Genome-Wide Identification of SRS Gene Family in Wheat and Expression Analysis Under Abiotic Stress
Abstract
1. Introduction
2. Results
2.1. Identification and Physicochemical Properties of Wheat SRS Genes
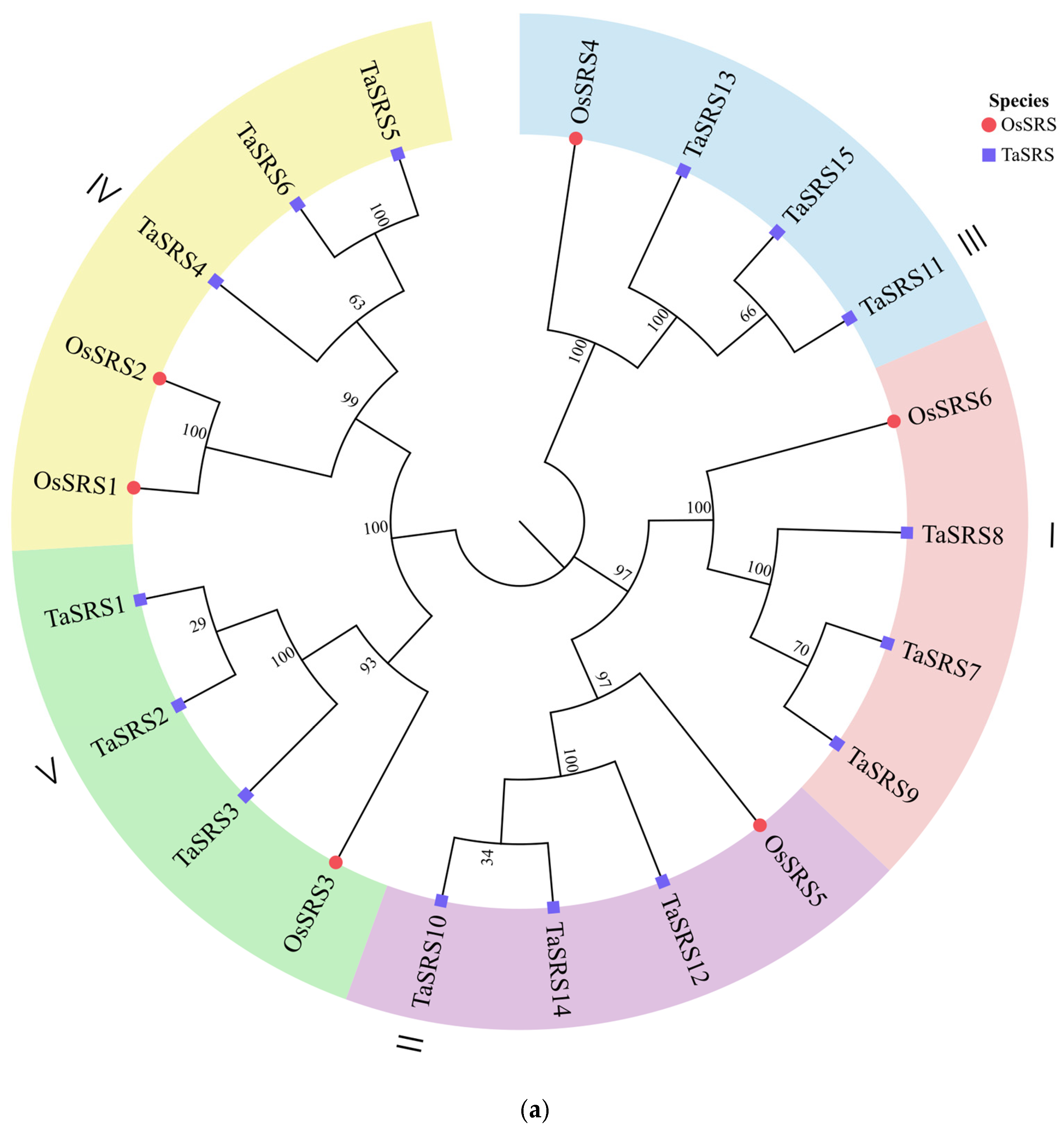
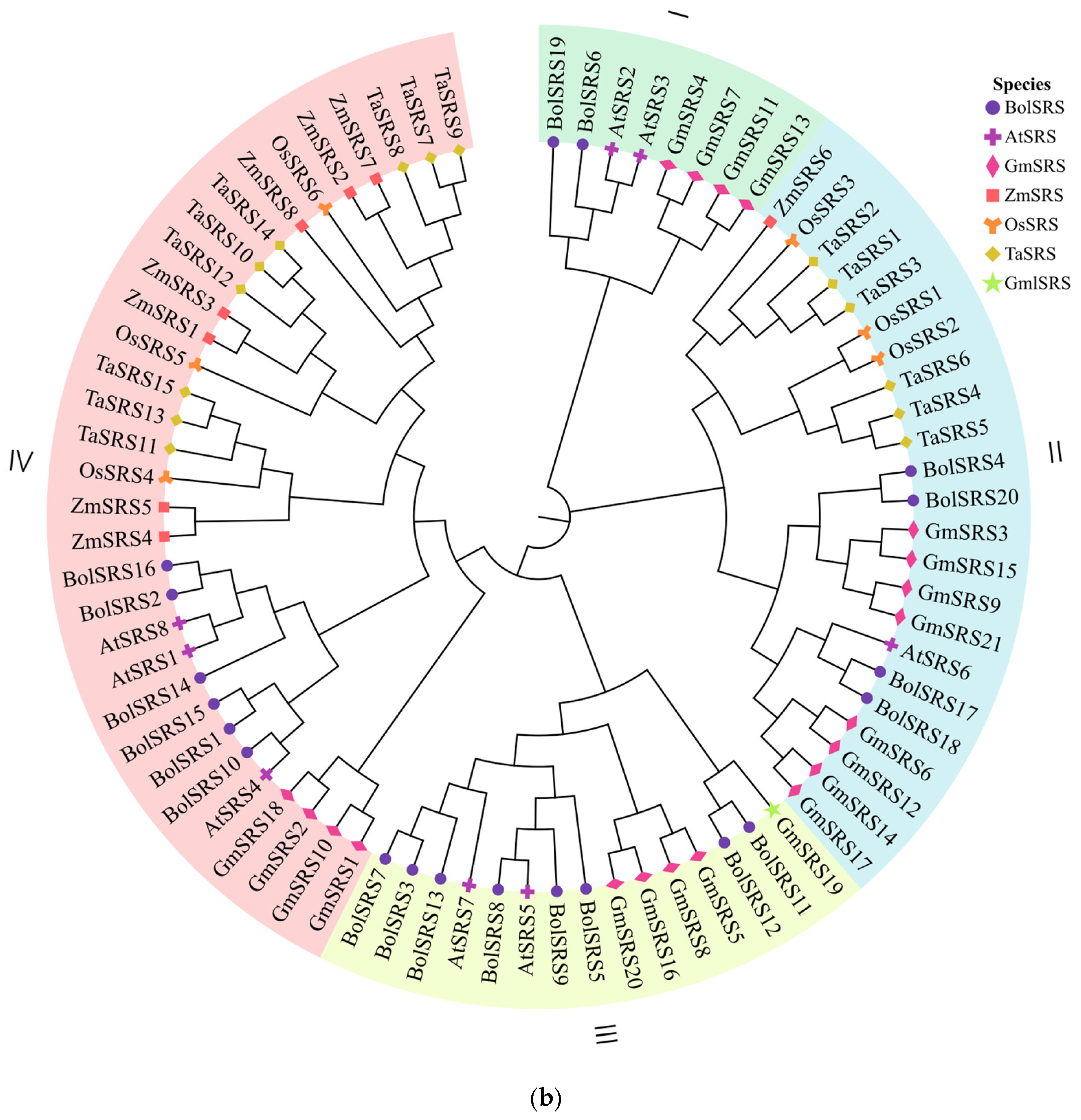
2.2. Analysis of Conserved Motif and Phylogenetic of Wheat SRS Gene Family
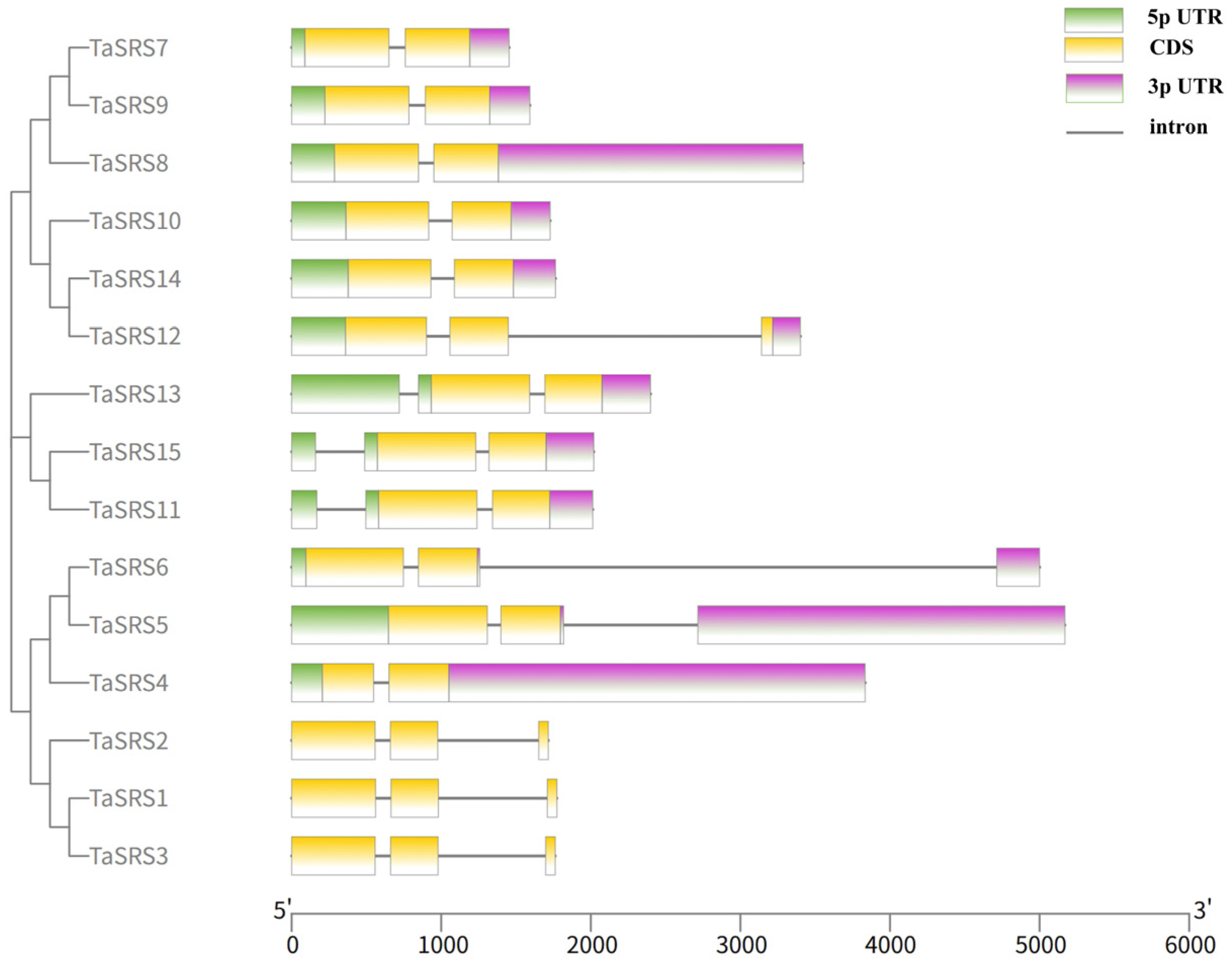
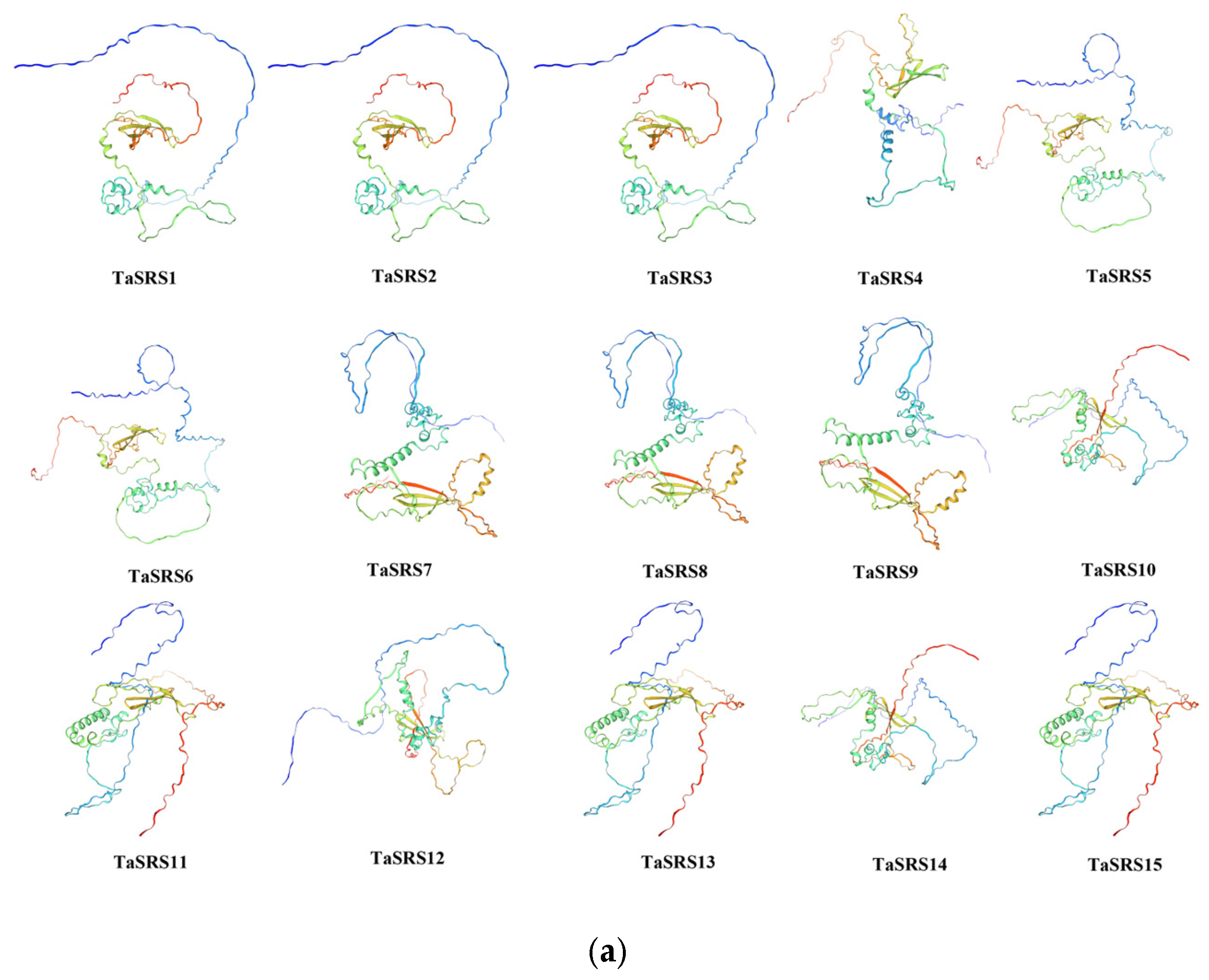
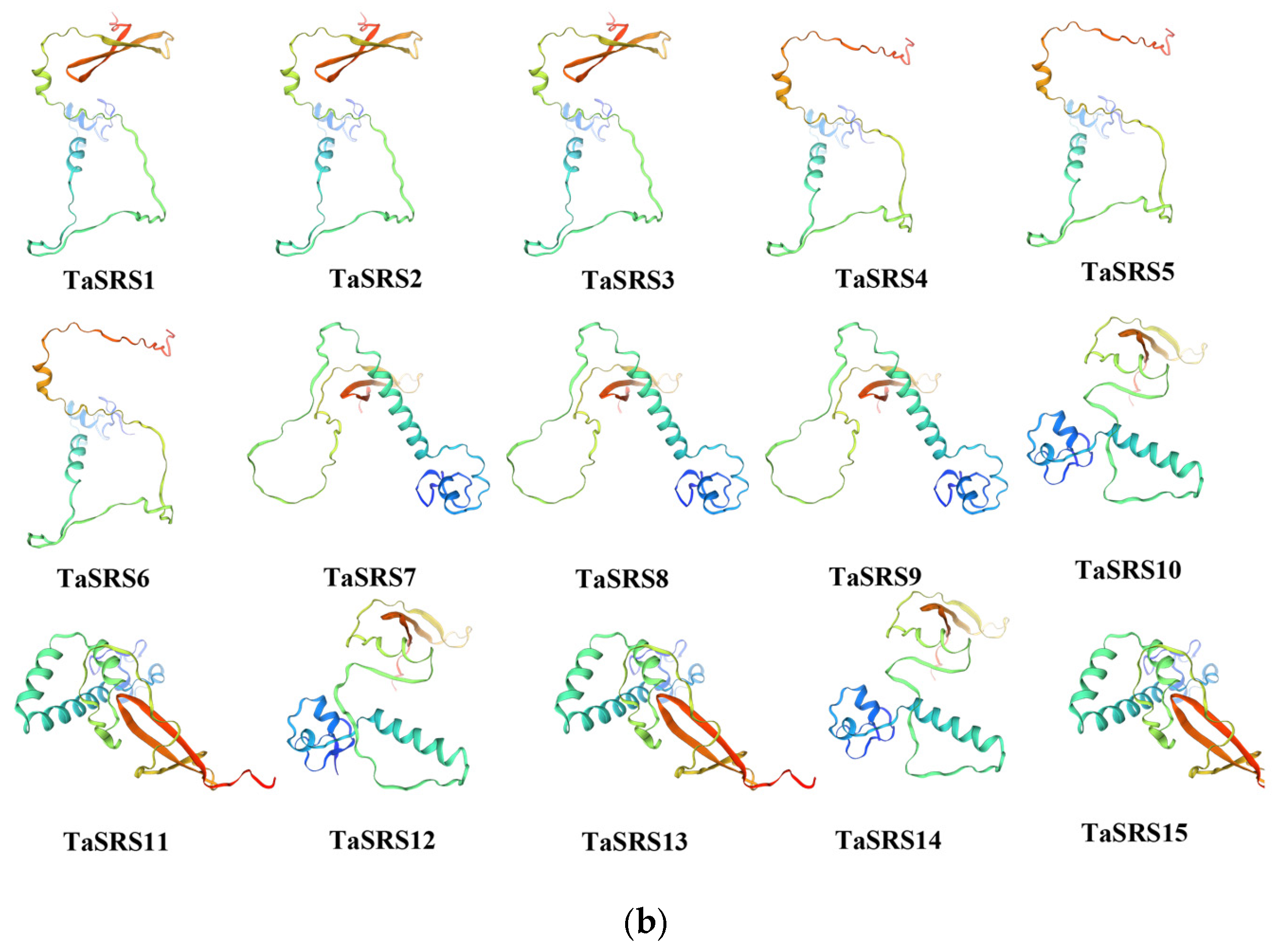
2.3. Analysis of Wheat SRS Gene Structure and Prediction of Protein Tertiary Structure
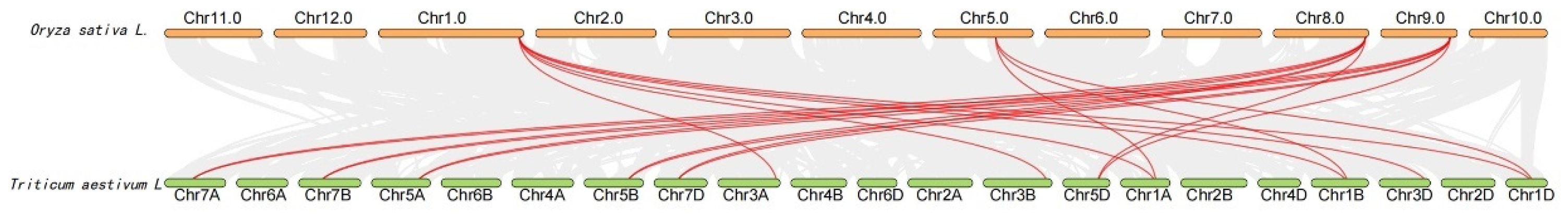
2.4. Analysis of Chromosome Localization and Collinearity of Wheat SRS Gene
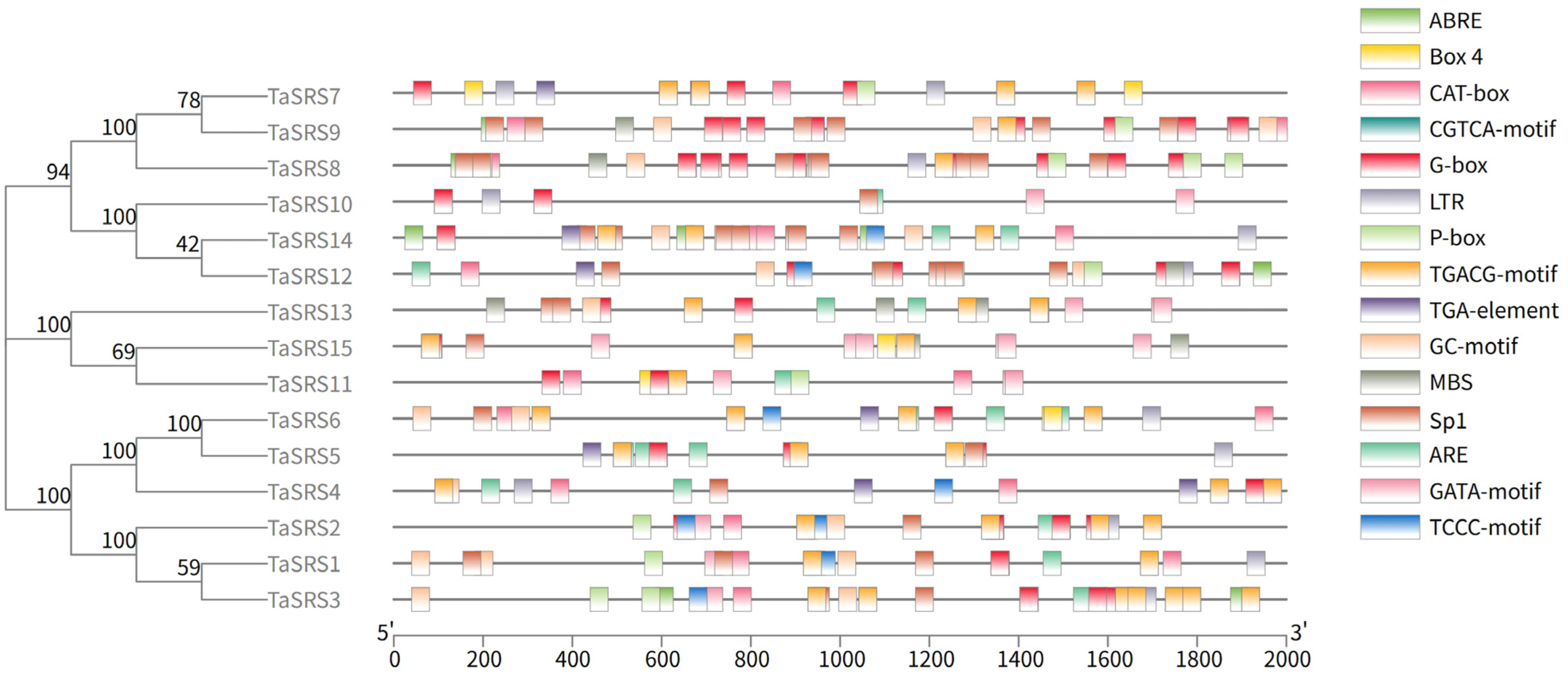
2.5. Analysis of Cis-Acting Elements of Wheat SRS Gene Family
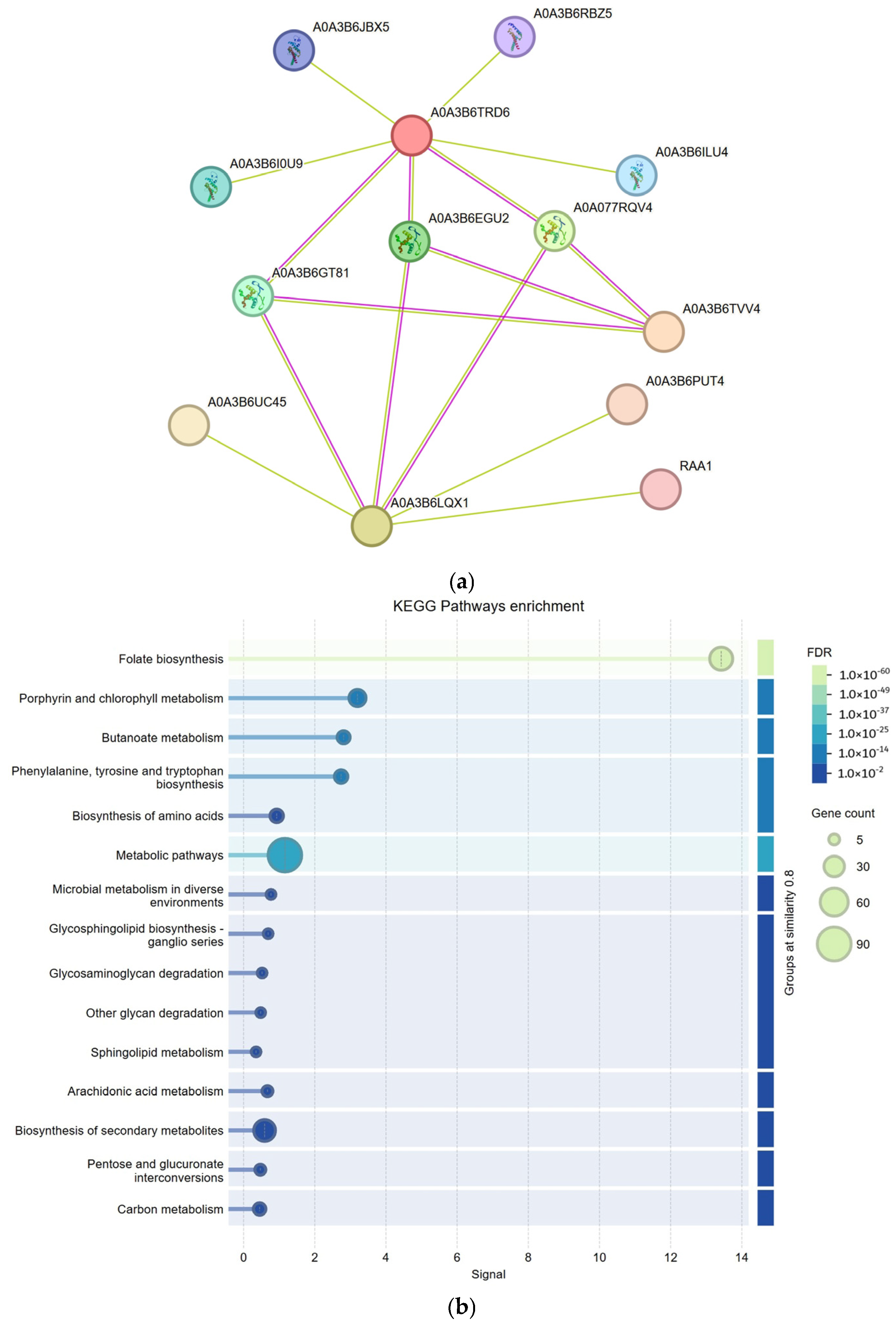
2.6. Analysis of the Gene Regulatory Network of the TaSRS Gene Family in Wheat
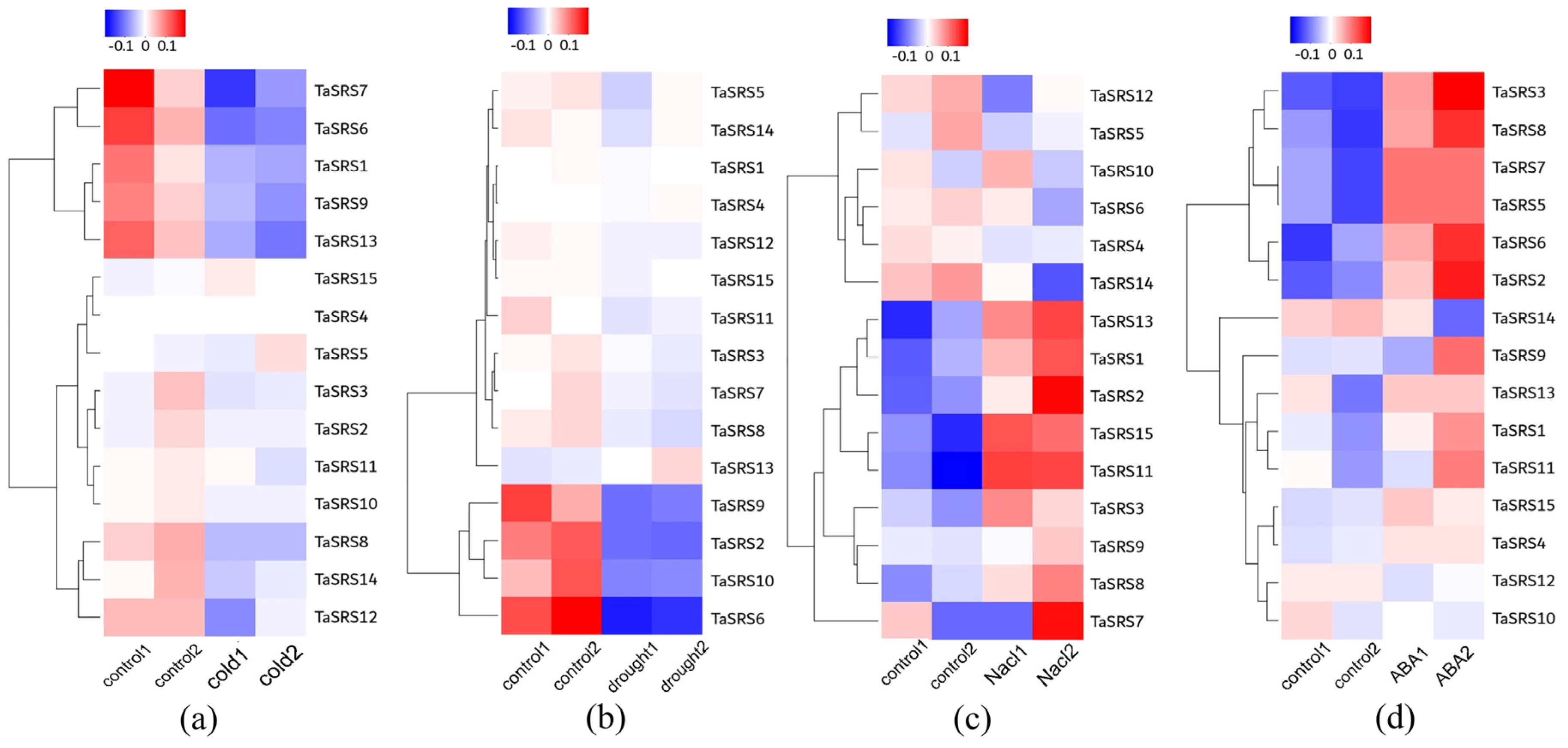
2.7. The Expression Profile of the Wheat TaSRS Gene in Response to Abiotic Stress
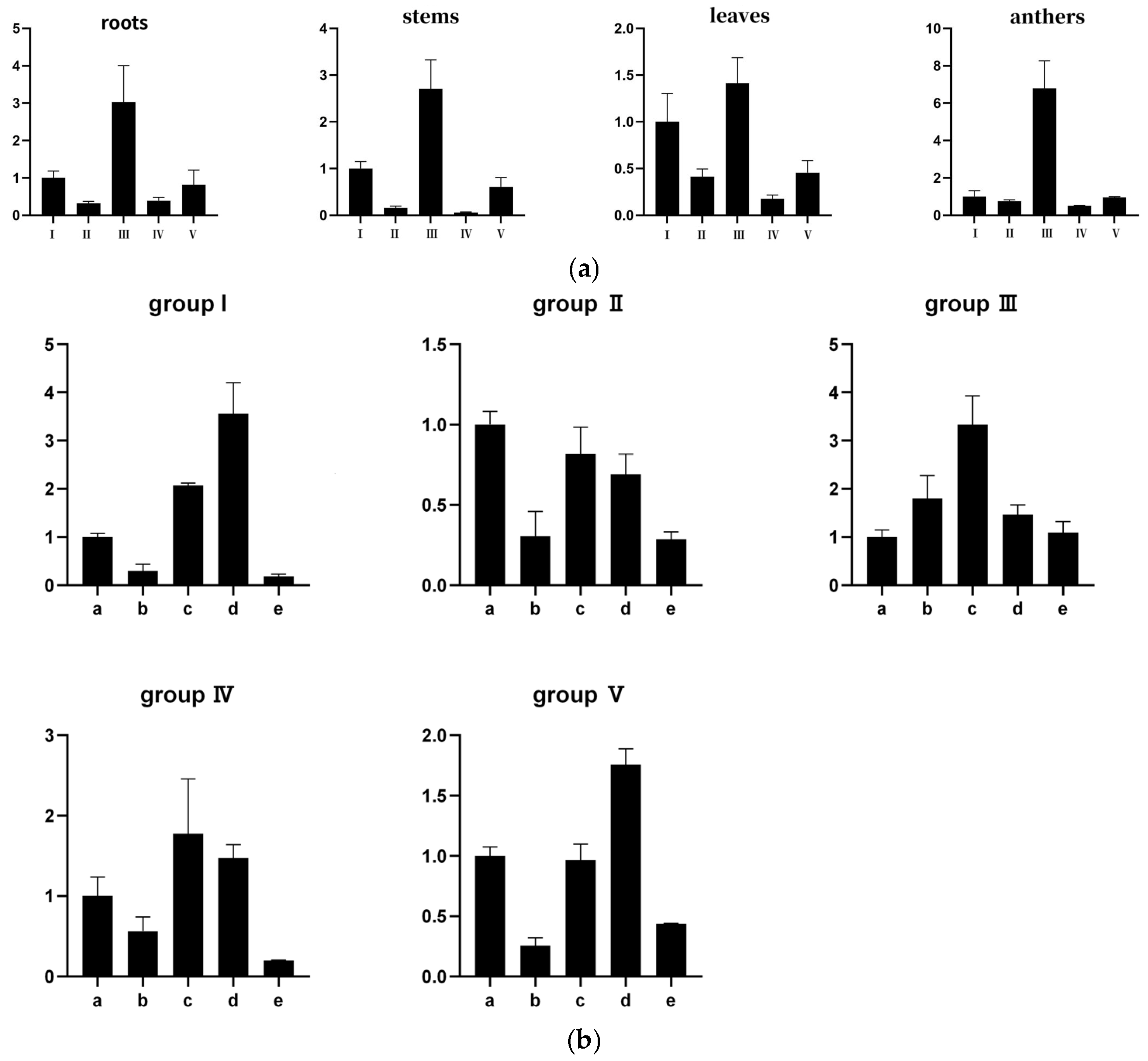
2.8. Expression of Wheat SRS Gene Family Under Different Tissue, Abiotic Stress and Hormone Treatment
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Identification of Wheat SRS Gene Family Members
4.3. Analysis of the Conserved Motifs and Phylogenetic Evolution of the SRS Gene Family
4.4. Analysis of the Structure of Wheat SRS Gene and Three-Dimensional Structure of Wheat SRS Protein
4.5. Analysis of Chromosome Localization and Synteny of the SRS Gene
4.6. Analysis of Cis-Acting Elements of SRS Gene Family in Wheat
4.7. TaSRS Gene Family Regulatory Network Analysis in Wheat
4.8. The Expression Profile of the Wheat TaSRS Gene in Response to Abiotic Stress
4.9. Quantitative Expression Analysis of SRS Gene Family in Wheat
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef] [PubMed]
- Fridborg, I.; Kuusk, S.; Robertson, M.; Sundberg, E. The Arabidopsis protein SHI represses gibberellin responses in Arabidopsis and barley. Plant Physiol. 2001, 127, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.-P.; Song, X.-Y.; Guo, L.-L.; Zhang, X.-Z.; Zheng, W.-J. Genome-Wide Analysis of the Shi-Related Sequence Family and Functional Identification of GmSRS18 Involving in Drought and Salt Stresses in Soybean. Int. J. Mol. Sci. 2020, 21, 1810. [Google Scholar] [CrossRef] [PubMed]
- Lovering, R.; Hanson, I.M.; Borden, K.L.; Martin, S.; O’Reilly, N.J.; Evan, G.I.; Rahman, D.; Pappin, D.J.; Trowsdale, J.; Freemont, P.S. Identification and preliminary characterization of a protein motif related to the zinc finger. Proc. Natl. Acad. Sci. USA 1993, 90, 2112–2116. [Google Scholar] [CrossRef]
- Hu, M.; Xie, M.; Cui, X.; Huang, J.; Cheng, X.; Liu, L.; Yan, S.; Liu, S.; Tong, C. Characterization and Potential Function Analysis of the SRS Gene Family in Brassica napus. Genes 2023, 14, 1421. [Google Scholar] [CrossRef]
- Klug, A. Zinc finger peptides for the regulation of gene expression. J. Mol. Biol. 1999, 293, 215–218. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, B.; Wang, X.; Wei, X. Genome-wide identification, structural analysis and expression profiles of short internodes related sequence gene family in quinoa. Front. Genet. 2022, 13, 961925. [Google Scholar] [CrossRef]
- Youssef, H.M.; Eggert, K.; Koppolu, R.; Alqudah, A.M.; Poursarebani, N.; Fazeli, A.; Sakuma, S.; Tagiri, A.; Rutten, T.; Govind, G.; et al. VRS2 regulates hormone-mediated inflorescence patterning in barley. Nat. Genet. 2017, 49, 157–161. [Google Scholar] [CrossRef]
- Gomariz-Fernández, A.; Sánchez-Gerschon, V.; Fourquin, C.; Ferrándiz, C. The Role of SHI/STY/SRS Genes in Organ Growth and Carpel Development Is Conserved in the Distant Eudicot Species Arabidopsis thaliana and Nicotiana benthamiana. Front. Plant Sci. 2017, 8, 814. [Google Scholar] [CrossRef]
- Lu, W.; Wang, Y.; Shi, Y.; Liang, Q.; Lu, X.; Su, D.; Xu, X.; Pirrello, J.; Gao, Y.; Huang, B.; et al. Identification of SRS transcription factor family in Solanum lycopersicum, and functional characterization of their responses to hormones and abiotic stresses. BMC Plant Biol. 2023, 23, 495. [Google Scholar] [CrossRef]
- Eklund, D.M.; StÅldal, V.; Valsecchi, I.; Cierlik, I.; Eriksson, C.; Hiratsu, K.; Ohme-Takagi, M.; Sundström, J.F.; Thelander, M.; Ezcurra, I.; et al. The Arabidopsis thaliana STYLISH1 Protein Acts as a Transcriptional Activator Regulating Auxin Biosynthesis. Plant Cell 2010, 22, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.; Zhang, W.; Cheng, X.; Hu, F.; Ye, Z.; Cao, J. Molecular evolutionary analysis of the SHI/STY gene family in land plants: A focus on the Brassica species. Front. Plant Sci. 2022, 13, 958964. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Yadav, S.; Singh, A.; Mahima, M.; Singh, A.; Gautam, V.; Sarkar, A.K. Auxin signaling modulates LATERAL ROOT PRIMORDIUM1 (LRP1) expression during lateral root development in Arabidopsis. Plant J. Cell Mol. Biol. 2020, 101, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Fridborg, I.; Kuusk, S.; Moritz, T.; Sundberg, E. The Arabidopsis dwarf mutant shi exhibits reduced gibberellin responses conferred by overexpression of a new putative zinc finger protein. Plant Cell 1999, 11, 1019–1031. [Google Scholar] [CrossRef]
- Ståldal, V.; Sundberg, E. The role of auxin in style development and apical-basal patterning of the Arabidopsis thaliana gynoecium. Plant Signal. Behav. 2009, 4, 83–85. [Google Scholar] [CrossRef]
- Kim, S.-G.; Lee, S.; Kim, Y.-S.; Yun, D.-J.; Woo, J.-C.; Park, C.-M. Activation tagging of an Arabidopsis SHI-RELATED SEQUENCE gene produces abnormal anther dehiscence and floral development. Plant Mol. Biol. 2010, 74, 337–351. [Google Scholar] [CrossRef]
- Kuusk, S.; Sohlberg, J.J.; Eklund, D.M.; Sundberg, E. Functionally redundant SHI family genes regulate Arabidopsis gynoecium development in a dose-dependent manner. Plant J. 2006, 47, 99–111. [Google Scholar] [CrossRef]
- Yuan, T.-T.; Xu, H.-H.; Zhang, Q.; Zhang, L.-Y.; Lu, Y.-T. The COP1 Target SHI-RELATED SEQUENCE5 Directly Activates Photomorphogenesis-Promoting Genes. Plant Cell 2018, 30, 2368–2382. [Google Scholar] [CrossRef]
- Duan, E.; Wang, Y.; Li, X.; Lin, Q.; Zhang, T.; Wang, Y.; Zhou, C.; Zhang, H.; Jiang, L.; Wang, J.; et al. OsSHI1 Regulates Plant Architecture Through Modulating the Transcriptional Activity of IPA1 in Rice. Plant Cell 2019, 31, 1026–1042. [Google Scholar] [CrossRef]
- Ma, B.; Nian, L.; Ain, N.U.; Liu, X.; Yang, Y.; Zhu, X.; Haider, F.U.; Lv, Y.; Bai, P.; Zhang, X.; et al. Genome-Wide Identification and Expression Profiling of the SRS Gene Family in Melilotus albus Reveals Functions in Various Stress Conditions. Plants 2022, 11, 3101. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Shi, H.; Yang, Y.; Feng, X.; Chen, X.; Xiao, F.; Lin, H.; Guo, Y. Insights into plant salt stress signaling and tolerance. J. Genet. Genom. 2024, 51, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Ma, J.; Li, J.; Yin, B.; Fu, C. Advances of salt stress-responsive transcription factors in plants. Chin. J. Biotechnol. 2022, 38, 50–65. [Google Scholar]
- Liu, H.; Tang, X.; Zhang, N.; Li, S.; Si, H. Role of bZIP Transcription Factors in Plant Salt Stress. Int. J. Mol. Sci. 2023, 24, 7893. [Google Scholar] [CrossRef]
- Delgado, C.; Mora-Poblete, F.; Ahmar, S.; Chen, J.-T.; Figueroa, C.R. Jasmonates and Plant Salt Stress: Molecular Players, Physiological Effects, and Improving Tolerance by Using Genome-Associated Tools. Int. J. Mol. Sci. 2021, 22, 3082. [Google Scholar] [CrossRef]
- Singh, A.; Roychoudhury, A. Gene regulation at transcriptional and post-transcriptional levels to combat salt stress in plants. Physiol. Plant. 2021, 173, 1556–1572. [Google Scholar] [CrossRef]
- Danquah, A.; de Zelicourt, A.; Colcombet, J.; Hirt, H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 2014, 32, 40–52. [Google Scholar] [CrossRef]
- Li, S.; Liu, S.; Zhang, Q.; Cui, M.; Zhao, M.; Li, N.; Wang, S.; Wu, R.; Zhang, L.; Cao, Y.; et al. The interaction of ABA and ROS in plant growth and stress resistances. Front. Plant Sci. 2022, 13, 1050132. [Google Scholar] [CrossRef]
- Brookbank, B.P.; Patel, J.; Gazzarrini, S.; Nambara, E. Role of Basal ABA in Plant Growth and Development. Genes 2021, 12, 1936. [Google Scholar] [CrossRef]
- Hong, J.H.; Seah, S.W.; Xu, J. The root of ABA action in environmental stress response. Plant Cell Rep. 2013, 32, 971–983. [Google Scholar] [CrossRef]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef]
- Zhao, Q.; Guan, X.; Zhou, L.; Asad, M.A.; Xu, Y.; Pan, G.; Cheng, F. ABA-triggered ROS burst in rice developing anthers is critical for tapetal programmed cell death induction and heat stress-induced pollen abortion. Plant Cell Environ. 2023, 46, 1453–1471. [Google Scholar] [CrossRef]
- Tian, W.; Zheng, Y.; Wang, W.; Wang, D.; Tilley, M.; Zhang, G.; He, Z.; Li, Y. A comprehensive review of wheat phytochemicals: From farm to fork and beyond. Compr. Rev. Food Sci. Food Saf. 2022, 21, 2274–2308. [Google Scholar] [CrossRef]
- Dinu, M.; Whittaker, A.; Pagliai, G.; Benedettelli, S.; Sofi, F. Ancient wheat species and human health: Biochemical and clinical implications. J. Nutr. Biochem. 2018, 52, 1–9. [Google Scholar] [CrossRef]
- Reynolds, M.; Foulkes, J.; Furbank, R.; Griffiths, S.; King, J.; Murchie, E.; Parry, M.; Slafer, G. Achieving yield gains in wheat. Plant Cell Environ. 2012, 35, 1799–1823. [Google Scholar] [CrossRef]
- Casiraghi, M.C.; Pagani, M.A.; Erba, D.; Marti, A.; Cecchini, C.; D’Egidio, M.G. Quality and nutritional properties of pasta products enriched with immature wheat grain. Int. J. Food Sci. Nutr. 2013, 64, 544–550. [Google Scholar] [CrossRef]
- Yin, Y.; Yuan, C.; Zhang, Y.; Li, S.; Bai, B.; Wu, L.; Ren, Y.; Singh, R.P.; Lan, C. Genetic analysis of stripe rust resistance in the common wheat line Kfa/2*Kachu under a Chinese rust environment. Theor. Appl. Genet. 2023, 136, 185. [Google Scholar] [CrossRef]
- Dekomah, S.D.; Wang, Y.; Qin, T.; Xu, D.; Sun, C.; Yao, P.; Liu, Y.; Bi, Z.; Bai, J. Identification and Expression Analysis of Calcium-Dependent Protein Kinases Gene Family in Potato Under Drought Stress. Front. Genet. 2022, 13, 874397. [Google Scholar] [CrossRef]
- Han, Z.; Liu, Y.; Deng, X.; Liu, D.; Liu, Y.; Hu, Y.; Yan, Y. Genome-wide identification and expression analysis of expansin gene family in common wheat (Triticum aestivum L.). BMC Genom. 2019, 20, 101. [Google Scholar] [CrossRef]
- Priest, H.D.; Fox, S.E.; Rowley, E.R.; Murray, J.R.; Michael, T.P.; Mockler, T.C. Analysis of global gene expression in Brachypodium distachyon reveals extensive network plasticity in response to abiotic stress. PLoS ONE 2014, 9, e87499. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, Y.; Chen, J.; Lin, H.; Zhao, M.; Peng, H.; Liu, L.; Yuan, G.; Zhang, S.; Zhang, Z.; et al. Genome expression profile analysis reveals important transcripts in maize roots responding to the stress of heavy metal Pb. Physiol. Plant. 2013, 147, 270–282. [Google Scholar] [CrossRef]
- Zawaski, C.; Kadmiel, M.; Ma, C.; Gai, Y.; Jiang, X.; Strauss, S.H.; Busov, V.B. SHORT INTERNODES-like genes regulate shoot growth and xylem proliferation in Populus. New Phytol 2011, 191, 678–691. [Google Scholar] [CrossRef]
- Huang, H.; Song, J.; Feng, Y.; Zheng, L.; Chen, Y.; Luo, K. Genome-Wide Identification and Expression Analysis of the SHI-Related Sequence Family in Cassava. Genes 2023, 14, 870. [Google Scholar] [CrossRef]
- Yang, Y.W.; Lai, K.N.; Tai, P.Y.; Ma, D.P.; Li, W.H. Molecular phylogenetic studies of Brassica, rorippa, arabidopsis and allied genera based on the internal transcribed spacer region of 18S–25S rDNA. Mol. Phylogenetics Evol. 1999, 13, 455–462. [Google Scholar] [CrossRef]
- Smith, D.L.; Fedoroff, N.V. LRP1, a gene expressed in lateral and adventitious root primordia of arabidopsis. Plant Cell 1995, 7, 735. [Google Scholar]
- Eklund, D.M.; Cierlik, I.; Ståldal, V.; Claes, A.R.; Vestman, D.; Chandler, J.; Sundberg, E. Expression of Arabidopsis SHORT INTERNODES/STYLISH family genes in auxin biosynthesis zones of aerial organs is dependent on a GCC box-like regulatory element. Plant Physiol. 2011, 157, 2069–2080. [Google Scholar] [CrossRef]
- Sun, C.; Yu, L.; Zhang, S.; Gu, Q.; Wang, M. Genome-wide characterization of the SHORT INTER-NODES/STYLISH and Shi-Related Sequence family in Gossypium hirsutum and functional identification of GhSRS21 under salt stress. Front. Plant Sci. 2022, 13, 1078083. [Google Scholar] [CrossRef]
- Chapman, E.J.; Estelle, M. Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 2009, 43, 265–285. [Google Scholar] [CrossRef]
- He, B.; Shi, P.; Lv, Y.; Gao, Z.; Chen, G. Gene coexpression network analysis reveals the role of SRS genes in senescence leaf of maize (Zea mays L.). J. Genet. 2020, 99, 3. [Google Scholar] [CrossRef]
- Yang, J.; Xu, P.; Yu, D. Genome-Wide Identification and Characterization of the SHI-Related Sequence Gene Family in Rice. Evol. Bioinform. 2020, 16, 1176934320941495. [Google Scholar] [CrossRef]
- Du, W.; Yang, J.; Li, Q.; He, C.; Pang, Y. Identification and Characterization of Abiotic Stress–Responsive NF-YB Family Genes in Medicago. Int. J. Mol. Sci. 2022, 23, 6906. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chu, Z. Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium distachyon. BMC Genom. 2015, 16, 227. [Google Scholar] [CrossRef] [PubMed]
- Bechtold, U.; Field, B. Molecular mechanisms controlling plant growth during abiotic stress. J. Exp. Bot. 2018, 69, 2753–2758. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, C.; Chen, H.; Wang, P.; Wang, P.; Song, C.; Zhang, X.; Wang, D. Multi-gene co-expression can improve comprehensive resistance to multiple abiotic stresses in Brassica napus L. Plant Sci. 2018, 274, 410–419. [Google Scholar] [CrossRef]


| Gene Name | Gene ID | Length (aa) | Length (bp) | Molecular Weight | Isoelectric Point | Exon Number | Group |
|---|---|---|---|---|---|---|---|
| TaSRS1 | TraesCS1A02G241400 | 314 | 945 | 32,544.10 | 8.89 | 3 | V |
| TaSRS2 | TraesCS1B02G253200 | 313 | 942 | 32,580.13 | 8.89 | 3 | V |
| TaSRS3 | TraesCS1D02G241300 | 313 | 942 | 32,449.04 | 8.89 | 3 | V |
| TaSRS4 | TraesCS3A02G491500 | 247 | 744 | 25,092.11 | 8.43 | 2 | IV |
| TaSRS5 | TraesCS3B02G552400 | 352 | 1059 | 35,484.59 | 7.72 | 2 | IV |
| TaSRS6 | TraesCS3D02G497300 | 348 | 1047 | 35,259.38 | 7.70 | 2 | IV |
| TaSRS7 | TraesCS5A02G319200 | 330 | 993 | 33,920.42 | 8.96 | 2 | I |
| TaSRS8 | TraesCS5B02G319600 | 330 | 993 | 33,924.41 | 8.96 | 2 | I |
| TaSRS9 | TraesCS5D02G325400 | 330 | 993 | 33,920.42 | 8.96 | 2 | I |
| TaSRS10 | TraesCS7A02G270900 | 315 | 948 | 32,500.40 | 8.16 | 2 | II |
| TaSRS11 | TraesCS7A02G406300 | 346 | 1041 | 34,382.91 | 8.68 | 2 | III |
| TaSRS12 | TraesCS7B02G169200 | 335 | 1008 | 34,636.99 | 7.71 | 3 | II |
| TaSRS13 | TraesCS7B02G306300 | 346 | 1041 | 34,468.96 | 8.41 | 2 | III |
| TaSRS14 | TraesCS7D02G271400 | 315 | 948 | 32,386.30 | 8.16 | 2 | II |
| TaSRS15 | TraesCS7D02G400000 | 346 | 1041 | 34,428.93 | 8.68 | 2 | III |
| Name | Gene | Forward Primer Sequences (5′→3′) | Reverse Primer Sequences (5′→3′) |
|---|---|---|---|
| group I | TaSRS7, TaSRS8, TaSRS9 | CTGCTCCACCCACGTCAAGTCCA | AGACGCACGCATCGGAACACC |
| group II | TaSRS10, TaSRS12, TaSRS14 | CAGACCACCGTCAGCATCG | GTGGCCGTGGAAGAATGG |
| group III | TaSRS11, TaSRS13, TaSRS15 | GTCGTGTTGGGCTTCTCGC | GACCCAGGTGCTCTTGACG |
| group IV | TaSRS4, TaSRS5, TaSRS6 | GCACGTCAAGAGCACCTGGGTC | ACGCCCTGGTCGTAGAGGAAGC |
| group V | TaSRS1, TaSRS2, TaSRS3 | CGCCATCCAGTTCTGGCAATC | TCCTGACGTGCGTGGTGCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Y.; Chang, Q.; Li, C.; Wu, K.; Wang, Y.; Guo, C.; Shu, Y.; Bai, Y. Genome-Wide Identification of SRS Gene Family in Wheat and Expression Analysis Under Abiotic Stress. Int. J. Mol. Sci. 2025, 26, 6289. https://doi.org/10.3390/ijms26136289
Yu Y, Chang Q, Li C, Wu K, Wang Y, Guo C, Shu Y, Bai Y. Genome-Wide Identification of SRS Gene Family in Wheat and Expression Analysis Under Abiotic Stress. International Journal of Molecular Sciences. 2025; 26(13):6289. https://doi.org/10.3390/ijms26136289
Chicago/Turabian StyleYu, Yanan, Qihang Chang, Chunyue Li, Kaiyue Wu, Yanyan Wang, Changhong Guo, Yongjun Shu, and Yan Bai. 2025. "Genome-Wide Identification of SRS Gene Family in Wheat and Expression Analysis Under Abiotic Stress" International Journal of Molecular Sciences 26, no. 13: 6289. https://doi.org/10.3390/ijms26136289
APA StyleYu, Y., Chang, Q., Li, C., Wu, K., Wang, Y., Guo, C., Shu, Y., & Bai, Y. (2025). Genome-Wide Identification of SRS Gene Family in Wheat and Expression Analysis Under Abiotic Stress. International Journal of Molecular Sciences, 26(13), 6289. https://doi.org/10.3390/ijms26136289






