Structural Insights into the Regulatory Mechanisms of the Toxic Activity of Sofic in Anti-Phage Defense Systems
Abstract
1. Introduction
2. Results
2.1. The Small Protein AS1 Counteracts the Toxic Effects of Sofic in E. coli
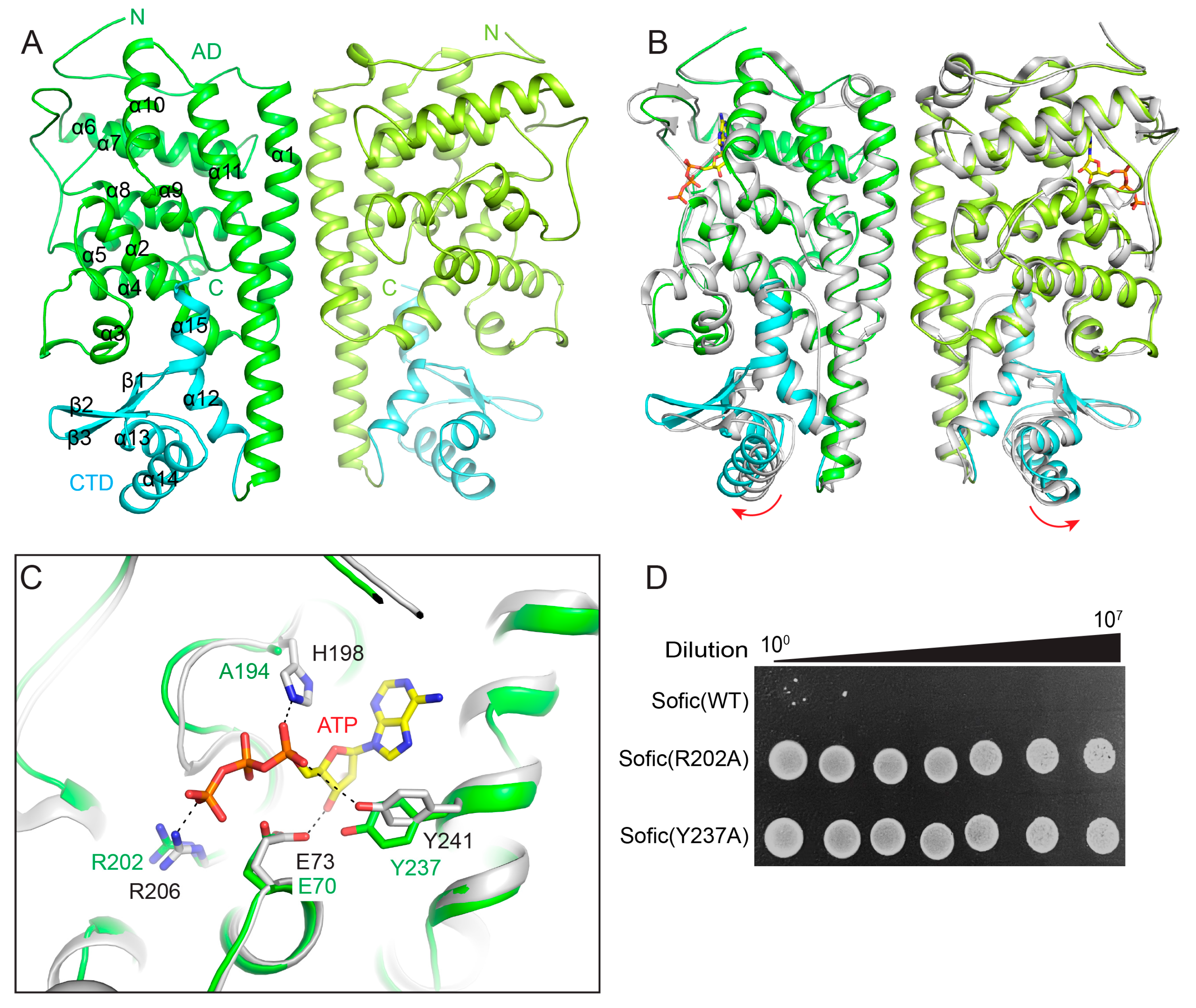
2.2. The Crystal Structure of Apo Sofic
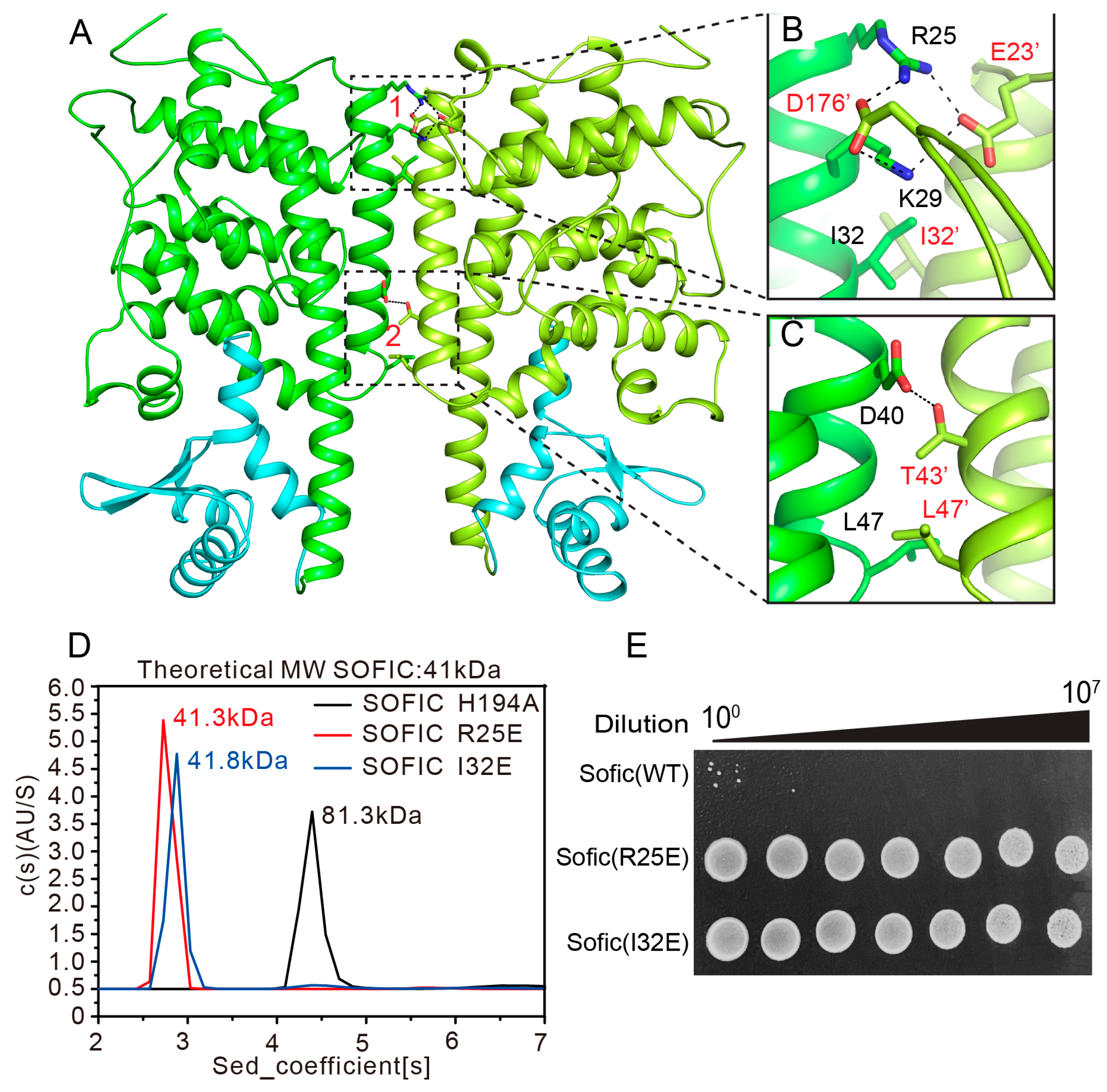
2.3. Dimerization of Sofic Is Required for Its Toxic Activity
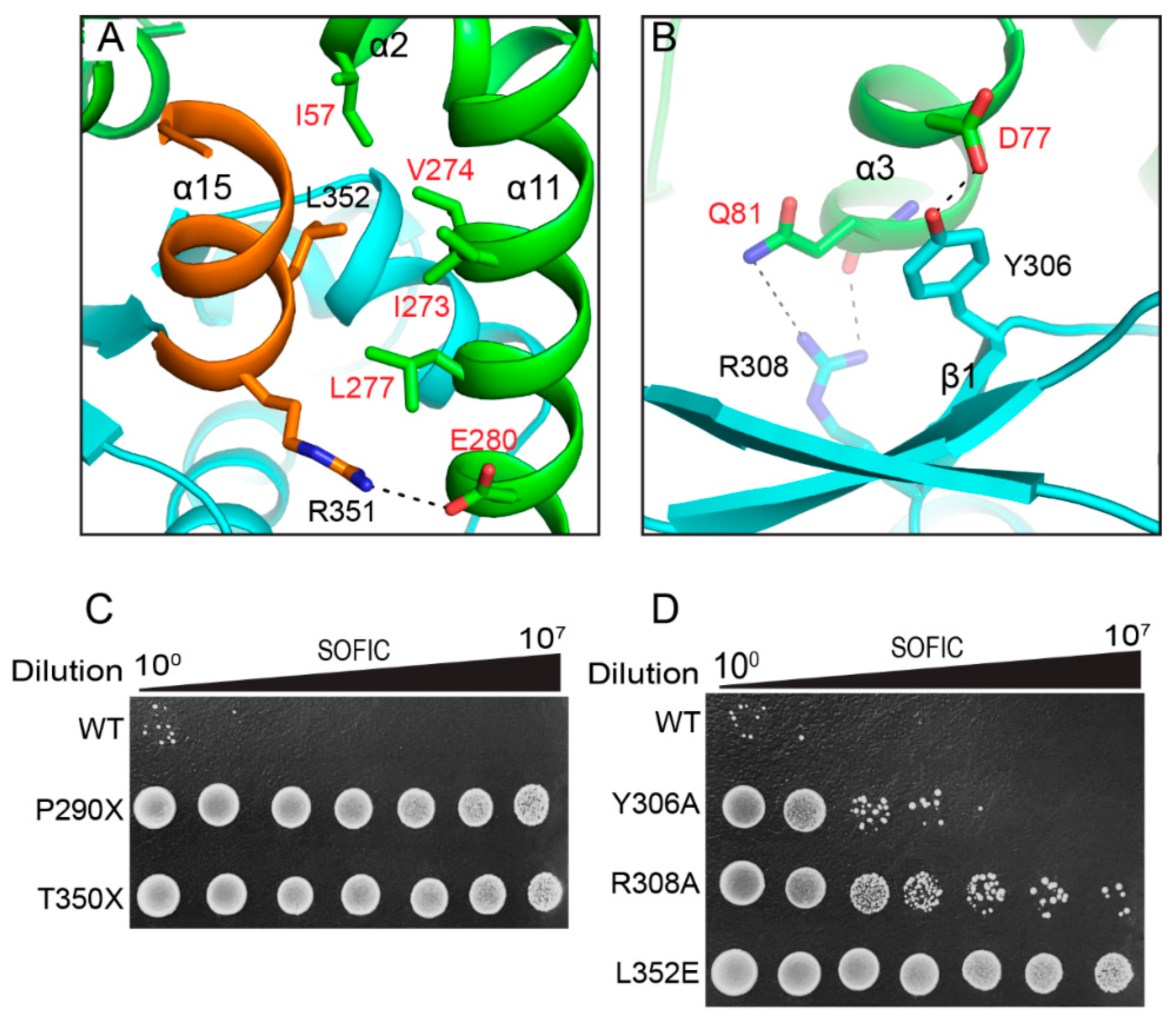
2.4. The C-Terminal Domain of EcSofic Regulates Its Toxic Activity
3. Discussion
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Georjon, H.; Bernheim, A. The highly diverse antiphage defence systems of bacteria. Nat. Rev. Microbiol. 2023, 21, 686–700. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Altae-Tran, H.; Bohning, F.; Makarova, K.S.; Segel, M.; Schmid-Burgk, J.L.; Koob, J.; Wolf, Y.I.; Koonin, E.V.; Zhang, F. Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 2020, 369, 1077–1084. [Google Scholar] [CrossRef] [PubMed]
- Doron, S.; Melamed, S.; Ofir, G.; Leavitt, A.; Lopatina, A.; Keren, M.; Amitai, G.; Sorek, R. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 2018, 359, eaar4120. [Google Scholar] [CrossRef]
- Patel, P.H.; Taylor, V.L.; Zhang, C.; Getz, L.J.; Fitzpatrick, A.D.; Davidson, A.R.; Maxwell, K.L. Anti-phage defence through inhibition of virion assembly. Nat. Commun. 2024, 15, 1644. [Google Scholar] [CrossRef]
- Loeff, L.; Walter, A.; Rosalen, G.T.; Jinek, M. DNA end sensing and cleavage by the Shedu anti-phage defense system. Cell 2025, 188, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Bernheim, A.; Sorek, R. The pan-immune system of bacteria: Antiviral defence as a community resource. Nat. Rev. Microbiol. 2020, 18, 113–119. [Google Scholar] [CrossRef]
- Lopatina, A.; Tal, N.; Sorek, R. Abortive Infection: Bacterial Suicide as an Antiviral Immune Strategy. Annu. Rev. Virol. 2020, 7, 371–384. [Google Scholar] [CrossRef]
- Shen, Z.; Lin, Q.; Yang, X.Y.; Fosuah, E.; Fu, T.M. Assembly-mediated activation of the SIR2-HerA supramolecular complex for anti-phage defense. Mol. Cell 2023, 83, 4586–4599. [Google Scholar] [CrossRef]
- Tamulaitiene, G.; Sabonis, D.; Sasnauskas, G.; Ruksenaite, A.; Silanskas, A.; Avraham, C.; Ofir, G.; Sorek, R.; Zaremba, M.; Siksnys, V. Activation of Thoeris antiviral system via SIR2 effector filament assembly. Nature 2024, 627, 431–436. [Google Scholar] [CrossRef]
- Wang, R.; Xu, Q.; Wu, Z.; Li, J.; Guo, H.; Liao, T.; Shi, Y.; Yuan, L.; Gao, H.; Yang, R.; et al. The structural basis of the activation and inhibition of DSR2 NADase by phage proteins. Nat. Commun. 2024, 15, 6185. [Google Scholar] [CrossRef]
- Garb, J.; Lopatina, A.; Bernheim, A.; Zaremba, M.; Siksnys, V.; Melamed, S.; Leavitt, A.; Millman, A.; Amitai, G.; Sorek, R. Multiple phage resistance systems inhibit infection via SIR2-dependent NAD(+) depletion. Nat. Microbiol. 2022, 7, 1849–1856. [Google Scholar] [CrossRef]
- Hobbs, S.J.; Kranzusch, P.J. Nucleotide Immune Signaling in CBASS, Pycsar, Thoeris, and CRISPR Antiphage Defense. Annu. Rev. Microbiol. 2024, 78, 255–276. [Google Scholar] [CrossRef]
- LeRoux, M.; Srikant, S.; Teodoro, G.I.C.; Zhang, T.; Littlehale, M.L.; Doron, S.; Badiee, M.; Leung, A.K.L.; Sorek, R.; Laub, M.T. The DarTG toxin-antitoxin system provides phage defence by ADP-ribosylating viral DNA. Nat. Microbiol. 2022, 7, 1028–1040. [Google Scholar] [CrossRef]
- Jurenas, D.; Fraikin, N.; Goormaghtigh, F.; Van Melderen, L. Biology and evolution of bacterial toxin-antitoxin systems. Nat. Rev. Microbiol. 2022, 20, 335–350. [Google Scholar] [CrossRef]
- Kelly, A.; Arrowsmith, T.J.; Went, S.C.; Blower, T.R. Toxin-antitoxin systems as mediators of phage defence and the implications for abortive infection. Curr. Opin. Microbiol. 2023, 73, 102293. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Goepfert, A.; Stanger, F.V.; Harms, A.; Schmidt, A.; Schirmer, T.; Dehio, C. Adenylylation control by intra- or intermolecular active-site obstruction in Fic proteins. Nature 2012, 482, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Roy, C.R.; Cherfils, J. Structure and function of Fic proteins. Nat. Rev. Microbiol. 2015, 13, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Worby, C.A.; Mattoo, S.; Kruger, R.P.; Corbeil, L.B.; Koller, A.; Mendez, J.C.; Zekarias, B.; Lazar, C.; Dixon, J.E. The fic domain: Regulation of cell signaling by adenylylation. Mol. Cell 2009, 34, 93–103. [Google Scholar] [CrossRef]
- Luong, P.; Kinch, L.N.; Brautigam, C.A.; Grishin, N.V.; Tomchick, D.R.; Orth, K. Kinetic and structural insights into the mechanism of AMPylation by VopS Fic domain. J. Biol. Chem. 2010, 285, 20155–20163. [Google Scholar] [CrossRef]
- Xiao, J.; Worby, C.A.; Mattoo, S.; Sankaran, B.; Dixon, J.E. Structural basis of Fic-mediated adenylylation. Nat. Struct. Mol. Biol. 2010, 17, 1004–1010. [Google Scholar] [CrossRef]
- Yarbrough, M.L.; Li, Y.; Kinch, L.N.; Grishin, N.V.; Ball, H.L.; Orth, K. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 2009, 323, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Goepfert, A.; Stanger, F.V.; Dehio, C.; Schirmer, T. Conserved inhibitory mechanism and competent ATP binding mode for adenylyltransferases with Fic fold. PLoS ONE 2013, 8, e64901. [Google Scholar] [CrossRef]
- Das, D.; Krishna, S.S.; McMullan, D.; Miller, M.D.; Xu, Q.; Abdubek, P.; Acosta, C.; Astakhova, T.; Axelrod, H.L.; Burra, P.; et al. Crystal structure of the Fic (Filamentation induced by cAMP) family protein SO4266 (gi|24375750) from Shewanella oneidensis MR-1 at 1.6 A resolution. Proteins 2009, 75, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Millman, A.; Melamed, S.; Leavitt, A.; Doron, S.; Bernheim, A.; Hor, J.; Garb, J.; Bechon, N.; Brandis, A.; Lopatina, A.; et al. An expanded arsenal of immune systems that protect bacteria from phages. Cell Host. Microbe. 2022, 30, 1556–1569. [Google Scholar] [CrossRef]
- Stokar-Avihail, A.; Fedorenko, T.; Hor, J.; Garb, J.; Leavitt, A.; Millman, A.; Shulman, G.; Wojtania, N.; Melamed, S.; Amitai, G.; et al. Discovery of phage determinants that confer sensitivity to bacterial immune systems. Cell 2023, 186, 1863–1876. [Google Scholar] [CrossRef]
- Jdeed, G.; Morozova, V.V.; Tikunova, N.V. Genome Analysis of Anti-Phage Defense Systems and Defense Islands in Stenotrophomonas maltophilia: Preservation and Variability. Viruses 2024, 16, 1903. [Google Scholar] [CrossRef]
- Beavogui, A.; Lacroix, A.; Wiart, N.; Poulain, J.; Delmont, T.O.; Paoli, L.; Wincker, P.; Oliveira, P.H. The defensome of complex bacterial communities. Nat. Commun. 2024, 15, 2146. [Google Scholar] [CrossRef]
- Sprenger, H.; Kienesberger, S.; Pertschy, B.; Poltl, L.; Konrad, B.; Bhutada, P.; Vorkapic, D.; Atzmuller, D.; Feist, F.; Hogenauer, C.; et al. Fic Proteins of Campylobacter fetus subsp. venerealis Form a Network of Functional Toxin-Antitoxin Systems. Front. Microbiol. 2017, 8, 1965. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.H.; McCloskey, A.; Chen, F.R.; Nakayasu, E.S.; Zhang, L.Q.; Luo, Z.Q. Fic Proteins Inhibit the Activity of Topoisomerase IV by AMPylation in Diverse Bacteria. Front. Microbiol. 2020, 11, 2084. [Google Scholar] [CrossRef] [PubMed]
- Minor, W.; Cymborowski, M.; Otwinowski, Z.; Chruszcz, M. HKL-3000: The integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta. Crystallogr. D. Biol. Crystallogr. 2006, 62, 859–866. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta. Crystallogr. D. Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J. Solving structures of protein complexes by molecular replacement with Phaser. Acta. Crystallogr. D. Biol. Crystallogr. 2007, 63, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta. Crystallogr. D. Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta. Crystallogr. D. Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B., 3rd; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta. Crystallogr. D. Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef]

| Structure | EcSofic |
|---|---|
| PDB ID | 9VJC |
| Data collection | |
| Space group | P1211 |
| Cell dimensions | |
| a, b, c (Å) | 68.595, 58.234, 102.863 |
| α, β, γ (º) | 90, 93.57, 90 |
| Resolution (Å) | 55.39–2.283 (2.364–2.283) * |
| Rpim (%) | 6.0 (28.0) |
| I/σI | 11.5 (3.0) |
| CC (1/2) | 0.995(0.922) |
| Completeness (%) | 94.36 (86.80) |
| Refinement | |
| No. of reflections | 35,092 (3222) |
| Rwork/Rfree (%) | 0.2117 (0.2767)/0.2666 (0.3527) |
| Ramachandran | |
| Favored (%) | 99.72 |
| Outlier (%) | 0.28 |
| R. m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 0.97 |
| Average B-factor | 31.09 |
| Protein | 30.90 |
| Solvent | 34.26 |
| Number of non-hydrogen atoms | 6125 |
| Protein | 5774 |
| Ligand | 0 |
| Solvent | 351 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Z.; Chen, G.; He, L.; Guo, H.; Yuan, R.; Su, H.; Xie, Z.; Li, F. Structural Insights into the Regulatory Mechanisms of the Toxic Activity of Sofic in Anti-Phage Defense Systems. Int. J. Mol. Sci. 2025, 26, 6074. https://doi.org/10.3390/ijms26136074
Wu Z, Chen G, He L, Guo H, Yuan R, Su H, Xie Z, Li F. Structural Insights into the Regulatory Mechanisms of the Toxic Activity of Sofic in Anti-Phage Defense Systems. International Journal of Molecular Sciences. 2025; 26(13):6074. https://doi.org/10.3390/ijms26136074
Chicago/Turabian StyleWu, Zhuoxi, Guodong Chen, Libang He, Hao Guo, Ruifang Yuan, Huiling Su, Zhenyang Xie, and Faxiang Li. 2025. "Structural Insights into the Regulatory Mechanisms of the Toxic Activity of Sofic in Anti-Phage Defense Systems" International Journal of Molecular Sciences 26, no. 13: 6074. https://doi.org/10.3390/ijms26136074
APA StyleWu, Z., Chen, G., He, L., Guo, H., Yuan, R., Su, H., Xie, Z., & Li, F. (2025). Structural Insights into the Regulatory Mechanisms of the Toxic Activity of Sofic in Anti-Phage Defense Systems. International Journal of Molecular Sciences, 26(13), 6074. https://doi.org/10.3390/ijms26136074






