Expression of WNT Family Genes in Mesenchymal Stromal Cells of the Hematopoietic Niche in Patients with Different Responses to Multiple Myeloma Treatment
Abstract
1. Introduction
2. Results
2.1. Differential Expression of WNT Family Genes and CTNNB1 Gene in the PCs of MM Patients
2.2. Quantification of WNT and β-Catenin Protein Levels in the BM of MM Patients
2.3. Distribution of WNT and β-Catenin Proteins in the BM of MM Patients
2.4. Transcriptional Activity of WNT Family Genes and the CTNNB1 Gene in BM MSCs Cultures from MM Patients
3. Discussion
Limitations of the Study
4. Materials and Methods
4.1. Ethical Guidelines
4.2. Bioinformatics
4.3. Patients
4.4. Collection of Trephine Biopsies and Aspirates
4.5. Cell Cultures and Lines
4.6. Oligonucleotides
4.7. RNA Isolation and cDNA Synthesis
4.8. Real-Time PCR
4.9. Immunohistochemistry (IHC)
4.10. Multiplex Fluorescence IHC
4.11. Microscopy
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BM | bone marrow |
| MM | multiple myeloma |
| UT | primary patients |
| NR | patients without response to treatment |
| PoCR | patients with complete or partial response |
| HD | healthy donors |
| MSCs | mesenchymal stromal cells |
| PC | plasma cells |
| TME | tumor microenvironment |
References
- Bessmeltsev, S.S.; Abdulkadyrov, K.M. Multiple Myeloma. A Guide for Physicians; MK: Moscow, Russia, 2016. [Google Scholar]
- van de Donk, N.W.C.J.; Pawlyn, C.; Yong, K.L. Multiple Myeloma. Lancet 2021, 397, 410–427. [Google Scholar] [CrossRef] [PubMed]
- Bessmeltsev, S.S. Multiple Myeloma (Management of Newly Diagnosed Patients): Literature Review and Our Own Data. Part II. Clin. Oncohematol. 2013, 6, 379–414. [Google Scholar]
- Meads, M.B.; Gatenby, R.A.; Dalton, W.S. Environment-Mediated Drug Resistance: A Major Contributor to Minimal Residual Disease. Nat. Rev. Cancer 2009, 9, 665–674. [Google Scholar] [CrossRef]
- de Jong, M.M.E.; Kellermayer, Z.; Papazian, N.; Tahri, S.; Hofste op Bruinink, D.; Hoogenboezem, R.; Sanders, M.A.; van de Woestijne, P.C.; Bos, P.K.; Khandanpour, C.; et al. The Multiple Myeloma Microenvironment Is Defined by an Inflammatory Stromal Cell Landscape. Nat. Immunol. 2021, 22, 769–780. [Google Scholar] [CrossRef]
- Noll, J.E.; Williams, S.A.; Purton, L.E.; Zannettino, A.C.W. Tug of War in the Haematopoietic Stem Cell Niche: Do Myeloma Plasma Cells Compete for the HSC Niche? Blood Cancer J. 2012, 2, e91. [Google Scholar] [CrossRef]
- Friedenstein, A.J.; Chailakhjan, R.K.; Lalykina, K.S. The development of fibroblast colonies in monolayer cultures of guinea-pig bone marrow and spleen cells. Cell Prolif. 1970, 3, 393–403. [Google Scholar] [CrossRef]
- Méndez-Ferrer, S.; Bonnet, D.; Steensma, D.P.; Hasserjian, R.P.; Ghobrial, I.M.; Gribben, J.G.; Andreeff, M.; Krause, D.S. Bone Marrow Niches in Haematological Malignancies. Nat. Rev. Cancer 2020, 20, 285–298. [Google Scholar] [CrossRef]
- Méndez-Ferrer, S.; Michurina, T.V.; Ferraro, F.; Mazloom, A.R.; MacArthur, B.D.; Lira, S.A.; Scadden, D.T.; Ma’ayan, A.; Enikolopov, G.N.; Frenette, P.S. Mesenchymal and Haematopoietic Stem Cells Form a Unique Bone Marrow Niche. Nature 2010, 466, 829–834. [Google Scholar] [CrossRef]
- Dazzi, F.; Ramasamy, R.; Glennie, S.; Jones, S.P.; Roberts, I. The Role of Mesenchymal Stem Cells in Haemopoiesis. Blood Rev. 2006, 20, 161–171. [Google Scholar] [CrossRef]
- Fröbel, J.; Landspersky, T.; Percin, G.; Schreck, C.; Rahmig, S.; Ori, A.; Nowak, D.; Essers, M.; Waskow, C.; Oostendorp, R.A.J. The Hematopoietic Bone Marrow Niche Ecosystem. Front. Cell Dev. Biol. 2021, 9, 1–19. [Google Scholar] [CrossRef]
- Somaiah, C.; Kumar, A.; Sharma, R.; Sharma, A.; Anand, T.; Bhattacharyya, J.; Das, D.; Deka Talukdar, S.; Jaganathan, B.G. Mesenchymal Stem Cells Show Functional Defect and Decreased Anti-Cancer Effect after Exposure to Chemotherapeutic Drugs. J. Biomed. Sci. 2018, 25, 5. [Google Scholar] [CrossRef]
- Enukashvily, N.I.; Semenova, N.; Chubar, A.V.; Ostromyshenskii, D.I.; Gushcha, E.A.; Gritsaev, S.; Bessmeltsev, S.S.; Rugal, V.I.; Prikhodko, E.M.; Kostroma, I.; et al. Pericentromeric Non-Coding DNA Transcription Is Associated with Niche Impairment in Patients with Ineffective or Partially Effective Multiple Myeloma Treatment. Int. J. Mol. Sci. 2022, 23, 3359. [Google Scholar] [CrossRef] [PubMed]
- Moschetta, M.; Kawano, Y.; Sacco, A.; Belotti, A.; Ribolla, R.; Chiarini, M.; Giustini, V.; Bertoli, D.; Sottini, A.; Valotti, M.; et al. Bone Marrow Stroma and Vascular Contributions to Myeloma Bone Homing. Curr. Osteoporos. Rep. 2017, 15, 499–506. [Google Scholar] [CrossRef]
- Edwards, C.M.; Zhuang, J.; Mundy, G.R. The Pathogenesis of the Bone Disease of Multiple Myeloma. Bone 2008, 42, 1007–1013. [Google Scholar] [CrossRef] [PubMed]
- Giannakoulas, N.; Ntanasis-Stathopoulos, I.; Terpos, E. The Role of Marrow Microenvironment in the Growth and Development of Malignant Plasma Cells in Multiple Myeloma. Int. J. Mol. Sci. 2021, 22, 4462. [Google Scholar] [CrossRef]
- Weißbach, S.; Heredia-Guerrero, S.C.; Barnsteiner, S.; Großhans, L.; Bodem, J.; Starz, H.; Langer, C.; Appenzeller, S.; Knop, S.; Steinbrunn, T.; et al. Exon-4 Mutations in KRAS Affect MEK/ERK and PI3K/AKT Signaling in Human Multiple Myeloma Cell Lines. Cancers 2020, 12, 455. [Google Scholar] [CrossRef]
- Ramakrishnan, V.; Kumar, S. PI3K/AKT/MTOR Pathway in Multiple Myeloma: From Basic Biology to Clinical Promise. Leuk. Lymphoma 2018, 59, 2524–2534. [Google Scholar] [CrossRef]
- Li, Z.-W.; Chen, H.; Campbell, R.A.; Bonavida, B.; Berenson, J.R. NF-KappaB in the Pathogenesis and Treatment of Multiple Myeloma. Curr. Opin. Hematol. 2008, 15, 391–399. [Google Scholar] [CrossRef]
- van Andel, H.; Kocemba, K.A.; Spaargaren, M.; Pals, S.T. Aberrant Wnt Signaling in Multiple Myeloma: Molecular Mechanisms and Targeting Options. Leukemia 2019, 33, 1063–1075. [Google Scholar] [CrossRef]
- Spaan, I.; Raymakers, R.A.; van de Stolpe, A.; Peperzak, V. Wnt Signaling in Multiple Myeloma: A Central Player in Disease with Therapeutic Potential. J. Hematol. Oncol. 2018, 11, 67. [Google Scholar] [CrossRef]
- Nemeth, M.J.; Mak, K.K.; Yang, Y.; Bodine, D.M. β-Catenin Expression in the Bone Marrow Microenvironment Is Required for Long-Term Maintenance of Primitive Hematopoietic Cells. Stem Cells 2009, 27, 1109–1119. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gaur, T.; Lengner, C.J.; Hovhannisyan, H.; Bhat, R.A.; Bodine, P.V.N.; Komm, B.S.; Javed, A.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; et al. Canonical WNT Signaling Promotes Osteogenesis by Directly Stimulating Runx2 Gene Expression. J. Biol. Chem. 2005, 280, 33132–33140. [Google Scholar] [CrossRef] [PubMed]
- Sugimura, R.; He, X.C.; Venkatraman, A.; Arai, F.; Box, A.; Semerad, C.; Haug, J.S.; Peng, L.; Zhong, X.; Suda, T.; et al. Noncanonical Wnt Signaling Maintains Hematopoietic Stem Cells in the Niche. Cell 2012, 150, 351–365. [Google Scholar] [CrossRef] [PubMed]
- Derksen, P.W.B.; Tjin, E.; Meijer, H.P.; Klok, M.D.; Mac Gillavry, H.D.; van Oers, M.H.J.; Lokhorst, H.M.; Bloem, A.C.; Clevers, H.; Nusse, R.; et al. Illegitimate WNT Signaling Promotes Proliferation of Multiple Myeloma Cells. Proc. Natl. Acad. Sci. USA 2004, 101, 6122–6127. [Google Scholar] [CrossRef]
- Qiang, Y.-W.; Chen, Y.; Stephens, O.; Brown, N.; Chen, B.; Epstein, J.; Barlogie, B.; Shaughnessy, J.D. Myeloma-Derived Dickkopf-1 Disrupts Wnt-Regulated Osteoprotegerin and RANKL Production by Osteoblasts: A Potential Mechanism Underlying Osteolytic Bone Lesions in Multiple Myeloma. Blood 2008, 112, 196–207. [Google Scholar] [CrossRef]
- García-Ortiz, A.; Rodríguez-García, Y.; Encinas, J.; Maroto-Martín, E.; Castellano, E.; Teixidó, J.; Martínez-López, J. The Role of Tumor Microenvironment in Multiple Myeloma Development and Progression. Cancers 2021, 13, 217. [Google Scholar] [CrossRef]
- Qiang, Y.-W.; Hu, B.; Chen, Y.; Zhong, Y.; Shi, B.; Barlogie, B.; Shaughnessy, J.D. Bortezomib Induces Osteoblast Differentiation via Wnt-Independent Activation of β-Catenin/TCF Signaling. Blood 2009, 113, 4319–4330. [Google Scholar] [CrossRef]
- Qin, K.; Yu, M.; Fan, J.; Wang, H.; Zhao, P.; Zhao, G.; Zeng, W.; Chen, C.; Wang, Y.; Wang, A.; et al. Canonical and Noncanonical Wnt Signaling: Multilayered Mediators, Signaling Mechanisms and Major Signaling Crosstalk. Genes. Dis. 2024, 11, 103–134. [Google Scholar] [CrossRef]
- Ling, L.; Nurcombe, V.; Cool, S.M. Wnt Signaling Controls the Fate of Mesenchymal Stem Cells. Gene 2009, 433, 1–7. [Google Scholar] [CrossRef]
- Chen, X.; Shi, C.; Cao, H.; Chen, L.; Hou, J.; Xiang, Z.; Hu, K.; Han, X. The Hedgehog and Wnt/β-Catenin System Machinery Mediate Myofibroblast Differentiation of LR-MSCs in Pulmonary Fibrogenesis. Cell Death Dis. 2018, 9, 639. [Google Scholar] [CrossRef]
- Behrens, J.; von Kries, J.P.; Kühl, M.; Bruhn, L.; Wedlich, D.; Grosschedl, R.; Birchmeier, W. Functional Interaction of β-Catenin with the Transcription Factor LEF-1. Nature 1996, 382, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Doumpas, N.; Lampart, F.; Robinson, M.D.; Lentini, A.; Nestor, C.E.; Cantù, C.; Basler, K. TCF/LEF Dependent and Independent Transcriptional Regulation of Wnt/Β-catenin Target Genes. EMBO J. 2019, 38, e98873. [Google Scholar] [CrossRef] [PubMed]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt Signaling in Cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef] [PubMed]
- Moskalev, E.A.; Luckert, K.; Vorobjev, I.A.; Mastitsky, S.E.; Gladkikh, A.A.; Stephan, A.; Schrenk, M.; Kaplanov, K.D.; Kalashnikova, O.B.; Pötz, O.; et al. Concurrent Epigenetic Silencing of Wnt/β-Catenin Pathway Inhibitor Genes in B Cell Chronic Lymphocytic Leukaemia. BMC Cancer 2012, 12, 213. [Google Scholar] [CrossRef]
- Chim, C.S.; Pang, R.; Fung, T.K.; Choi, C.L.; Liang, R. Epigenetic Dysregulation of Wnt Signaling Pathway in Multiple Myeloma. Leukemia 2007, 21, 2527–2536. [Google Scholar] [CrossRef]
- Maiso, P.; Mogollón, P.; Ocio, E.M.; Garayoa, M. Bone Marrow Mesenchymal Stromal Cells in Multiple Myeloma: Their Role as Active Contributors to Myeloma Progression. Cancers 2021, 13, 2542. [Google Scholar] [CrossRef]
- Petinati, N.A.; Bigildeev, A.E.; Karpenko, D.S.; Sats, N.V.; Kapranov, N.M.; Davydova, Y.O.; Fastova, E.A.; Magomedova, A.U.; Kravchenko, S.K.; Arapidi, G.P.; et al. Humoral Effect of a B-Cell Tumor on the Bone Marrow Multipotent Mesenchymal Stromal Cells. Biochemistry 2021, 86, 207–216. [Google Scholar] [CrossRef]
- Bessmeltsev, S.S. Multiple Myeloma (Pathogenesis, Clinical Features, Diagnosis, Differential Diagnosis). Part I. Clin. Oncohematol. 2013, 6, 237–257. [Google Scholar]
- Qiang, Y.-W.; Endo, Y.; Rubin, J.S.; Rudikoff, S. Wnt Signaling in B-Cell Neoplasia. Oncogene 2003, 22, 1536–1545. [Google Scholar] [CrossRef]
- Qiang, Y.-W.; Shaughnessy, J.D.; Yaccoby, S. Wnt3a Signaling within Bone Inhibits Multiple Myeloma Bone Disease and Tumor Growth. Blood 2008, 112, 374–382. [Google Scholar] [CrossRef][Green Version]
- Boyd, K.D.; Ross, F.M.; Chiecchio, L.; Dagrada, G.P.; Konn, Z.J.; Tapper, W.J.; Walker, B.A.; Wardell, C.P.; Gregory, W.M.; Szubert, A.J.; et al. A Novel Prognostic Model in Myeloma Based on Co-Segregating Adverse FISH Lesions and the ISS: Analysis of Patients Treated in the MRC Myeloma IX Trial. Leukemia 2012, 26, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Mahtouk, K.; Moreaux, J.; Hose, D.; Rème, T.; Meißner, T.; Jourdan, M.; Rossi, J.F.; Pals, S.T.; Goldschmidt, H.; Klein, B. Growth Factors in Multiple Myeloma: A Comprehensive Analysis of Their Expression in Tumor Cells and Bone Marrow Environment Using Affymetrix Microarrays. BMC Cancer 2010, 10, 198. [Google Scholar] [CrossRef]
- Available online: Https://Www.Proteinatlas.Org/ENSG00000169884-WNT10B/Cell+line (accessed on 25 June 2025).
- Kobune, M.; Chiba, H.; Kato, J.; Kato, K.; Nakamura, K.; Kawano, Y.; Takada, K.; Takimoto, R.; Takayama, T.; Hamada, H.; et al. Wnt3/RhoA/ROCK Signaling Pathway Is Involved in Adhesion-Mediated Drug Resistance of Multiple Myeloma in an Autocrine Mechanism. Mol. Cancer Ther. 2007, 6, 1774–1784. [Google Scholar] [CrossRef]
- Bjorklund, C.C.; Ma, W.; Wang, Z.-Q.; Davis, R.E.; Kuhn, D.J.; Kornblau, S.M.; Wang, M.; Shah, J.J.; Orlowski, R.Z. Evidence of a Role for Activation of Wnt/β-Catenin Signaling in the Resistance of Plasma Cells to Lenalidomide. J. Biol. Chem. 2011, 286, 11009–11020. [Google Scholar] [CrossRef] [PubMed]
- Bolzoni, M.; Donofrio, G.; Storti, P.; Guasco, D.; Toscani, D.; Lazzaretti, M.; Bonomini, S.; Agnelli, L.; Capocefalo, A.; Dalla Palma, B.; et al. Myeloma Cells Inhibit Non-Canonical Wnt Co-Receptor Ror2 Expression in Human Bone Marrow Osteoprogenitor Cells: Effect of Wnt5a/Ror2 Pathway Activation on the Osteogenic Differentiation Impairment Induced by Myeloma Cells. Leukemia 2013, 27, 451–463. [Google Scholar] [CrossRef]
- Fu, H.-D.; Wang, B.-K.; Wan, Z.-Q.; Lin, H.; Chang, M.-L.; Han, G.-L. Wnt5a Mediated Canonical Wnt Signaling Pathway Activation in Orthodontic Tooth Movement: Possible Role in the Tension Force-Induced Bone Formation. J. Mol. Histol. 2016, 47, 455–466. [Google Scholar] [CrossRef]
- Stevens, J.R.; Miranda-Carboni, G.A.; Singer, M.A.; Brugger, S.M.; Lyons, K.M.; Lane, T.F. Wnt10b Deficiency Results in Age-Dependent Loss of Bone Mass and Progressive Reduction of Mesenchymal Progenitor Cells. J. Bone Miner. Res. 2010, 25, 2138–2147. [Google Scholar] [CrossRef]
- Tazaki, Y.; Sugitani, K.; Ogai, K.; Kobayashi, I.; Kawasaki, H.; Aoyama, T.; Suzuki, N.; Tabuchi, Y.; Hattori, A.; Kitamura, K. RANKL, Ephrin-Eph and Wnt10b Are Key Intercellular Communication Molecules Regulating Bone Remodeling in Autologous Transplanted Goldfish Scales. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2018, 225, 46–58. [Google Scholar] [CrossRef]
- Zhao, X.; Ma, Y.; Luo, J.; Xu, K.; Tian, P.; Lu, C.; Song, J. Blocking the WNT/β-Catenin Pathway in Cancer Treatment:Pharmacological Targets and Drug Therapeutic Potential. Heliyon 2024, 10, e35989. [Google Scholar] [CrossRef]
- Geng, W.; Guo, X.; Zhang, L.; Ma, Y.; Wang, L.; Liu, Z.; Ji, H.; Xiong, Y. Resveratrol Inhibits Proliferation, Migration and Invasion of Multiple Myeloma Cells via NEAT1-Mediated Wnt/β-Catenin Signaling Pathway. Biomed. Pharmacother. 2018, 107, 484–494. [Google Scholar] [CrossRef]
- Kulig, P.; Milczarek, S.; Bakinowska, E.; Szalewska, L.; Baumert, B.; Machaliński, B. Lenalidomide in Multiple Myeloma: Review of Resistance Mechanisms, Current Treatment Strategies and Future Perspectives. Cancers 2023, 15, 963. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, Y.; Tang, J.; Qin, S.; Shen, X.; He, S.; Ju, S. CAM-DR: Mechanisms, Roles and Clinical Application in Tumors. Front. Cell Dev. Biol. 2021, 9, 698047. [Google Scholar] [CrossRef] [PubMed]
- Okoye, U.C.; Malbon, C.C.; Wang, H. Wnt and Frizzled RNA Expression in Human Mesenchymal and Embryonic (H7) Stem Cells. J. Mol. Signal 2008, 3, 16. [Google Scholar] [CrossRef]
- Zeng, F.-R.; Zhou, X.-Y.; Zeng, L.-G.; Sun, J.-C.; He, F.; Mo, W.; Wen, Y.; Wang, S.-Y.; Liu, Q.; Guo, L.-L. Identification of Key Genes and Pathway Related to Chemoresistance of Small Cell Lung Cancer through an Integrative Bioinformatics Analysis. Ann. Transl. Med. 2022, 10, 968. [Google Scholar] [CrossRef]
- Niu, Q.; Liu, Z.; Gao, J.; Wang, Q. MiR-338-3p Enhances Ovarian Cancer Cell Sensitivity to Cisplatin by Downregulating WNT2B. Yonsei Med. J. 2019, 60, 1146. [Google Scholar] [CrossRef]
- General Assembly of the World Medical Association. World Medical Association Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects. J. Am. Coll. Dent. 2014, 81, 14–18. [Google Scholar]
- Emde-Rajaratnam, M.; Beck, S.; Benes, V.; Salwender, H.; Bertsch, U.; Scheid, C.; Hänel, M.; Weisel, K.; Hielscher, T.; Raab, M.S.; et al. RNA-Sequencing Based First Choice of Treatment and Determination of Risk in Multiple Myeloma. Front. Immunol. 2023, 14, 1286700. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- BabrahamBioinformatics. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 27 October 2024).
- Homo Sapiens Genome Assembly GRCh38.P14. Available online: https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_000001405.40/ (accessed on 25 October 2024).
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Homo Sapiens Genome Assembly GRCh38.P14. Available online: https://ftp.ncbi.nlm.nih.gov/genomes/all/GCA/000/001/405/GCA_000001405.29_GRCh38.p14/ (accessed on 25 October 2024).
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, M.-C.; Konaté, M.M.; Chen, L.; Das, B.; Karlovich, C.; Williams, P.M.; Evrard, Y.A.; Doroshow, J.H.; McShane, L.M. TPM, FPKM, or Normalized Counts? A Comparative Study of Quantification Measures for the Analysis of RNA-Seq Data from the NCI Patient-Derived Models Repository. J. Transl. Med. 2021, 19, 269. [Google Scholar] [CrossRef] [PubMed]
- RDocumantation. Pheatmap: A Function to Draw Clustered Heatmaps. Available online: https://www.rdocumentation.org/packages/pheatmap/versions/1.0.13/topics/pheatmap (accessed on 28 October 2024).
- Mendeleeva, L.P.; Votyakova, O.M.; Pokrovskaya, O.S.; Rekhtina, I.G.; Darskaya, E.I.; Galtseva, I.V.; Kaplanov, K.D.; Motorin, D.V.; Samoylova, O.S.; Semochkin, S.V.; et al. National Clinical Recommendations on Diagnosis and Treatment of Multiple Myeloma. Hematol. Transfusiology Russ. J. (Gematol. I Transfusiologiya) 2016, 61, 1–24. [Google Scholar] [CrossRef]
- Ivanovic, Z. Hypoxia or in Situ Normoxia: The Stem Cell Paradigm. J. Cell Physiol. 2009, 219, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Nawata, M.; Wakitani, S. Expression Profiles and Functional Analyses of Wnt-Related Genes in Human Joint Disorders. Am. J. Pathol. 2005, 167, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Dobrynin, M.A.; Korchagina, N.M.; Prjibelski, A.D.; Shafranskaya, D.; Ostromyshenskii, D.I.; Shunkina, K.; Stepanova, I.; Kotova, A.V.; Podgornaya, O.I.; Enukashvily, N.I. Human Pericentromeric Tandemly Repeated DNA Is Transcribed at the End of Oocyte Maturation and Is Associated with Membraneless Mitochondria-Associated Structures. Sci. Rep. 2020, 10, 19634. [Google Scholar] [CrossRef]
- Faget, L.; Hnasko, T.S. Tyramide Signal Amplification for Immunofluorescent Enhancement. Methods Mol. Biol. 2015, 1318, 161–172. [Google Scholar]
- Stritt, M.; Stalder, A.K.; Vezzali, E. Orbit Image Analysis: An Open-Source Whole Slide Image Analysis Tool. PLoS Comput. Biol. 2020, 16, e1007313. [Google Scholar] [CrossRef]
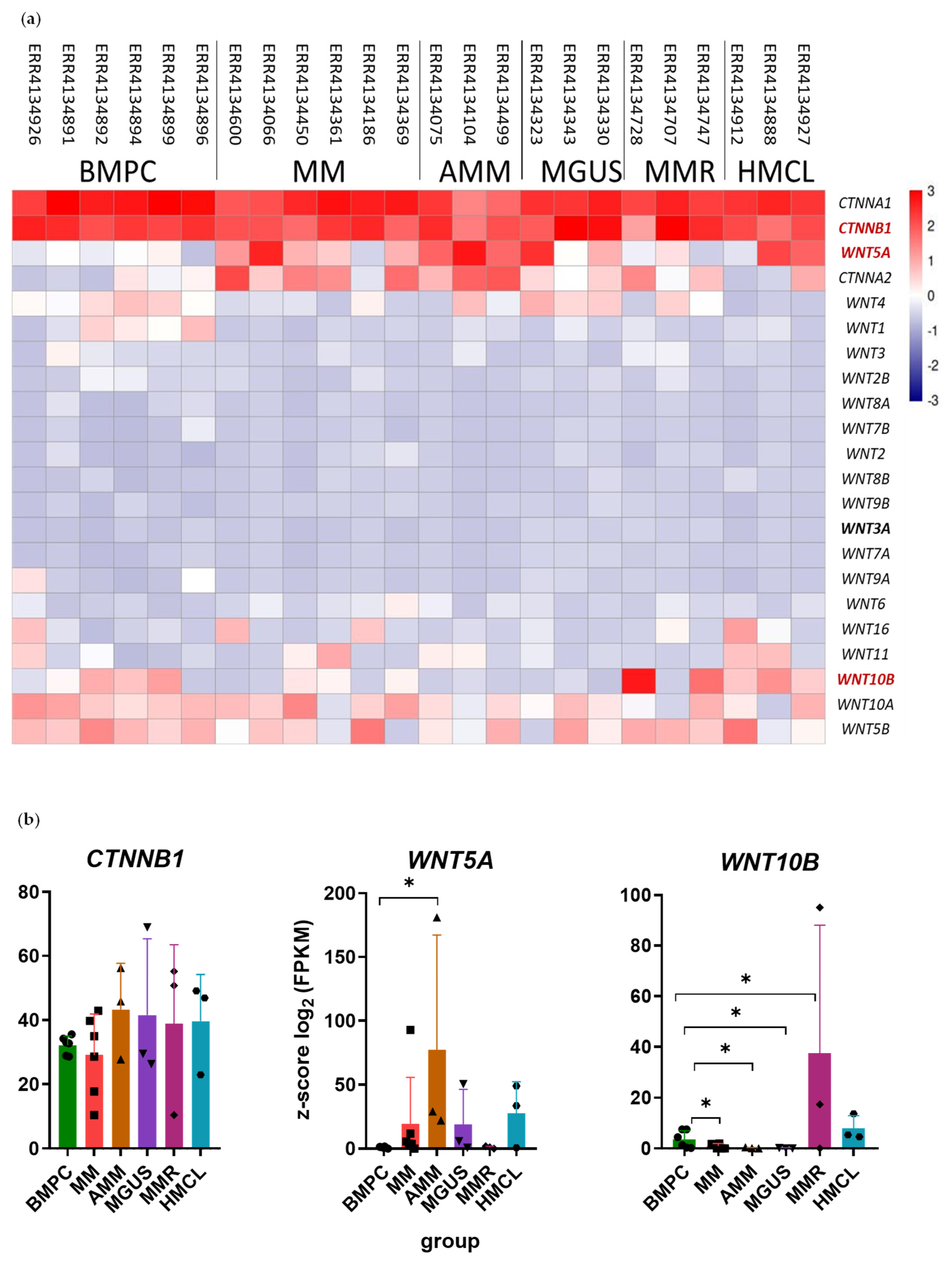

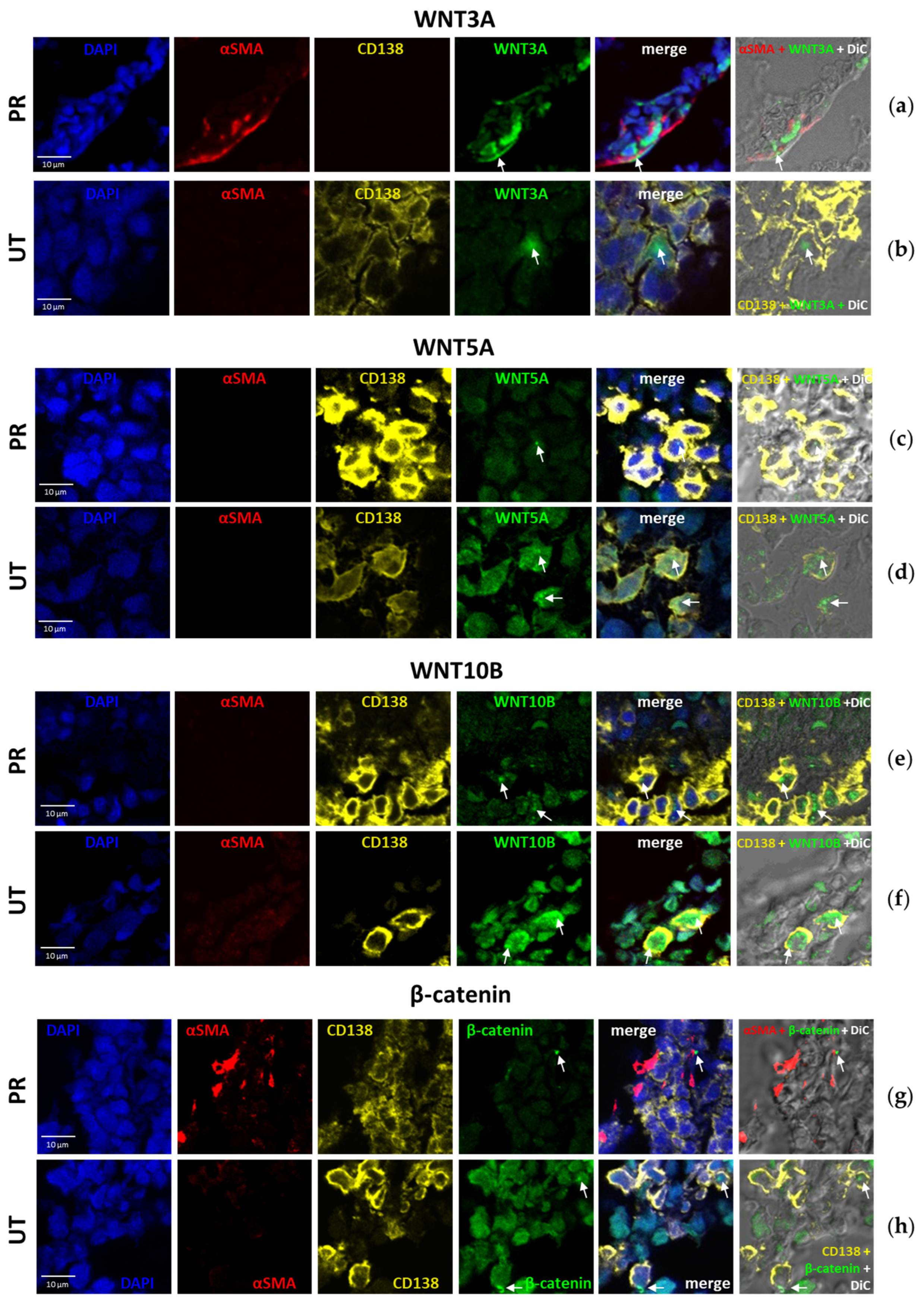

| UT * | PoCR | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MSCs | PCs | Others | MSCs | PCs | Others | |||||||
| Stained Cells, % | Signals per 3 Fields of View | Stained Cells, % | Signals per 3 Fields of View | Stained Cells, % | Signals per 3 Fields of View | Stained Cells, % | Signals per 3 Fields of View | Stained Cells, % | Signals per 3 Fields of View | Stained Cells, % | Signals per 3 Fields of View | |
| WNT3A | 11.75 ± 7.27 | 10 | 17.09 ± 18.92 | 94 | 0 | 41 | 36.23 ± 5.7 | 223 | 60.97 ± 10.7 | 15 | 0 | 356 |
| WNT5A | 5.09 ± 4.49 | 19 | 1.24 ± 2.56 | 77 | 0 | 39 | 3.94 ± 2.77 | 5 | 2.72 ± 0.59 | 4 | 0 | 2 |
| WNT10B | 21.5 ± 5.58 | 6 | 15.74 ± 4.92 | 12 | 0 | 28 | - | 1 | 21.64 ± 2.67 | 46 | 0 | 122 |
| β-catenin | 26.68 ± 8.21 | 35 | 1.24 ± 2.02 | 22 | 0 | 49 | 54.55 | 6 | 5.10 ± 2.62 | 30 | 0 | 159 |
| Group | Age | Type of Response * | M-Protein Type | BM Infiltration, % (Pre-Treatment) | Myelogram, % PCs (Pre- and Post-Treatment) | Karyotype | Therapy ** |
|---|---|---|---|---|---|---|---|
| PoCR PCs < 10% (n = 6) | 65 | VGPR | IgG kappa | 40 | Before: 43.6 After: 0 | ? | 4 CVD courses, 6 DaraRd courses |
| 61 | VGPR | IgG lambda | 20–30 | After: 2.2 | 13q14 and 13q34 deletions | 3 CVD courses, autoHSCT | |
| 59 | PR | IgG kappa | 60–70 | Before: 8.8 After: 5.6 | ? | 3 CVD courses, 1 RVD course, autoHSCT | |
| 69 | PR | IgA kappa | 70–80 | ? | ? | 5 VRD courses | |
| 64 | PR | IgA kappa | Single cells | After: 2.8 | No abnormalities | autoHSCT | |
| 54 | SD | - (non-secretory MM) | 10 | After: 6 | ? | 4 CVD courses | |
| NR (n = 1) | 61 | SD | IgA kappa | 50 | After: 14.4 | No abnormalities | 3 VCD courses, autoHSCT |
| UT (n = 8) (Trephine biopsies: n = 9 (2 trephine biopsies were taken from one patient)) | 83 | - | IgG lambda | 15 | ? | ? | - |
| 83 | - | IgG lambda | 30–40 | 46.8 | ? | - | |
| 72 | - | IgA kappa | 30–40 | 21.6 | ? | - | |
| 71 | - | IgG kappa | 95 | 42.6 | 17p/TP53 deletion | - | |
| 58 | - | IgA kappa | 70–80 | ? | ? | - | |
| 60 | - | ? | 90 | ? | ? | - | |
| 70 | - | IgG lambda | 80–90 | 36.4 | ? | - | |
| 56 | - | - (non-secretory MM) | 80 | 6 | ? | - |
| Group | Age | Type of Response * | M-Protein Type | BM Infiltration, % (Post-Treatment) | Myelogram, % PCs (Post-Treatment) | Karyotype | Therapy ** |
|---|---|---|---|---|---|---|---|
| PoCR PC < 10% (n = 3) | 61 | CR | IgG lambda | 1–2 | 1 | No abnormalities | 5 VCD courses, autoHSCT |
| 59 | CR | IgG kappa | 1–2 | 2.6 | No abnormalities | 6 VD courses | |
| 59 | VGPR | IgG lambda | 1–2 | 2.8 | No abnormalities | VRD, autoHSCT | |
| NR (n = 3) | 50 | SD | IgG kappa | 50 | 47 | 14q32 monosomy, 13q14 and 13q34 deletions, TP53/17p13 deletion or monosomy | 6 VD courses, autoHSCT |
| 61 | SD | IgA kappa | 50 | 14.4 | No abnormalities | 3 VCD courses, autoHSCT | |
| 70 | SD | IgG lambda | 90 | 82.4 | Y chromosome loss | 2 VCD courses | |
| UT (n = 3) | 64 | - | ? | ? | 66 | ? | - |
| ? | - | IgG kappa | ? | ? | ? | - | |
| ? | - | ? | ? | ? | ? | - |
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| WNT3 | 5′-GGAGAAGCGGAAGGAAAAATG-3′ | 5′-GCACGTCGTAGATGCGAATACA-3′ |
| WNT3A | 5′-CCTGCACTCCATCCAGCTACA-3′ | 5′-GACCTCTCTTCCTACCTTTCCCTTA-3′ |
| WNT5A | 5′-GAAATGCGTGTTGGGTTGAA-3′ | 5′-ATGCCCTCTCCACAAAGTGAA-3′ |
| WNT5B | 5′-CTGCCTTTCCAGCGAGAATT-3′ | 5′-AGGTCAAATGGCCCCCTTT-3′ |
| WNT7B | 5′-CCCGGCAAGTTCTCTTTCTTC-3′ | 5′-GGCGTAGCTTTTCTGTGTCCAT-3′ |
| WNT8B | 5′-TCCCAGAAAAACTGAGGAAACTG-3′ | 5′-AACCTCTGCCTCTAGGAACCAA-3′ |
| WNT10B | 5′-CTTTTCAGCCCTTTGCTCTGAT-3′ | 5′-CCCCTAAAGCTGTTTCCAGGTA-3′ |
| WNT2B (previously WNT13) | 5′-TGCCAAGGAGAAGAGGCTTAAG-3′ | 5′-GTGCGACCACAGCGGTTATT-3′ |
| WNT9A (previously WNT14) | 5′-CTTAAGTACAGCAGCAAGTTCGTCAA-3′ | 5′-CCACGAGGTTGTTGTGGAAGT-3′ |
| WNT9B (previously WNT15) | 5′-CAGGTGCTGAAACTGCGCTAT-3′ | 5′-GCCCAAGGCCTCATTGGT-3′ |
| CTNNB1 | 5′-CTGCTGTTTTGTTCCGAATGTC-3′ | 5′-CCATTGGCTCTGTTCTGAAGAGA-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belik, L.A.; Enukashvily, N.I.; Semenova, N.Y.; Ostromyshenskii, D.I.; Motyko, E.V.; Kirienko, A.N.; Kustova, D.V.; Bessmeltsev, S.S.; Sidorkevich, S.V.; Martynkevich, I.S. Expression of WNT Family Genes in Mesenchymal Stromal Cells of the Hematopoietic Niche in Patients with Different Responses to Multiple Myeloma Treatment. Int. J. Mol. Sci. 2025, 26, 6236. https://doi.org/10.3390/ijms26136236
Belik LA, Enukashvily NI, Semenova NY, Ostromyshenskii DI, Motyko EV, Kirienko AN, Kustova DV, Bessmeltsev SS, Sidorkevich SV, Martynkevich IS. Expression of WNT Family Genes in Mesenchymal Stromal Cells of the Hematopoietic Niche in Patients with Different Responses to Multiple Myeloma Treatment. International Journal of Molecular Sciences. 2025; 26(13):6236. https://doi.org/10.3390/ijms26136236
Chicago/Turabian StyleBelik, Liubov A., Natella I. Enukashvily, Natalia Y. Semenova, Dmitrii I. Ostromyshenskii, Ekaterina V. Motyko, Anna N. Kirienko, Daria V. Kustova, Stanislav S. Bessmeltsev, Sergey V. Sidorkevich, and Irina S. Martynkevich. 2025. "Expression of WNT Family Genes in Mesenchymal Stromal Cells of the Hematopoietic Niche in Patients with Different Responses to Multiple Myeloma Treatment" International Journal of Molecular Sciences 26, no. 13: 6236. https://doi.org/10.3390/ijms26136236
APA StyleBelik, L. A., Enukashvily, N. I., Semenova, N. Y., Ostromyshenskii, D. I., Motyko, E. V., Kirienko, A. N., Kustova, D. V., Bessmeltsev, S. S., Sidorkevich, S. V., & Martynkevich, I. S. (2025). Expression of WNT Family Genes in Mesenchymal Stromal Cells of the Hematopoietic Niche in Patients with Different Responses to Multiple Myeloma Treatment. International Journal of Molecular Sciences, 26(13), 6236. https://doi.org/10.3390/ijms26136236







