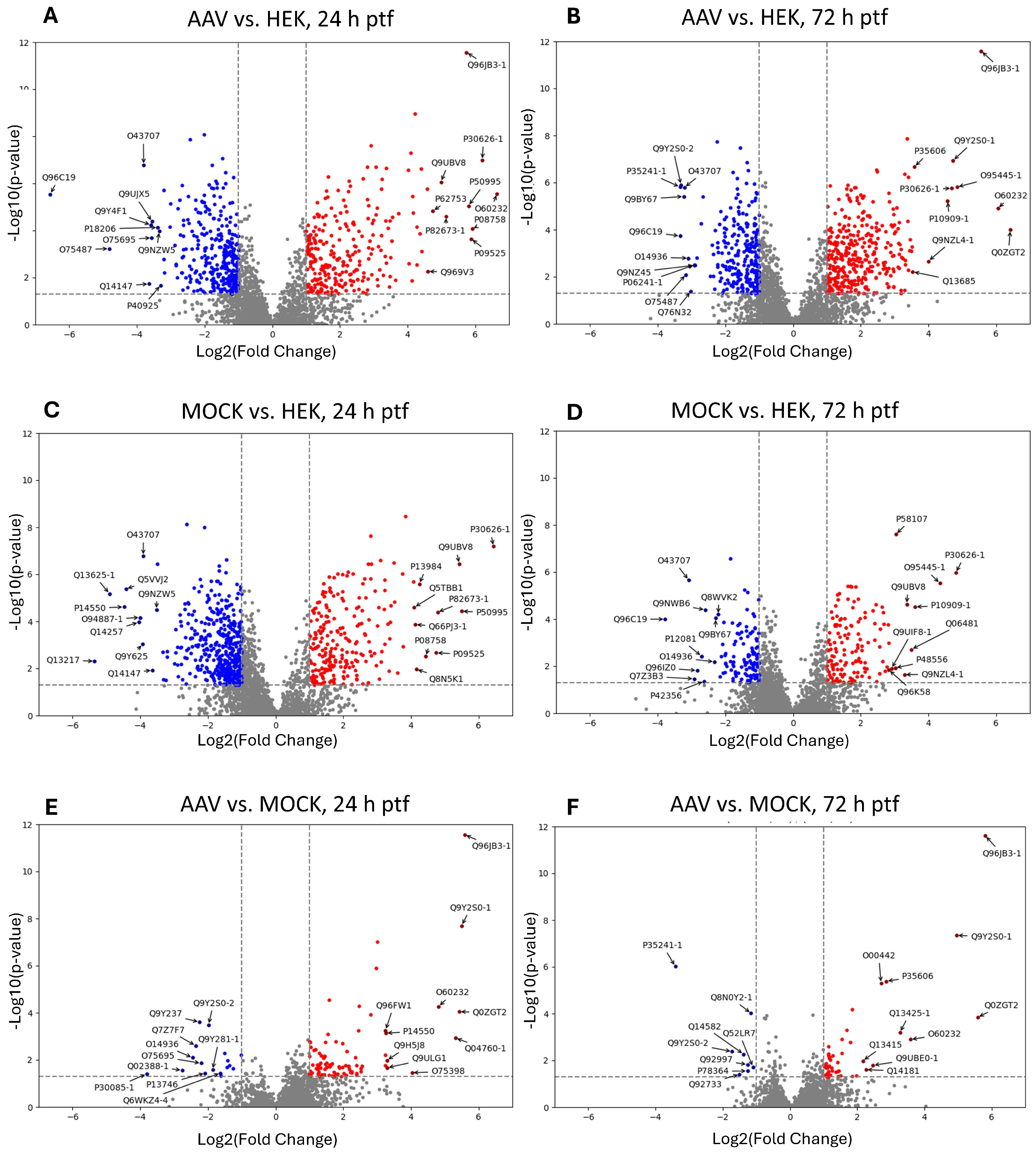
2.2. Identification of Top Regulated HEK-293 Proteins
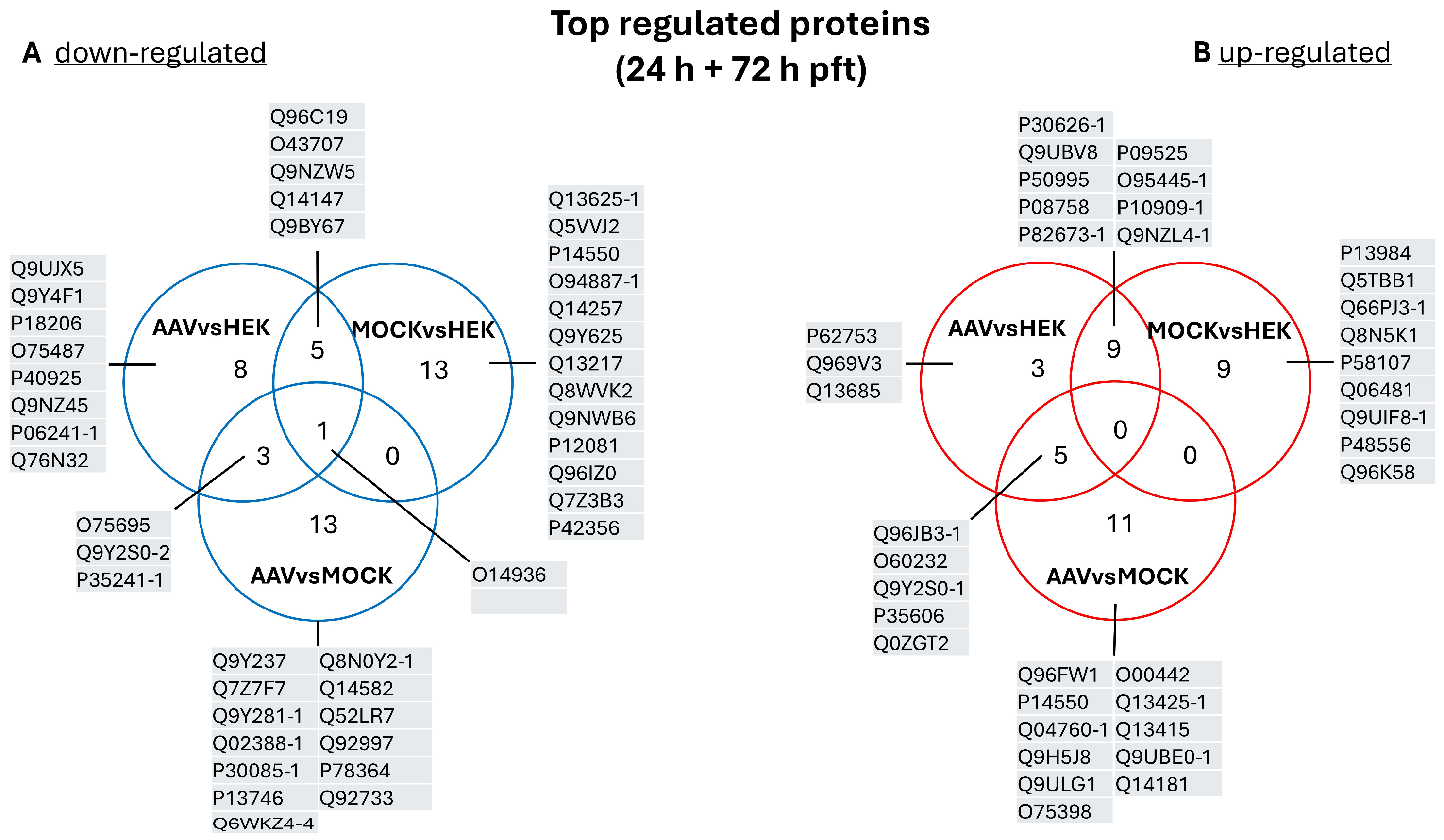
2.3. Comparison of Top Regulated Proteins
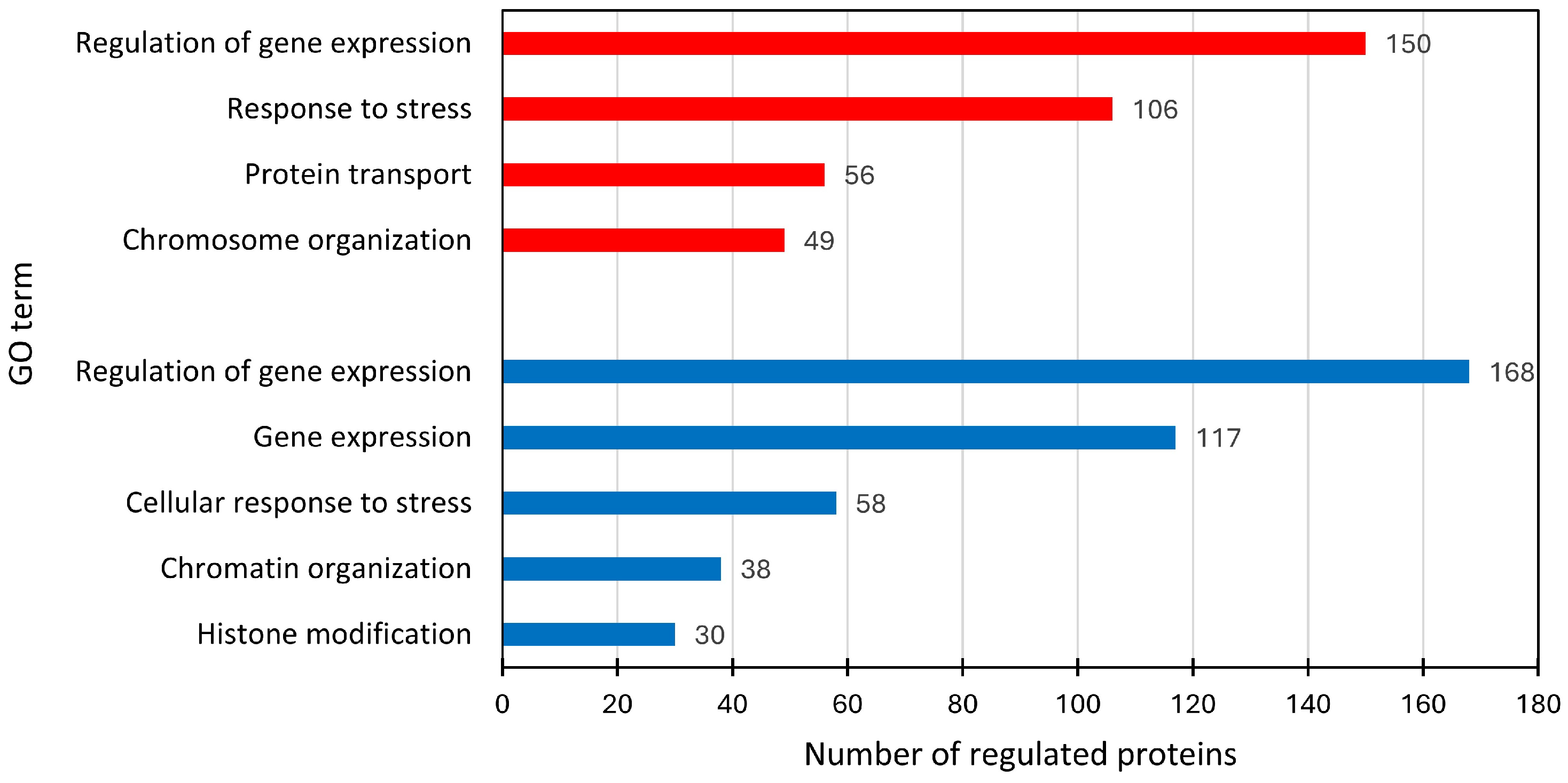
2.4. Comparison of Significantly Regulated Proteins and Respective Functional Groups
2.5. Up-Regulated Proteins in the Context of the GO Term ‘Response to Virus’
2.6. rAAV-Production Specific Response in HEK-293 Cells
2.7. Identified Proteins with Context to Adenoviral Proteins
3. Discussion
3.1. Protein Identification and Sample Variability
3.2. Comparison of Top Regulated Proteins
3.3. Comparison of Significantly Regulated Proteins and Respective Functional Groups
3.4. Up-Regulated Proteins in the Context of the GO Term ‘Response to Virus’
3.4.1. Clusterin (CLUS)
3.4.2. Deoxynucleoside Triphosphate Triphosphohydrolase SAMHD1 (SAMHD1)
3.4.3. Schlafen Family Member 11 (SLFN11)
3.4.4. Histone-Lysine N-Methyltransferase SETD2 (SETD2)
3.4.5. 2′,5′-Phosphodiesterase 12 (PDE12)
3.4.6. ATP-Dependent RNA Helicase DDX3X (DDX3X)
3.4.7. Heat Shock Protein Beta-1 (HSPB1)
3.4.8. Tripartite Motif-Containing Protein 6 (TRIM6)
3.4.9. Zinc Finger CCHC Domain-Containing Protein 3 (ZCCHC3)
3.4.10. Heat Shock Protein HSP 90-Alpha (HSP90AA1)
3.5. rAAV-Production-Specific Response in HEK-293 Cells
3.6. Identified Proteins with Context to Adenoviral Proteins
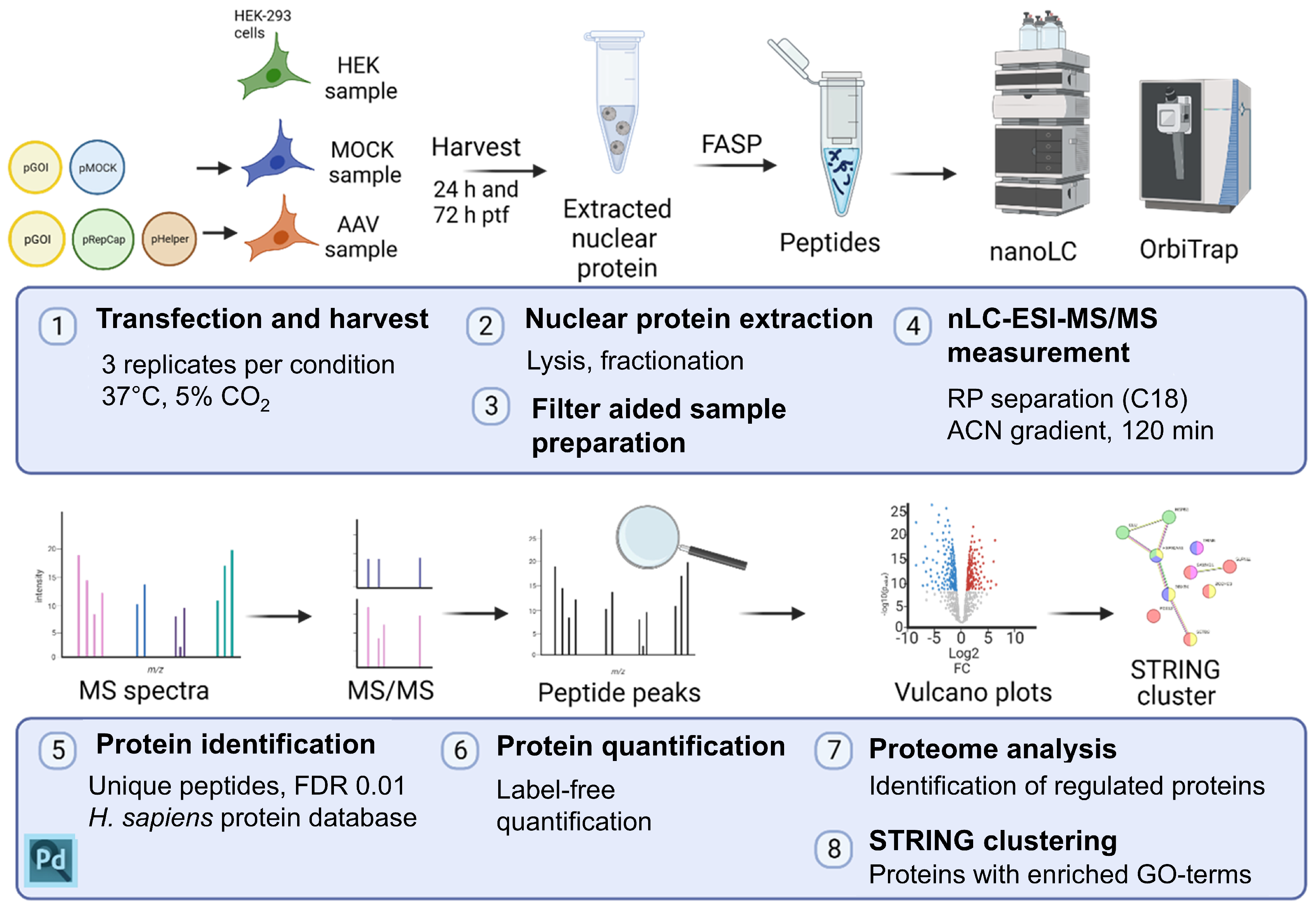
4. Materials and Methods
4.1. Cell Culture of HEK-293 Cells
4.2. Transfection of HEK-293 Cells
4.3. Fluorescence Microscopy
4.4. Protein Fractionation into Nuclear Fraction
4.5. Filter-Aided Sample Preparation (FASP) for Mass Spectrometry
4.6. Nano-Liquid Chromatography–Orbitrap Mass Spectrometry Measurement
4.7. Data Analysis: Protein Identification and Label-Free Quantification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| rAAV | recombinant adeno-associated virus |
| HEK | HEK samples (untransfected) |
| MOCK | MOCK samples (mock-transfected) |
| AAV | AAV samples (triple-transfected with rAAV2 production plasmids) |
| ptf | post-transfection |
| ITR | inverted terminal repeats |
| LFQ | label-free quantification |
| PCA | principle component analysis |
| GO | gene ontology |
References
- Glybera. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/glybera (accessed on 8 July 2024).
- Luxturna. Available online: https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/luxturna (accessed on 11 July 2024).
- Zolgensma. Available online: https://www.fda.gov/vaccines-blood-biologics/zolgensma (accessed on 8 July 2024).
- Hemgenix. Available online: https://www.fda.gov/vaccines-blood-biologics/vaccines/hemgenix (accessed on 9 July 2024).
- Upstaza. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/upstaza (accessed on 15 January 2025).
- Roctavian. Available online: https://www.fda.gov/vaccines-blood-biologics/roctavian (accessed on 8 July 2024).
- Elevidys. Available online: https://www.fda.gov/vaccines-blood-biologics/tissue-tissue-products/elevidys (accessed on 9 July 2024).
- Beqvez. Available online: https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/beqvez (accessed on 11 July 2024).
- Russell, W.C.; Graham, F.L.; Smiley, J.; Nairn, R. Characteristics of a Human Cell Line Transformed by DNA from Human Adenovirus Type 5. J. Gen. Virol. 1977, 36, 59–72. [Google Scholar] [CrossRef]
- Berk, A.J. Recent Lessons in Gene Expression, Cell Cycle Control, and Cell Biology from Adenovirus. Oncogene 2005, 24, 7673–7685. [Google Scholar] [CrossRef] [PubMed]
- Aponte-Ubillus, J.J.; Barajas, D.; Sterling, H.; Aghajanirefah, A.; Bardliving, C.; Peltier, J.; Shamlou, P.; Roy, M.; Gold, D. Proteome Profiling and Vector Yield Optimization in a Recombinant Adeno-associated Virus-producing Yeast Model. Microbiologyopen 2020, 9, e1136. [Google Scholar] [CrossRef] [PubMed]
- Handyside, B.; Ismail, A.M.; Zhang, L.; Yates, B.; Xie, L.; Sihn, C.-R.; Murphy, R.; Bouwman, T.; Kim, C.K.; De Angelis, R.; et al. Vector Genome Loss and Epigenetic Modifications Mediate Decline in Transgene Expression of AAV5 Vectors Produced in Mammalian and Insect Cells. Mol. Ther. 2022, 30, 3570–3586. [Google Scholar] [CrossRef]
- Merten, O.-W. Development of Stable Packaging and Producer Cell Lines for the Production of AAV Vectors. Microorganisms 2024, 12, 384. [Google Scholar] [CrossRef]
- Cao, T.M.; Chen, D.; Barnard, G.C.; Shen, A. Recombinant Adeno-associated Virus Production Evaluation in Chinese Hamster Ovary Cells. Biotechnol. Bioeng. 2024, 121, 395–402. [Google Scholar] [CrossRef]
- Le, D.T.; Radukic, M.T.; Müller, K.M. Adeno-Associated Virus Capsid Protein Expression in Escherichia Coli and Chemically Defined Capsid Assembly. Sci. Rep. 2019, 9, 18631. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Radukic, M.T.; Teschner, K.; Becker, L.; Müller, K.M. Synthesis and Concomitant Assembly of Adeno-Associated Virus-like Particles in Escherichia Coli. ACS Synth. Biol. 2022, 11, 3601–3607. [Google Scholar] [CrossRef]
- Strasser, L.; Boi, S.; Guapo, F.; Donohue, N.; Barron, N.; Rainbow-Fletcher, A.; Bones, J. Proteomic Landscape of Adeno-Associated Virus (AAV)-Producing HEK293 Cells. Int. J. Mol. Sci. 2021, 22, 11499. [Google Scholar] [CrossRef]
- Chung, C.-H.; Murphy, C.M.; Wingate, V.P.; Pavlicek, J.W.; Nakashima, R.; Wei, W.; McCarty, D.; Rabinowitz, J.; Barton, E. Production of rAAV by Plasmid Transfection Induces Antiviral and Inflammatory Responses in Suspension HEK293 Cells. Mol. Ther.—Methods Clin. Dev. 2023, 28, 272–283. [Google Scholar] [CrossRef]
- Wang, Y.; Fu, Q.; Lee, Y.S.; Sha, S.; Yoon, S. Transcriptomic Features Reveal Molecular Signatures Associated with Recombinant Adeno-associated Virus Production in HEK293 Cells. Biotechnol. Prog. 2023, 39, e3346. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Lee, Z.; Hu, W. Multi-omics Kinetic Analysis of Recombinant Adeno-associated Virus Production by Plasmid Transfection of HEK293 Cells. Biotechnol. Prog. 2024, 40, e3428. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.; Lu, M.; Cai, W.; Hu, W.-S. Comparative Transcriptomic and Proteomic Kinetic Analysis of Adeno-Associated Virus Production Systems. Appl. Microbiol. Biotechnol. 2024, 108, 385. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.T.; Tan, E.; Kok, Y.J.; Ng, S.K.; Bi, X. Temporal Insights into Molecular and Cellular Responses during rAAV Production in HEK293T Cells. Mol. Ther.—Methods Clin. Dev. 2024, 32, 101278. [Google Scholar] [CrossRef]
- Wang, Y.; Fu, Q.; Park, S.Y.; Lee, Y.S.; Park, S.-Y.; Lee, D.-Y.; Yoon, S. Decoding Cellular Mechanism of Recombinant Adeno-Associated Virus (rAAV) and Engineering Host-Cell Factories toward Intensified Viral Vector Manufacturing. Biotechnol. Adv. 2024, 71, 108322. [Google Scholar] [CrossRef]
- Gurazada, S.G.R.; Kennedy, H.M.; Braatz, R.D.; Mehrman, S.J.; Polson, S.W.; Rombel, I.T. HEK-Omics: The Promise of Omics to Optimize HEK293 for Recombinant Adeno-Associated Virus (rAAV) Gene Therapy Manufacturing. Biotechnol. Adv. 2025, 79, 108506. [Google Scholar] [CrossRef]
- Wistuba, A.; Kern, A.; Weger, S.; Grimm, D.; Kleinschmidt, J.A. Subcellular Compartmentalization of Adeno-Associated Virus Type 2 Assembly. J. Virol. 1997, 71, 1341–1352. [Google Scholar] [CrossRef]
- Maurer, A.C.; Weitzman, M.D. Adeno-Associated Virus Genome Interactions Important for Vector Production and Transduction. Hum. Gene Ther. 2020, 31, 499–511. [Google Scholar] [CrossRef]
- Golm, S.K.; Hübner, W.; Müller, K.M. Fluorescence Microscopy in Adeno-Associated Virus Research. Viruses 2023, 15, 1174. [Google Scholar] [CrossRef]
- Lv, Y.; Qi, J.; Babon, J.J.; Cao, L.; Fan, G.; Lang, J.; Zhang, J.; Mi, P.; Kobe, B.; Wang, F. The JAK-STAT Pathway: From Structural Biology to Cytokine Engineering. Signal Transduct. Target. Ther. 2024, 9, 221. [Google Scholar] [CrossRef]
- Saxena, N.K.; Vertino, P.M.; Anania, F.A.; Sharma, D. Leptin-Induced Growth Stimulation of Breast Cancer Cells Involves Recruitment of Histone Acetyltransferases and Mediator Complex to CYCLIN D1 Promoter via Activation of Stat3. J. Biol. Chem. 2007, 282, 13316–13325. [Google Scholar] [CrossRef] [PubMed]
- Vo, N.; Goodman, R.H. CREB-Binding Protein and P300 in Transcriptional Regulation. J. Biol. Chem. 2001, 276, 13505–13508. [Google Scholar] [CrossRef] [PubMed]
- Cohen, H.Y.; Lavu, S.; Bitterman, K.J.; Hekking, B.; Imahiyerobo, T.A.; Miller, C.; Frye, R.; Ploegh, H.; Kessler, B.M.; Sinclair, D.A. Acetylation of the C Terminus of Ku70 by CBP and PCAF Controls Bax-Mediated Apoptosis. Mol. Cell 2004, 13, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Sui, H.; Chen, Q.; Imamichi, T. Cytoplasmic-translocated Ku70 Senses Intracellular DNA and Mediates Interferon-lambda1 Induction. Immunology 2021, 163, 323–337. [Google Scholar] [CrossRef]
- Nash, K.; Chen, W.; Salganik, M.; Muzyczka, N. Identification of Cellular Proteins That Interact with the Adeno-Associated Virus Rep Protein. J. Virol. 2009, 83, 454–469. [Google Scholar] [CrossRef]
- Schwartz, R.A.; Carson, C.T.; Schuberth, C.; Weitzman, M.D. Adeno-Associated Virus Replication Induces a DNA Damage Response Coordinated by DNA-Dependent Protein Kinase. J. Virol. 2009, 83, 6269–6278. [Google Scholar] [CrossRef]
- Collaco, R.F.; Bevington, J.M.; Bhrigu, V.; Kalman-Maltese, V.; Trempe, J.P. Adeno-Associated Virus and Adenovirus Coinfection Induces a Cellular DNA Damage and Repair Response via Redundant Phosphatidylinositol 3-like Kinase Pathways. Virology 2009, 392, 24–33. [Google Scholar] [CrossRef]
- Ning, K.; Kuz, C.A.; Cheng, F.; Feng, Z.; Yan, Z.; Qiu, J. Adeno-Associated Virus Monoinfection Induces a DNA Damage Response and DNA Repair That Contributes to Viral DNA Replication. mBio 2023, 14, e03528-22. [Google Scholar] [CrossRef]
- Bresch, A.-M.; Yerich, N.; Wang, R.; Sperry, A.O. The PP1 Regulator PPP1R2 Coordinately Regulates AURKA and PP1 to Control Centrosome Phosphorylation and Maintain Central Spindle Architecture. BMC Mol. Cell Biol. 2020, 21, 84. [Google Scholar] [CrossRef]
- Rebelo, S.; Santos, M.; Martins, F.; Da Cruz E Silva, E.F.; Da Cruz E Silva, O.A.B. Protein Phosphatase 1 Is a Key Player in Nuclear Events. Cell. Signal. 2015, 27, 2589–2598. [Google Scholar] [CrossRef]
- Willems, E.; Dedobbeleer, M.; Digregorio, M.; Lombard, A.; Lumapat, P.N.; Rogister, B. The Functional Diversity of Aurora Kinases: A Comprehensive Review. Cell Div. 2018, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Jian, C.; Xu, J.; Huang, A.Y.; Xi, J.; Hu, K.; Wei, L.; Cheng, H.; Wang, X. Identification of EFHD1 as a Novel Ca2+ Sensor for Mitoflash Activation. Cell Calcium 2016, 59, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Mun, S.A.; Park, J.; Park, K.R.; Lee, Y.; Kang, J.Y.; Park, T.; Jin, M.; Yang, J.; Jun, C.-D.; Eom, S.H. Structural and Biochemical Characterization of EFhd1/Swiprosin-2, an Actin-Binding Protein in Mitochondria. Front. Cell Dev. Biol. 2021, 8, 628222. [Google Scholar] [CrossRef] [PubMed]
- Deltour, S.; Pinte, S.; Guérardel, C.; Leprince, D. Characterization of HRG22, a Human Homologue of the Putative Tumor Suppressor Gene HIC1. Biochem. Biophys. Res. Commun. 2001, 287, 427–434. [Google Scholar] [CrossRef]
- Huang, P.; Peslak, S.A.; Shehu, V.; Keller, C.A.; Giardine, B.; Shi, J.; Hardison, R.C.; Blobel, G.A.; Khandros, E. Let-7 miRNAs Repress HIC2 to Regulate BCL11A Transcription and Hemoglobin Switching. Blood 2024, 143, 1980–1991. [Google Scholar] [CrossRef]
- Lu, P.; Shangguan, W.; Zhao, Q. HIC2 Promotes Cell Cycle Transitions by Upregulating CDK1 Expression in Glioblastoma. Asian J. Surg. 2023, 46, 4536–4538. [Google Scholar] [CrossRef]
- Deltour, S.; Pinte, S.; Guerardel, C.; Wasylyk, B.; Leprince, D. The Human Candidate Tumor Suppressor Gene HIC1 Recruits CtBP through a Degenerate GLDLSKK Motif. Mol. Cell. Biol. 2002, 22, 4890–4901. [Google Scholar] [CrossRef]
- Misiaszek, A.D.; Girbig, M.; Grötsch, H.; Baudin, F.; Murciano, B.; Lafita, A.; Müller, C.W. Cryo-EM Structures of Human RNA Polymerase I. Nat. Struct. Mol. Biol. 2021, 28, 997–1008. [Google Scholar] [CrossRef]
- Zhou, S.; Van Bortle, K. The Pol III Transcriptome: Basic Features, Recurrent Patterns, and Emerging Roles in Cancer. WIREs RNA 2023, 14, e1782. [Google Scholar] [CrossRef]
- Tian, Y.; Sun, F.; Zhong, Y.; Huang, W.; Wang, G.; Liu, C.; Xiao, Y.; Wu, J.; Mu, L. Expression and Clinical Significance of POLR1D in Colorectal Cancer. Oncology 2020, 98, 138–145. [Google Scholar] [CrossRef]
- Zhou, Q.; Perakis, S.O.; Ulz, P.; Mohan, S.; Riedl, J.M.; Talakic, E.; Lax, S.; Tötsch, M.; Hoefler, G.; Bauernhofer, T.; et al. Cell-Free DNA Analysis Reveals POLR1D-Mediated Resistance to Bevacizumab in Colorectal Cancer. Genome Med. 2020, 12, 20. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Hakuno, F.; Northcott, P.; Pessin, J.E.; Adcock, M.R. Nexilin, a Cardiomyopathy-Associated F-Actin Binding Protein, Binds and Regulates IRS1 Signaling in Skeletal Muscle Cells. PLoS ONE 2013, 8, e55634. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Liu, C.; Liao, H.-K.; Zhang, R.; Yuan, B.; Yang, H.; Li, R.; Zhu, S.; Fang, X.; Rodriguez Esteban, C.; et al. In Vivo Rescue of Genetic Dilated Cardiomyopathy by Systemic Delivery of Nexilin. Genome Biol. 2024, 25, 135. [Google Scholar] [CrossRef] [PubMed]
- Nonnenmacher, M.; Weber, T. Adeno-Associated Virus 2 Infection Requires Endocytosis through the CLIC/GEEC Pathway. Cell Host Microbe 2011, 10, 563–576. [Google Scholar] [CrossRef]
- Wang, H.; Li, Z.; Wang, J.; Sun, K.; Cui, Q.; Song, L.; Zou, Y.; Wang, X.; Liu, X.; Hui, R.; et al. Mutations in NEXN, a Z-Disc Gene, Are Associated with Hypertrophic Cardiomyopathy. Am. J. Hum. Genet. 2010, 87, 687–693. [Google Scholar] [CrossRef]
- Kreycy, N.; Gotzian, C.; Fleming, T.; Flechtenmacher, C.; Grabe, N.; Plinkert, P.; Hess, J.; Zaoui, K. Glyoxalase 1 Expression Is Associated with an Unfavorable Prognosis of Oropharyngeal Squamous Cell Carcinoma. BMC Cancer 2017, 17, 382. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, X.; Liu, L.; Cai, D.; Gou, S.; Hao, S.; Li, Y.; Shen, J.; Chen, Y.; Zhao, Y.; et al. GLO1 Regulates Hepatocellular Carcinoma Proliferation and Migration through the Cell Cycle Pathway. BMC Cancer 2024, 24, 1297. [Google Scholar] [CrossRef]
- Maki, M.; Kitaura, Y.; Satoh, H.; Ohkouchi, S.; Shibata, H. Structures, Functions and Molecular Evolution of the Penta-EF-Hand Ca2+-Binding Proteins. Biochim. Biophys. Acta BBA—Proteins Proteom. 2002, 1600, 51–60. [Google Scholar] [CrossRef]
- Kitaura, Y.; Matsumoto, S.; Satoh, H.; Hitomi, K.; Maki, M. Peflin and ALG-2, Members of the Penta-EF-Hand Protein Family, Form a Heterodimer That Dissociates in a Ca2+-Dependent Manner. J. Biol. Chem. 2001, 276, 14053–14058. [Google Scholar] [CrossRef]
- Satoh, H.; Shibata, H.; Nakano, Y.; Kitaura, Y.; Maki, M. ALG-2 Interacts with the Amino-Terminal Domain of Annexin XI in a Ca2+-Dependent Manner. Biochem. Biophys. Res. Commun. 2002, 291, 1166–1172. [Google Scholar] [CrossRef]
- Hayes, M.J.; Rescher, U.; Gerke, V.; Moss, S.E. Annexin–Actin Interactions. Traffic 2004, 5, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Maki, M.; Suzuki, H.; Shibata, H. Structure and Function of ALG-2, a Penta-EF-Hand Calcium-Dependent Adaptor Protein. Sci. China Life Sci. 2011, 54, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Shibata, H.; Kanadome, T.; Sugiura, H.; Yokoyama, T.; Yamamuro, M.; Moss, S.E.; Maki, M. A New Role for Annexin A11 in the Early Secretory Pathway via Stabilizing Sec31A Protein at the Endoplasmic Reticulum Exit Sites (ERES). J. Biol. Chem. 2015, 290, 4981–4993. [Google Scholar] [CrossRef] [PubMed]
- Rayl, M.; Truitt, M.; Held, A.; Sargeant, J.; Thorsen, K.; Hay, J.C. Penta-EF-Hand Protein Peflin Is a Negative Regulator of ER-To-Golgi Transport. PLoS ONE 2016, 11, e0157227. [Google Scholar] [CrossRef]
- Perdreau-Dahl, H.; Progida, C.; Barfeld, S.J.; Guldsten, H.; Thiede, B.; Arntzen, M.; Bakke, O.; Mills, I.G.; Krauss, S.; Morth, J.P. Sjögren Syndrome/Scleroderma Autoantigen 1 Is a Direct Tankyrase Binding Partner in Cancer Cells. Commun. Biol. 2020, 3, 123. [Google Scholar] [CrossRef]
- Ferrer-Acosta, Y.N.; Rodriguez Cruz, E.; Del, C.; Vaquer, A.; E Vega, I. Functional and Structural Analysis of the Conserved EFhd2 Protein. Protein Pept. Lett. 2013, 20, 573–583. [Google Scholar] [CrossRef]
- Lehne, F.; Pokrant, T.; Parbin, S.; Salinas, G.; Großhans, J.; Rust, K.; Faix, J.; Bogdan, S. Calcium Bursts Allow Rapid Reorganization of EFhD2/Swip-1 Cross-Linked Actin Networks in Epithelial Wound Closure. Nat. Commun. 2022, 13, 2492. [Google Scholar] [CrossRef]
- Hornbruch-Freitag, C.; Griemert, B.; Buttgereit, D.; Renkawitz-Pohl, R. Drosophila Swiprosin-1/EFHD2 Accumulates at the Prefusion Complex Stage during Drosophila Myoblast Fusion. J. Cell Sci. 2011, 124, 3266–3278. [Google Scholar] [CrossRef]
- Lehne, F.; Bogdan, S. Swip-1 Promotes Exocytosis of Glue Granules in the Exocrine Drosophila Salivary Gland. J. Cell Sci. 2023, 136, jcs260366. [Google Scholar] [CrossRef]
- Mun, S.A.; Park, J.; Kang, J.Y.; Park, T.; Jin, M.; Yang, J.; Eom, S.H. Structural and Biochemical Insights into Zn2+ -Bound EF-Hand Proteins, EFhd1 and EFhd2. IUCrJ 2023, 10, 233–245. [Google Scholar] [CrossRef]
- Tworig, J.; Grafton, F.; Hörer, M.; Reid, C.A.; Mandegar, M.A. Transcriptomics-Informed Pharmacology Identifies Epigenetic and Cell Cycle Regulators as Enhancers of AAV Production. bioRxiv 2024. [Google Scholar] [CrossRef]
- Kadam, R.; Harish, M.; Dalvi, K.; Teni, T. Novel Nucleolar Localization of Clusterin and Its Associated Functions in Human Oral Cancers: An in Vitro and in Silico Analysis. Cell Biochem. Funct. 2021, 39, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Razin, S.V.; Gavrilov, A.A.; Iarovaia, O.V. Modification of Nuclear Compartments and the 3D Genome in the Course of a Viral Infection. Acta Naturae 2020, 12, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Fraefel, C.; Bittermann, A.G.; Büeler, H.; Heid, I.; Bächi, T.; Ackermann, M. Spatial and Temporal Organization of Adeno-Associated Virus DNA Replication in Live Cells. J. Virol. 2004, 78, 389–398. [Google Scholar] [CrossRef][Green Version]
- Carvalho, T.; Seeler, J.S.; Ohman, K.; Jordan, P.; Pettersson, U.; Akusjärvi, G.; Carmo-Fonseca, M.; Dejean, A. Targeting of Adenovirus E1A and E4-ORF3 Proteins to Nuclear Matrix-Associated PML Bodies. J. Cell Biol. 1995, 131, 45–56. [Google Scholar] [CrossRef]
- Müller, S.; Dejean, A. Viral Immediate-Early Proteins Abrogate the Modification by SUMO-1 of PML and Sp100 Proteins, Correlating with Nuclear Body Disruption. J. Virol. 1999, 73, 5137–5143. [Google Scholar] [CrossRef]
- Zhang, K.; Lv, D.-W.; Li, R. Conserved Herpesvirus Protein Kinases Target SAMHD1 to Facilitate Virus Replication. Cell Rep. 2019, 28, 449–459.e5. [Google Scholar] [CrossRef]
- Chamontin, C.; Bossis, G.; Nisole, S.; Arhel, N.J.; Maarifi, G. Regulation of Viral Restriction by Post-Translational Modifications. Viruses 2021, 13, 2197. [Google Scholar] [CrossRef]
- Valdez, F.; Salvador, J.; Palermo, P.M.; Mohl, J.E.; Hanley, K.A.; Watts, D.; Llano, M. Schlafen 11 Restricts Flavivirus Replication. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Nightingale, K.; Potts, M.; Hunter, L.M.; Fielding, C.A.; Zerbe, C.M.; Fletcher-Etherington, A.; Nobre, L.; Wang, E.C.Y.; Strang, B.L.; Houghton, J.W.; et al. Human Cytomegalovirus Protein RL1 Degrades the Antiviral Factor SLFN11 via Recruitment of the CRL4 E3 Ubiquitin Ligase Complex. Proc. Natl. Acad. Sci. USA 2022, 119, e2108173119. [Google Scholar] [CrossRef]
- Mu, Y.; Lou, J.; Srivastava, M.; Zhao, B.; Feng, X.; Liu, T.; Chen, J.; Huang, J. SLFN 11 Inhibits Checkpoint Maintenance and Homologous Recombination Repair. EMBO Rep. 2016, 17, 94–109. [Google Scholar] [CrossRef] [PubMed]
- Murai, J.; Tang, S.-W.; Leo, E.; Baechler, S.A.; Redon, C.E.; Zhang, H.; Al Abo, M.; Rajapakse, V.N.; Nakamura, E.; Jenkins, L.M.M.; et al. SLFN11 Blocks Stressed Replication Forks Independently of ATR. Mol. Cell 2018, 69, 371–384.e6. [Google Scholar] [CrossRef] [PubMed]
- Murai, J.; Zhang, H.; Pongor, L.; Tang, S.-W.; Jo, U.; Moribe, F.; Ma, Y.; Tomita, M.; Pommier, Y. Chromatin Remodeling and Immediate Early Gene Activation by SLFN11 in Response to Replication Stress. Cell Rep. 2020, 30, 4137–4151.e6. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Kao, E.; Gao, X.; Sandig, H.; Limmer, K.; Pavon-Eternod, M.; Jones, T.E.; Landry, S.; Pan, T.; Weitzman, M.D.; et al. Codon-Usage-Based Inhibition of HIV Protein Synthesis by Human Schlafen 11. Nature 2012, 491, 125–128. [Google Scholar] [CrossRef]
- Chen, K.; Liu, J.; Liu, S.; Xia, M.; Zhang, X.; Han, D.; Jiang, Y.; Wang, C.; Cao, X. Methyltransferase SETD2-Mediated Methylation of STAT1 Is Critical for Interferon Antiviral Activity. Cell 2017, 170, 492–506.e14. [Google Scholar] [CrossRef]
- Zhang, Y.; Shang, L.; Zhang, J.; Liu, Y.; Jin, C.; Zhao, Y.; Lei, X.; Wang, W.; Xiao, X.; Zhang, X.; et al. An Antibody-Based Proximity Labeling Map Reveals Mechanisms of SARS-CoV-2 Inhibition of Antiviral Immunity. Cell Chem. Biol. 2022, 29, 5–18.e6. [Google Scholar] [CrossRef]
- De Almeida, S.F.; Grosso, A.R.; Koch, F.; Fenouil, R.; Carvalho, S.; Andrade, J.; Levezinho, H.; Gut, M.; Eick, D.; Gut, I.; et al. Splicing Enhances Recruitment of Methyltransferase HYPB/Setd2 and Methylation of Histone H3 Lys36. Nat. Struct. Mol. Biol. 2011, 18, 977–983. [Google Scholar] [CrossRef]
- Rorbach, J.; Nicholls, T.J.J.; Minczuk, M. PDE12 Removes Mitochondrial RNA Poly(A) Tails and Controls Translation in Human Mitochondria. Nucleic Acids Res. 2011, 39, 7750–7763. [Google Scholar] [CrossRef]
- Wood, E.R.; Bledsoe, R.; Chai, J.; Daka, P.; Deng, H.; Ding, Y.; Harris-Gurley, S.; Kryn, L.H.; Nartey, E.; Nichols, J.; et al. The Role of Phosphodiesterase 12 (PDE12) as a Negative Regulator of the Innate Immune Response and the Discovery of Antiviral Inhibitors. J. Biol. Chem. 2015, 290, 19681–19696. [Google Scholar] [CrossRef]
- Khadivjam, B.; Stegen, C.; Hogue-Racine, M.-A.; El Bilali, N.; Döhner, K.; Sodeik, B.; Lippé, R. The ATP-Dependent RNA Helicase DDX3X Modulates Herpes Simplex Virus 1 Gene Expression. J. Virol. 2017, 91, e02411-16. [Google Scholar] [CrossRef]
- Atkinson, S.C.; Heaton, S.M.; Audsley, M.D.; Kleifeld, O.; Borg, N.A. TRIM25 and DEAD-Box RNA Helicase DDX3X Cooperate to Regulate RIG-I-Mediated Antiviral Immunity. Int. J. Mol. Sci. 2021, 22, 9094. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Cheng, Y.; Wang, H.; Yan, Y.; Sun, J. Chicken DDX3X Activates IFN-b via the chSTING-chIRF7-IFN-b Signaling Axis. Front. Immunol. 2018, 10, 822. [Google Scholar] [CrossRef] [PubMed]
- Pene, V.; Li, Q.; Sodroski, C.; Hsu, C.-S.; Liang, T.J. Dynamic Interaction of Stress Granules, DDX3X, and IKK-α Mediates Multiple Functions in Hepatitis C Virus Infection. J Virol 2015, 89, 5462–5477. [Google Scholar] [CrossRef] [PubMed]
- Yedavalli, V.S.R.K.; Zhang, N.; Cai, H.; Zhang, P.; Starost, M.F.; Hosmane, R.S.; Jeang, K.-T. Ring Expanded Nucleoside Analogues Inhibit RNA Helicase and Intracellular Human Immunodeficiency Virus Type 1 Replication. J. Med. Chem. 2008, 51, 5043–5051. [Google Scholar] [CrossRef]
- Bruey, J.-M.; Ducasse, C.; Bonniaud, P.; Ravagnan, L.; Susin, S.A.; Diaz-Latoud, C.; Gurbuxani, S.; Arrigo, A.-P.; Kroemer, G.; Solary, E.; et al. Hsp27 Negatively Regulates Cell Death by Interacting with Cytochrome c. Nat. Cell Biol. 2000, 2, 645–652. [Google Scholar] [CrossRef]
- Sakamoto, H.; Mashima, T.; Yamamoto, K.; Tsuruo, T. Modulation of Heat-Shock Protein 27 (Hsp27) Anti-Apoptotic Activity by Methylglyoxal Modification. J. Biol. Chem. 2002, 277, 45770–45775. [Google Scholar] [CrossRef]
- Havasi, A.; Li, Z.; Wang, Z.; Martin, J.L.; Botla, V.; Ruchalski, K.; Schwartz, J.H.; Borkan, S.C. Hsp27 Inhibits Bax Activation and Apoptosis via a Phosphatidylinositol 3-Kinase-Dependent Mechanism. J. Biol. Chem. 2008, 283, 12305–12313. [Google Scholar] [CrossRef]
- Giraldo, M.I.; Hage, A.; Van Tol, S.; Rajsbaum, R. TRIM Proteins in Host Defense and Viral Pathogenesis. Curr. Clin. Microbiol. Rep. 2020, 7, 101–114. [Google Scholar] [CrossRef]
- Bharaj, P.; Atkins, C.; Luthra, P.; Giraldo, M.I.; Dawes, B.E.; Miorin, L.; Johnson, J.R.; Krogan, N.J.; Basler, C.F.; Freiberg, A.N.; et al. The Host E3-Ubiquitin Ligase TRIM6 Ubiquitinates the Ebola Virus VP35 Protein and Promotes Virus Replication. J. Virol. 2017, 91, e00833-17. [Google Scholar] [CrossRef]
- Van Tol, S.; Kalveram, B.; Ilinykh, P.A.; Ronk, A.; Huang, K.; Aguilera-Aguirre, L.; Bharaj, P.; Hage, A.; Atkins, C.; Giraldo, M.I.; et al. Ubiquitination of Ebola Virus VP35 at Lysine 309 Regulates Viral Transcription and Assembly. PLoS Pathog. 2022, 18, e1010532. [Google Scholar] [CrossRef]
- Lian, H.; Wei, J.; Zang, R.; Ye, W.; Yang, Q.; Zhang, X.-N.; Chen, Y.-D.; Fu, Y.-Z.; Hu, M.-M.; Lei, C.-Q.; et al. ZCCHC3 Is a Co-Sensor of cGAS for dsDNA Recognition in Innate Immune Response. Nat. Commun. 2018, 9, 3349. [Google Scholar] [CrossRef] [PubMed]
- Lian, H.; Zang, R.; Wei, J.; Ye, W.; Hu, M.-M.; Chen, Y.-D.; Zhang, X.-N.; Guo, Y.; Lei, C.-Q.; Yang, Q.; et al. The Zinc-Finger Protein ZCCHC3 Binds RNA and Facilitates Viral RNA Sensing and Activation of the RIG-I-like Receptors. Immunity 2018, 49, 438–448.e5. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, Z.; Wang, S.; Tong, G.; Chen, K.; Zhao, Y. Proteomic Analysis Reveals Zinc-Finger CCHC-Type Containing Protein 3 as a Factor Inhibiting Virus Infection by Promoting Innate Signaling. Virus Res. 2022, 319, 198876. [Google Scholar] [CrossRef] [PubMed]
- Hoter, A.; El-Sabban, M.E.; Naim, H.Y. The HSP90 Family: Structure, Regulation, Function, and Implications in Health and Disease. Int. J. Mol. Sci. 2018, 19, 2560. [Google Scholar] [CrossRef]
- Silbermann, L.-M.; Vermeer, B.; Schmid, S.; Tych, K. The Known Unknowns of the Hsp90 Chaperone. eLife 2024, 13, e102666. [Google Scholar] [CrossRef]
- Lubkowska, A.; Pluta, W.; Strońska, A.; Lalko, A. Role of Heat Shock Proteins (HSP70 and HSP90) in Viral Infection. Int. J. Mol. Sci. 2021, 22, 9366. [Google Scholar] [CrossRef]
- Dietmair, S.; Hodson, M.P.; Quek, L.-E.; Timmins, N.E.; Gray, P.; Nielsen, L.K. A Multi-Omics Analysis of Recombinant Protein Production in Hek293 Cells. PLoS ONE 2012, 7, e43394. [Google Scholar] [CrossRef]
- Li, M.; Shu, H.-B. Dephosphorylation of cGAS by PPP6C Impairs Its Substrate Binding Activity and Innate Antiviral Response. Protein Cell 2020, 11, 584–599. [Google Scholar] [CrossRef]
- Ni, G.; Ma, Z.; Wong, J.P.; Zhang, Z.; Cousins, E.; Major, M.B.; Damania, B. PPP6C Negatively Regulates STING-Dependent Innate Immune Responses. mBio 2020, 11, e01728-20. [Google Scholar] [CrossRef]
- Oshiumi, H. Recent Advances and Contradictions in the Study of the Individual Roles of Ubiquitin Ligases That Regulate RIG-I-Like Receptor-Mediated Antiviral Innate Immune Responses. Front. Immunol. 2020, 11, 1296. [Google Scholar] [CrossRef]
- Li, S.; Brignole, C.; Marcellus, R.; Thirlwell, S.; Binda, O.; McQuoid, M.J.; Ashby, D.; Chan, H.; Zhang, Z.; Miron, M.-J.; et al. The Adenovirus E4orf4 Protein Induces G2/M Arrest and Cell Death by Blocking Protein Phosphatase 2A Activity Regulated by the B55 Subunit. J. Virol. 2009, 83, 8340–8352. [Google Scholar] [CrossRef] [PubMed]
- Kleinberger, T.; Shenk, T. Adenovirus E4orf4 Protein Binds to Protein Phosphatase 2A, and the Complex Down Regulates ElA-Enhanced junB Transcription. J. Virol. 1993, 67, 7556–7560. [Google Scholar] [CrossRef] [PubMed]
- Ben-Israel, H.; Sharf, R.; Rechavi, G.; Kleinberger, T. Adenovirus E4orf4 Protein Downregulates MYC Expression through Interaction with the PP2A-B55 Subunit. J. Virol. 2008, 82, 9381–9388. [Google Scholar] [CrossRef] [PubMed]
- Greer, A.E.; Hearing, P.; Ketner, G. The Adenovirus E4 11 k Protein Binds and Relocalizes the Cytoplasmic P-Body Component Ddx6 to Aggresomes. Virology 2011, 417, 161–168. [Google Scholar] [CrossRef]
- Huang, J.-H.; Ku, W.-C.; Chen, Y.-C.; Chang, Y.-L.; Chu, C.-Y. Dual Mechanisms Regulate the Nucleocytoplasmic Localization of Human DDX6. Sci. Rep. 2017, 7, 42853. [Google Scholar] [CrossRef]
- Okumura, F.; Joo-Okumura, A.; Nakatsukasa, K.; Kamura, T. The Role of Cullin 5-Containing Ubiquitin Ligases. Cell Div. 2016, 11, 1. [Google Scholar] [CrossRef]
- Stracker, T.H.; Petrini, J.H.J. The MRE11 Complex: Starting from the Ends. Nat. Rev. Mol. Cell Biol. 2011, 12, 90–103. [Google Scholar] [CrossRef]
- Radukic, M.T.; Le, D.T.; Krassuski, T.; Borchert, P.; Leach, D.R.F.; Müller, K.M. Degradation and Stable Maintenance of AAV Inverted Terminal Repeats in E. coli. bioRxiv 2024. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal Sample Preparation Method for Proteome Analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bandla, C.; Kundu, D.J.; Kamatchinathan, S.; Bai, J.; Hewapathirana, S.; John, N.S.; Prakash, A.; Walzer, M.; Wang, S.; et al. The PRIDE Database at 20 Years: 2025 Update. Nucleic Acids Res. 2025, 53, D543–D553. [Google Scholar] [CrossRef]


| Group | Protein | UniProt | Fold Change | |||||
|---|---|---|---|---|---|---|---|---|
| AAV/HEK | MOCK/HEK | AAV/MOCK | ||||||
| 24 h | 72 h | 24 h | 72 h | 24 h | 72 h | |||
| AAV vs. HEK | Glypican-4 | O75487 | 0.04 * | 0.11 * | 0.13 * | 0.19 | 0.26 | 0.59 |
| Tyrosine-protein kinase Fyn | P06241-1 | 0.11 * | 0.13 * | 0.13 * | 0.29 | 0.84 | 0.46 | |
| Vinculin | P18206 | 0.10 * | 0.39 * | 0.25 * | 0.45 | 0.39 | 0.87 | |
| Malate dehydrogenase, cytoplasmic | P40925 | 0.10 * | 0.27 | 0.19 | 0.43 | 0.54 | 0.61 | |
| Centrosomal protein of 68 kDa | Q76N32 | 3.17 | 0.12 * | 0.53 | 0.28 | 5.96 | 0.44 | |
| CDGSH iron-sulfur domain-containing protein 1 | Q9NZ45 | 0.69 | 0.13 * | 0.71 | 0.22 | 0.97 | 0.59 | |
| Anaphase-promoting complex subunit 4 | Q9UJX5 | 0.09 * | 0.58 | 0.14 * | 0.41 | 0.6 | 1.44 | |
| FERM, RhoGEF, and pleckstrin domain-containing protein 1 | Q9Y4F1 | 0.08 * | 0.39 | 0.22 * | 0.39 | 0.38 | 1.01 | |
| MOCK vs. HEK | FERM, RhoGEF, and pleckstrin domain-containing protein 2 | O94887-1 | 0.21 | 0.42 | 0.06 * | 0.24 * | 3.43 | 1.78 |
| Histidine-tRNA ligase | P12081 | 0.43 | 0.35 | 0.42 | 0.15 * | 1.01 | 2.26 | |
| Alcohol dehydrogenase [NADP(+)] | P14550 | 0.43 | 1.41 | 0.05 * | 1.07 | 9.56 | 1.32 | |
| Phosphatidylinositol 4-kinase alpha | P42356 | 1.3 | 0.57 | 0.29 | 0.16 * | 4.58 | 3.54 | |
| DnaJ homolog subfamily C member 3 | Q13217 | 0.17 | 1.75 | 0.03 * | 0.96 | 6.74 | 1.83 | |
| Apoptosis-stimulating of p53 protein 2 | Q13625-1 | 0.17 * | 0.36 | 0.03 * | 0.53 | 5.09 | 0.68 | |
| Reticulocalbin-2 | Q14257 | 0.16 * | 0.88 | 0.06 * | 0.25 * | 2.6 | 3.54 | |
| Histone H2A deubiquitinase MYSM1 | Q5VVJ2 | 0.06 | 0.26 | 0.05 * | 0.43 | 1.29 | 0.61 | |
| KAT8 regulatory NSL complex subunit 1 | Q7Z3B3 | 0.24 | 0.27 | 0.14 * | 0.13 * | 1.77 | 2.04 | |
| U4/U6.U5 small nuclear ribonucleoprotein 27 kDa protein | Q8WVK2 | 0.7 | 0.15 * | 0.43 * | 0.22 * | 1.63 | 0.7 | |
| PRKC apoptosis WT1 regulator protein | Q96IZ0 | 1.34 | 0.38 | 0.86 | 0.14 * | 1.55 | 2.7 | |
| Arginine- and glutamate-rich protein 1 | Q9NWB6 | 0.54 | 0.24 * | 0.50 | 0.17 * | 1.07 | 1.45 | |
| Glypican-6 | Q9Y625 | 0.14 * | 0.4 | 0.07 * | 0.57 | 2.11 | 0.7 | |
| AAV vs. MOCK | HLA class I histocompatibility antigen, A-11 alpha chain | P13746 | 0.35 | 1.25 | 1.46 | 1.46 | 0.24 * | 0.85 |
| UMP-CMP kinase | P30085-1 | 0.29 | 1.49 | 4.06 | 0.67 | 0.07 * | 2.24 | |
| Polyhomeotic-like protein 1 | P78364 | 0.30 * | 0.26 * | 0.57 | 0.63 | 0.52 | 0.42 * | |
| Collagen alpha-1(VII) chain | Q02388-1 | 0.31 | 0.9 | 2.12 | 1.38 | 0.15 * | 0.65 | |
| Max dimerization protein 4 | Q14582 | 0.12 * | 0.25 * | 0.35 * | 0.65 | 0.35 * | 0.38 * | |
| Enhancer of polycomb homolog 2 | Q52LR7 | 0.29 * | 0.22 * | 0.34 * | 0.47 | 0.85 | 0.47 * | |
| Rab11 family-interacting protein 1 | Q6WKZ4-4 | 0.36 | 1.94 | 1.13 | 1.94 | 0.32 * | 1.00 | |
| 39S ribosomal protein L55, mitochondrial | Q7Z7F7 | 2.03 | 2.69 | 10.41 * | 1.77 | 0.20 * | 1.52 | |
| Zinc finger protein 444 | Q8N0Y2-1 | 0.76 | 0.45 * | 1.23 | 1 | 0.62 | 0.45 * | |
| Proline-rich protein PRCC | Q92733 | 0.25 * | 0.19 * | 0.27 * | 0.54 | 0.93 | 0.35 * | |
| segment polarity protein disheveled homolog DVL-3 | Q92997 | 2.61 | 0.69 | 4.32 * | 1.66 | 0.60 | 0.42 * | |
| Peptidyl-prolyl cis-trans isomerase NIMA-interacting 4 | Q9Y237 | 0.15 * | 0.82 | 0.72 | 0.8 | 0.21 * | 1.02 | |
| Cofilin-2 | Q9Y281-1 | 0.67 | 0.81 | 2.44 | 1.21 | 0.28 * | 0.67 | |
| AAV vs. HEK and MOCK vs. HEK | Alpha-actinin-4 | O43707 | 0.07 * | 0.11 * | 0.07 * | 0.12 * | 1.08 | 0.92 |
| Probable ATP-dependent RNA helicase DHX34 | Q14147 | 0.08 * | 0.07 | 0.08 * | 0.13 | 1 | 0.52 | |
| EF-hand domain-containing protein D2 | Q96C19 | 0.01 * | 0.10 * | 0.01 | 0.07 * | 1.19 | 1.35 | |
| Cell adhesion molecule 1 | Q9BY67 | 0.14 * | 0.11 * | 0.16 * | 0.21 * | 0.92 | 0.52 | |
| MAGUK p55 subfamily member 6 | Q9NZW5 | 0.10 * | 0.21 * | 0.09 * | 0.25 * | 1.13 | 0.86 | |
| AAV vs. HEK and AAV vs. MOCK | Protein XRP2 | O75695 | 0.09 * | 0.20 * | 0.39 | 0.43 | 0.22 * | 0.47 |
| Radixin | P35241-1 | 1.13 | 0.10 * | 1.13 | 1.05 | 1.00 | 0.09 * | |
| Isoform 2 of DNA-directed RNA polymerases I and III subunit RPAC2 | Q9Y2S0-2 P0DPB5 | 0.25 * | 0.10 * | 0.98 | 0.33 * | 0.25 * | 0.30 * | |
| AAV vs. MOCK and MOCK vs. HEK and AAV vs. HEK | Peripheral plasma membrane protein CASK | O14936 | 0.14 * | 0.12 * | 0.76 | 0.20 * | 0.18 * | 0.59 |
| Group | Protein | UniProt | Fold Change | |||||
|---|---|---|---|---|---|---|---|---|
| AAV/HEK | MOCK/HEK | AAV/MOCK | ||||||
| 24 h | 72 h | 24 h | 72 h | 24 h | 72 h | |||
| AAV vs. HEK | 40S ribosomal protein S6 | P62753 | 26.9 * | 4.1 * | 5.98 * | 3.44 | 4.5 | 1.18 |
| Angio-associated migratory cell protein | Q13685 | 1.5 | 11.6 * | 0.91 | 4.31 | 1.69 | 2.7 | |
| Nicalin, BOS complex subunit NCLN | Q969V3 | 24.2 * | 1.8 | 6.48 | 1.66 | 3.73 | 1.11 | |
| MOCK vs. HEK | General transcription factor IIF subunit 2 | P13984 | 23.93 * | 8.74 * | 19.1 * | 6.5 * | 1.26 | 1.35 |
| 26S proteasome non-ATPase regulatory subunit 8 | P48556 | 1.11 | 11.46 * | 1.1 | 8.1 * | 0.98 | 1.41 | |
| Epiplakin | P58107 | 18.55 * | 10.31 * | 14.4 * | 8.2 * | 1.29 | 1.26 | |
| Amyloid-like protein 2 | Q06481 | 2.29 | 8.78 * | 5.2 * | 11.3 * | 0.44 | 0.78 | |
| Ribonuclease h2 subunit b | Q5TBB1 | 17.89 * | 10.44 * | 17.2 * | 7.0 * | 1.04 | 1.5 | |
| ADP-ribosylation factor-like protein 6-interacting protein 4 | Q66PJ3-1 | 19.43 * | 1.14 | 17.5 * | 1.2 | 1.11 | 0.94 | |
| CDGSH iron-sulfur domain-containing protein 2 | Q8N5K1 | 17.4 | 1.19 | 17.8 * | 0.7 | 0.97 | 1.63 | |
| Zinc finger protein 668 | Q96K58 | 1.3 | 4.21 | 1.8 | 7.1 * | 0.71 | 0.59 | |
| Bromodomain adjacent to zinc finger domain protein 2B | Q9UIF8-1 | 12.49 | 10.62 * | 12.2 * | 7.5 * | 1.03 | 1.42 | |
| AAV vs. MOCK | RNA 3′-terminal phosphate cyclase | O00442 | 0.01 | 2.84 * | 0.26 | 0.43 * | 0.01 | 6.5 * |
| Deformed epidermal autoregulatory factor 1 homolog | O75398 | 3.46 | 0.64 | 0.21 | 0.57 | 16.3 * | 1.1 | |
| Alcohol dehydrogenase [NADP(+)] | P14550 | 0.43 | 1.41 | 0.04 * | 1.07 | 9.6 * | 1.3 | |
| Lactoylglutathione lyase | Q04760-1 | 4.09 | 1.02 | 0.1 | 0.58 | 40.0 * | 1.8 | |
| Origin recognition complex subunit 1 | Q13415 | 9.73 | 1.52 | 2.33 | 0.34 | 4.2 | 4.5 * | |
| Beta-2-syntrophin | Q13425-1 | 0.35 | 3.86 | 0.51 | 0.4 | 0.7 | 9.6 * | |
| DNA polymerase alpha subunit B | Q14181 | 0.31 | 3.05 | 0.01 | 0.64 | ≥100 | 4.8 * | |
| Ubiquitin thioesterase otub1 | Q96FW1 | 3.9 | 2.13 | 0.41 | 0.66 | 9.5 * | 3.2 | |
| TATA box-binding protein-associated factor RNA polymerase I subunit D | Q9H5J8 | 19.23 * | 0.78 | 1.97 | 0.74 | 9.8 * | 1.0 | |
| SUMO-activating enzyme subunit 1 | Q9UBE0-1 | 1.07 | 2.78 | 0.34 | 0.5 | 3.2 | 5.5 * | |
| DNA helicase ino80 | Q9ULG1 | 4.21 | 1.38 | 0.43 | 0.96 | 9.9 * | 1.4 | |
| AAV vs. HEK and MOCK vs. HEK | Apolipoprotein M | O95445-1 | 4.2 * | 28.8 * | 4.2 * | 20.3 * | 1.02 | 1.42 |
| Annexin A5 | P08758 | 60.0 * | 4.4 | 21.6 * | 6.4 | 2.77 | 0.68 | |
| Annexin A4 | P09525 | 59.1 * | 4.8 | 26.8 * | 3.6 | 2.2 | 1.33 | |
| Clusterin | P10909-1 | ≥100 | 23.7 * | ≥100 | 12.2 * | 1.78 | 1.95 | |
| Sorcin | P30626-1 | 74.1 * | 25.9 * | 86.7 * | 28.1 * | 0.85 | 0.92 | |
| Annexin A11 | P50995 | 55.9 * | 6.7 | 45.2 * | 6.2 | 1.23 | 1.08 | |
| 28S ribosomal protein S35, mitochondrial | P82673-1 | 35.4 * | 3.3 | 27.7 * | 2.9 | 1.27 | 1.14 | |
| Hsp70-binding protein 1 | Q9NZL4-1 | 1.7 | 16.0 * | 1.9 | 9.8 * | 0.93 | 1.63 | |
| Peflin | Q9UBV8 | 31.8 * | 7.5 * | 43.2 * | 10.3 * | 0.73 | 0.72 | |
| AAV vs. HEK and AAV vs. MOCK | ZNRD2 | O60232 | ≥100 * | 66.7 * | 3.54 | 5.54 | 28.3 * | 12.0 * |
| Coatomer subunit beta | P35606 | 17.1 * | 12.0 * | 2.19 | 1.64 | 7.8 * | 7.3 * | |
| Nexilin | Q0ZGT2 | 21.3 * | 85.8 * | 0.49 | 1.77 | 43.2 * | 48.3 * | |
| Hypermethylated in cancer 2 protein | Q96JB3-1 | 53.3 * | 47.2 * | 1.11 | 0.84 | 48.1 * | 55.9 * | |
| DNA-directed RNA polymerases I and III subunit RPAC2 | Q9Y2S0-1 P0DPB6 | 21.1 * | 26.5 * | 0.467 | 0.851 | 45.2 * | 31.1 * | |
| UniProt Accession | Description | Protein | Gene Symbol | Fold Change AAV vs. HEK | Fold Change MOCK vs. HEK | Fold Change AAV vs. MOCK | |||
|---|---|---|---|---|---|---|---|---|---|
| 24 h | 72 h | 24 h | 72 h | 24 h | 72 h | ||||
| P10909-1 | Clusterin | CLUS | CLU | 4.9 * | 20.9 ** | 4.2 | 9.2 ** | 1.2 | 2.3 |
| Q9Y3Z3 | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1 | SAMH1 | SAMHD1 | 4.2 * | 10.8 ** | 1.0 | 4.0 * | 4.2 * | 2.7 |
| Q7Z7L1 | Schlafen family member 11 | SLN11 | SLFN11 | 8.1 * | 6.2 ** | 4.3 * | 2.5 | 1.9 | 2.5 |
| Q9BYW2 | Histone-lysine N-methyltransferase SETD2 | SETD2 | SETD2 | 1.4 | 4.3 ** | 0.6 | 1.5 | 2.2 | 2.9 |
| Q6L8Q7-1 | 2′,5′-phosphodiesterase 12 | PDE12 | PDE12 | n/a *** | 3.2 * | n/a | 1.3 | n/a | 2.4 * |
| O00571 | ATP-dependent RNA helicase DDX3X | DDX3X | DDX3X | 2.7 ** | 2.6 ** | 2.0 ** | 2.2 ** | 1.3 | 1.2 |
| P04792 | Heat shock protein beta-1 | HSPB1 | HSPB1 | 0.9 | 2.5 * | 1.9 | 9.9 * | 0.9 | 1.2 |
| Q9NUD5 | Zinc finger CCHC domain-containing protein 3 | ZCHC3 | ZCCHC3 | 4.8 ** | 2.3 * | 5.9 ** | 1.8 | 0.8 | 1.2 |
| Q9C030 | Tripartite motif-containing protein 6 | TRIM6 | TRIM6 | 0.7 | 2.1 * | 0.8 | 1.8 | 0.9 | 1.2 |
| P07900 | Heat shock protein HSP 90-alpha | HSP90AA1 | HSP90AA1 | 0.6 | 2.0 ** | 0.6 | 1.2 | 1.1 | 1.7 * |
| UniProt Accession | Description | Protein | Gene Symbol | Fold Change AAV vs. HEK | Fold Change MOCK vs. HEK | Fold Change AAV vs. MOCK | |||
|---|---|---|---|---|---|---|---|---|---|
| 24 h | 72 h | 24 h | 72 h | 24 h | 72 h | ||||
| O00743 | Serine/threonine-protein phosphatase 6 catalytic subunit | PPP6C | PPP6C | 1.1 | 2.8 * | 0.8 | 1.1 | 1.3 | 2.6 |
| Q9NUD5 | Zinc finger CCHC domain-containing protein 3 | ZCHC3 | ZCCHC3 | 4.8 ** | 2.3 * | 5.9 ** | 1.8 | 0.8 | 1.2 |
| UniProt Accession | Description | Protein | Gene Symbol | Fold Change AAV vs. HEK | Fold Change MOCK vs. HEK | Fold Change AAV vs. MOCK | |||
|---|---|---|---|---|---|---|---|---|---|
| 24 h | 72 h | 24 h | 72 h | 24 h | 72 h | ||||
| P63151 | Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform | 2ABA | PPP2R2A | 1.3 | 2.6 * | 0.6 | 1.3 | 2.2 * | 2 |
| P30154 | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform | 2AAB | PPP2R1B | 0.7 | 0.6 | 0.3 ** | 0.6 | 2.3 * | 0.9 |
| P01106 | Myc proto-oncogene protein | MYC | MYC | 0.6 | 0.7 | 0.7 | 0.9 | 0.9 | 0.8 |
| P26196 | Probable ATP-dependent RNA helicase DDX6 | DDX6 | DDX6 | 2.1 ** | 1.9 ** | 1.6 * | 1.5 | 1.3 | 1.3 |
| P04637 | Cellular tumor antigen p53 | P53 | TP53 | 0.8 | 0.6 ** | 0.9 | 0.8 | 0.9 | 0.7 |
| Q92878 | DNA repair protein Rad50 | RAD50 | RAD50 | 0.4 * | 0.2 ** | 0.2 | 0.6 | 1.7 | 0.3 * |
| Q93034 | Cullin-5 | CUL5 | CUL5 | 2.7 * | 5.4 ** | 1.6 | 2 | 1.7 | 2.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Golm, S.K.; Hoffrogge, R.; Müller, K.M. Nuclear Fraction Proteome Analyses During rAAV Production of AAV2-Plasmid-Transfected HEK-293 Cells. Int. J. Mol. Sci. 2025, 26, 6315. https://doi.org/10.3390/ijms26136315
Golm SK, Hoffrogge R, Müller KM. Nuclear Fraction Proteome Analyses During rAAV Production of AAV2-Plasmid-Transfected HEK-293 Cells. International Journal of Molecular Sciences. 2025; 26(13):6315. https://doi.org/10.3390/ijms26136315
Chicago/Turabian StyleGolm, Susanne K., Raimund Hoffrogge, and Kristian M. Müller. 2025. "Nuclear Fraction Proteome Analyses During rAAV Production of AAV2-Plasmid-Transfected HEK-293 Cells" International Journal of Molecular Sciences 26, no. 13: 6315. https://doi.org/10.3390/ijms26136315
APA StyleGolm, S. K., Hoffrogge, R., & Müller, K. M. (2025). Nuclear Fraction Proteome Analyses During rAAV Production of AAV2-Plasmid-Transfected HEK-293 Cells. International Journal of Molecular Sciences, 26(13), 6315. https://doi.org/10.3390/ijms26136315






