Repeatome Dynamics and Sex Chromosome Differentiation in the XY and XY1Y2 Systems of the Fish Hoplias malabaricus (Teleostei; Characiformes)
Abstract
1. Introduction
2. Results
2.1. Satellitome Characterization
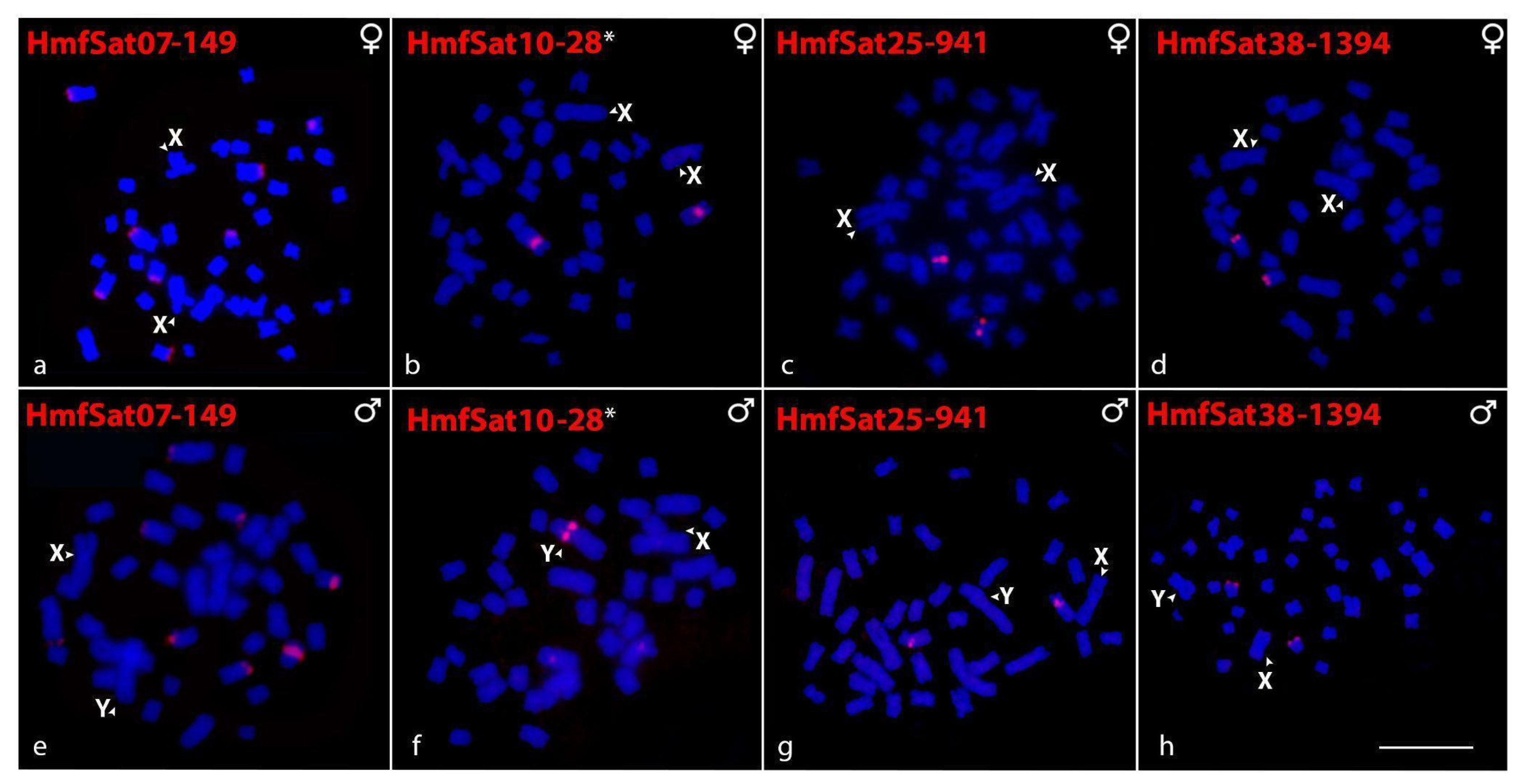
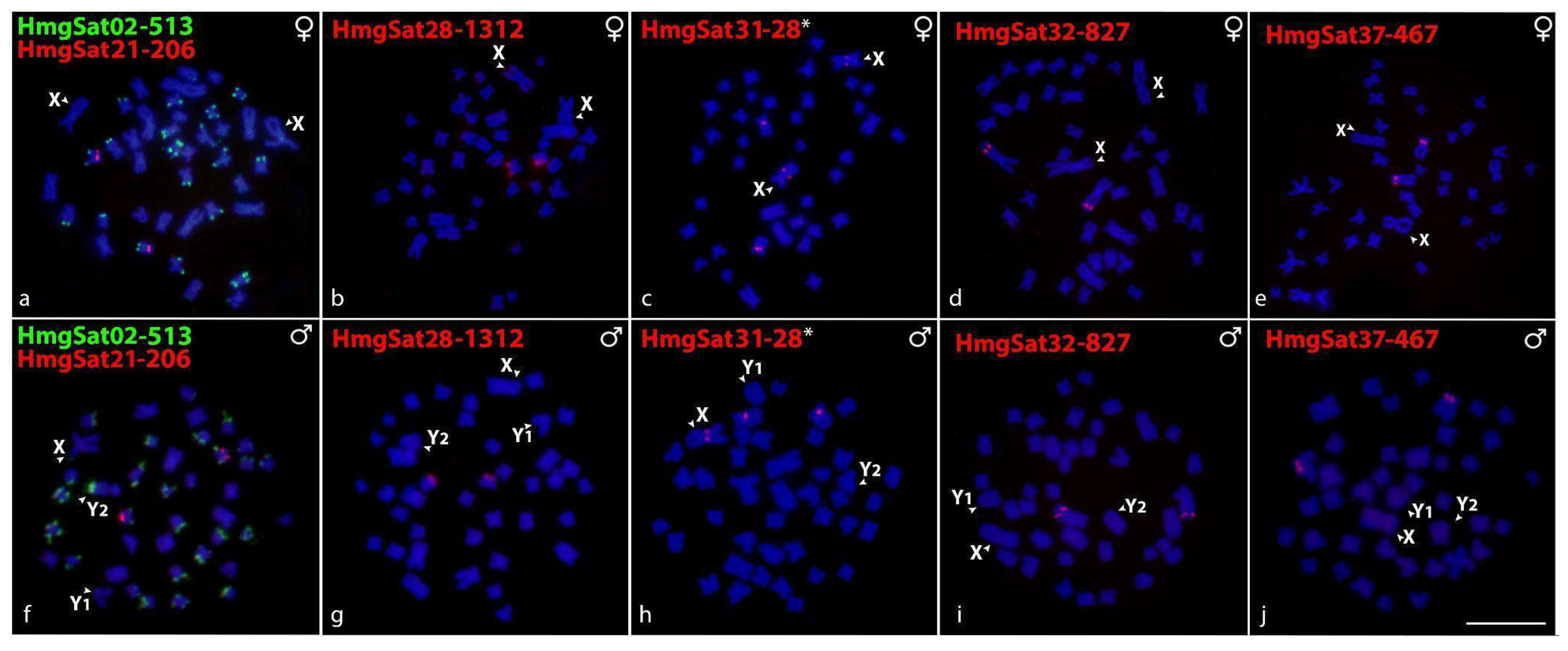
2.2. Chromosomal Distribution of HmfSatDNAs and HmgSatDNAs
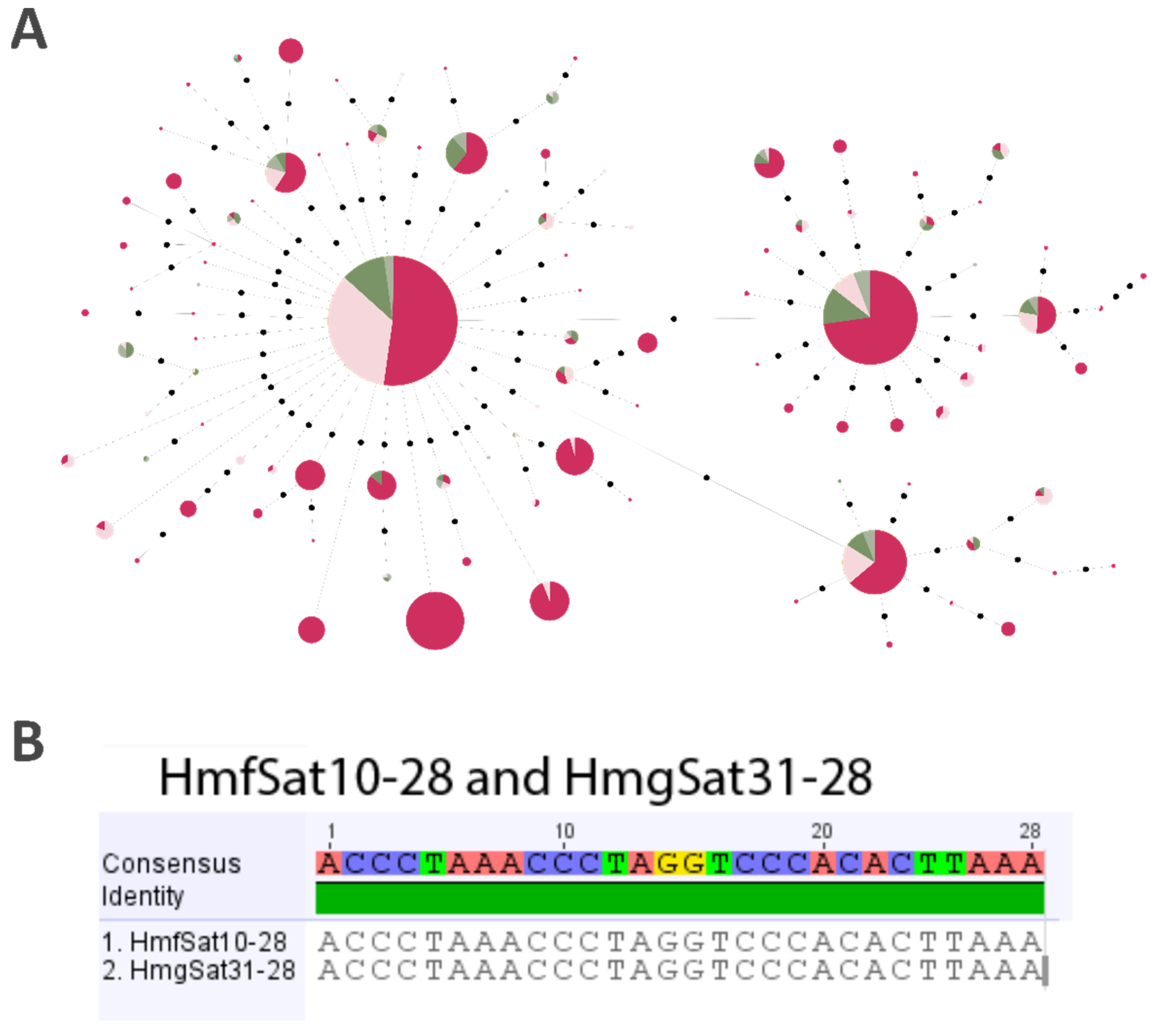
2.3. Minimum Spanning Tree (MST)
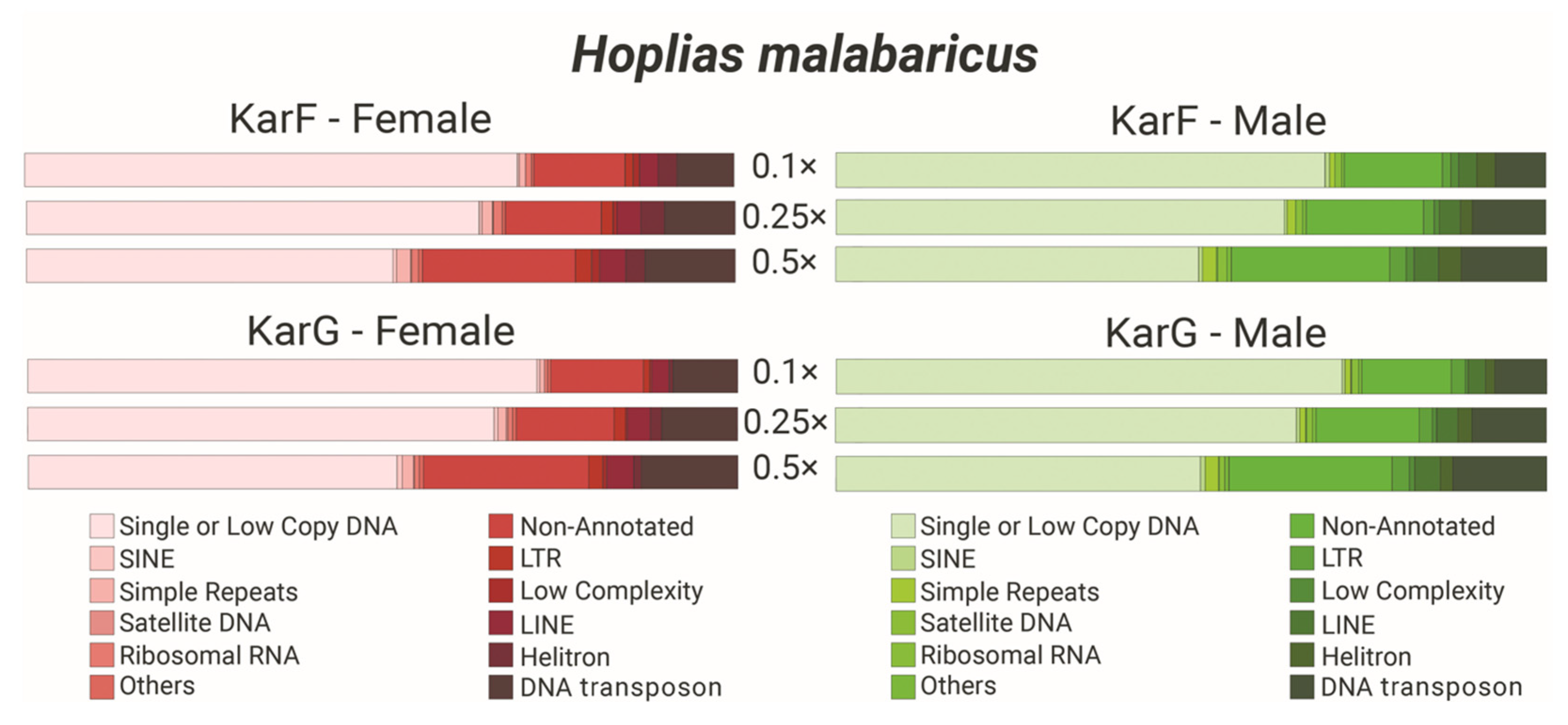
2.4. Repeatome Composition of H. malabaricus Karyomorphs F and G
2.5. Sex-Linked Markers
3. Discussion
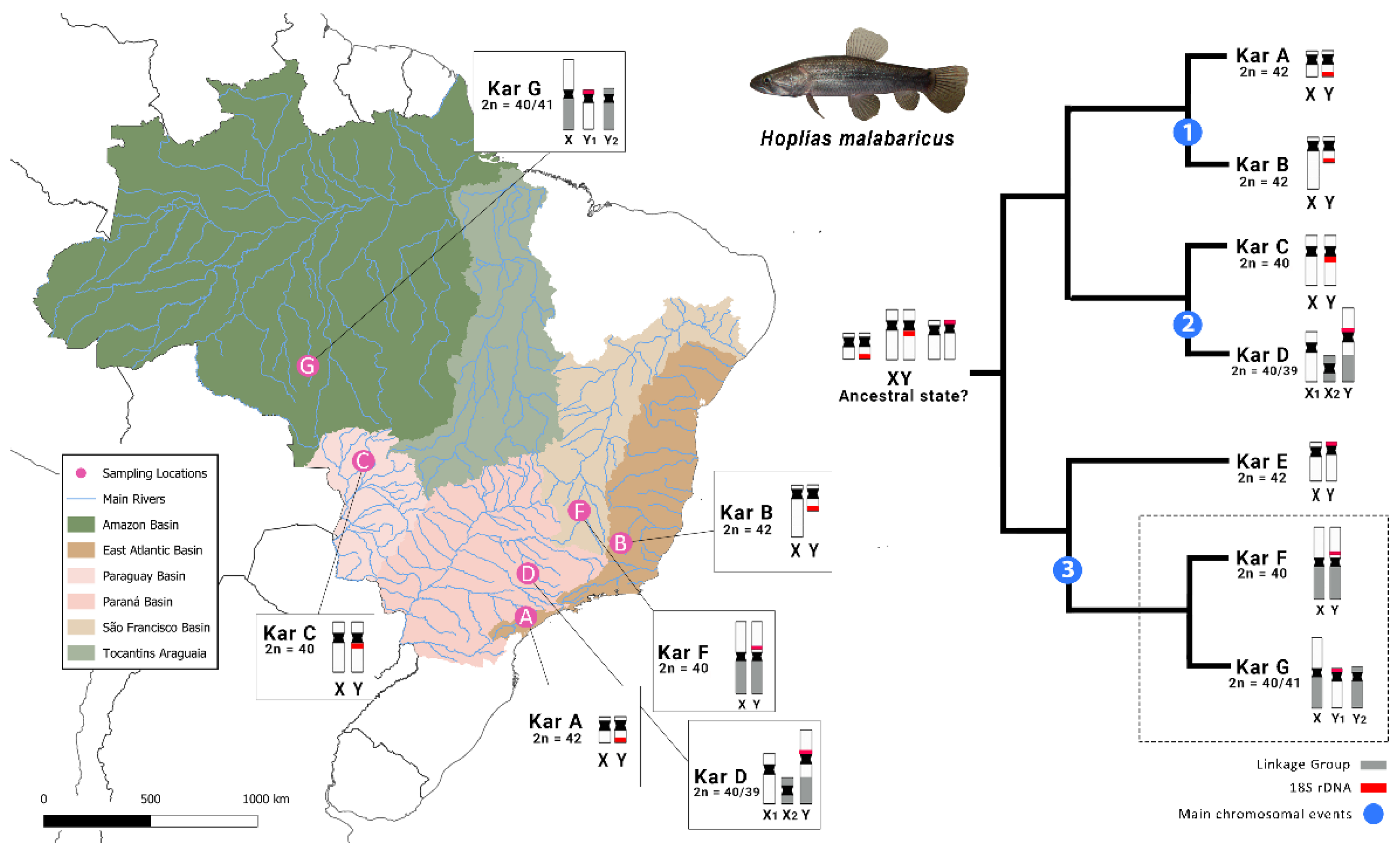
3.1. Different Evolutionary Paths of Related Sex Chromosome Systems
3.2. Insights on Sex Chromosome Differentiation Through Repetitive DNAs and Putative Sex-Linked Markers
4. Materials and Methods
4.1. Chromosome Preparation, DNA Extraction, and Low-Coverage Genome Sequencing
4.2. Characterization of the Satellitome
4.3. Satellite Amplification, Labeling, and Fluorescence In Situ Hybridization (FISH)
4.4. Repeatome Analysis
4.5. Genotyping by Sequencing (GBS) and Sex-Linked Markers Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Kar | Karyomorph |
| 2n | Diploid number |
| satDNA | Satellite DNA |
| TE | Transposable Element |
| SNP | Single Nucleotide Polymorphism |
| RADseq | Restriction Site-associated DNA Sequencing |
| ICMBIO | Chico Mendes Institute of Biodiversity Conservation |
| ISBIO | Brazilian Biodiversity Information and Authorization System |
| CEUA | Ethics Committee on Animal Experimentation |
| FISH | Fluorescence in situ Hybridization |
| DarT-Seq | Diversity Arrays Technology Sequencing |
| SLM | Sex-linked Marker |
| RFLP | Restriction Fragment Length Polymorphism |
| AFLP | Amplified Fragment Length Polymorphism |
| RFX4 | Regulatory Factor X 4 |
| TF | Transcription Factor |
| DBD | DNA-binding Domain |
References
- Nelson, J.S.; Grande, T.C.; Wilson, M.V. Fishes of the World; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar] [CrossRef]
- Fricke, R.; Eschmeyer, W.N.; Fong, J.D. Eschmeyer’s Catalog of Fishes: Genera/Species by Family/Subfamily. 2024. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/SpeciesByFamily.asp (accessed on 21 May 2024).
- Sember, A.; Nguyen, P.; Perez, M.F.; Altmanová, M.; Ráb, P.; Cioffi, M.D.B. Multiple sex chromosomes in teleost fishes from a cytogenetic perspective: State of the art and future challenges. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021, 376, 20200098. [Google Scholar] [CrossRef]
- Graves, J.A.M.; Shetty, S. Sex from W to Z: Evolution of vertebrate sex chromosomes and sex-determining genes. J. Exp. Zool. 2001, 290, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Schartl, M.; Schmid, M.; Nanda, I. Dynamics of vertebrate sex chromosome evolution: From equal size to giants and dwarfs. Chromosoma 2016, 125, 553–571. [Google Scholar] [CrossRef] [PubMed]
- Bertollo, L.A.C.; Born, G.G.; Dergam, J.A.; Fenocchio, A.S.; Moreira-Filho, O. A biodiversity approach in the neotropical Erythrinidae fish, Hoplias malabaricus: Karyotypic survey, geographic distribution of cytotypes, and cytotaxonomic considerations. Chromosome Res. 2000, 8, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, M.B.; Martins, C.; Vicari, M.R.; Rebordinos, L.; Bertollo, L.A.C. Differentiation of the XY sex chromosomes in the fish Hoplias malabaricus (Characiformes, Erythrinidae): Unusual accumulation of repetitive sequences on the X chromosome. Sex. Dev. 2010, 4, 176–185. [Google Scholar] [CrossRef]
- Souza, F.H.S.S.; Perez, M.F.; Ferreira, P.H.; Bertollo, L.A.C.; Ezaz, T.; Charlesworth, D.; Cioffi, M.B. Multiple karyotype differences between populations of the Hoplias malabaricus (Teleostei; Characiformes), a species complex in the gray area of the speciation process. Heredity 2024, 133, 216–226. [Google Scholar] [CrossRef]
- Born, G.G.; Bertollo, L.A.C. An XX/XY sex chromosome system in a fish species, Hoplias malabaricus, with a polymorphic NOR-bearing X chromosome. Chromosome Res. 2000, 8, 111–118. [Google Scholar] [CrossRef]
- de Oliveira, E.A.; Sember, A.; Bertollo, L.A.C.; Yano, C.F.; Ezaz, T.; Moreira-Filho, O.; Hatanaka, T.; Trifonov, V.; Liehr, T.; Al-Rikabi, A.B.H.; et al. Tracking the evolutionary pathway of sex chromosomes among fishes: Characterizing the unique XX/XY1Y2 system in Hoplias malabaricus (Teleostei, Characiformes). Chromosoma 2018, 127, 115–128. [Google Scholar] [CrossRef]
- Freitas, N.L.; Al-Rikabi, A.B.; Bertollo, L.A.C.; Ezaz, T.; Yano, C.F.; de Oliveira, E.A.; Hatanaka, T.; Cioffi, M.B. Early stages of XY sex chromosomes differentiation in the fish Hoplias malabaricus (Characiformes, Erythrinidae) revealed by DNA repeats accumulation. Curr. Genomics. 2018, 127, 115–128. [Google Scholar] [CrossRef]
- Deakin, J.E.; Potter, S.; O’Neill, R.; Ruiz-Herrera, A.; Cioffi, M.B.; Eldridge, M.D.; Fukui, K.; Graves, J.A.M.; Griffin, D.; Grutzner, F.; et al. Chromosomics: Bridging the gap between genomes and chromosomes. Genes 2019, 10, 627. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 2016, 6, 28333. [Google Scholar] [CrossRef] [PubMed]
- Utsunomia, R.; Silva, D.M.; Ruiz-Ruano, F.J.; Goes, C.A.G.; Melo, S.; Ramos, L.P.; Oliveira, C.; Porto-Foresti, F.; Foresti, F.; Hashimoto, D.T. Satellitome landscape analysis of Megaleporinus macrocephalus (Teleostei, Anostomidae) reveals intense accumulation of satellite sequences on the heteromorphic sex chromosome. Sci. Rep. 2019, 9, 5856. [Google Scholar] [CrossRef]
- Kretschmer, R.; Goes, C.A.G.; Bertollo, L.A.C.; Ezaz, T.; Porto-Foresti, F.; Toma, G.A.; Utsunomia, R.; Cioffi, M.B. Satellitome analysis illuminates the evolution of ZW sex chromosomes of Triportheidae fishes (Teleostei: Characiformes). Chromosoma 2022, 131, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Goes, C.A.G.; Dos Santos, R.Z.; Aguiar, W.R.C.; Alves, D.C.; Silva, D.M.; Foresti, F.; Oliveira, C.; Utsunomia, R.; Porto-Foresti, F. Revealing the satellite DNA history in Psalidodon and Astyanax characid fish by comparative satellitomics. Front. Genet. 2022, 13, 884072. [Google Scholar] [CrossRef]
- Crepaldi, C.; Martí, E.; Gonçalves, E.M.; Martí, D.A.; Parise-Maltempi, P.P. Genomic differences between the sexes in a fish species seen through satellite DNAs. Front. Genet. 2021, 12, 728670. [Google Scholar] [CrossRef]
- Lisachov, A.; Nguyen, D.H.M.; Panthum, T.; Ahmad, S.F.; Singchat, W.; Ponjarat, J.; Jaisamut, K.; Srisapoome, P.; Duengkae, P.; Hatachote, S.; et al. Emerging importance of bighead catfish (Clarias macrocephalus) and North African catfish (C. gariepinus) as a bioresource and their genomic perspective. Aquaculture 2023, 573, 739585. [Google Scholar] [CrossRef]
- Faber-Hammond, J.J.; Phillips, R.B.; Brown, K.H. Comparative Analysis of the Shared Sex-Determination Region (SDR) among Salmonid Fishes. Genome Bio. Evol. 2015, 7, 1972–1987. [Google Scholar] [CrossRef]
- Crepaldi, C.; Cabral-de-Mello, D.C.; Parise-Maltempi, P.P. Comparative analysis of transposable elements dynamics in fish with different sex chromosome systems. Genome 2024, 67, 339–350. [Google Scholar] [CrossRef]
- Schemberger, M.O.; Nascimento, V.D.; Coan, R.; Ramos, É.; Nogaroto, V.; Ziemniczak, K.; Valente, G.T.; Moreira-Filho, O.; Martins, C.; Vicari, M.R. DNA transposon invasion and microsatellite accumulation guide W chromosome differentiation in a Neotropical fish genome. Chromosoma 2019, 128, 547–560. [Google Scholar] [CrossRef]
- Palmer, D.H.; Rogers, T.F.; Dean, R.; Wright, A.E. How to identify sex chromosomes and their turnover. Mol. Ecol. 2019, 28, 4709–4724. [Google Scholar] [CrossRef]
- Baird, N.A.; Etter, P.D.; Atwood, T.S.; Currey, M.C.; Shiver, A.L.; Lewis, Z.A.; Selker, E.U.; Cresko, W.A.; Johnson, E.A. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 2008, 3, e3376. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.L.; Rodríguez Marí, A.; Braasch, I.; Amores, A.; Hohenlohe, P.; Batzel, P.; Postlethwait, J.H. Multiple sex-associated regions and a putative sex chromosome in zebrafish revealed by RAD mapping and population genomics. PLoS ONE 2012, 7, e40701. [Google Scholar] [CrossRef]
- Palaiokostas, C.; Bekaert, M.; Khan, M.G.; Taggart, J.B.; Gharbi, K.; McAndrew, B.J.; Penman, D.J. Mapping and validation of the major sex-determining region in Nile tilapia (Oreochromis niloticus L.) using RAD sequencing. PLoS ONE 2013, 8, e68389. [Google Scholar] [CrossRef]
- Palaiokostas, C.; Bekaert, M.; Davie, A.; Cowan, M.E.; Oral, M.; Taggart, J.B.; Gharbi, K.; McAndrew, B.J.; Penman, D.J.; Migaud, H. Mapping the sex determination locus in the Atlantic halibut (Hippoglossus hippoglossus) using RAD sequencing. BMC Genom. 2013, 14, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Palaiokostas, C.; Bekaert, M.; Taggart, J.B.; Gharbi, K.; McAndrew, B.J.; Chatain, B.; Penman, D.J.; Vandeputte, M. A new SNP-based vision of the genetics of sex determination in European sea bass (Dicentrarchus labrax). Genet. Sel. Evol. 2015, 47, 68. [Google Scholar] [CrossRef]
- Barchi, L.; Lanteri, S.; Portis, E.; Acquadro, A.; Valè, G.; Toppino, L.; Rotino, G.L. Identification of SNP and SSR markers in eggplant using RAD tag sequencing. BMC Genom. 2011, 12, 1–9. [Google Scholar] [CrossRef]
- Kafkas, S.; Khodaeiaminjan, M.; Güney, M.; Kafkas, E. Identification of sex-linked SNP markers using RAD sequencing suggests ZW/ZZ sex determination in Pistacia vera L. BMC Genom. 2015, 16, 1–11. [Google Scholar] [CrossRef]
- Kilian, B.; Graner, A. NGS technologies for analyzing germplasm diversity in genebanks. Brief. Funct. Genom. 2012, 11, 38–50. [Google Scholar] [CrossRef]
- Meyer, K.D. DART-seq: An antibody-free method for global m6A detection. Nat. Methods 2019, 16, 1275–1280. [Google Scholar] [CrossRef]
- Pereira, W.J.; Pappas, M.d.C.R.; Grattapaglia, D.; Pappas, G.J.J. A cost-effective approach to DNA methylation detection by Methyl Sensitive DArT sequencing. PLoS ONE 2020, 15, e0233800. [Google Scholar] [CrossRef]
- Lambert, M.R.; Skelly, D.K.; Ezaz, T. Sex-linked markers in the North American green frog (Rana clamitans) developed using DArTseq provide early insight into sex chromosome evolution. BMC Genom. 2016, 17, 844. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Muramoto, J.; Christian, L.; Atkin, N.B. Diploid-tetraploid relationship among old-world members of the fish family Cyprinidae. Chromosoma 1967, 23, 1–9. [Google Scholar] [CrossRef]
- Charlesworth, B.; Crispin, Y.J.; Charlesworth, D. The evolutionary dynamics of sexually antagonistic mutations in pseudoautosomal regions of sex chromosomes. Evolution 2014, 68, 1339–1350. [Google Scholar] [CrossRef] [PubMed]
- Bertollo, L.A.C.; Takahashi, C.S.; Moreira-Filho, O. Multiple sex chromosomes in the genus Hoplias (Pisces: Erythrinidae). Cytologia 1983, 48, 1–12. [Google Scholar] [CrossRef]
- Cioffi, M.B.; Camacho, J.P.M.; Bertollo, L.A.C. Repetitive DNAs and differentiation of sex chromosomes in neotropical fishes. Cytogenet. Genome Res. 2011, 132, 188–194. [Google Scholar] [CrossRef]
- Cioffi, M.B.; Liehr, T.; Trifonov, V.; Molina, W.F.; Bertollo, L.A.C. Independent sex chromosome evolution in lower vertebrates: A molecular cytogenetic overview in the Erythrinidae fish family. Cytogenet. Genome Res. 2013, 141, 186–194. [Google Scholar] [CrossRef]
- Toma, G.A.; Sember, A.; Goes, C.A.G.; Kretschmer, R.; Porto-Foresti, F.; Bertollo, L.A.C.; Liehr, T.; Utsunomia, R.; Cioffi, M.B. Satellite DNAs and the evolution of the multiple X1X2Y sex chromosomes in the wolf fish Hoplias malabaricus (Teleostei; Characiformes). Sci. Rep. 2024, 14, 20402. [Google Scholar] [CrossRef]
- Sember, A.; Bertollo, L.A.C.; Ráb, P.; Yano, C.F.; Hatanaka, T.; De Oliveira, E.A.; Cioffi, M.B. Sex chromosome evolution and genomic divergence in the fish Hoplias malabaricus (Characiformes, Erythrinidae). Front. Genet. 2018, 9, 71. [Google Scholar] [CrossRef]
- Charlesworth, D. The timing of genetic degeneration of sex chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021, 376, 20200093. [Google Scholar] [CrossRef]
- Kratochvíl, L.; Stöck, M.; Rovatsos, M.; Bullejos, M.; Herpin, A.; Jeffries, D.L.; Peichel, C.L.; Perrin, N.; Valenzuela, N.; Pokorná, M.J. Expanding the classical paradigm: What we have learnt from vertebrates about sex chromosome evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021, 376, 20200097. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Jurka, J. The Long Terminal Repeat of an Endogenous Retrovirus Induces Alternative Splicing and Encodes an Additional Carboxy-Terminal Sequence in the Human Leptin Receptor. J. Mol. Evol. 1999, 48, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.H.; Choo, K.H.A. Evolutionary dynamics of transposable elements at the centromere. Trends Genet. 2004, 20, 611–616. [Google Scholar] [CrossRef] [PubMed]
- Deon, G.A.; Dos Santos, R.Z.; Sassi, F.D.M.C.; Moreira-Filho, O.; Vicari, M.R.; Porto-Foresti, F.; Utsunomia, R.; Cioffi, M.B. The role of satellite DNAs in the chromosomal rearrangements and the evolution of the rare XY1Y2 sex system in Harttia (Siluriformes: Loricariidae). J. Hered. 2024, 115, 541–551. [Google Scholar] [CrossRef]
- Sopniewski, J.; Shams, F.; Scheele, B.C.; Kefford, B.J.; Ezaz, T. Identifying sex-linked markers in Litoria aurea: A novel approach to understanding sex chromosome evolution in an amphibian. Sci. Rep. 2019, 9, 16591. [Google Scholar] [CrossRef]
- Gelaw, Y.M.; Eleblu, J.S.; Ofori, K.; Fenta, B.A.; Mukankusi, C.; Emam, E.A.; Offei, S. High-density DArTSeq SNP markers revealed wide genetic diversity and structured population in common bean (Phaseolus vulgaris L.) germplasm in Ethiopia. Mol. Biol. Rep. 2023, 50, 6739–6751. [Google Scholar] [CrossRef]
- Baloch, F.S.; Alsaleh, A.; Shahid, M.Q.; Çiftçi, V.; Sáenz de Miera, L.E.; Aasim, M.; Nadeem, M.A.; Aktaş, H.; Özkan, H.; Hatipoğlu, R. A whole genome DArTseq and SNP analysis for genetic diversity assessment in durum wheat from central fertile crescent. PLoS ONE 2017, 12, e0167821. [Google Scholar] [CrossRef] [PubMed]
- Hill, P.L.; Burridge, C.P.; Ezaz, T.; Wapstra, E. Conservation of sex-linked markers among conspecific populations of a viviparous skink, Niveoscincus ocellatus, exhibiting genetic and temperature-dependent sex determination. Genome Biol. Evol. 2018, 10, 1079–1087. [Google Scholar] [CrossRef]
- Kimura, M. Random Genetic Drift in Multi-Allelic Locus. Evolution 1955, 9, 419–435. [Google Scholar] [CrossRef]
- Gajiwala, K.S.; Chen, H.; Cornille, F.; Roques, B.P.; Reith, W.; Mach, B.; Burley, S.K. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature 2000, 403, 916–921. [Google Scholar] [CrossRef]
- Zaim, J.; Speina, E.; Kierzek, A.M. Identification of new genes regulated by the Crt1 transcription factor, an effector of the DNA damage checkpoint pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 28–37. [Google Scholar] [CrossRef]
- Garg, A.; Futcher, B.; Leatherwood, J. A new transcription factor for mitosis: In Schizosaccharomyces pombe, the RFX transcription factor Sak1 works with forkhead factors to regulate mitotic expression. Nucleic Acids Res. 2015, 43, 6874–6888. [Google Scholar] [CrossRef]
- Reith, W.; Mach, B. The bare lymphocyte syndrome and the regulation of MHC expression. Annu. Rev. Immunol. 2001, 19, 331–373. [Google Scholar] [CrossRef]
- He, S.; Song, Y.; Zhang, S.; Yuan, Z.; Yang, L.; Fang, C.; Seim, I.; Liu, S.; Liu, Q.; Wang, C.; et al. Genetic Innovations Underpin Morphological Diversity and Radiation in Teleost Fish. Res. Sq. 2023. [Google Scholar] [CrossRef]
- Morotomi-Yano, K.; Yano, K.I.; Saito, H.; Sun, Z.; Iwama, A.; Miki, Y. Human regulatory factor X 4 (RFX4) is a testis-specific dimeric DNA-binding protein that cooperates with other humanRFX members. J. Biol. Chem. 2002, 277, 836–842. [Google Scholar] [CrossRef]
- Sugiaman-Trapman, D.; Vitezic, M.; Jouhilahti, E.M.; Mathelier, A.; Lauter, G.; Misra, S.; Daub, C.O.; Kere, J.; Swoboda, P. Characterization of the human RFX transcription factor family by regulatory and target gene analysis. BMC Genom. 2018, 19, 1–15. [Google Scholar] [CrossRef]
- Zhu, Z.; Younas, L.; Zhou, Q. Evolution and regulation of animal sex chromosomes. Nat. Rev. Genet. 2025, 26, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Guarracino, A.; Buonaiuto, S.; de Lima, L.G.; Potapova, T.; Rhie, A.; Koren, S.; Rubinstein, B.; Fischer, C.; Human Pangenome Reference Consortium; Gerton, J.L.; et al. Recombination between heterologous human acrocentric chromosomes. Nature 2023, 617, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Bertollo, L.A.C.; Cioffi, M.B.; Moreira-Filho, O. Direct chromosome preparation from freshwater teleost fishes. In Fish Cytogenetic Techniques (Chondrichthyans and Teleosts), 1st ed.; Ozouf-Costaz, C., Pisano, E., Foresti, F., Toledo, L.F.A., Eds.; CRC Press: Boca Raton, FL, USA, 2015; pp. 21–26. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Novák, P.; Ávila Robledillo, L.; Koblížková, A.; Vrbová, I.; Neumann, P.; Macas, J. TAREAN: A computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res. 2017, 45, e111. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Smit, A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. 2013–2015. Available online: http://www.repeatmasker.org (accessed on 20 April 2024).
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, M.; Sousa, A.; Ramirez, M.; Francisco, A.P.; Carriço, J.A.; Vaz, C. PHYLOViZ 2.0: Providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 2017, 33, 128–129. [Google Scholar] [CrossRef]
- Morgulis, A.; Coulouris, G.; Raytselis, Y.; Madden, T.L.; Agarwala, R.; Schäffer, A.A. Database indexing for production MegaBLAST searches. Bioinformatics 2008, 24, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Sassi, F.M.C.; Toma, G.A.; Cioffi, M.B. FISH-in fish chromosomes. In Cytogenetics and Molecular Cytogenetics, 1st ed.; Liehr, T., Ed.; CRC Press: Boca Raton, FL, USA, 2022; Volume 1, pp. 281–297. [Google Scholar]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Goubert, C.; Modolo, L.; Vieira, C.; ValienteMoro, C.; Mavingui, P.; Boulesteix, M. De novo assembly and annotation of the Asian tiger mosquito (Aedes albopictus) repeatome with dnaPipeTE from raw genomic reads and comparative analysis with the yellow fever mosquito (Aedes aegypti). Genome Bio. Evol. 2015, 7, 1192–1205. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://www.R-project.org/ (accessed on 4 June 2024).
- Eaton, D.A. PyRAD: Assembly of de novo RADseq loci for phylogenetic analyses. Bioinformatics 2014, 30, 1844–1849. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; Ugene Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
| Karyomorph | N | SmRUL | LgRUL | M% | F% |
|---|---|---|---|---|---|
| KarF | 56 | 28 (HmfSat10) | 2944 (HmfSat08) | 6.1 | 5.3 |
| KarG | 45 | 21 (HmgSat41) | 1380 (HmgSat27) | 7.7 | 7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Oliveira, M.P.B.; Deon, G.A.; de Menezes Cavalcante Sassi, F.; de Souza, F.H.S.; Goes, C.A.G.; Utsunomia, R.; Porto-Foresti, F.; Vidal, J.A.D.; da Silva, A.B.; Ezaz, T.; et al. Repeatome Dynamics and Sex Chromosome Differentiation in the XY and XY1Y2 Systems of the Fish Hoplias malabaricus (Teleostei; Characiformes). Int. J. Mol. Sci. 2025, 26, 6039. https://doi.org/10.3390/ijms26136039
de Oliveira MPB, Deon GA, de Menezes Cavalcante Sassi F, de Souza FHS, Goes CAG, Utsunomia R, Porto-Foresti F, Vidal JAD, da Silva AB, Ezaz T, et al. Repeatome Dynamics and Sex Chromosome Differentiation in the XY and XY1Y2 Systems of the Fish Hoplias malabaricus (Teleostei; Characiformes). International Journal of Molecular Sciences. 2025; 26(13):6039. https://doi.org/10.3390/ijms26136039
Chicago/Turabian Stylede Oliveira, Mariannah Pravatti Barcellos, Geize Aparecida Deon, Francisco de Menezes Cavalcante Sassi, Fernando Henrique Santos de Souza, Caio Augusto Gomes Goes, Ricardo Utsunomia, Fábio Porto-Foresti, Jhon Alex Dziechciarz Vidal, Amanda Bueno da Silva, Tariq Ezaz, and et al. 2025. "Repeatome Dynamics and Sex Chromosome Differentiation in the XY and XY1Y2 Systems of the Fish Hoplias malabaricus (Teleostei; Characiformes)" International Journal of Molecular Sciences 26, no. 13: 6039. https://doi.org/10.3390/ijms26136039
APA Stylede Oliveira, M. P. B., Deon, G. A., de Menezes Cavalcante Sassi, F., de Souza, F. H. S., Goes, C. A. G., Utsunomia, R., Porto-Foresti, F., Vidal, J. A. D., da Silva, A. B., Ezaz, T., Liehr, T., & de Bello Cioffi, M. (2025). Repeatome Dynamics and Sex Chromosome Differentiation in the XY and XY1Y2 Systems of the Fish Hoplias malabaricus (Teleostei; Characiformes). International Journal of Molecular Sciences, 26(13), 6039. https://doi.org/10.3390/ijms26136039









