Epigenetic Remodeling of Regulatory Regions by Indicaxanthin Suggests a Shift in Cell Identity Programs in Colorectal Cancer Cells
Abstract
1. Introduction
2. Results and Discussion
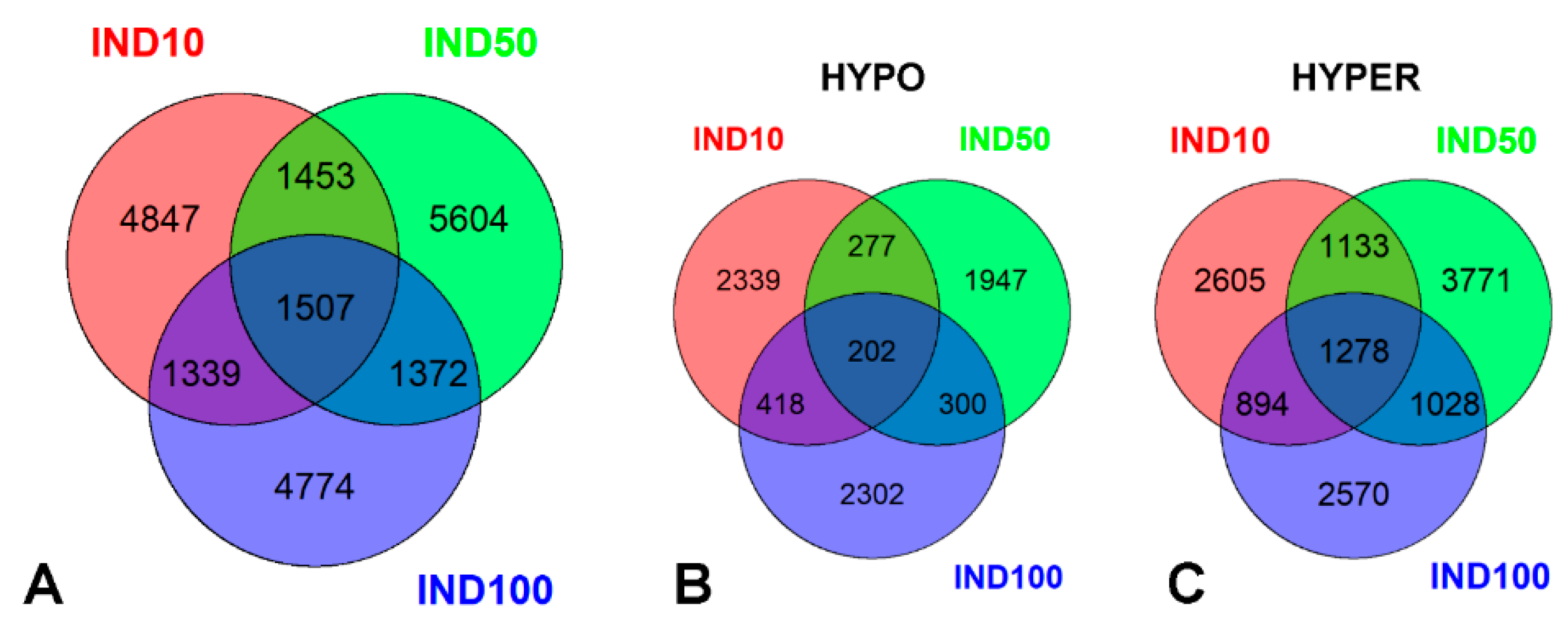
2.1. Indicaxanthin-Induced Methylation Changes in Caco-2 Cells
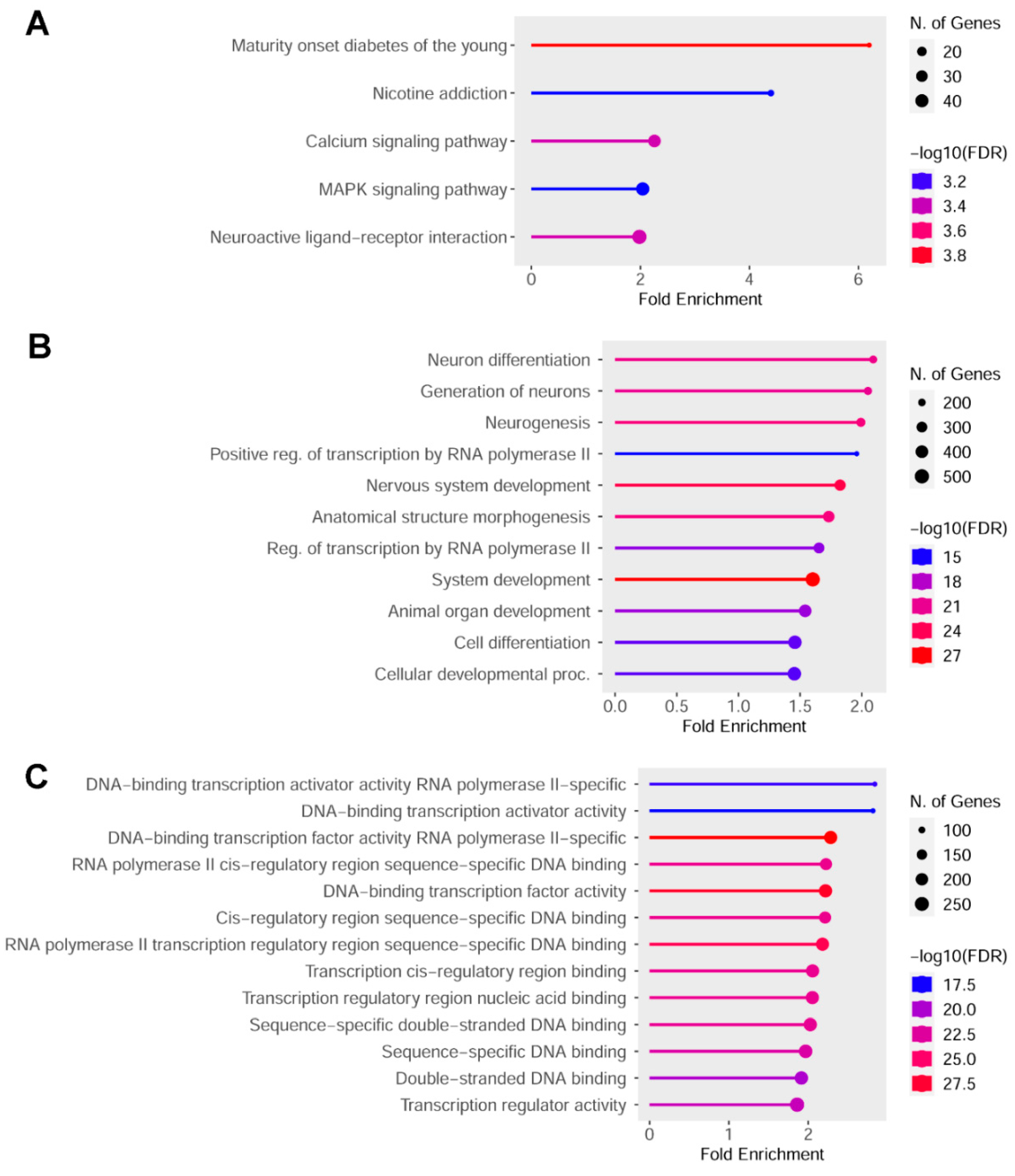
2.2. Epigenetic Impact of Indicaxanthin on Promoter and Regulatory Regions
2.3. Top Hypomethylated and Hypermethylated Targets Induced by Indicaxanthin
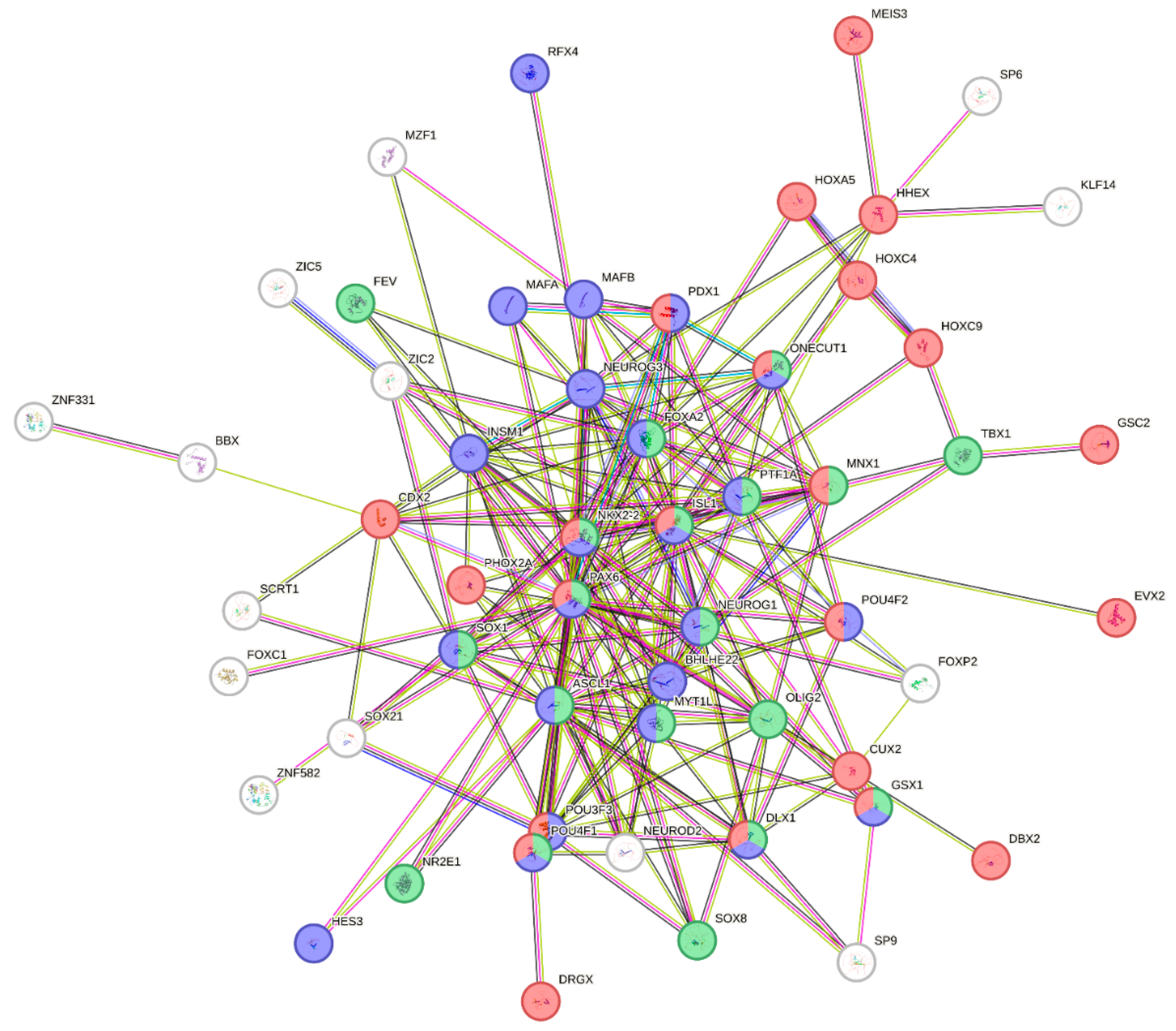
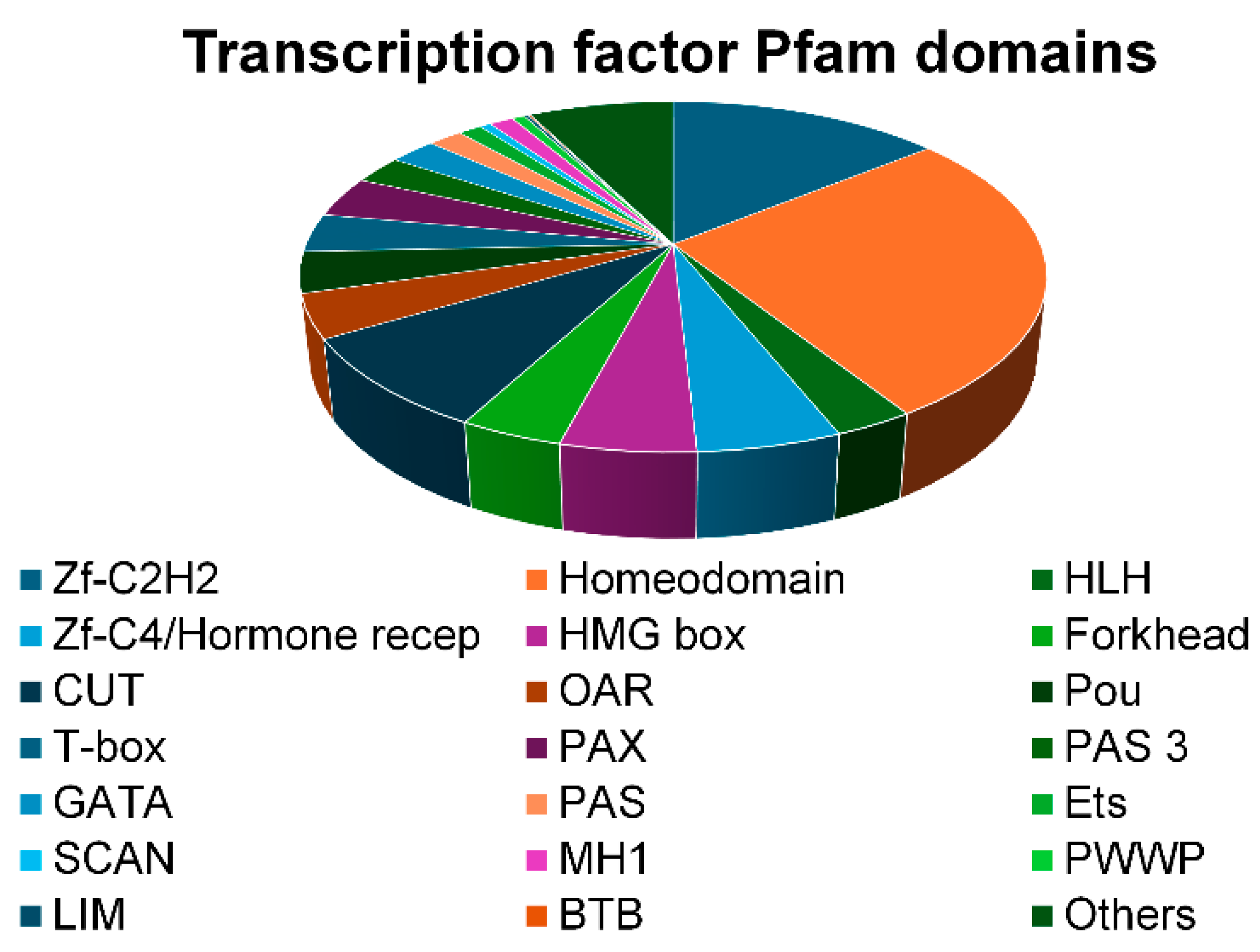
2.4. Indicaxanthin Targeted the Epigenetic Landscape of Several Transcription Factors
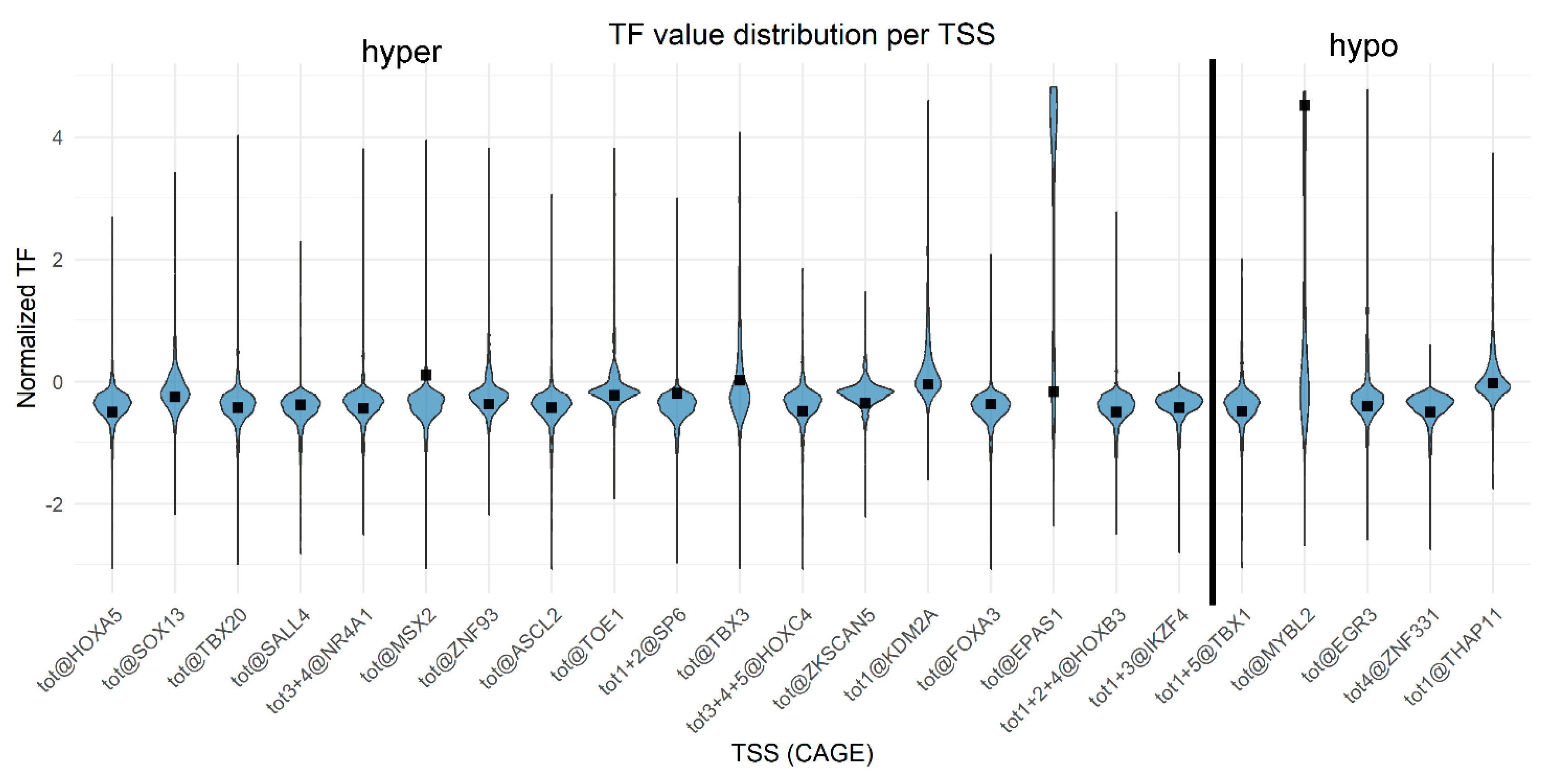
2.5. Analysis of Cell Type Signature (Transcription Factor TSS Expression)
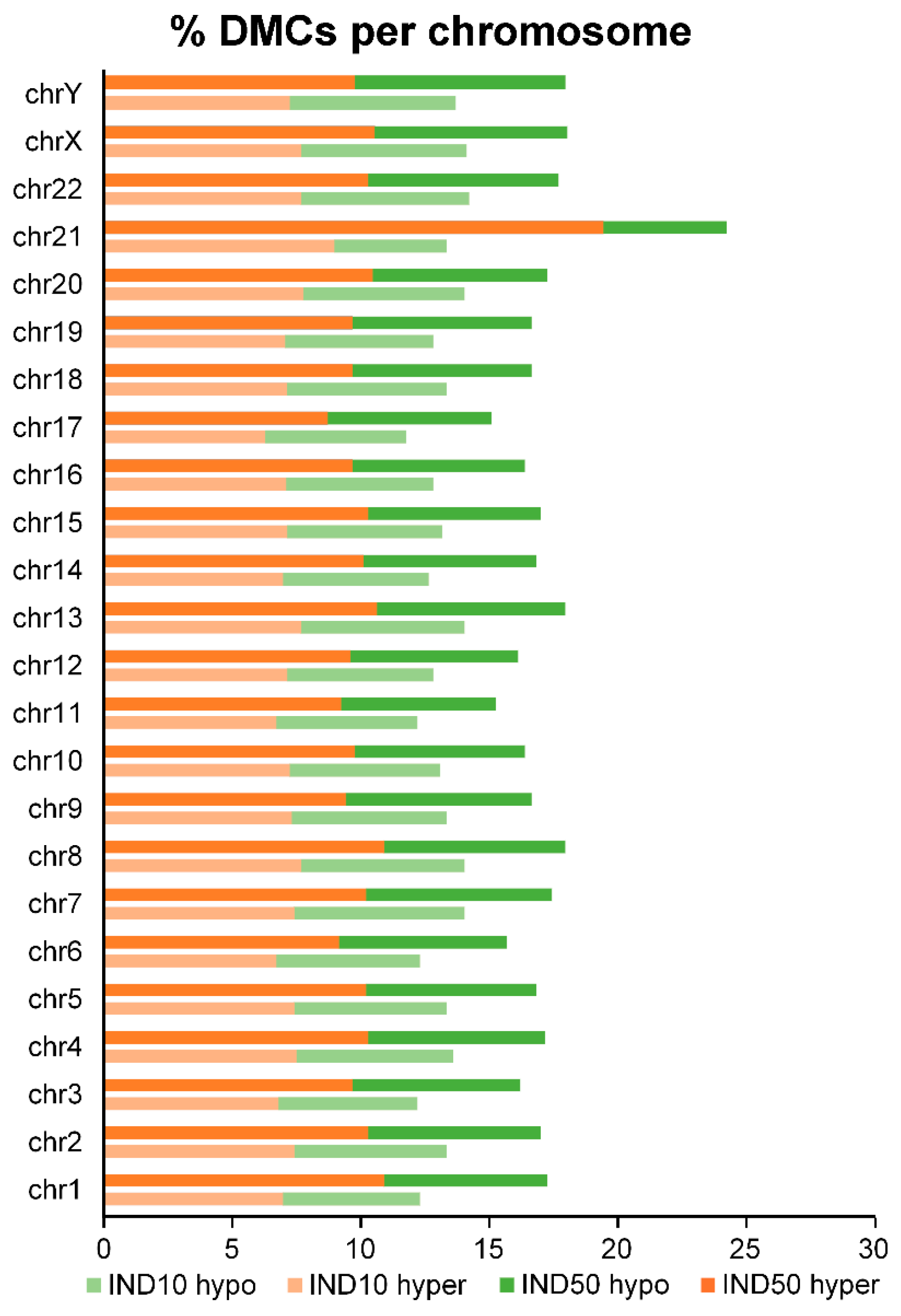
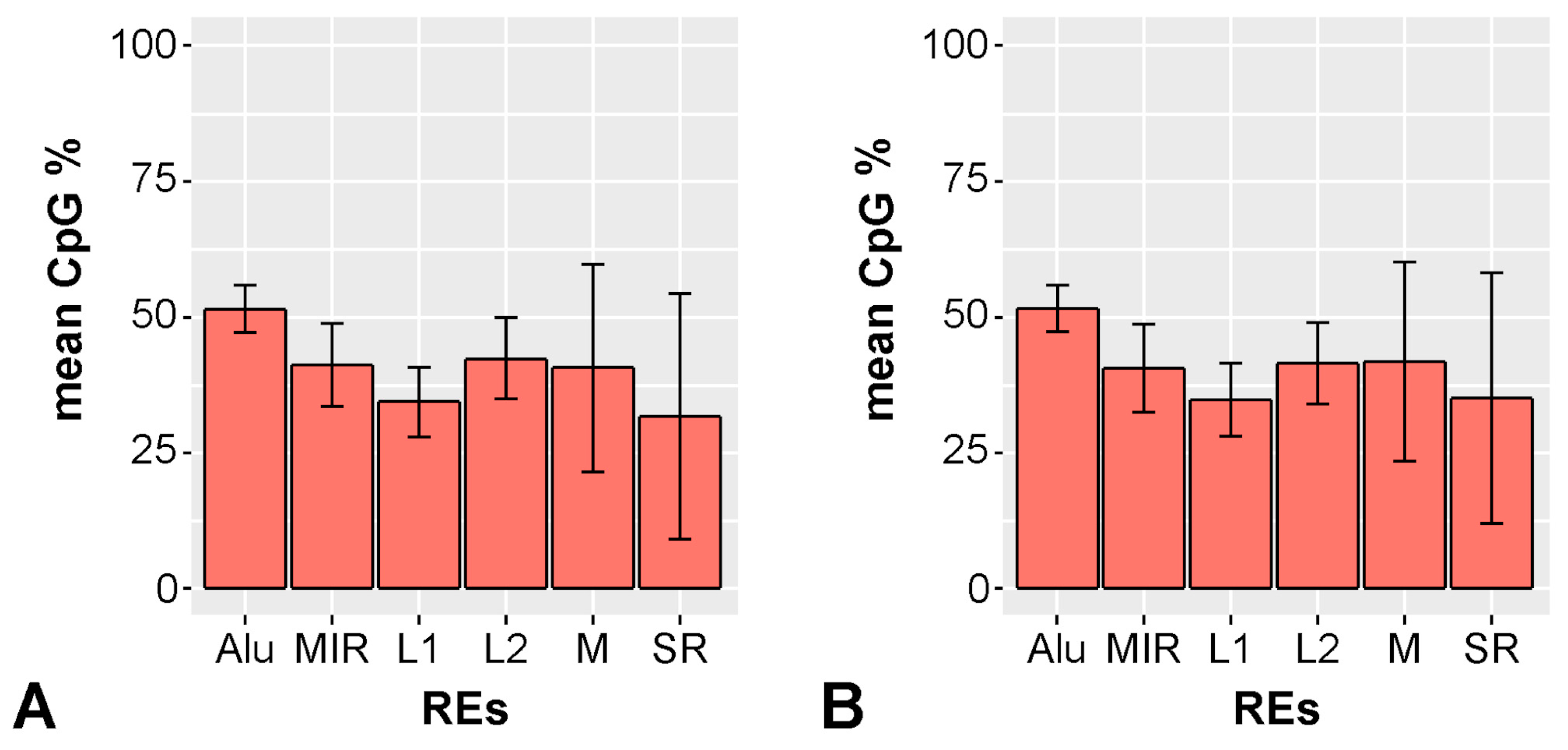
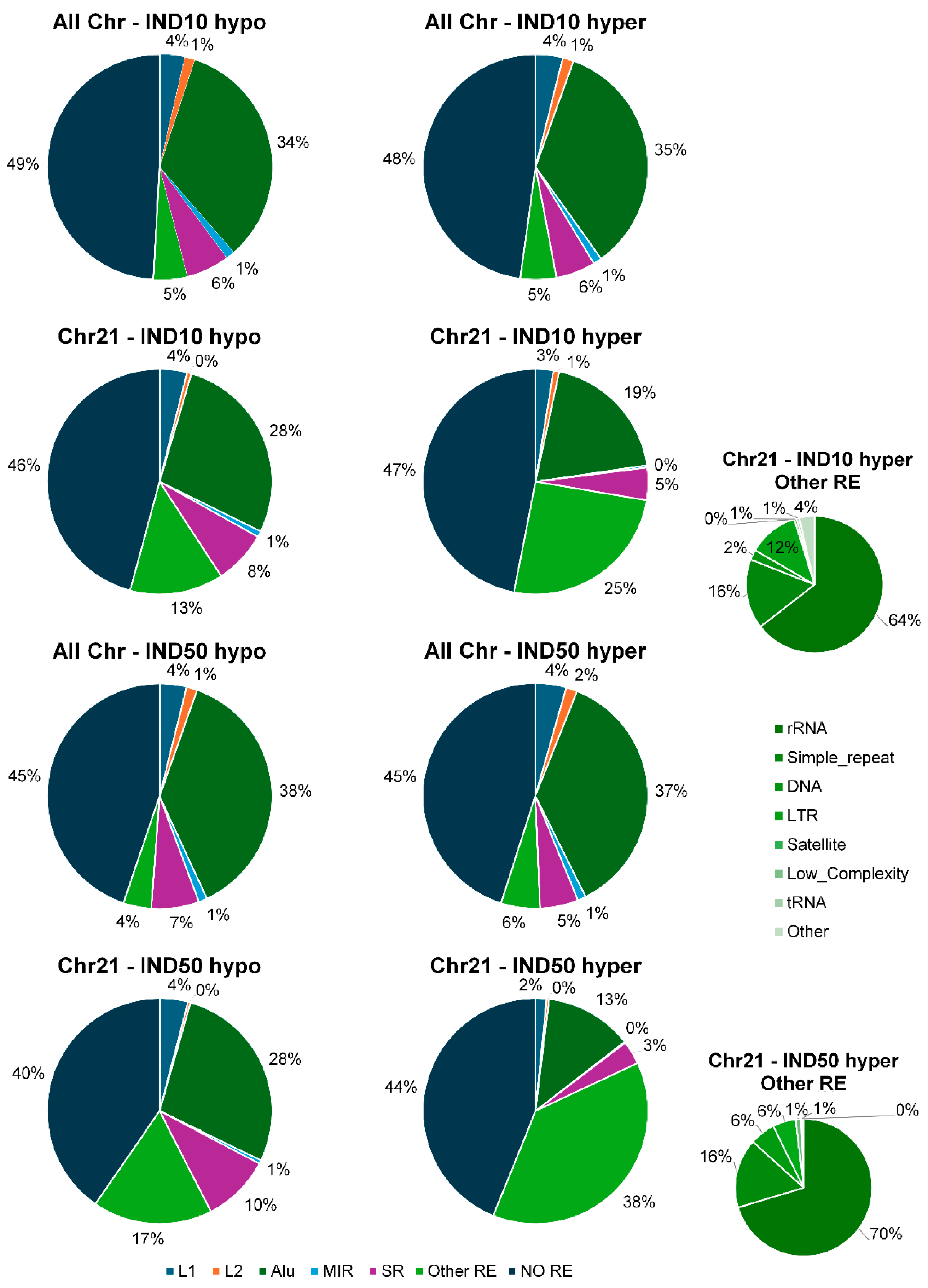
2.6. Indicaxanthin Influences Methylation Dynamics Within Repetitive Genomic Regions
3. Materials and Methods
3.1. Cell Culture and Treatments
3.2. Reduced Representation Bisulfite Sequencing (RRBS) and Differential Methylation Analysis
3.3. Functional Analysis of Differentially Methylated Regions
3.4. Statistical Analysis
3.5. Transcription Factor Analysis
3.6. Analysis of Cell Type Signature
- Maximum effect: minimum TF value for hypermethylated TSSs and maximum TF value for hypomethylated TSSs.
- Medium effect: for hypermethylated TSSs, TF = TF_Caco-2—(median − minimum)/4; for hypomethylated TSSs, TF = TF_ Caco-2 + (maximum − median)/4.
- Low effect: for hypermethylated TSSs, TF = TF_ Caco-2—(median − minimum)/8; for hypomethylated TSSs, TF = TF_ Caco-2 + (maximum − median)/8.
3.7. Methylation Analysis of Repetitive Elements
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Naselli, F.; Volpes, S.; Cardinale, P.S.; Micheli, S.; Cicio, A.; Scoglio, G.D.; Chiarelli, R.; Zizzo, M.G.; Picone, P.; Caradonna, F.; et al. First Evidence of Epigenetic Modulation of Human Gene Methylation by Microalga Aphanizomenon Flos-Aquae (AFA) in Inflammation-Related Pathways in Intestinal Cells. Clin. Epigenet. 2025, 17, 44. [Google Scholar] [CrossRef] [PubMed]
- Volpes, S.; Cruciata, I.; Ceraulo, F.; Schimmenti, C.; Naselli, F.; Pinna, C.; Mauro, M.; Picone, P.; Dallavalle, S.; Nuzzo, D.; et al. Nutritional Epigenomic and DNA-Damage Modulation Effect of Natural Stilbenoids. Sci. Rep. 2023, 13, 658. [Google Scholar] [CrossRef]
- Lakshminarasimhan, R.; Liang, G. The Role of DNA Methylation in Cancer. In DNA Methyltransferases—Role and Function; Jeltsch, A., Jurkowska, R.Z., Eds.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2016; Volume 945, pp. 151–172. ISBN 978-3-319-43622-7. [Google Scholar]
- Artursson, P.; Palm, K.; Luthman, K. Caco-2 Monolayers in Experimental and Theoretical Predictions of Drug transport. Adv. Drug Deliv. Rev. 1996, 22, 67–84, Erratum in Adv. Drug Deliv. Rev. 2001, 46, 27–43. [Google Scholar] [CrossRef]
- Tesoriere, L.; Gentile, C.; Angileri, F.; Attanzio, A.; Tutone, M.; Allegra, M.; Livrea, M.A. Trans-Epithelial Transport of the Betalain Pigments Indicaxanthin and Betanin across Caco-2 Cell Monolayers and Influence of Food Matrix. Eur. J. Nutr. 2013, 52, 1077–1087. [Google Scholar] [CrossRef]
- Mannino, G.; Caradonna, F.; Cruciata, I.; Lauria, A.; Perrone, A.; Gentile, C. Melatonin Reduces Inflammatory Response in Human Intestinal Epithelial Cells Stimulated by Interleukin-1β. J. Pineal Res. 2019, 67, e12598. [Google Scholar] [CrossRef]
- Lechner, J.F.; Stoner, G.D. Red Beetroot and Betalains as Cancer Chemopreventative Agents. Molecules 2019, 24, 1602. [Google Scholar] [CrossRef]
- Tesoriere, L.; Fazzari, M.; Angileri, F.; Gentile, C.; Livrea, M.A. In Vitro Digestion of Betalainic Foods. Stability and Bioaccessibility of Betaxanthins and Betacyanins and Antioxidative Potential of Food Digesta. J. Agric. Food Chem. 2008, 56, 10487–10492. [Google Scholar] [CrossRef]
- Tesoriere, L.; Allegra, M.; Gentile, C.; Livrea, M.A. Betacyanins as Phenol Antioxidants. Chemistry and Mechanistic Aspects of the Lipoperoxyl Radical-Scavenging Activity in Solution and Liposomes. Free Radic. Res. 2009, 43, 706–717. [Google Scholar] [CrossRef]
- Tesoriere, L.; Attanzio, A.; Allegra, M.; Gentile, C.; Livrea, M.A. Indicaxanthin Inhibits NADPH Oxidase (NOX)-1 Activation and NF-κB-Dependent Release of Inflammatory Mediators and Prevents the Increase of Epithelial Permeability in IL-1β-Exposed Caco-2 Cells. Br. J. Nutr. 2014, 111, 415–423. [Google Scholar] [CrossRef]
- Naselli, F.; Belshaw, N.J.; Gentile, C.; Tutone, M.; Tesoriere, L.; Livrea, M.A.; Caradonna, F. Phytochemical Indicaxanthin Inhibits Colon Cancer Cell Growth and Affects the DNA Methylation Status by Influencing Epigenetically Modifying Enzyme Expression and Activity. J. Nutr. Nutr. 2015, 8, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Naselli, F.; Tesoriere, L.; Caradonna, F.; Bellavia, D.; Attanzio, A.; Gentile, C.; Livrea, M.A. Anti-Proliferative and pro-Apoptotic Activity of Whole Extract and Isolated Indicaxanthin from Opuntia Ficus-Indica Associated with Re-Activation of the Onco-Suppressor p16INK4a Gene in Human Colorectal Carcinoma (Caco-2) Cells. Biochem. Biophys. Res. Commun. 2014, 450, 652–658. [Google Scholar] [CrossRef]
- Ragusa, M.A.; Naselli, F.; Cruciata, I.; Volpes, S.; Schimmenti, C.; Serio, G.; Mauro, M.; Librizzi, M.; Luparello, C.; Chiarelli, R.; et al. Indicaxanthin Induces Autophagy in Intestinal Epithelial Cancer Cells by Epigenetic Mechanisms Involving DNA Methylation. Nutrients 2023, 15, 3495. [Google Scholar] [CrossRef] [PubMed]
- Milagro, F.I.; Mansego, M.L.; De Miguel, C.; Martínez, J.A. Dietary Factors, Epigenetic Modifications and Obesity Outcomes: Progresses and Perspectives. Mol. Asp. Med. 2013, 34, 782–812. [Google Scholar] [CrossRef]
- Hardy, T.M.; Tollefsbol, T.O. Epigenetic Diet: Impact on the Epigenome and Cancer. Epigenomics 2011, 3, 503–518. [Google Scholar] [CrossRef] [PubMed]
- Mathers, J.C. Nutrigenomics in the Modern Era. Proc. Nutr. Soc. 2017, 76, 265–275. [Google Scholar] [CrossRef]
- Ferguson, L.R. Nutrigenomics Approaches to Functional Foods. J. Am. Diet. Assoc. 2009, 109, 452–458. [Google Scholar] [CrossRef]
- Ingham, P.W. Hedgehog Signaling. In Current Topics in Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2022; Volume 149, pp. 1–58. ISBN 978-0-12-817097-7. [Google Scholar]
- Jiang, J. Hedgehog Signaling Mechanism and Role in Cancer. Semin. Cancer Biol. 2022, 85, 107–122. [Google Scholar] [CrossRef]
- Chatel, G.; Ganeff, C.; Boussif, N.; Delacroix, L.; Briquet, A.; Nolens, G.; Winkler, R. Hedgehog Signaling Pathway Is Inactive in Colorectal Cancer Cell Lines. Int. J. Cancer 2007, 121, 2622–2627. [Google Scholar] [CrossRef]
- Lehner, F.; Kulik, U.; Klempnauer, J.; Borlak, J. The Hepatocyte Nuclear Factor 6 (HNF6) and FOXA2 Are Key Regulators in Colorectal Liver Metastases. FASEB J. 2007, 21, 1445–1462. [Google Scholar] [CrossRef]
- Wingelhofer, B.; Neubauer, H.A.; Valent, P.; Han, X.; Constantinescu, S.N.; Gunning, P.T.; Müller, M.; Moriggl, R. Implications of STAT3 and STAT5 Signaling on Gene Regulation and Chromatin Remodeling in Hematopoietic Cancer. Leukemia 2018, 32, 1713–1726. [Google Scholar] [CrossRef]
- Brew, K.; Dinakarpandian, D.; Nagase, H. Tissue Inhibitors of Metalloproteinases: Evolution, Structure and Function. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 2000, 1477, 267–283. [Google Scholar] [CrossRef]
- Makarenkova, H.P.; Meech, R. Barx Homeobox Family in Muscle Development and Regeneration. In International Review of Cell and Molecular Biology; Elsevier: Amsterdam, The Netherlands, 2012; Volume 297, pp. 117–173. ISBN 978-0-12-394308-8. [Google Scholar]
- Li, B.; Huang, Q.; Wei, G.-H. The Role of HOX Transcription Factors in Cancer Predisposition and Progression. Cancers 2019, 11, 528. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, B.; Meglio, P.D.; Gialitakis, M.; Duarte, J.H. The Aryl Hydrocarbon Receptor: Multitasking in the Immune System. Annu. Rev. Immunol. 2014, 32, 403–432. [Google Scholar] [CrossRef]
- Sondermann, N.C.; Faßbender, S.; Hartung, F.; Hätälä, A.M.; Rolfes, K.M.; Vogel, C.F.A.; Haarmann-Stemmann, T. Functions of the Aryl Hydrocarbon Receptor (AHR) beyond the Canonical AHR/ARNT Signaling Pathway. Biochem. Pharmacol. 2023, 208, 115371. [Google Scholar] [CrossRef]
- Tischkau, S.A. Mechanisms of Circadian Clock Interactions with Aryl Hydrocarbon Receptor Signalling. Eur. J. Neurosci. 2020, 51, 379–395. [Google Scholar] [CrossRef] [PubMed]
- Ullah, K.; Wu, R. Hypoxia-Inducible Factor Regulates Endothelial Metabolism in Cardiovascular Disease. Front. Physiol. 2021, 12, 670653. [Google Scholar] [CrossRef]
- Larigot, L.; Juricek, L.; Dairou, J.; Coumoul, X. AhR Signaling Pathways and Regulatory Functions. Biochim. Open 2018, 7, 1–9. [Google Scholar] [CrossRef]
- Thong-asa, W.; Puenpha, K.; Lairaksa, T.; Saengjinda, S. Neuroprotective Effects of Betanin in Mice with Cerebral Ischemia-Reperfusion Injury. Exp. Anim. 2023, 72, 336–345. [Google Scholar] [CrossRef]
- Allegra, M.; Tutone, M.; Tesoriere, L.; Attanzio, A.; Culletta, G.; Almerico, A.M. Evaluation of the IKKβ Binding of Indicaxanthin by Induced-Fit Docking, Binding Pose Metadynamics, and Molecular Dynamics. Front. Pharmacol. 2021, 12, 701568. [Google Scholar] [CrossRef]
- Blake, J.A.; Ziman, M.R. Pax Genes: Regulators of Lineage Specification and Progenitor Cell Maintenance. Development 2014, 141, 737–751. [Google Scholar] [CrossRef] [PubMed]
- Pingault, V.; Zerad, L.; Bertani-Torres, W.; Bondurand, N. SOX10: 20 Years of Phenotypic Plurality and Current Understanding of Its Developmental Function. J. Med. Genet. 2022, 59, 105–114. [Google Scholar] [CrossRef]
- Bondurand, N.; Sham, M.H. The Role of SOX10 during Enteric Nervous System Development. Dev. Biol. 2013, 382, 330–343. [Google Scholar] [CrossRef]
- Hu, N.; Strobl-Mazzulla, P.H.; Simoes-Costa, M.; Sánchez-Vásquez, E.; Bronner, M.E. DNA Methyltransferase 3B Regulates Duration of Neural Crest Production via Repression of Sox10. Proc. Natl. Acad. Sci. USA 2014, 111, 17911–17916. [Google Scholar] [CrossRef] [PubMed]
- Herbarth, B.; Pingault, V.; Bondurand, N.; Kuhlbrodt, K.; Hermans-Borgmeyer, I.; Puliti, A.; Lemort, N.; Goossens, M.; Wegner, M. Mutation of the Sry-Related Sox10 Gene in Dominant Megacolon, a Mouse Model for Human Hirschsprung Disease. Proc. Natl. Acad. Sci. USA 1998, 95, 5161–5165. [Google Scholar] [CrossRef]
- Jackstadt, R.; Röh, S.; Neumann, J.; Jung, P.; Hoffmann, R.; Horst, D.; Berens, C.; Bornkamm, G.W.; Kirchner, T.; Menssen, A.; et al. AP4 Is a Mediator of Epithelial–Mesenchymal Transition and Metastasis in Colorectal Cancer. J. Exp. Med. 2013, 210, 1331–1350. [Google Scholar] [CrossRef]
- Gaudet, F.; Hodgson, J.G.; Eden, A.; Jackson-Grusby, L.; Dausman, J.; Gray, J.W.; Leonhardt, H.; Jaenisch, R. Induction of Tumors in Mice by Genomic Hypomethylation. Science 2003, 300, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Widschwendter, M.; Jiang, G.; Woods, C.; Müller, H.M.; Fiegl, H.; Goebel, G.; Marth, C.; Müller-Holzner, E.; Zeimet, A.G.; Laird, P.W.; et al. DNA Hypomethylation and Ovarian Cancer Biology. Cancer Res. 2004, 64, 4472–4480. [Google Scholar] [CrossRef] [PubMed]
- Jasinska, A.; Krzyzosiak, W.J. Repetitive Sequences That Shape the Human Transcriptome. FEBS Lett. 2004, 567, 136–141. [Google Scholar] [CrossRef]
- Su, M.; Han, D.; Boyd-Kirkup, J.; Yu, X.; Han, J.-D.J. Evolution of Alu Elements toward Enhancers. Cell Rep. 2014, 7, 376–385. [Google Scholar] [CrossRef]
- Elliott, G.; Hong, C.; Xing, X.; Zhou, X.; Li, D.; Coarfa, C.; Bell, R.J.A.; Maire, C.L.; Ligon, K.L.; Sigaroudinia, M.; et al. Intermediate DNA Methylation Is a Conserved Signature of Genome Regulation. Nat. Commun. 2015, 6, 6363. [Google Scholar] [CrossRef]
- Maurano, M.T.; Wang, H.; John, S.; Shafer, A.; Canfield, T.; Lee, K.; Stamatoyannopoulos, J.A. Role of DNA Methylation in Modulating Transcription Factor Occupancy. Cell Rep. 2015, 12, 1184–1195. [Google Scholar] [CrossRef] [PubMed]
- Schübeler, D. Function and Information Content of DNA Methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef]
- Englander, E.W.; Howard, B.H. Nucleosome Positioning by Human Alu Elements in Chromatin. J. Biol. Chem. 1995, 270, 10091–10096. [Google Scholar] [CrossRef]
- Salih, F.; Salih, B.; Kogan, S.; Trifonov, E.N. Epigenetic Nucleosomes: Alu Sequences and CG as Nucleosome Positioning Element. J. Biomol. Struct. Dyn. 2008, 26, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Rollins, R.A.; Haghighi, F.; Edwards, J.R.; Das, R.; Zhang, M.Q.; Ju, J.; Bestor, T.H. Large-Scale Structure of Genomic Methylation Patterns. Genome Res. 2006, 16, 157–163. [Google Scholar] [CrossRef]
- Edwards, J.R.; O’Donnell, A.H.; Rollins, R.A.; Peckham, H.E.; Lee, C.; Milekic, M.H.; Chanrion, B.; Fu, Y.; Su, T.; Hibshoosh, H.; et al. Chromatin and Sequence Features That Define the Fine and Gross Structure of Genomic Methylation Patterns. Genome Res. 2010, 20, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Bártová, E.; Horáková, A.H.; Uhlířová, R.; Raška, I.; Galiová, G.; Orlova, D.; Kozubek, S. Structure and Epigenetics of Nucleoli in Comparison with Non-Nucleolar Compartments. J. Histochem. Cytochem. 2010, 58, 391–403. [Google Scholar] [CrossRef]
- McStay, B.; Grummt, I. The Epigenetics of rRNA Genes: From Molecular to Chromosome Biology. Annu. Rev. Cell Dev. Biol. 2008, 24, 131–157. [Google Scholar] [CrossRef]
- Pelletier, J.; Thomas, G.; Volarević, S. Ribosome Biogenesis in Cancer: New Players and Therapeutic Avenues. Nat. Rev. Cancer 2018, 18, 51–63. [Google Scholar] [CrossRef]
- Dannheisig, D.P.; Schimansky, A.; Donow, C.; Pfister, A.S. Nucleolar Stress Functions Upstream to Stimulate Expression of Autophagy Regulators. Cancers 2021, 13, 6220. [Google Scholar] [CrossRef] [PubMed]
- Pecoraro, A.; Pagano, M.; Russo, G.; Russo, A. Role of Autophagy in Cancer Cell Response to Nucleolar and Endoplasmic Reticulum Stress. Int. J. Mol. Sci. 2020, 21, 7334. [Google Scholar] [CrossRef]
- Pfister, A.S. An Update on Nucleolar Stress: The Transcriptional Control of Autophagy. Cells 2023, 12, 2071. [Google Scholar] [CrossRef] [PubMed]
- Butera, D.; Tesoriere, L.; Di Gaudio, F.; Bongiorno, A.; Allegra, M.; Pintaudi, A.M.; Kohen, R.; Livrea, M.A. Antioxidant Activities of Sicilian Prickly Pear (Opuntia Ficus Indica) Fruit Extracts and Reducing Properties of Its Betalains: Betanin and Indicaxanthin. J. Agric. Food Chem. 2002, 50, 6895–6901. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, J.; Wu, H.; Liu, S.; Wang, J.; Wu, B.; Huang, S.; Li, N.; Wang, J.; Zhang, X. Systematic Assessment of Reduced Representation Bisulfite Sequencing to Human Blood Samples: A Promising Method for Large-Sample-Scale Epigenomic Studies. J. Biotechnol. 2012, 157, 1–6. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Abugessaisa, I.; Shimoji, H.; Sahin, S.; Kondo, A.; Harshbarger, J.; Lizio, M.; Hayashizaki, Y.; Carninci, P.; Forrest, A.; Kasukawa, T.; et al. FANTOM5 Transcriptome Catalog of Cellular States Based on Semantic MediaWiki. Database 2016, 2016, baw105. [Google Scholar] [CrossRef][Green Version]


| tot | % (tot) | Hyper | % (Hyper) | Hypo | % (Hyper) | |
|---|---|---|---|---|---|---|
| Genecode.prom | 1761 | 54 | 800 | 53 | 961 | 55 |
| CGI | 1706 | 52 | 775 | 52 | 931 | 53 |
| CGI shores | 652 | 20 | 324 | 22 | 328 | 19 |
| CCRE | 2512 | 77 | 1113 | 74 | 1399 | 80 |
| DHS | 1380 | 42 | 588 | 39 | 792 | 45 |
| CTCF | 235 | 7 | 96 | 6 | 139 | 8 |
| H3K4me1 | 616 | 19 | 281 | 19 | 335 | 19 |
| H3K4me3 | 1570 | 48 | 696 | 46 | 874 | 50 |
| H3K9me3 | 42 | 1 | 22 | 1 | 20 | 1 |
| H3K36me3 | 2 | 0 | 2 | 0 | 0 | 0 |
| TSS.CAGE | 612 | 19 | 283 | 19 | 329 | 19 |
| CAGE.prom | 2060 | 63 | 927 | 62 | 1133 | 65 |
| Type | % | Type | % | Type | % |
|---|---|---|---|---|---|
| protein_coding | 61.6 | misc_RNA | 2.0 | rRNA | 1.0 |
| lncRNA | 29.3 | TEC | 1.4 | snoRNA | 0.3 |
| miRNA | 2.3 | snRNA | 1.3 | other | 0.5 |
| Gene | diff.meth IND50 | Type | Description | Tumor Function |
|---|---|---|---|---|
| ACY3 | −70.8 | coding | aminoacylase 3 | |
| C19orf73 | −82.9 | coding | chromosome 19 open reading frame 73 | |
| CABIN1 | −74.6 | lncRNA | calcineurin binding protein 1 | Potential oncogene |
| DIO3 | −81 | coding | iodothyronine deiodinase 3 | |
| DSCAM | −80 | coding | DS cell adhesion molecule | |
| FXYD7 | −78.1 | coding | FXYD domain containing ion transport regulator 7 | |
| LINC01551 | −72.5 | lncRNA | long intergenic non-protein coding RNA 1551 | |
| ONECUT1 | −96 | coding | one cut homeobox 1 | tumor suppressor |
| PROK2 | −73.3 | coding | prokineticin 2 | |
| RAP1GAP2 | −71.2 | coding | RAP1 GTPase activating protein 2 | tumor suppressor |
| SHC2 | −80.2 | coding | SHC adaptor protein 2 | oncogene |
| SHH | −88.6 | coding | sonic hedgehog signaling molecule | oncogene |
| ZNF525 | −100 | coding | zinc finger protein 525 |
| Gene | diff.meth IND50 | Type | Description |
|---|---|---|---|
| BARX2 | 86.3 | coding | BARX homeobox 2 |
| CD72 | 83.3 | coding | CD72 molecule |
| CHFR | 91.6 | coding | checkpoint with forkhead and ring finger domains |
| CYP1B1-AS1 | 92.7 | lncRNA | CYP1B1 antisense RNA 1 |
| EFCAB6 | 84.6 | coding | EF-hand calcium binding domain 6 |
| MSX2 | 95.2 | coding | msh homeobox 2 |
| NAPRT | 88.8 | coding | nicotinate phosphoribosyltransferase |
| PTGIS | 100 | coding | prostaglandin I2 synthase |
| RHBDL1 | 90.9 | coding | rhomboid like 1 |
| RNU6-765P | 85.5 | snRNA | RNA, U6 small nuclear 765, pseudogene |
| STAT5A | 89.5 | coding | signal transducer and activator of transcription 5A |
| TIMP2 | 85.3 | coding | TIMP metallopeptidase inhibitor 2 |
| TMEM200C | 85.2 | coding | transmembrane protein 200C |
| UGDH | 81 | coding | UDP-glucose 6-dehydrogenase |
| ZKSCAN5 | 93.9 | coding | zinc finger with KRAB and SCAN domains 5 |
| Enrichment FDR | nGenes | Pathway Genes | Fold Enrichment | Pathways |
|---|---|---|---|---|
| 9.4 × 10−6 | 12 | 27 | 6.5 | SOX10 target gene |
| 1.2 × 10−5 | 61 | 440 | 2 | TFAP4 target gene |
| 2.2 × 10−8 | 127 | 1041 | 1.8 | PAX5 target gene |
| 7.9 × 10−7 | 127 | 1107 | 1.7 | AHR target gene |
| 3.1 × 10−5 | 108 | 970 | 1.6 | NFKB1 target gene |
| 5.3 × 10−9 | 194 | 1757 | 1.6 | ARNT target gene |
| Gene | Entrez Gene | Diff Meth Prom | Meth.Diff IND50 |
|---|---|---|---|
| HYPER | |||
| ASCL2 | 430 | all | 62.1 |
| EPAS1 | 2034 | all | 62.1 |
| FOXA3 | 3171 | all | 44.8 |
| HOXA5 | 3202 | all | 55.1 |
| HOXB3 | 3213 | p1+p2+p4@HOXB3 | 37 |
| HOXC4 | 3221 | p3@HOXC4 | 39.8 |
| IKZF4 | 64375 | p1+p3@IKZF4 | 42 |
| KDM2A | 22992 | p1@KDM2A | 41.1 |
| MSX2 | 4488 | all | 95.2 |
| NR4A1 | 3164 | p3@NR4A1 | 67.7 |
| SALL4 | 57167 | all | 35.5 |
| SOX13 | 9580 | all | 27.6 |
| SP6 | 80320 | p1+p2@SP6 | 34 |
| TBX20 | 57057 | all | 25.1 |
| TBX3 | 6926 | all | 56.5 |
| TOE1 | 114034 | all | 29.2 |
| ZKSCAN5 | 23660 | all | 93.9 |
| ZNF93 | 81931 | all | 58.9 |
| HYPO | |||
| TBX1 | 6899 | p1+p5@TBX1 | -32.3 |
| MYBL2 | 4605 | all | -39.9 |
| EGR3 | 1960 | all | -38.9 |
| ZNF331 | 55422 | p4+p7@ZNF331 | -98 |
| THAP11 | 57215 | all | -29 |
| IND10 Hyper | IND10 Hypo | IND50 Hyper | IND50 Hypo | |
|---|---|---|---|---|
| All DMC | 99707 | 80526 | 92256 | 60594 |
| All DMC in RE | 52043 | 40970 | 50691 | 33452 |
| Chr21 DMC | 1918 | 939 | 3178 | 808 |
| Chr21 DMC in RE | 1018 | 509 | 1784 | 482 |
| Track | CLASS | Family | Genome (%) | Chr21 (%) |
|---|---|---|---|---|
| RepeatMasker | LINE | L1 | 17.7 | 15.6 |
| RepeatMasker | LINE | L2 | 3.5 | 2.3 |
| RepeatMasker | SINE | Alu | 10.6 | 8.8 |
| RepeatMasker | SINE | MIR | 2.8 | 1.6 |
| RepeatMasker | Other | Other | 17.4 | 23.3 |
| RepeatMasker | All repeats | - | 52 | 51.6 |
| Simple Repeats | simpleRepeat | - | 4.8 | 9.2 |
| RepeatMasker | Simple_repeat | Simple_repeat | 1.3 | 1.8 |
| Track | Table or Experiment ID |
|---|---|
| Genes | AllGencodev39 and RefSeq All (ncbiRefSeq) |
| CpG Islands | cpgIslandExt |
| ENCODE cCRE | encodeCcreCombined (2024) |
| TSS CAGE | hg38_liftover+new_CAGE_peaks_phase1and2 |
| Dnase HS (Caco2) | wgEncodeRegDnaseUwCaco2Peak |
| ChIP-seq CTCF (Caco2) | ENCFF990ZZT |
| ChIP-seq H3K36me3 (Caco2) | ENCFF479TQU |
| ChIP-seq H3K27me3 (Caco2) | ENCFF649WIT |
| ChIP-seq H3K4me3 (Caco2) | ENCFF642BGI |
| ChIP-seq H3K4me1 (Caco2) | ENCFF095KMQ |
| ChIP-seq H3K9me3 (Caco2) | ENCFF945SLD |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ragusa, M.A.; Gentile, C.; Nicosia, A.; Costa, S.; Volpes, S.; Greco, L.; Naselli, F.; Caradonna, F. Epigenetic Remodeling of Regulatory Regions by Indicaxanthin Suggests a Shift in Cell Identity Programs in Colorectal Cancer Cells. Int. J. Mol. Sci. 2025, 26, 6072. https://doi.org/10.3390/ijms26136072
Ragusa MA, Gentile C, Nicosia A, Costa S, Volpes S, Greco L, Naselli F, Caradonna F. Epigenetic Remodeling of Regulatory Regions by Indicaxanthin Suggests a Shift in Cell Identity Programs in Colorectal Cancer Cells. International Journal of Molecular Sciences. 2025; 26(13):6072. https://doi.org/10.3390/ijms26136072
Chicago/Turabian StyleRagusa, Maria Antonietta, Carla Gentile, Aldo Nicosia, Salvatore Costa, Sara Volpes, Laura Greco, Flores Naselli, and Fabio Caradonna. 2025. "Epigenetic Remodeling of Regulatory Regions by Indicaxanthin Suggests a Shift in Cell Identity Programs in Colorectal Cancer Cells" International Journal of Molecular Sciences 26, no. 13: 6072. https://doi.org/10.3390/ijms26136072
APA StyleRagusa, M. A., Gentile, C., Nicosia, A., Costa, S., Volpes, S., Greco, L., Naselli, F., & Caradonna, F. (2025). Epigenetic Remodeling of Regulatory Regions by Indicaxanthin Suggests a Shift in Cell Identity Programs in Colorectal Cancer Cells. International Journal of Molecular Sciences, 26(13), 6072. https://doi.org/10.3390/ijms26136072









