Proteomic Analysis of the Low Molecular Mass Fraction of Newly Diagnosed and Recurrent Glioblastoma CUSA Fluid: A Pilot Investigation of the Peptidomic Profile
Abstract
1. Introduction
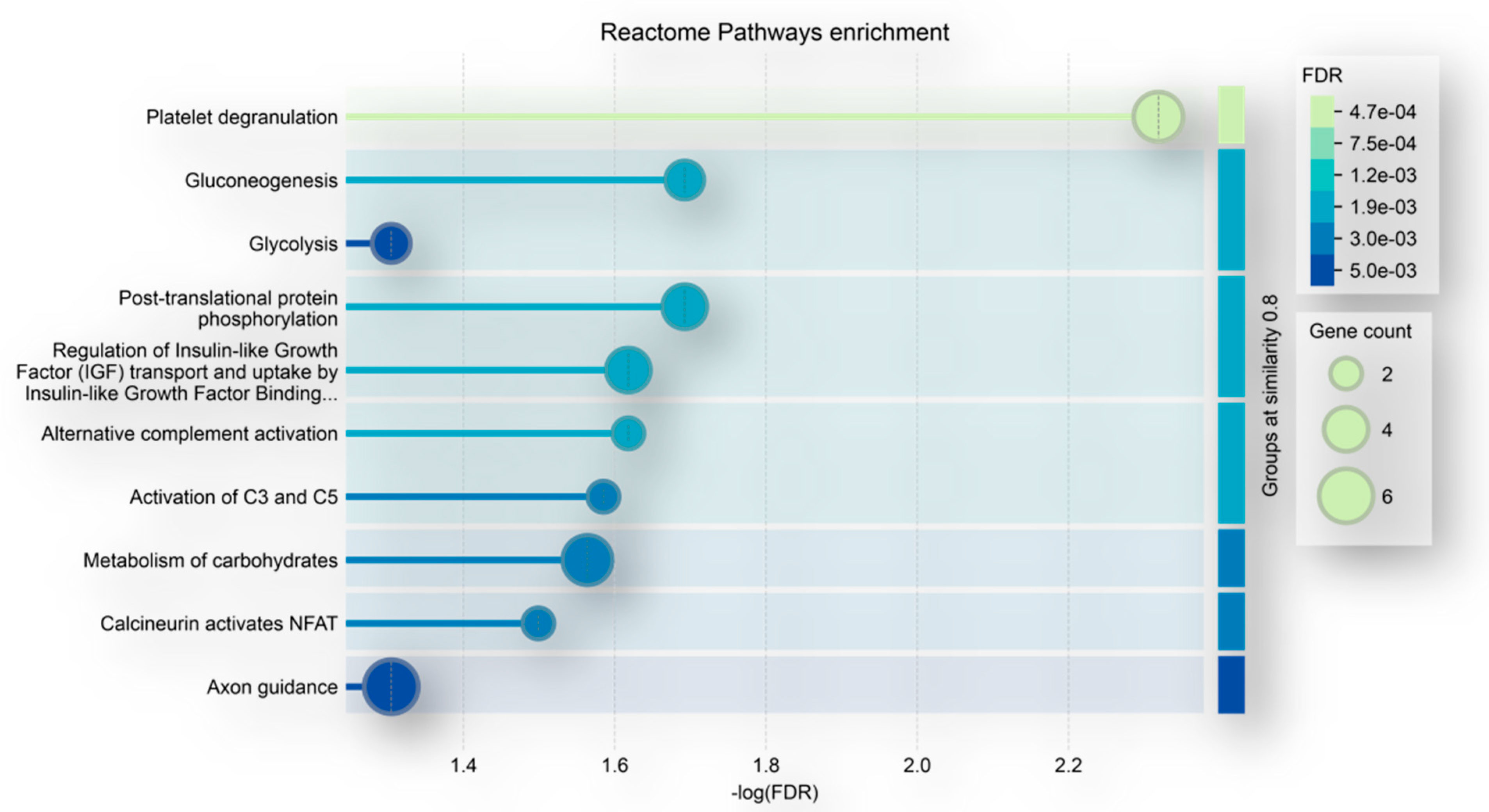
2. Results and Discussion
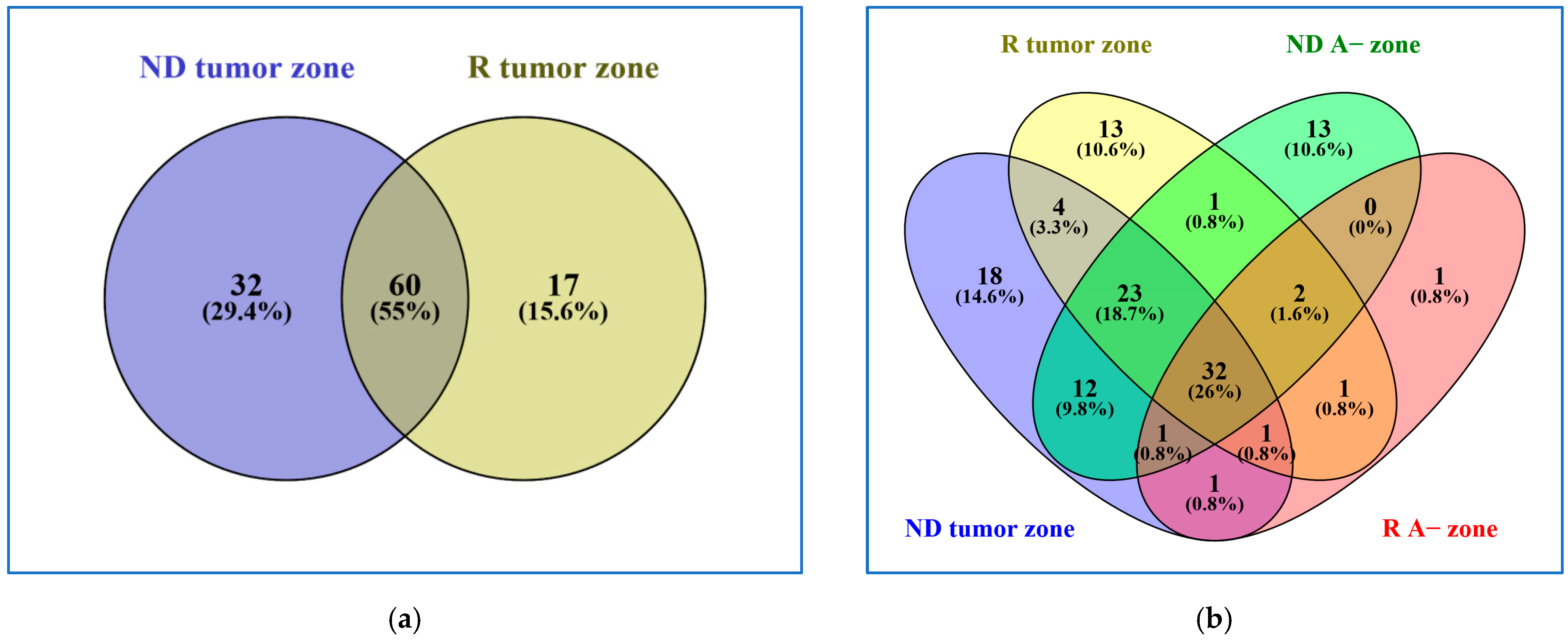
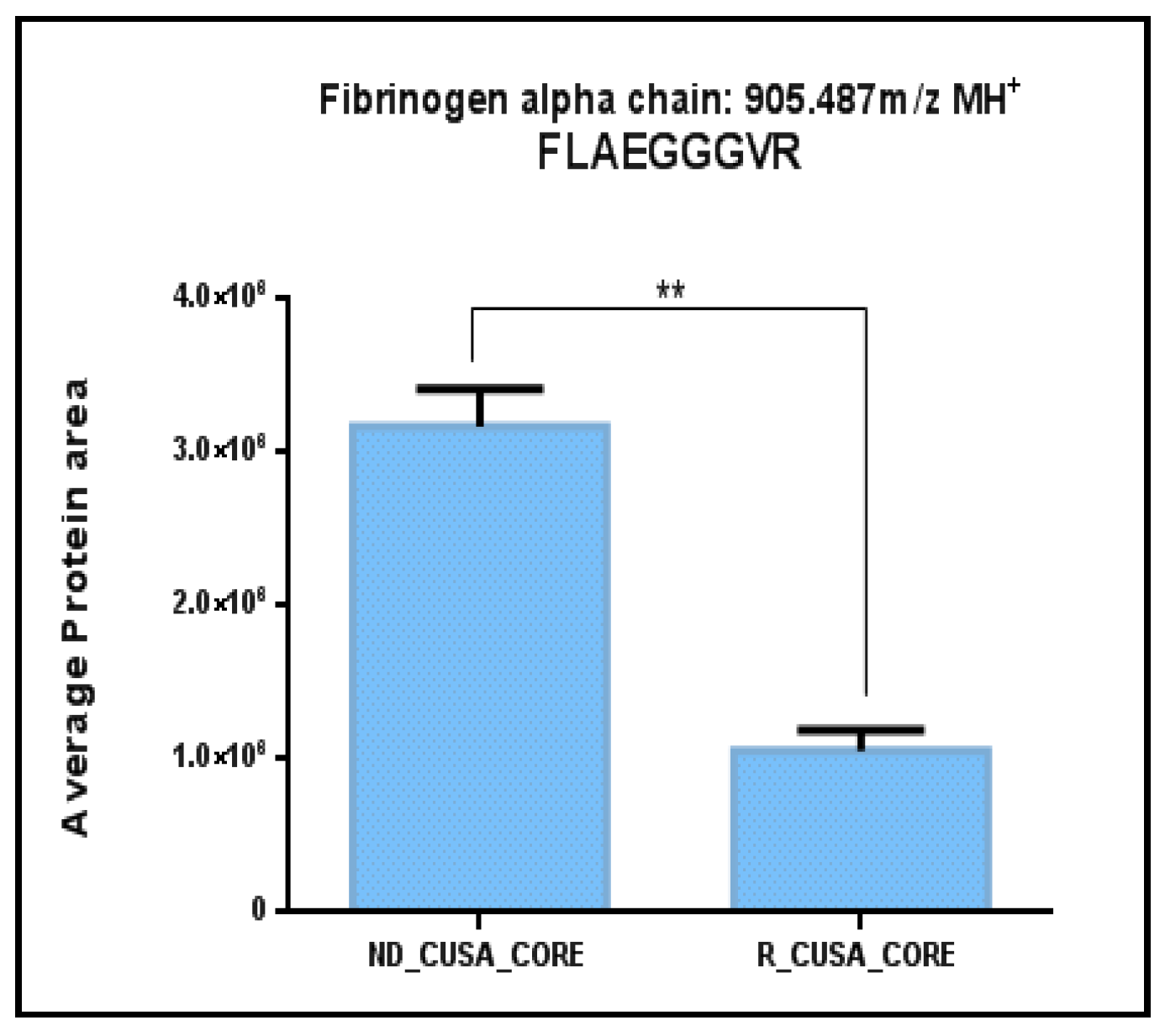
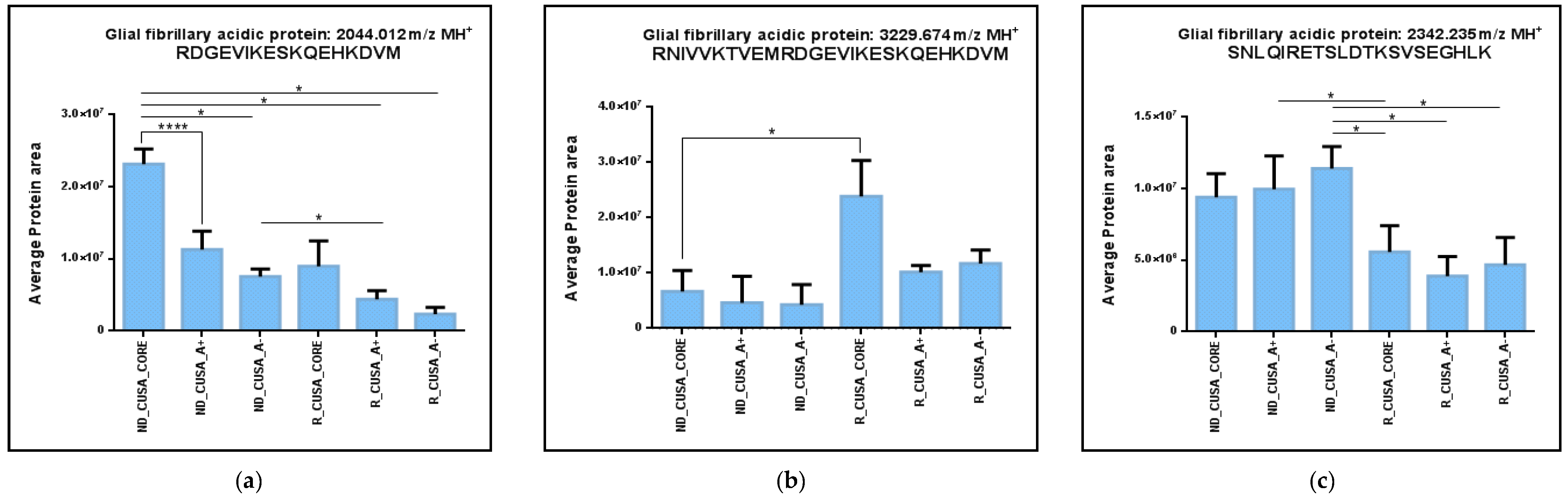
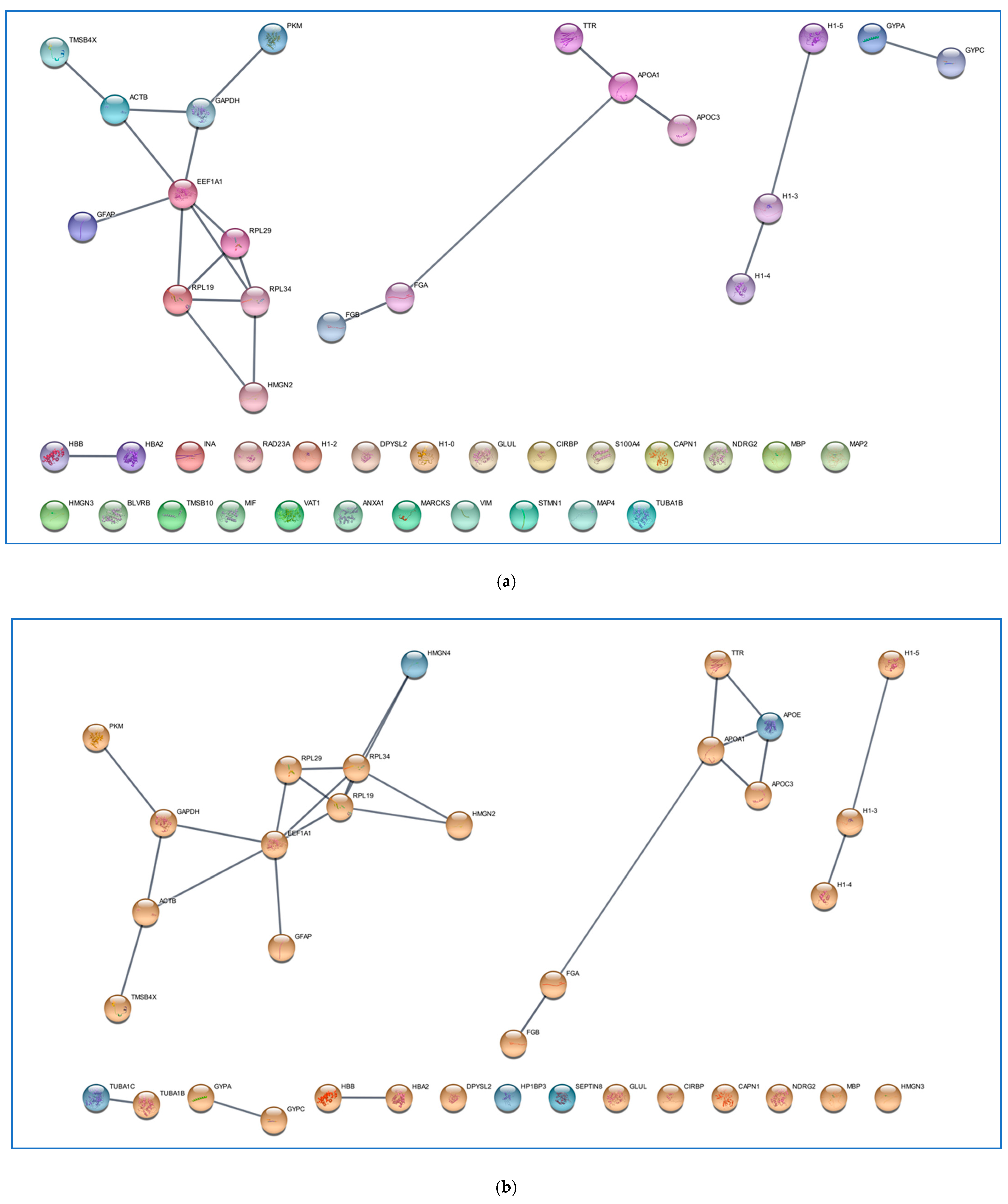
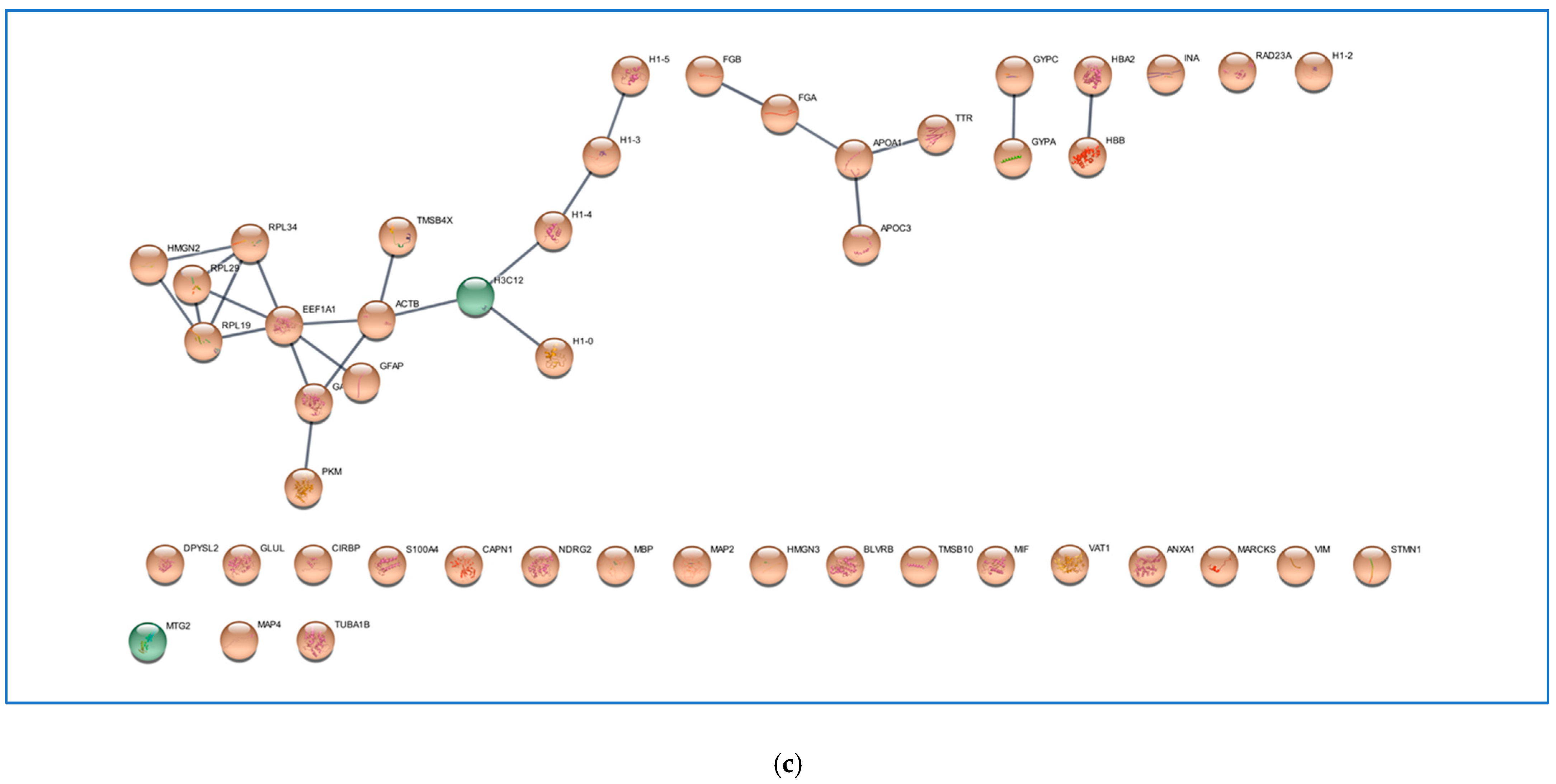
2.1. Comparative Analysis of the ND and R GBM Tumor CORE Zones
2.2. Analysis of the GBM CUSA A+ Zone: The ND and R GBM Exclusive and Shared Peptides
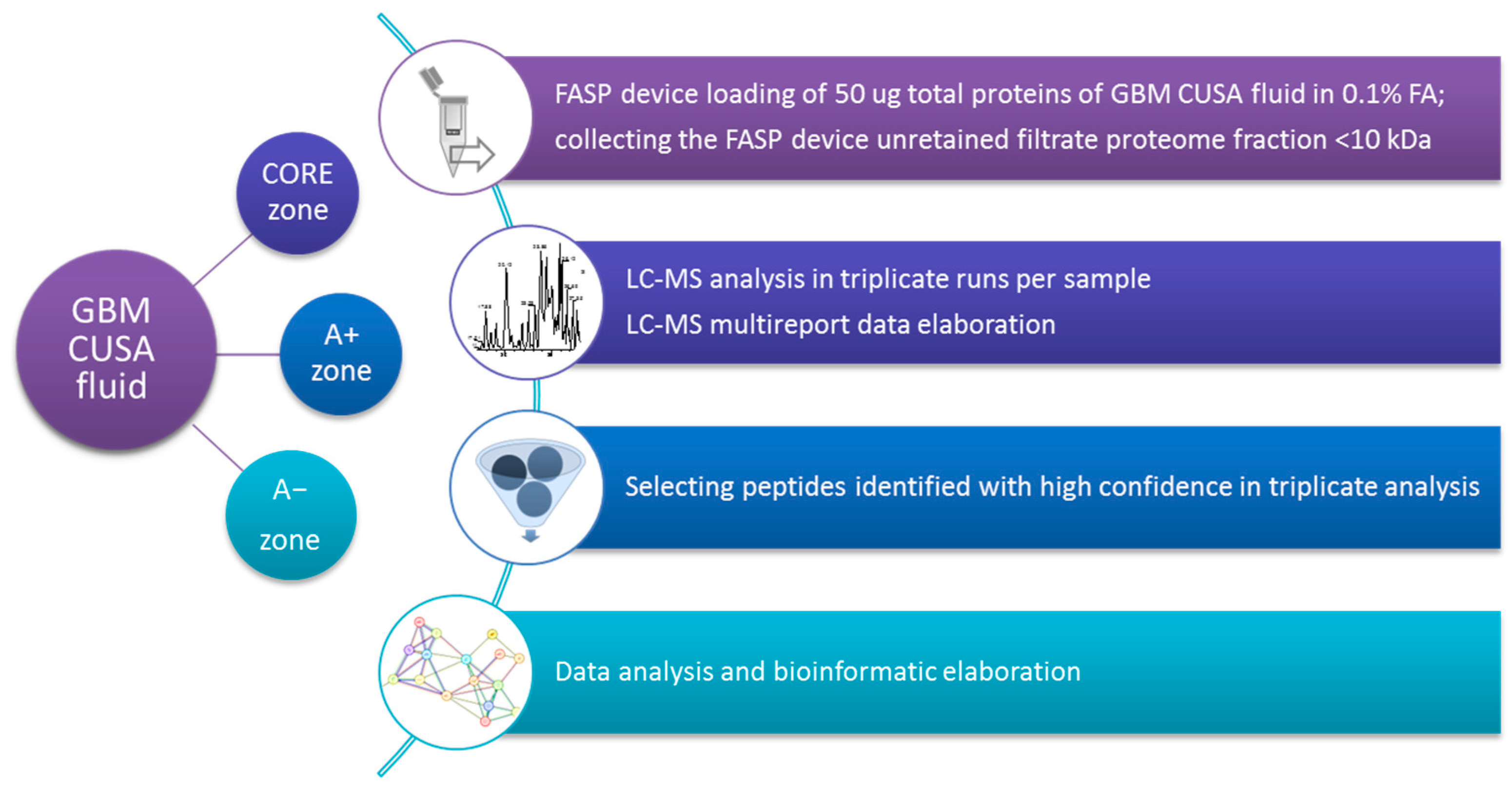
3. Materials and Methods
3.1. Chemicals
3.2. Sample Collection and Pretreatment
3.3. LC-MS Peptidomic Analysis
3.4. LC-MS Data Elaboration and Bioinformatics Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GBM | Glioblastoma multiforme |
| ND GBM | Newly diagnosed GBM |
| R GBM | Recurrent GBM |
| CUSA | Cavitron Ultrasonic Surgical Aspirator |
| FASP | Filter-Aided Sample Preparation |
| LC | Liquid Chromatography |
| MS | Mass spectrometry |
| CNS | Central nervous system |
| FA | Formic acid |
| 5-ALA | 5-aminolevulinic acid |
| ACN | acetonitrile |
| PIC | Protease inhibitor cocktail |
| FDR | False Discovery Rate |
| HPA | Human Protein Atlas |
References
- Lan, Z.; Li, X.; Zhang, X. Glioblastoma: An Update in Pathology, Molecular Mechanisms and Biomarkers. Int. J. Mol. Sci. 2024, 25, 3040. [Google Scholar] [CrossRef] [PubMed]
- Stoyanov, G.S.; Lyutfi, E.; Georgieva, R.; Georgiev, R.; Dzhenkov, D.L.; Petkova, L.; Ivanov, B.D.; Kaprelyan, A.; Ghenev, P. Reclassification of Glioblastoma Multiforme According to the 2021 World Health Organization Classification of Central Nervous System Tumors: A Single Institution Report and Practical Significance. Cureus 2022, 14, 21822. [Google Scholar] [CrossRef] [PubMed]
- Mathew, E.N.; Berry, B.C.; Yang, H.W.; Carroll, R.S.; Johnson, M.D. Delivering Therapeutics to Glioblastoma: Overcoming Biological Constraints. Int. J. Mol. Sci. 2022, 23, 1711. [Google Scholar] [CrossRef]
- Bikfalvi, A.; Da Costa, C.A.; Avril, T.; Barnier, J.V.; Bauchet, L.; Brisson, L.; Cartron, P.F.; Castel, H.; Chevet, E.; Chneiweiss, H.; et al. Challenges in glioblastoma research: Focus on the tumor microenvironment. Trends Cancer 2023, 9, 9. [Google Scholar] [CrossRef]
- Kim, M.; Ladomersky, E.; Mozny, A.; Kocherginsky, M.; O’Shea, K.; Reinstein, Z.Z.; Zhai, L.; Bell, A.; Lauing, K.L.; Bollu, L.; et al. Glioblastoma as an age-related neurological disorder in adults. Neurooncol. Adv. 2021, 3, 125. [Google Scholar] [CrossRef]
- Hadjipanayis, C.G.; Stummer, W. 5-ALA and FDA approval for glioma surgery. J. Neurooncol. 2019, 141, 479. [Google Scholar] [CrossRef]
- Hishii, M.; Matsumoto, T.; Arai, H. Diagnosis and Treatment of Early-Stage Glioblastoma. Asian J. Neurosurg. 2019, 14, 589. [Google Scholar] [CrossRef]
- Hellinger, R.; Sigurdsson, A.; Wu, W.; Romanova, E.V.; Li, L.; Sweedler, J.V.; Süssmuth, R.D.; Gruber, C.W. Peptidomics. Nat. Rev. Methods Primers 2023, 3, 25. [Google Scholar] [CrossRef]
- Foreman, R.E.; George, A.L.; Reimann, F.; Gribble, F.M.; Kay, R.G. Peptidomics: A Review of Clinical Applications and Methodologies. J. Proteome Res. 2021, 20, 3782. [Google Scholar] [CrossRef]
- Chong, C.; Coukos, G.; Bassani-Sternberg, M. Identification of tumor antigens with immunopeptidomics. Nat. Biotechnol. 2022, 40, 175. [Google Scholar] [CrossRef]
- Wang, S.; Mao, C.; Liu, S. Peptides encoded by noncoding genes: Challenges and perspectives. Signal Transduct. Target. Ther. 2019, 4, 57. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Zhang, J.; Wie, M.; Liu, B.; Dong, K. Emerging role of long noncoding RNA-encoded micropeptides in cancer. Cancer Cell Int. 2020, 20, 506. [Google Scholar] [CrossRef] [PubMed]
- La Rocca, G.; Simboli, G.A.; Vincenzoni, F.; Rossetti, D.V.; Urbani, A.; Ius, T.; Della Pepa, G.M.; Olivi, A.; Sabatino, G.; Desiderio, C. Glioblastoma CUSA Fluid Protein Profiling: A Comparative Investigation of the Core and Peripheral Tumor Zones. Cancers 2020, 13, 30. [Google Scholar] [CrossRef]
- Moresi, F.; Rossetti, D.V.; Vincenzoni, F.; Simboli, G.A.; La Rocca, G.; Olivi, A.; Urbani, A.; Sabatino, G.; Desiderio, C. Investigating Glioblastoma Multiforme Sub-Proteomes: A Computational Study of CUSA Fluid Proteomic Data. Int. J. Mol. Sci. 2022, 23, 2058. [Google Scholar] [CrossRef]
- Iavarone, F.; Desiderio, C.; Vitali, A.; Messana, I.; Martelli, C.; Castagnola, M.; Cabras, T. Cryptides: Latent peptides everywhere. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 246. [Google Scholar] [CrossRef]
- Muntiu, A.; Papait, A.; Vincenzoni, F.; Rossetti, D.V.; Romele, P.; Cargnoni, A.; Silini, A.; Parolini, O.; Desiderio, C. Proteomic analysis of the human amniotic mesenchymal stromal cell secretome by integrated approaches via filter-aided sample preparation. J. Proteom. 2025, 310, 105339. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef]
- Dobra, G.; Gyukity-Sebestyén, E.; Bukva, M.; Harmati, M.; Nagy, V.; Szabó, Z.; Pankotai, T.; Klekner, Á.; Buzás, K. MMP-9 as Prognostic Marker for Brain Tumours: A Comparative Study on Serum-Derived Small Extracellular Vesicles. Cancers 2023, 15, 712. [Google Scholar] [CrossRef]
- Xue, Q.; Cao, L.; Chen, X.Y.; Zhao, J.; Gao, L.; Li, S.Z.; Fei, Z. High expression of MMP9 in glioma affects cell proliferation and is associated with patient survival rates. Oncol. Lett. 2017, 13, 1325. [Google Scholar] [CrossRef]
- Chen, X.C.; Wei, X.T.; Guan, J.H.; Shu, H.; Chen, D. EGF stimulates glioblastoma metastasis by induction of matrix metalloproteinase-9 in an EGFR-dependent mechanism. Oncotarget 2017, 8, 65969. [Google Scholar] [CrossRef]
- Choe, G.; Park, J.K.; Jouben-Steele, L.; Kremen, T.J.; Liau, L.M.; Vinters, H.V.; Cloughesy, T.F.; Mischel, P.S. Active matrix metalloproteinase 9 expression is associated with primary glioblastoma subtype. Clin. Cancer Res. 2002, 8, 2894. [Google Scholar] [PubMed]
- Slovacek, H.; Khanna, R.; Poredos, P.; Jezovnik, M.; Hoppensteadt, D.; Fareed, J.; Hopkinson, W. Interrelationship of Osteopontin, MMP-9 and ADAMTS4 in Patients With Osteoarthritis Undergoing Total Joint Arthroplasty. Clin. Appl. Thromb. Hemost. 2020, 26, 1076029620964864. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, Y.; Oishi, T.; Yamashita, A.; Murata, Y.; Yamamoto, T.; Takashima, I.; Isa, T.; Higo, N. Neuronal and microglial localization of secreted phosphoprotein 1 (osteopontin) in intact and damaged motor cortex of macaques. Brain Res. 2019, 1714, 52. [Google Scholar] [CrossRef] [PubMed]
- Wormser, U.; Sintov, A.; Vinceti, M.; Mandrioli, J.; Brodsky, B.; Proscura, E.; Finkelstein, Y. Unique cerebrospinal fluid peptides: Potential amyotrophic lateral sclerosis biomarkers and etiological factors. Explor. Neuroprot. Ther. 2023, 3, 435. [Google Scholar] [CrossRef]
- Zhang, H.; Zhou, Y.; Cui, B.; Liu, Z.; Shen, H. Novel insights into astrocyte-mediated signaling of proliferation.; invasion and tumor immune microenvironment in glioblastoma. Biomed. Pharmacother. 2020, 126, 110086. [Google Scholar] [CrossRef]
- Osswald, M.; Jung, E.; Sahm, F.; Solecki, G.; Venkataramani, V.; Blaes, J.; Weil, S.; Horstmann, H.; Wiestler, B.; Syed, M.; et al. Brain tumour cells interconnect to a functional and resistant network. Nature 2015, 528, 93. [Google Scholar] [CrossRef]
- Semenzato, M.; Scorrano, L. Mind the GAP (43) for mitochondria transfer to glioblastomas. Nat. Cancer 2023, 4, 588. [Google Scholar] [CrossRef]
- Baum, C.E.; Price, D.K.; Figg, W.D. Sarcosine as a potential prostate cancer biomarker and therapeutic target. Cancer Biol. Ther. 2010, 9, 341. [Google Scholar] [CrossRef]
- Botta, G.; Perruolo, G.; Libertini, S.; Cassese, A.; Abagnale, A.; Beguinot, F.; Formisano, P.; Portella, G. PED/PEA-15 modulates coxsackievirus-adenovirus receptor expression and adenoviral infectivity via ERK-mediated signals in glioma cells. Hum. Gene Ther. 2010, 21, 1067. [Google Scholar] [CrossRef]
- Eckert, A.; Böck, B.C.; Tagscherer, K.E.; Haas, T.L.; Grund, K.; Sykora, J.; Herold-Mende, C.; Ehemann, V.; Hollstein, M.; Chneiweiss, H.; et al. The PEA-15/PED protein protects glioblastoma cells from glucose deprivation-induced apoptosis via the ERK/MAP kinase pathway. Oncogene 2008, 27, 1155. [Google Scholar] [CrossRef]
- Wei, Y. On the Quest of Cellular Functions of PEA-15 and the Therapeutic Opportunities. Pharmaceuticals 2015, 8, 455. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Z.; Liu, X.N.; Fan, R.C.; Jia, Y.P.; Zhang, Q.K.; Gao, X.Q.; Wang, Y.Q.; Yang, M.Q.; Ji, L.Z.; Zhou, Y.Q.; et al. Identification of pimavanserin tartrate as a potent Ca2+-calcineurin-NFAT pathway inhibitor for glioblastoma therapy. Acta Pharmacol. Sin. 2021, 42, 1860. [Google Scholar] [CrossRef] [PubMed]
- Campanella, R.; Guarnaccia, L.; Cordiglieri, C.; Trombetta, E.; Caroli, M.; Carrabba, G.; La Verde, N.; Rampini, P.; Gaudino, C.; Costa, A.; et al. Tumor-Educated Platelets and Angiogenesis in Glioblastoma: Another Brick in the Wall for Novel Prognostic and Targetable Biomarkers, Changing the Vision from a Localized Tumor to a Systemic Pathology. Cells 2020, 9, 294. [Google Scholar] [CrossRef]
- Martins, F.; Van der Kellen, D.; Gonçalves, L.G.; Serpa, J. Metabolic Profiles Point Out Metabolic Pathways Pivotal in Two Glioblastoma (GBM) Cell Lines, U251 and U-87MG. Biomedicines 2023, 11, 2041. [Google Scholar] [CrossRef]
- Pi, Y.; Fang, C.L.; Su, Z.Y. Protein phosphorylation: A potential target in glioma development. Ibrain 2022, 8, 176. [Google Scholar] [CrossRef]
- Pienkowski, T.; Kowalczyk, T.; Cysewski, D.; Kretowski, A.; Ciborowski, M. Glioma and post-translational modifications: A complex relationship. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 189009. [Google Scholar] [CrossRef]
- Lewitt, M.S.; Boyd, G.W. The Role of Insulin-Like Growth Factors and Insulin-Like Growth Factor-Binding Proteins in the Nervous System. Biochem. Insights 2019, 12, 1178626419842176. [Google Scholar] [CrossRef]
- Zhu, H.; Yu, X.; Zhang, S.; Shu, K. Targeting the Complement Pathway in Malignant Glioma Microenvironments. Front. Cell Dev. Biol. 2021, 9, 657472. [Google Scholar] [CrossRef]
- Chokshi, C.R.; Shaikh, M.V.; Brakel, B.; Rossotti, M.A.; Tieu, D.; Maich, W.; Anand, A.; Chafe, S.C.; Zhai, K.; Suk, Y.; et al. Targeting axonal guidance dependencies in glioblastoma with ROBO1 CAR T cells. Nat. Med. 2024, 30, 2936. [Google Scholar] [CrossRef]
- Muntiu, A.; Moresi, F.; Vincenzoni, F.; Rossetti, D.V.; Iavarone, F.; Messana, I.; Castagnola, M.; La Rocca, G.; Mazzucchi, E.; Olivi, A.; et al. Proteomic Profiling of Pre- and Post-Surgery Saliva of Glioblastoma Patients: A Pilot Investigation. Int. J. Mol. Sci. 2024, 25, 12984. [Google Scholar] [CrossRef]
- Rossetti, D.V.; Inserra, I.; Nesticò, A.; Vincenzoni, F.; Iavarone, F.; Messana, I.; Castagnola, M.; Massimi, L.; Tamburrini, G.; Caldarelli, M.; et al. Pediatric Brain Tumors: Signatures from the Intact Proteome. Int. J. Mol. Sci. 2022, 23, 3196. [Google Scholar] [CrossRef] [PubMed]
- Inserra, I.; Iavarone, F.; Martelli, C.; D’Angelo, L.; Delfino, D.; Rossetti, D.V.; Tamburrini, G.; Massimi, L.; Caldarelli, M.; Di Rocco, C.; et al. Proteomic study of pilocytic astrocytoma pediatric brain tumor intracystic fluid. J. Proteome Res. 2014, 13, 4594. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Huang, Y.; Li, H.; Luo, Y.; Zuo, Z.; Ren, J.; Xie, Y. SPENCER: A comprehensive database for small peptides encoded by noncoding RNAs in cancer patients. Nucleic Acids Res. 2022, 50, D1373. [Google Scholar] [CrossRef] [PubMed]
- Uceda-Castro, R.; Van Asperen, J.V.; Vennin, C.; Sluijs, J.A.; Van Bodegraven, E.J.; Margarido, A.S.; Robe, P.A.J.; Van Rheenen, J.; Hol, E.M. GFAP splice variants fine-tune glioma cell invasion and tumour dynamics by modulating migration persistence. Sci. Rep. 2022, 12, 424. [Google Scholar] [CrossRef]
- Simone, L.; Capobianco, D.L.; Di Palma, F.; Binda, E.; Legnani, F.G.; Vescovi, A.L.; Svelto, M.; Pisani, F. GFAP serves as a structural element of tunneling nanotubes between glioblastoma cells and could play a role in the intercellular transfer of mitochondria. Front. Cell Dev. Biol. 2023, 11, 1221671. [Google Scholar] [CrossRef]
- Marcu, A.; Bichmann, L.; Kuchenbecker, L.; Kowalewski, D.J.; Freudenmann, L.K.; Backert, L.; Mühlenbruch, L.; Szolek, A.; Lübke, M.; Wagner, P.; et al. HLA Ligand Atlas: A benign reference of HLA-presented peptides to improve T-cell-based cancer immunotherapy. J. Immunother. Cancer 2021, 9, 2071. [Google Scholar] [CrossRef]
- Wang, Y.; He, J.; Lian, S.; Zeng, Y.; He, S.; Xu, J.; Luo, L.; Yang, W.; Jiang, J. Targeting Metabolic-Redox Nexus to Regulate Drug Resistance: From Mechanism to Tumor Therapy. Antioxidants 2024, 13, 828. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, L.; Dong, X.; Liu, L.; Huo, L.; Chen, H. High Expression of Vimentin is Associated With Progression and a Poor Outcome in Glioblastoma. Appl. Immunohistochem. Mol. Morphol. 2018, 26, 337. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, S.; Chen, Y.; Ma, W.; Lu, S.; He, L.; Chen, J.; Chen, X.; Zhang, X.; Shi, Y.; et al. Vimentin promotes glioma progression and maintains glioma cell resistance to oxidative phosphorylation inhibition. Cell. Oncol. 2023, 46, 1791. [Google Scholar] [CrossRef]
- Wei, L.; Shao, N.; Peng, Y.; Zhou, P. Inhibition of Cathepsin S Restores TGF-β-induced Epithelial-to-mesenchymal Transition and Tight Junction Turnover in Glioblastoma Cells. J. Cancer 2021, 12, 1592. [Google Scholar] [CrossRef]
- Martelli, C.; D’Angelo, L.; Barba, M.; Baranzini, M.; Inserra, I.; Iavarone, F.; Vincenzoni, F.; Tamburrini, G.; Massimi, L.; Rocco, C.D.; et al. Top-down proteomic characterization of DAOY medulloblastoma tumor cell line. EuPA Open Proteom. 2016, 12, 13. [Google Scholar] [CrossRef] [PubMed]
- Attilio, P.J.; Flora, M.; Kamnaksh, A.; Bradshaw, D.J.; Agoston, D.; Mueller, G.P. The Effects of Blast Exposure on Protein Deimination in the Brain. Oxid. Med. Cell. Longev. 2017, 8398072. [Google Scholar] [CrossRef] [PubMed]
- Hjalmarsson, C.; Bjerke, M.; Andersson, B.; Blennow, K.; Zetterberg, H.; Åberg, N.D.; Olsson, B.; Eckerström, C.; Bokemark, L.; Wallin, A. Neuronal and glia-related biomarkers in cerebrospinal fluid of patients with acute ischemic stroke. J. Cent. Nerv. Syst. Dis. 2014, 6, 51. [Google Scholar] [CrossRef] [PubMed]
- Zavialova, M.G.; Shevchenko, V.E.; Nikolaev, E.N.; Zgoda, V.G. Is myelin basic protein a potential biomarker of brain cancer? Eur. J. Mass Spectrom. 2017, 23, 192–196. [Google Scholar] [CrossRef]
- Azimi, A.; Kaufman, K.L.; Ali, M.; Arthur, J.; Kossard, S.; Fernandez-Penas, P. Differential proteomic analysis of actinic keratosis, Bowen’s disease and cutaneous squamous cell carcinoma by label-free LC-MS/MS. J. Dermatol. Sci. 2018, 91, 69. [Google Scholar] [CrossRef]
- Byrling, J.; Kristl, T.; Hu, D.; Pla, I.; Sanchez, A.; Sasor, A.; Andersson, R.; Marko-Varga, G.; Andersson, B. Mass spectrometry-based analysis of formalin-fixed, paraffin-embedded distal cholangiocarcinoma identifies stromal thrombospondin-2 as a potential prognostic marker. J. Transl. Med. 2020, 18, 343. [Google Scholar] [CrossRef]
- Tong, J.; Helmy, M.; Cavalli, F.M.; Jin, L.; St-Germain, J.; Karisch, R.; Taylor, P.; Minden, M.D.; Taylor, M.D.; Neel, B.G.; et al. Integrated analysis of proteome, phosphotyrosine-proteome, tyrosine-kinome, and tyrosine-phosphatome in acute myeloid leukemia. Proteomics 2017, 17, 10. [Google Scholar] [CrossRef]
- Lan, Y.L.; Wang, X.; Lou, J.C.; Ma, X.C.; Zhang, B. The potential roles of aquaporin 4 in malignant gliomas. Oncotarget 2017, 8, 32345. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Guadagnin, G.; Cappello, P.; Novelli, F. Post-Translational Modifications in Tumor-Associated Antigens as a Platform for Novel Immuno-Oncology Therapies. Cancers 2022, 15, 138. [Google Scholar] [CrossRef]
- Tatari, N.; Khan, S.; Livingstone, J.; Zhai, K.; Mckenna, D.; Ignatchenko, V.; Chokshi, C.; Gwynne, W.D.; Singh, M.; Revill, S.; et al. The proteomic landscape of glioblastoma recurrence reveals novel and targetable immunoregulatory drivers. Acta Neuropathol. 2022, 144, 1127. [Google Scholar] [CrossRef]
- Kvartsberg, H.; Portelius, E.; Andreasson, U.; Brinkmalm, G.; Hellwig, K.; Lelental, N.; Kornhuber, J.; Hansson, O.; Minthon, L.; Spitzer, P.; et al. Characterization of the postsynaptic protein neurogranin in paired cerebrospinal fluid and plasma samples from Alzheimer’s disease patients and healthy controls. Alzheimers Res. Ther. 2015, 7, 40. [Google Scholar] [CrossRef] [PubMed]
- Öhrfelt, A.; Dumurgier, J.; Zetterberg, H.; Vrillon, A.; Ashton, N.J.; Kvartsberg, H.; Bouaziz-Amar, E.; Hugon, J.; Paquet, C.; Blennow, K. Full-length and C-terminal neurogranin in Alzheimer’s disease cerebrospinal fluid analyzed by novel ultrasensitive immunoassays. Alzheimers Res. Ther. 2020, 12, 168. [Google Scholar] [CrossRef] [PubMed]
- Kvartsberg, H.; Duits, F.H.; Ingelsson, M.; Andreasen, N.; Öhrfelt, A.; Andersson, K.; Brinkmalm, G.; Lannfelt, L.; Minthon, L.; Hansson, O.; et al. Cerebrospinal fluid levels of the synaptic protein neurogranin correlates with cognitive decline in prodromal Alzheimer’s disease. Alzheimers Dement. 2015, 11, 1180. [Google Scholar] [CrossRef]
- Portelius, E.; Zetterberg, H.; Skillbäck, T.; Törnqvist, U.; Andreasson, U.; Trojanowski, J.Q.; Weiner, M.W.; Shaw, L.M.; Mattsson, N.; Blennow, K. Alzheimer’s Disease Neuroimaging Initiative. Cerebrospinal fluid neurogranin: Relation to cognition and neurodegeneration in Alzheimer’s disease. Brain 2015, 138, 3373. [Google Scholar] [CrossRef]
- Feng, R.; Chen, M.; Luo, G.; Chen, Y.; Wang, J.; Xu, J.; Xu, M.; Zhong, P.; Wen, T. Potential glioblastoma biomarkers identified by mass spectroscopy and iTRAQ labeling. Genes Dis. 2022, 10, 1767. [Google Scholar] [CrossRef]
- Cesca, F.; Baldelli, P.; Valtorta, F.; Benfenati, F. The synapsins: Key actors of synapse function and plasticity. Prog. Neurobiol. 2010, 91, 313. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, X.; Zhu, X.L.; Hao, J.Q.; Bai, H.; Xiao, Y.C.; Wang, Z.Z.; Hao, C.Y.; Duan, H.B. Bioinformatics analysis of potential core genes for glioblastoma. Biosci. Rep. 2020, 40, BSR20201625. [Google Scholar] [CrossRef]
- Polat, B.; Wohlleben, G.; Kosmala, R.; Lisowski, D.; Mantel, F.; Lewitzki, V.; Löhr, M.; Blum, R.; Herud, P.; Flentje, M.; et al. Differences in stem cell marker and osteopontin expression in primary and recurrent glioblastoma. Cancer Cell Int. 2022, 22, 87. [Google Scholar] [CrossRef]
- Guadagno, E.; Borrelli, G.; Califano, M.; Calì, G.; Solari, D.; Del Basso De Caro, M. Immunohistochemical expression of stem cell markers CD44 and nestin in glioblastomas: Evaluation of their prognostic significance. Pathol. Res. Pract. 2016, 212, 825. [Google Scholar] [CrossRef]
- Jin, X.; Jin, X.; Jung, J.E.; Beck, S.; Kim, H. Cell surface Nestin is a biomarker for glioma stem cells. Biochem. Biophys. Res. Commun. 2013, 433, 496. [Google Scholar] [CrossRef]
- Prosniak, M.; Kenyon, L.C.; Hooper, D.C. Glioblastoma Contains Topologically Distinct Proliferative and Metabolically Defined Subpopulations of Nestin- and Glut1-Expressing Cells. J. Neuropathol. Exp. Neurol. 2021, 80, 674. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.; Lu, L.; Hu, Z.; Fei, Z.; Liu, M.; Wei, L.; Xu, J. Nestin Expression Is Associated with Poor Clinicopathological Features and Prognosis in Glioma Patients: An Association Study and Meta-analysis. Mol. Neurobiol. 2017, 54, 727. [Google Scholar] [CrossRef] [PubMed]
- Benecke, A.G.; Eilebrecht, S. RNA-Mediated Regulation of HMGA1 Function. Biomolecules 2015, 5, 943. [Google Scholar] [CrossRef] [PubMed]
- Reeves, R. Molecular biology of HMGA proteins: Hubs of nuclear function. Gene 2001, 277, 63. [Google Scholar] [CrossRef]
- Pang, B.; Fan, H.; Zhang, I.Y.; Liu, B.; Feng, B.; Meng, L.; Zhang, R.; Sadeghi, S.; Guo, H.; Pang, Q. HMGA1 expression in human gliomas and its correlation with tumor proliferation, invasion and angiogenesis. J. Neurooncol. 2012, 106, 543. [Google Scholar] [CrossRef]
- Liu, B.; Pang, B.; Liu, H.; Arakawa, Y.; Zhang, R.; Feng, B.; Zhong, P.; Murata, D.; Fan, H.; Xin, T.; et al. High mobility group A1 expression shows negative correlation with recurrence time in patients with glioblastoma multiforme. Pathol. Res. Pract. 2015, 211, 596. [Google Scholar] [CrossRef]
- Que, T.; Zheng, H.; Zeng, Y.; Liu, X.; Qi, G.; La, Q.; Liang, T.; Li, Z.; Yi, G.; Zhang, S.; et al. HMGA1 stimulates MYH9-dependent ubiquitination of GSK-3β via PI3K/Akt/c-Jun signaling to promote malignant progression and chemoresistance in gliomas. Cell Death Dis. 2022, 13, 164. [Google Scholar] [CrossRef]
- Hock, R.; Furusawa, T.; Ueda, T.; Bustin, M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007, 17, 72. [Google Scholar] [CrossRef]
- Fedele, M.; Battista, S.; Manfioletti, G.; Croce, C.M.; Giancotti, V.; Fusco, A. Role of the high mobility group A proteins in human lipomas. Carcinogenesis 2001, 22, 1583. [Google Scholar] [CrossRef]
- Shender, V.; Arapidi, G.; Butenko, I.; Anikanov, N.; Ivanova, O.; Govorun, V. Peptidome profiling dataset of ovarian cancer and non-cancer proximal fluids: Ascites and blood sera. Data Brief 2018, 22, 557. [Google Scholar] [CrossRef]
- Nowicki, M.O.; Hayes, J.L.; Chiocca, E.A.; Lawler, S.E. Proteomic Analysis Implicates Vimentin in Glioblastoma Cell Migration. Cancers 2019, 11, 466. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, S.S.; Garner, B.; Ooi, L. Understanding the Role of ApoE Fragments in Alzheimer’s Disease. Neurochem. Res. 2019, 44, 1297. [Google Scholar] [CrossRef] [PubMed]
- Hervás-Corpión, I.; Gallardo-Orihuela, A.; Catalina-Fernández, I.; Iglesias-Lozano, I.; Soto-Torres, O.; Geribaldi-Doldán, N.; Domínguez-García, S.; Luna-García, N.; Romero-García, R.; Mora-López, F.; et al. Potential Diagnostic Value of the Differential Expression of Histone H3 Variants between Low- and High-Grade Gliomas. Cancers 2021, 13, 5261. [Google Scholar] [CrossRef]
- Wu, G.; Broniscer, A.; McEachron, T.A.; Lu, C.; Paugh, B.S.; Becksfort, J.; Qu, C.; Ding, L.; Huether, R.; Parker, M.; et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012, 44, 251. [Google Scholar]
- Oliveros, J.C. (2007–2015) Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Version 2.1.0. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 25 March 2025).
- Uhlén, M.; Björling, E.; Agaton, C.; Szigyarto, C.A.; Amini, B.; Andersen, E.; Andersson, A.C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteom. 2005, 4, 1920. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, 2507. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Nastou, K.; Koutrouli, M.; Kirsch, R.; Mehryary, F.; Hachilif, R.; Hu, D.; Peluso, M.E.; Huang, Q.; Fang, T.; et al. The STRING database in 2025: Protein networks with directionality of regulation. Nucleic Acids Res. 2025, 53, D730–D737. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543. [Google Scholar] [CrossRef]


| Peptide Sequence § | Gene Name | Uniprot Accession * | m/z (MH+) Monoisotopic | PTM | Sequence Position (N- or C-Terminal) | A+ | A− |
|---|---|---|---|---|---|---|---|
| DAHKSEVAHRFKDLGEENFKALVL | ALB | P02768 | 2753.444 | - | 24–47 | - | - |
| VSVSEHTW | ANK1 | P16157 | 944.449 | - | 1755–1763 | - | - |
| VELGTQPATQ | APOA2 | P02652 | 1043.540 | - | 91–100 (C-Term) | - | - |
| MVDNWRPAQPLKNRQIKASFK | CA2 | P00918 | 2543.372 | M1 (Oxid.) | 240–260 (C-Term) | - | - |
| IEGVDAEDGHGPGEQQ | CFB | P00751 | 1637.710 | - | 242–257 | - | - |
| ITREEKPAVTAAPKK | CRYAB | P02511 | 1638.953 | - | 161–175 (C-Term) | - | - |
| FDQFFGEHL | CRYAB | P02511 | 1139.518 | - | 24–32 | - | - |
| DVELLKLE | FKBP1A | P62942 | 958.547 | - | 101–108 (C-Term) | - | - |
| ESARQDEGKEEEPEADQEHA | GAP43 | P17677 | 2283.961 | - | 219–238 (C-Term) | - | - |
| PDPAKSAPAP | H2BC17 | P23527 | 950.496 | - | 2–11 (N-Term) | - | - |
| GGGYDNYGGGNY | HNRNPA2B1 | P22626 | 1193.454 | - | 284–295 | - | - |
| AAGVEAAAEVAATEIK | HNRNPM | P52272 | 1542.807 | Acetyl (N-Term) | 2–17 (N-Term) | - | - |
| SDEFSLADALPEHSPAKTSAVS | MAPK1IP1L | Q8NDC0 | 2301.099 | Acetyl (N-Term) | 2–23 (N-Term) | - | - |
| EDMIRLGDLNEAGILR | MYO7A | Q13402 | 1814.945 | - | 68–83 | - | - |
| LEGKVLPGVDALSNI | PGK1 | P00558 | 1524.867 | - | 403–417 (C-Term) | - | - |
| AANFLLQQNFDED | RAD23B | P54727 | 1524.701 | - | 397–409 (C-Term) | - | - |
| KAPAQKVPAQKATGQKAAPAPKA | RPL14 | P50914 | 2257.317 | - | 173–194 | - | - |
| MEDLDQSPLVSSSDSPPRPQPAFKY | RTN4 | Q9NQC3 | 2833.341 | Acetyl (N-Term) | 1–25 (N-Term) | - | - |
| KTIAYGYIHDPSGGPVSLDFVKSGDYALERM | SARDH | Q9UL12 | 3402.681 | M31(Oxi) | 863–893 | - | - |
| LVRYTKKVPQVSTPTL | ALB | P02768 | 1830.091 | - | 432–447 | x | - |
| GEYKFQNALL | ALB | P02768 | 1182.618 | - | 423–432 | x | - |
| AEYGTLLQDLTNN | PEA15 | Q15121 | 1493.716 | Acetyl (N-Term) | 2–14 (N-Term) | x | - |
| EGKVLPGVDALSNI | PGK1 | P00558 | 1411.782 | - | 404–417 (C-Term) | x | - |
| AASGAEPQVLVQY | PTRHD1 | Q6GMV3 | 1374.695 | Acetyl (N-Term) | 16–28 | x | - |
| VDIINAKQ | TPI1 | P60174 | 900.518 | - | 242–249 (C-Term) | x | - |
| SPTVSQVTERSQD | ANK1 | P16157 | 1433.692 | - | 1686–1698 | x | x |
| SSKITHRIHWESASLLR | C3 | P01024 | 2021.109 | - | 1304–1320 | x | x |
| MVDNWRPAQPLKNRQIKASFK | CA2 | P00918 | 2527.375 | - | 240–260 (C-Term) | x | x |
| SSEPPPPPQPPTHQA | CARHSP1 | Q9Y2V2 | 1608.773 | Acetyl (N-Term) | 2–16 (N-Term) | x | x |
| SARQDEGKEEEPEADQEHA | GAP43 | P17677 | 2154.921 | - | 220–238 (C-Term) | x | x |
| SETPDQFMTADETRNLQ | CD44 | P16070 | 1982.878 | - | 718–734 | x | x |
| SYTLDSLGNPSA | NEFM | P07197 | 1266.589 | Acetyl (N-Term) | 2–13 (N-Term) | x | x |
| FVSNHAY | ALDOA | P04075 | 837.391 | - | 358–364 (C-Term) | - | x |
| FVELGTQPATQ | APOA2 | P02652 | 1190.607 | - | 90–100 (C-Term) | - | x |
| FVQMMTAK | CALM3 | P0DP25 | 955.477 | - | 142–149 (C-Term) | - | x |
| ASGVAVSDGVIKVF | CFL1 | P23528 | 1390.761 | Acetyl (N-Term) | 2–15 (N-Term) | - | x |
| DDIGDDDEDIDDI | EIF1AY | O14602 | 1464.554 | - | 132–144 (C-Term) | - | x |
| AGWNAYIDNL | PFN1 | P07737 | 1178.551 | Acetyl (N-Term) | 2–11 (N-Term) | - | x |
| ISHELDSASSEVN | SPP1 | P10451 | 1387.638 | - | 302–314 (C-Term) | - | x |
| AERPAEETGPQEEEGETAGEAPVSH | SPTB | P11277 | 2607.157 | - | 2082–2106 | - | x |
| Peptide Sequence § | Gene Name | Uniprot Accession * | m/z (MH+) Monoisotopic | PTM | Sequence Position (N- or C-Terminal) | A+ | A− | A+ | A− |
|---|---|---|---|---|---|---|---|---|---|
| ND | ND | R | R | ||||||
| VIKAVDKKAAGAGKVTKSAQKAQKA | EEF1A1 | P68104 | 2496.497 | - | 2–13 (N-Term) | - | - | - | - |
| FLAEGGGVR | FGA | P02671 | 905.487 | - | 27–35 | - | - | - | - |
| RNIVVKTVEMRDGEVIKE | GFAP | P14136 | 2115.164 | - | 406–423 | - | - | - | - |
| SKPHSEAGTAFIQTQQLH | PKM | P14618 | 2022.009 | Acetyl (N-Term) | 2–19 (N-Term) | - | - | - | - |
| KNPLPSKETIEQEKQAGES | TMSB4X | P62328 | 2113.084 | - | 26–44 (C-Term) | - | - | - | - |
| IENEEQEYVQTVK | ANXA1 | P04083 | 1608.778 | - | 14–26 | x | - | - | - |
| VGVIKAVDKKAAGAGKVTKSA | EEF1A1 | P68104 | 1998.209 | - | 435–455 | x | - | - | - |
| YDMNAANVGWNNSTFA | MIF | P14174 | 1790.75 | M3 (Oxi) | 100–115 (C-Term) | x | - | - | - |
| NAANVGWNNSTFA | MIF | P14174 | 1365.621 | - | 103–115 (C-Term) | x | - | - | - |
| TLPTKETIEQEKRSEIS | TMSB10 | P63313 | 1989.056 | - | 28–43 | x | - | - | - |
| KTETQEKNPLPSKETIEQEKQAG | TMSB4X | P62328 | 2613.355 | - | 20–42 | x | - | - | - |
| LSALEEYTKKLNTQ | APOA1 | P02647 | 1637.879 | - | 254–267 (C-Term) | - | x | - | - |
| RETSLDTKSVSEGHL | GFAP | P14136 | 1658.838 | Acetyl (N-Term) | 390–404 | - | x | - | - |
| IRETSLDTKSVSE | GFAP | P14136 | 1464.76 | - | 389–401 | - | x | - | - |
| ETSLDTKSVSEGHL | GFAP | P14136 | 1502.737 | - | 391–404 | - | x | - | - |
| ASVSTVLTSKY | HBA1; HBA2 | P69905 | 1155.627 | - | 131–141 | - | x | - | - |
| VLSPADKTNVKAAWGKVG | HBA1; HBA2 | P69905 | 1841.035 | - | 2–19 (N-Term) | - | x | - | - |
| IEEQKIVVK | RPL34 | P49207 | 1085.659 | - | 97–105 | - | x | - | - |
| TQEKNPLPSKETIEQEKQAGES | TMSB4X | P62328 | 2471.235 | - | 23–44 (C-Term) | - | x | - | - |
| GKVKVGVNGFGRIG | GAPDH | P04406 | 1387.82 | - | 2–15 (N-Term) | x | x | - | - |
| MRDGEVIKESKQEHKDVM | GFAP | P14136 | 2159.061 | - | 415–432 (C-Term) | x | x | - | - |
| PAAPAPAEKTPVKKKARK | H1-4 | P10412 | 1888.15 | - | 8–26 | x | x | - | - |
| SETAPAETATPAPVEKSPAK | H1-5 | P16401 | 2024.028 | Acetyl (N-Term) | 2–21 (N-Term) | x | x | - | - |
| VSTVLTSKY | HBA1; HBA2 | P69905 | 997.558 | - | 133–141 | x | x | - | - |
| AEAFDDVVGETVGKTD | MAP4 | P27816 | 1652.77 | - | 31–46 | x | x | - | - |
| YDMNAANVGWNNSTFA | MIF | P14174 | 1774.752 | - | 100–115 (C-Term) | x | x | - | - |
| AELQEVQITE | NDRG2 | Q9UN36 | 1201.597 | Acetyl (N-Term) | 2–11 (N-Term) | x | x | - | - |
| KEEIIKTLSKEEETK | RPL19 | P84098 | 1804.995 | - | 181–195 | x | x | - | - |
| AQAAAPASVPAQAPK | RPL29 | P47914 | 1377.753 | - | 135–149 | x | x | - | - |
| ASSDIQVKELEKRASGQAF | STMN1 | P16949 | 2106.087 | Acetyl (N-Term) | 2–20 (N-Term) | x | x | - | - |
| SDKPDMAEIEKFDKSKL | TMSB4X | P62328 | 2023.007 | Acetyl (N-Term) | 2–18 (N-Term) | x | x | - | - |
| SDKPDMAEIEKFDKS | TMSB4X | P62328 | 1781.83 | Acetyl (N-Term) | 2–16 (N-Term) | x | x | - | - |
| SDEREVAEAATGEDA | VAT1 | Q99536 | 1591.676 | Acetyl (N-Term) | 2–16 (N-Term) | x | x | - | - |
| TVETRDGQVINETSQHHDDLE | VIM | P08670 | 2423.117 | - | 446–466 (C-Term) | x | x | - | - |
| TVETRDGQVINETS | VIM | P08670 | 1548.756 | - | 446–459 | x | x | - | - |
| QIRETSLDTKSVSEGHLK | GFAP | P14136 | 2028.076 | - | 388–405 | - | - | x | x |
| GKRKASGPPVSELITK | H1-3 | P16402 | 1667.981 | - | 32–47 | - | - | x | x |
| LASVSTVLTSKYR | HBA1; HBA2 | P69905 | 1424.812 | - | 130–142 (C-Term) | - | - | x | x |
| ALLSPYSYSTTAVVTNPKE | TTR | P02766 | 2041.057 | - | 129–147 (C-Term) | - | - | x | x |
| GYTEHQVVSSDFNSDTH | GAPDH | P04406 | 1922.819 | - | 275–291 | x | - | x | - |
| ADKPDMGEIASFDKAKLKKTETQEKN | TMSB10 | P63313 | 2980.501 | Acetyl (N-Term) | 2–27 (N-Term) | x | - | x | - |
| DSYDSYATHNE | CIRBP | Q14011 | 1301.497 | - | 162–172 (C-Term) | x | x | x | - |
| EGDFLAEGGGVR | FGA | P02671 | 1206.577 | - | 24–35 | x | x | x | - |
| RDGEVIKESKQEHKDVM | GFAP | P14136 | 2028.02 | - | 416–432 (C-Term) | x | x | x | - |
| TENSTSAPAAKPK | H1-0 | P07305 | 1343.682 | Acetyl (N-Term) | 2–14 (N-Term) | x | x | x | - |
| PAAPAAPAPAEKTPVKKKAR | H1-4 | P10412 | 1999.184 | - | 6–25 | x | x | x | - |
| GAAKRKASGPPVSELITK | H1-4 | P10412 | 1810.054 | - | 29–46 | x | x | x | - |
| VLSPADKTNVKAAWGKVGAHAGEYGAEALER | HBA1; HBA2 | P69905 | 3195.662 | - | 2–32 (N-Term) | x | x | x | - |
| PAVHASLDKF | HBA1; HBA2 | P69905 | 1084.579 | - | 120–129 | x | x | x | - |
| HFDLSHGSAQVK | HBA1; HBA2 | P69905 | 1325.662 | - | 46–57 | x | x | x | - |
| SPKAEDGATPSPSNETPKK | MARCKS | P29966 | 1940.961 | - | 135–153 | x | x | x | - |
| SPKAEDGATPSPSNETPK | MARCKS | P29966 | 1812.866 | - | 135–152 | x | x | x | - |
| FEGFPDKQPR | S100A4 | P26447 | 1220.606 | - | 90–99 | x | x | x | - |
| LKKTETQEKNPLPSKETIEQEKQAGES | TMSB4X | P62328 | 3070.6 | - | 18–44 (C-Term) | x | x | x | - |
| TVETRDGQVINETSQ | VIM | P08670 | 1676.815 | - | 446–460 | x | x | x | - |
| IRETSLDTKSVSEGHL | GFAP | P14136 | 1771.925 | - | 389–404 | x | x | - | x |
| IKTVETRDGQVINETSQHHDDLE | VIM | P08670 | 2664.295 | - | 444–466 (C-Term) | x | x | - | x |
| IKTVETRDGQVINETSQ | VIM | P08670 | 1917.994 | - | 444–460 | x | x | - | x |
| EIENPETSDQ | GYPA | P02724 | 1161.493 | - | 141–150 (C-Term) | - | x | x | - |
| SETAPAETATPAPVEKSPAKKKAT | H1-5 | P16401 | 2452.305 | Acetyl (N-Term) | 2–25 (N-Term) | - | x | x | - |
| FLSFPTTKTY | HBA1; HBA2 | P69905 | 1204.628 | - | 34–43 | - | x | x | - |
| AEDVTAALAKQGL | MAP2 | P11137 | 1286.697 | - | 1815–1827 (C-Term) | - | x | x | - |
| SEEIITPVY | CAPN1 | P07384 | 1092.547 | Acetyl (N-Term) | 2–10 (N-Term) | - | x | - | x |
| APPGGRANITSLG | DPYSL2 | Q16555 | 1210.657 | - | 560–572 (C-Term) | - | x | - | x |
| VNDNEEGFFSA | FGB | P02675 | 1228.514 | - | 33–43 | - | x | - | x |
| SETAPAAPAAPAPAEKTPVKKK | H1-4 | P10412 | 2202.219 | Acetyl (N-Term) | 2–23 (N-Term) | - | x | - | x |
| ENPVVHFF | MBP | P02686 | 988.491 | - | 217–224 | - | x | - | x |
| DLEPTVIDEVRTGTY | TUBA1B | P68363 | 1707.847 | - | 69–83 | - | x | - | x |
| GEGDFLAEGGGVR | FGA | P02671 | 1263.596 | - | 23–35 | - | x | x | x |
| MRDGEVIKESKQEHKDVM | GFAP | P14136 | 2191.046 | M1(Oxi) M18 (Oxi) | 415–432 (C-Term) | - | x | x | x |
| LNETGDEPFQYKN | GLUL | P15104 | 1554.71 | - | 361–373 (C-Term) | - | x | x | x |
| SVEIENPETSDQ | GYPA | P02724 | 1347.594 | - | 139–150 (C-Term) | - | x | x | x |
| GDPALQDAGDSSRKEYFI | GYPC | P04921 | 1968.935 | - | 111–128 (C-Term) | - | x | x | x |
| SETAPAAPAAAPPAEKAPVKKKAA | H1-2 | P16403 | 2314.285 | Acetyl (N-Term) | 2–25 (N-Term) | - | x | x | x |
| ASVSTVLTSKYR | HBA1; HBA2 | P69905 | 1311.731 | - | 131–142 (C-Term) | - | x | x | x |
| SVSTVLTSKYR | HBA1; HBA2 | P69905 | 1240.693 | - | 132–142 (C-Term) | - | x | x | x |
| VHLTPEEKSAVTAL | HBB | P68871 | 1494.821 | - | 2–15 (N-Term) | - | x | x | x |
| GFKGVDAQGTLS | MBP | P02686 | 1179.603 | - | 274–285 | - | x | x | x |
| QDENPVVHFF | MBP | P02686 | 1231.577 | - | 215–224 | - | x | x | x |
| SLPQKSHGRTQDENPVVH | MBP | P02686 | 2029.02 | - | 205–222 | - | x | x | x |
| DDDIAALVVDNG | ACTB | P60709 | 1258.584 | Acetyl (N-Term) | 2–13 (N-Term) | x | x | x | x |
| DDDIAALVVDNGSG | ACTB | P60709 | 1402.637 | Acetyl (N-Term) | 2–15 (N-Term) | x | x | x | x |
| FIENEEQEYVQTVK | ANXA1 | P04083 | 1755.845 | - | 13–26 | x | x | x | x |
| SEAEDASLLSF | APOC3 | P02656 | 1168.539 | - | 21–31 (N-Term) | x | x | x | x |
| GKDQSGEVLSSV | AQP4 | P55087 | 1205.604 | - | 312–323 (C-Term) | x | x | x | x |
| LTTDEYDGHSTYPSHQYQ | BLVRB | P30043 | 2141.912 | - | 189–206 (C-Term) | x | x | x | x |
| SAKTSPAKQQAPPVRNLH | DPYSL2 | Q16555 | 1930.065 | - | 518–535 | x | x | x | x |
| ADSGEGDFLAEGGGVR | FGA | P02671 | 1536.695 | - | 20–35 | x | x | x | x |
| DSGEGDFLAEGGGVR | FGA | P02671 | 1465.658 | - | 21–35 | x | x | x | x |
| GDFLAEGGGVR | FGA | P02671 | 1077.535 | - | 25–35 | x | x | x | x |
| IRETSLDTKSVSEGHLK | GFAP | P14136 | 1900.017 | - | 389–405 | x | x | x | x |
| MRDGEVIKESKQEHKDVM | GFAP | P14136 | 2175.054 | M18(Oxi) | 415–432 (C-Term) | x | x | x | x |
| RDGEVIKESKQEHKDVM | GFAP | P14136 | 2044.012 | M17(Oxi) | 416–432 (C-Term) | x | x | x | x |
| RNIVVKTVEMRDGEVIKE | GFAP | P14136 | 2131.157 | M10(Oxi) | 406–423 | x | x | x | x |
| SNLQIRETSLDTKSVSEGHLK | GFAP | P14136 | 2342.235 | - | 375–405 | x | x | x | x |
| RNIVVKTVEMRDGEVIKESKQEHKDVM | GFAP | P14136 | 3229.674 | M10(Oxi) M27 (Oxi) | 406–432 (C-Term) | x | x | x | x |
| SLDTKSVSEGHLK | GFAP | P14136 | 1400.74 | - | 393–404 | x | x | x | x |
| SSVEIENPETSDQ | GYPA | P02724 | 1434.629 | - | 138–150 (C-Term) | x | x | x | x |
| VEIENPETSDQ | GYPA | P02724 | 1260.563 | - | 140–150 (C-Term) | x | x | x | x |
| SETAPAAPAAPAPAEKTPVK | H1-4 | P10412 | 1946.028 | Acetyl (N-Term) | 2–21 (N-Term) | x | x | x | x |
| AAKRKASGPPVSELITK | H1-4 | P10412 | 1753.035 | - | 30–46 | x | x | x | x |
| SETAPAAPAAPAPAEKTPVKKKAR | H1-4 | P10412 | 2429.354 | Acetyl (N-Term) | 2–25 (N-Term) | x | x | x | x |
| SETAPAAPAAPAPAEKTPVKKKARK | H1-4 | P10412 | 2557.452 | Acetyl (N-Term) | 2–26 (N-Term) | x | x | x | x |
| SETAPAAPAAPAPAEKTPVKKKA | H1-4 | P10412 | 2273.258 | Acetyl (N-Term) | 2–24 (N-Term) | x | x | x | x |
| SETAPAAPAAPAPAEKTPVKK | H1-4 | P10412 | 2074.126 | Acetyl (N-Term) | 2–22 (N-Term) | x | x | x | x |
| VLSPADKTNVKAAWGKVGAHAGEYGAEALERM | HBA1; HBA2 | P69905 | 3342.701 | M32(Oxi) | 2–33 (N-Term) | x | x | x | x |
| AAHLPAEFTPAVHASLDKF | HBA1; HBA2 | P69905 | 2022.049 | - | 111–128 | x | x | x | x |
| LASVSTVLTSKY | HBA1; HBA2 | P69905 | 1268.711 | - | 130–141 | x | x | x | x |
| FDLSHGSAQVK | HBA1; HBA2 | P69905 | 1188.604 | - | 47–57 | x | x | x | x |
| GKVNVDEVGGEALG | HBB | P68871 | 1343.684 | - | 17–30 | x | x | x | x |
| VHLTPEEKSAVT | HBB | P68871 | 1310.696 | - | 2–13 (N-Term) | x | x | x | x |
| LSAKPAPPKPEPKPK | HMGN2 | P05204 | 1584.946 | - | 28–42 | x | x | x | x |
| SARLSAKPAPPKPEPKPK | HMGN2, HMG17 | P05204 | 1899.117 | - | 25–42 | x | x | x | x |
| SARLSAKPAPPKPEPKPR | HMGN3 | Q15651 | 1927.124 | - | 28–44 | x | x | x | x |
| TEKSNIEETTISSQKI | INA | Q16352 | 1807.93 | - | 484–499 (C-Term) | x | x | x | x |
| YGSLPQKSHGRTQDENPVVH | MBP | P02686 | 2249.113 | - | 203–222 | x | x | x | x |
| TQDENPVVHFF | MBP | P02686 | 1332.623 | - | 214–224 | x | x | x | x |
| DTGILDSIGR | MBP | P02686 | 1046.55 | - | 168–177 | x | x | x | x |
| DTGILDSIGRF | MBP | P02686 | 1193.619 | - | 168–178 | x | x | x | x |
| GRTQDENPVVHF | MBP | P02686 | 1398.678 | - | 212–223 | x | x | x | x |
| GSLPQKSHGRTQDENPVVH | MBP | P02686 | 2086.043 | - | 204–222 | x | x | x | x |
| FLLSQNFDDE | RAD23A | P54725 | 1227.553 | - | 354–363 (C-Term) | x | x | x | x |
| KTETQEKNPLPSKETIEQEKQ | TMSB4X | P62328 | 2485.284 | - | 20–40 | x | x | x | x |
| KTETQEKNPLPSKETIEQEKQAGES | TMSB4X | P62328 | 2829.421 | - | 20–44 (C-Term) | x | x | x | x |
| SDKPDMAEIEKFDKS | TMSB4X | P62328 | 1797.826 | Acetyl (N-Term) | 2–16 (N-Term) | x | x | x | x |
| SDKPDMAEIEKFDKSK | TMSB4X | P62328 | 1925.919 | Acetyl (N-Term) | 2–17 (N-Term) | x | x | x | x |
| LLSPYSYSTTAVVTNPKE | TTR | P02766 | 1970.017 | - | 130–147 (C-Term) | x | x | x | x |
| VDLEPTVIDEVR | TUBA1B | P68363 | 1384.735 | - | 68–79 | x | x | x | x |
| DLEPTVIDEVR | TUBA1B | P68363 | 1285.667 | - | 69–79 | x | x | x | x |
| LIKTVETRDGQVINETSQHHDDLE | VIM | P08670 | 2777.378 | - | 443–466 (C-Term) | x | x | x | x |
| RDGQVINETSQHHDDLE | VIM | P08670 | 1992.905 | - | 450–466 (C-Term) | x | x | x | x |
| LIKTVETRDGQVINETSQ | VIM | P08670 | 2031.078 | - | 443–460 | x | x | x | x |
| Peptide Sequence § | Gene Name | Uniprot Accession * | m/z (MH+) Monoisotopic | PTM | Sequence Position (N- or C-Terminal) | A+ | A− |
|---|---|---|---|---|---|---|---|
| FSNKITPIQSKEAY | AK4 | P27144 | 1625.859 | - | 210–223 (C-Term) | - | - |
| GELAKHAVSEGTKAVTK | H2BK1 | A0A2R8Y619 | 1725.954 | - | 101–117 | - | - |
| RDNIQGITKPAIRR | H4C1, H4C2, H4C3, H4C4, H4C5 | P62805 | 1637.960 | - | 24–37 | - | - |
| FRDGDILGKYVD | HSPE1 | P61604 | 1397.711 | - | 91–102 (C-Term) | - | - |
| ATLKEKLIAPVA | LDHB | P07195 | 1295.797 | Acetyl (N-Term) | 2–13 (N-Term) | - | - |
| LEEENQESLR | NES | P48681 | 1246.596 | - | 882–893 | - | - |
| GPGPGGPGGAGVARGGAGGGP | NRGN | Q92686 | 1559.774 | - | 55–75 | - | - |
| ASGNARIGKPAPDFK | PRDX2 | P32119 | 1570.839 | Acetyl (N-Term) | 2–16 (N-Term) | - | - |
| IVKPVKVSAPRVGGK | RPL24 | P83731 | 1534.981 | - | 142–156 | - | - |
| IAKLEKAKAKELATKLG | RPL7A | P62424 | 1812.138 | - | 250–266 (C-Term) | - | - |
| SQDEVKAETIRSL | SYN1 | P17600 | 1475.774 | - | 682–695 | - | - |
| ANEDEAKAETIRSL | SYN2 | Q92777 | 1546.775 | - | 559–572 | - | - |
| TEPLPEKTQESL | TPD52 | P55327 | 1371.706 | - | 212–224 (C-Term) | - | - |
| SPKVSDTVVEPYN | TUBB | P07437 | 1434.715 | - | 172–184 | - | - |
| SHEKSFLVSGDN | GRINA | Q7Z429 | 1361.637 | Acetyl (N-Term) | 2–13 (N-Term) | x | - |
| KLEKEEEEGISQESSEEEQ | HMGA1 | P17096 | 2237.003 | - | 89–107 (C-Term) | x | - |
| VELQKEEAQKL | HNRNPU | Q00839 | 1314.729 | - | 655–665 | x | - |
| GPGGPGGAGVARGGAGGGP | NRGN | Q92686 | 1405.702 | - | 57–75 | x | - |
| DFGSLSNLQVTQP | RPS17 | P08708 | 1405.697 | - | 110–122 | x | - |
| EIVHIQAGQCG | TUBB | P07437 | 1154.566 | - | 3–13 | x | - |
| GGPGGAGVARGGAGGGPSGD | NRGN | Q92686 | 1510.707 | - | 59–78 (C-Term) | x | x |
| RKGPGPGGPGGAGVARGGAGGGPSGD | NRGN | Q92686 | 2103.056 | - | 53–78 (C-Term) | x | x |
| GGPGGAGVARGGAGGGP | NRGN | Q92686 | 1251.623 | - | 59–75 | x | x |
| RTGPPTTQQPRPSGPGPAGRPKP | SYN1 | P17600 | 2.337.261 | - | 612–634 | - | x |
| Peptide Sequence § | Gene Name | Uniprot Accession | m/z (MH+) Monoisotopic | PTM | Sequence Position (N- or C-terminal) | A+ ND | A + R |
|---|---|---|---|---|---|---|---|
| VDLEPTVIDEVRTGTY | TUBA1C | Q9BQE3 | 1806.915 | - | 68–83 | x | - |
| SDKTIGGGDDSFNT | TUBA1C | Q9BQE3 | 1413.618 | - | 38–51 | x | - |
| EQLITGKEDAANNY | TUBA1C | Q9BQE3 | 1565.749 | - | 90–103 | x | - |
| VDLEPTVIDEVR | TUBA1C | Q9BQE3 | 1384.737 | - | 68–79 | x | - |
| DLEPTVIDEVR | TUBA1C | Q9BQE3 | 1285.667 | - | 69–79 | x | - |
| SETGAGKHVPRAVF | TUBA1C | Q9BQE3 | 1455.774 | - | 54–67 | x | - |
| DLTEFQTNL | TUBA1C | Q9BQE3 | 1080.521 | - | 251–259 | x | - |
| KVEQAVETEPEPELRQQTE | APOE | P02649 | 2240.111 | - | 19–37 (N-term) | x | - |
| AATDLERFSNAEPEPR | SEPTIN8 | Q92599 | 1844.881 | Acetyl (N-Term) | 2–17 (N-term) | x | - |
| ATDTSQGELVHPK | HP1BP3 | Q5SSJ5 | 1424.707 | Acetyl (N-Term) | 2–14 (N-term) | x | - |
| SARLSAKPAPPKPEPRPKKASA | HMGN4 | O00479 | 2284.328 | - | 25–46 | x | - |
| EIRRYQKSTELLIR | H3C1 | P68431 | 1805.045 | - | 51–64 | - | x |
| YIAALGGAGGKGNRFFLAN | MTG2 | Q9H4K7 | 1897.015 | - | 180–198 | - | x |
| Peptide Sequence § | Gene Name | Uniprot Accession | m/z (MH+) Monoisotopic | PTM | Sequence Position (N- or C-Terminal) | A− ND | A− R |
|---|---|---|---|---|---|---|---|
| ADPGSDLFSVQALQ | SPTA1 | P02549 | 1447.712 | - | 1209–1222 | x | - |
| MVEEDEHEPKFEKF | SPTA1 | P02549 | 1793.809 | - | 2309–2322 | x | - |
| DPGSDLFSVQALQ | SPTA1 | P02549 | 1376.675 | - | 1210–1222 | x | - |
| TAEEIQERRQEVLT | SPTA1 | P02549 | 1701.884 | - | 20–33 | x | - |
| AQSKGNPEQTPVLKPEEEAPAPEVG | VTN | P04004 | 2602.311 | - | 104–128 | x | - |
| RDQKLSELDDRADALQ | VAMP3 | Q15836 | 1872.948 | - | 39–54 | x | - |
| EEEIAALVIDNG | ACTG1 | P63261 | 1314.644 | Acetyl (N-Term) | 2–13 (N-term) | x | - |
| AVGGEALGRL | HBD | P02042 | 942.539 | - | 23–32 | x | - |
| AEDMETKIKNY | COX6B1 | P14854 | 1383.653 | Acetyl (N-Term) | 2–12 (N-term) | x | - |
| SIQENISSL | PKLR | P30613 | 1032.522 | Acetyl (N-Term) | 2–10 (N-term) | x | - |
| TVATPLNQVANPNSAIFGGARPREEVVQKEQE | EIF4H | Q15056 | 3449.790 | - | 217–248 (C-term) | x | - |
| IEDVTPIPSDSTRRKGGRRGR | RPS14 | P62263 | 2353.286 | - | 129–149 | x | - |
| EEPAAHDTEATATDYHTT | SLC4A1 | P02730 | 1959.828 | - | 32–49 | x | - |
| ASTEGANNMPKQVEVRMHDSHLG | SLC1A2 | P43004 | 2550.182 | Acetyl (N-Term) | 2–24 (N-term) | x | - |
| WEVISDEHGIDPTGS | TUBB2A | Q13885 | 1641.745 | - | 21–35 | x | - |
| ADEVSASLAKQGL | MAPT | P10636 | 1288.678 | - | 746–758 (C-term) | x | - |
| TLADEVSASLAKQGL | MAPT | P10636 | 1502.813 | - | 744–758 (C-term) | x | - |
| EIRRYQKSTELLIR | H3-5 | Q6NXT2 | 1805.048 | - | 50–63 | - | x |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muntiu, A.; Vincenzoni, F.; Rossetti, D.V.; Urbani, A.; La Rocca, G.; Albanese, A.; Mazzucchi, E.; Olivi, A.; Sabatino, G.; Desiderio, C. Proteomic Analysis of the Low Molecular Mass Fraction of Newly Diagnosed and Recurrent Glioblastoma CUSA Fluid: A Pilot Investigation of the Peptidomic Profile. Int. J. Mol. Sci. 2025, 26, 6055. https://doi.org/10.3390/ijms26136055
Muntiu A, Vincenzoni F, Rossetti DV, Urbani A, La Rocca G, Albanese A, Mazzucchi E, Olivi A, Sabatino G, Desiderio C. Proteomic Analysis of the Low Molecular Mass Fraction of Newly Diagnosed and Recurrent Glioblastoma CUSA Fluid: A Pilot Investigation of the Peptidomic Profile. International Journal of Molecular Sciences. 2025; 26(13):6055. https://doi.org/10.3390/ijms26136055
Chicago/Turabian StyleMuntiu, Alexandra, Federica Vincenzoni, Diana Valeria Rossetti, Andrea Urbani, Giuseppe La Rocca, Alessio Albanese, Edoardo Mazzucchi, Alessandro Olivi, Giovanni Sabatino, and Claudia Desiderio. 2025. "Proteomic Analysis of the Low Molecular Mass Fraction of Newly Diagnosed and Recurrent Glioblastoma CUSA Fluid: A Pilot Investigation of the Peptidomic Profile" International Journal of Molecular Sciences 26, no. 13: 6055. https://doi.org/10.3390/ijms26136055
APA StyleMuntiu, A., Vincenzoni, F., Rossetti, D. V., Urbani, A., La Rocca, G., Albanese, A., Mazzucchi, E., Olivi, A., Sabatino, G., & Desiderio, C. (2025). Proteomic Analysis of the Low Molecular Mass Fraction of Newly Diagnosed and Recurrent Glioblastoma CUSA Fluid: A Pilot Investigation of the Peptidomic Profile. International Journal of Molecular Sciences, 26(13), 6055. https://doi.org/10.3390/ijms26136055









