Maltose and Maltotriose Transporters in Brewer’s Saccharomyces Yeasts: Polymorphic and Key Residues in Their Activity
Abstract
1. Introduction
2. Sugar Consumption in Brewing Wort
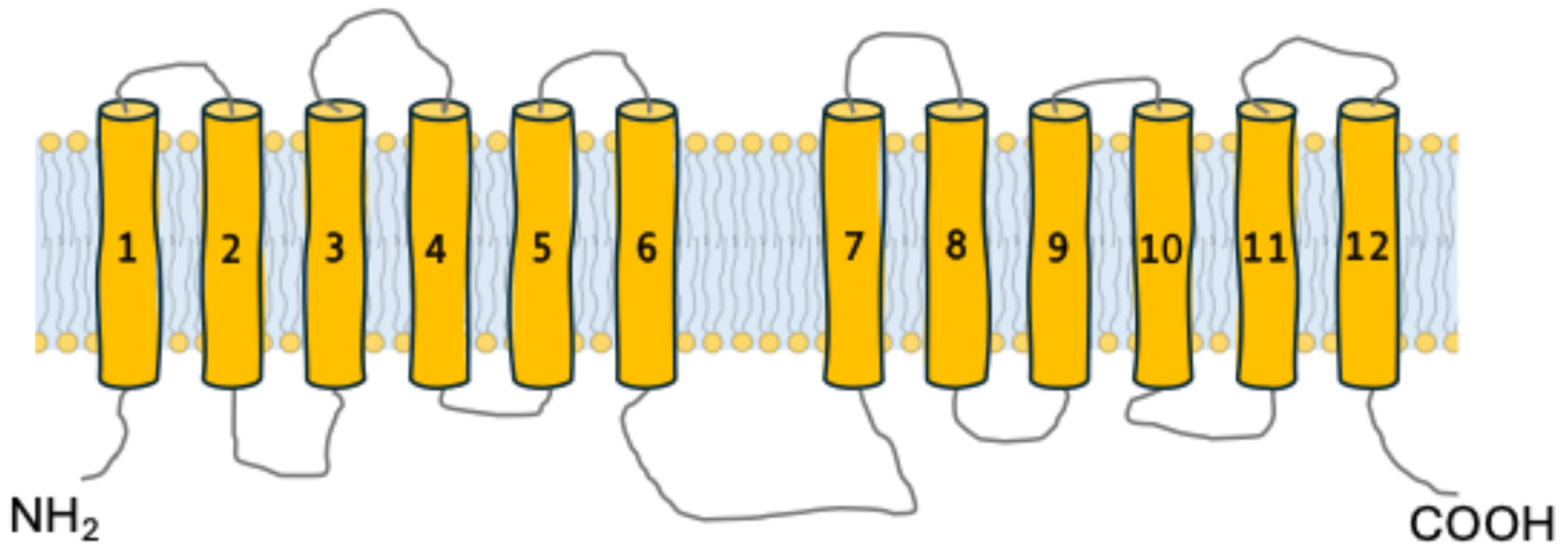
3. Common Structural Characteristics of α-Glucoside Transporters in Brewer’s Yeasts
4. MAL Loci
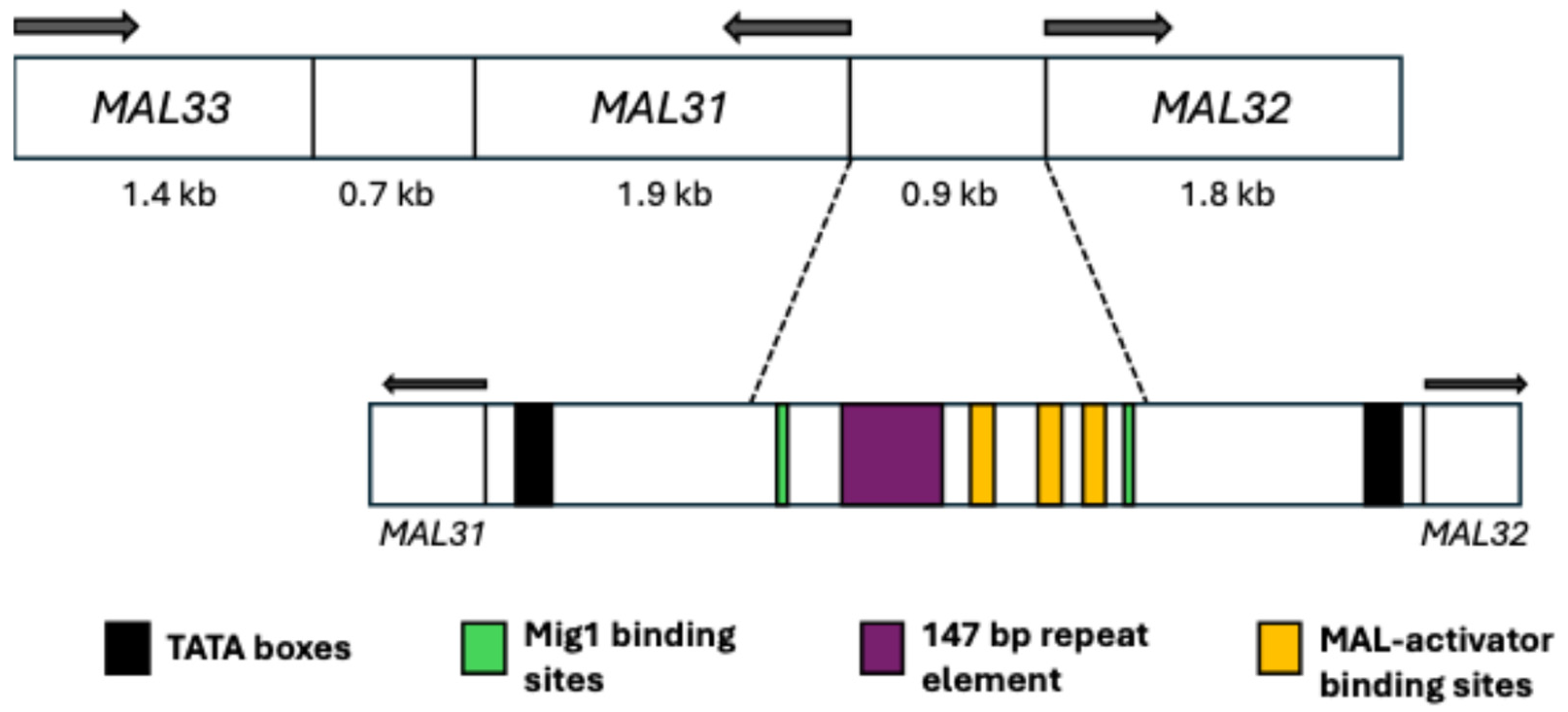
4.1. General Aspects
4.2. Regulation and Possible Evolutionary Origin of the MAL Loci
5. AGT1 Gene
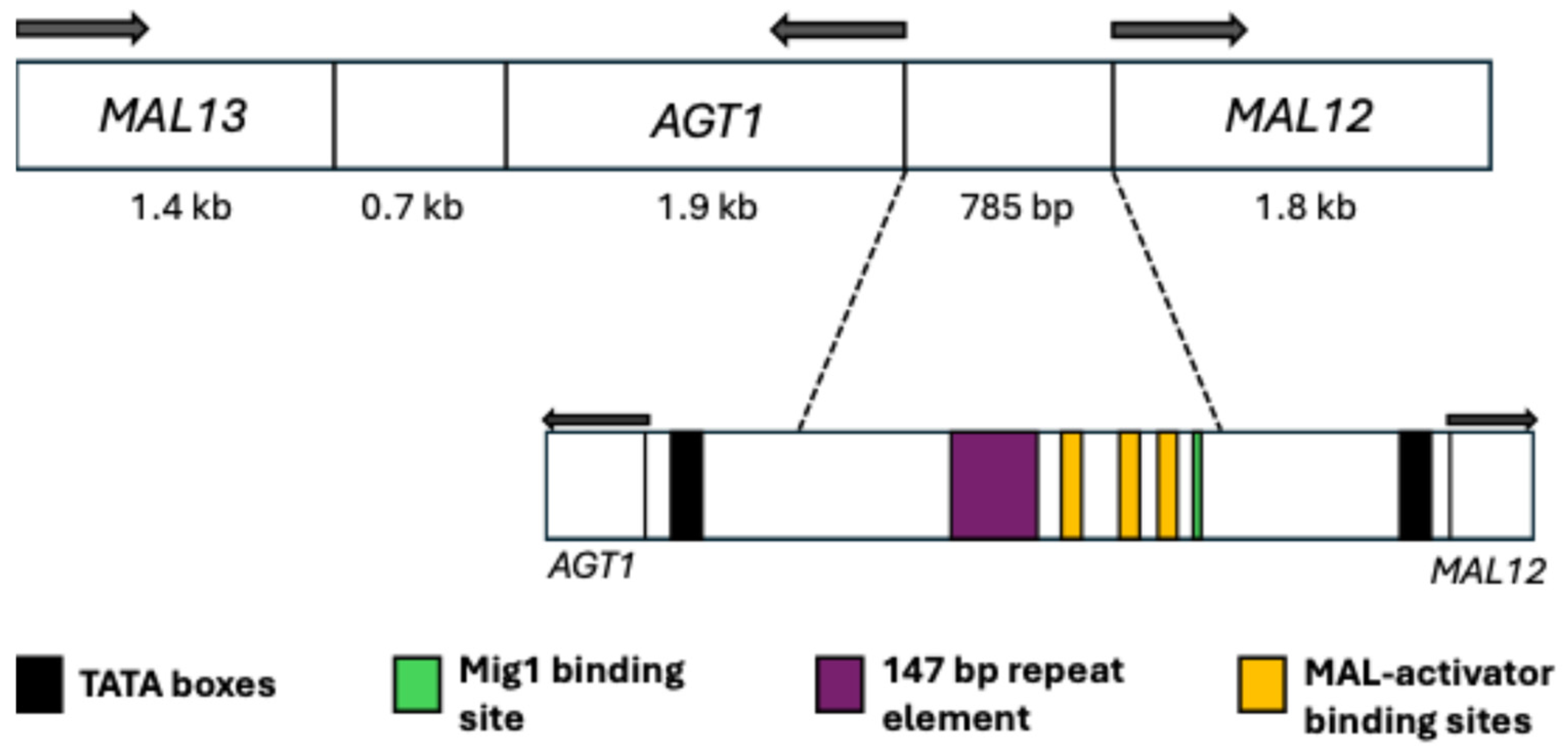
5.1. Main Elements
5.2. Regulation and Possible Evolutionary Origin of AGT1
6. MPHx Gene
6.1. General Features
6.2. Regulation and Possible Evolutionary Origin of MPHx
7. MTT1/MTY1 Gene
7.1. Overall Characteristics
7.2. Regulation and Possible Evolutionary Origin of MTT1
8. Other Reported Important Permeases
8.1. MALT434
8.2. SeMALT413
8.3. ScMALT#2 and ScMALT#5
9. Functional Divergence and Rapid Expansion of α-Glucoside Transporter Genes
10. Key and Polymorphic Residues Involved in α-Glucoside Transporters Activity
10.1. Polymorphic Residues in α-Glucoside Transporters
10.2. Key Residues Involved in Sugar Transport in α-Glucoside Transporters
10.3. Key Residues Reported in Post-Translational Regulation of α-Glucoside Transporters
11. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Johnson, E.A.; Echavarri-Erasun, C. Yeast Biotechnology. In The Yeasts; Elsevier: Amsterdam, The Netherlands, 2011; pp. 21–44. ISBN 978-0-444-52149-1. [Google Scholar]
- Dunn, B.; Sherlock, G. Reconstruction of the Genome Origins and Evolution of the Hybrid Lager Yeast Saccharomyces pastorianus. Genome Res. 2008, 18, 1610–1623. [Google Scholar] [CrossRef] [PubMed]
- Libkind, D.; Hittinger, C.T.; Valério, E.; Gonçalves, C.; Dover, J.; Johnston, M.; Gonçalves, P.; Sampaio, J.P. Microbe Domestication and the Identification of the Wild Genetic Stock of Lager-Brewing Yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 14539–14544. [Google Scholar] [CrossRef] [PubMed]
- Humia, B.V.; Santos, K.S.; Barbosa, A.M.; Sawata, M.; Mendonça, M.D.C.; Padilha, F.F. Beer Molecules and Its Sensory and Biological Properties: A Review. Molecules 2019, 24, 1568. [Google Scholar] [CrossRef] [PubMed]
- Petruzzi, L.; Rosaria Corbo, M.; Sinigaglia, M.; Bevilacqua, A. Brewer’s Yeast in Controlled and Uncontrolled Fermentations, with a Focus on Novel, Nonconventional, and Superior Strains. Food Rev. Int. 2016, 32, 341–363. [Google Scholar] [CrossRef]
- Londesborough, J. Fermentation of maltotriose by brewer’s and baker’s yeasts. Biotechnol. Lett. 2001, 23, 1995–2000. [Google Scholar] [CrossRef]
- Stewart, G. Saccharomyces Species in the Production of Beer. Beverages 2016, 2, 34. [Google Scholar] [CrossRef]
- Comitini, F.; Agarbati, A.; Canonico, L.; Ciani, M. Yeast Interactions and Molecular Mechanisms in Wine Fermentation: A Comprehensive Review. Int. J. Mol. Sci. 2021, 22, 7754. [Google Scholar] [CrossRef]
- Meneses, F.J.; Jiranek, V. Expression Patterns of Genes and Enzymes Involved in Sugar Catabolism in Industrial Saccharomyces cerevisiae Strains Displaying Novel Fermentation Characteristics. J. Inst. Brew. 2002, 108, 322–335. [Google Scholar] [CrossRef]
- Smit, A.; Moses, S.G.; Pretorius, I.S.; Cordero Otero, R.R. The Thr505 and Ser557 Residues of the AGT1-Encoded α-Glucoside Transporter Are Critical for Maltotriose Transport in Saccharomyces cerevisiae. J. Appl. Microbiol. 2008, 104, 1103–1111. [Google Scholar] [CrossRef]
- Trichez, D.; Knychala, M.M.; Figueiredo, C.M.; Alves, S.L.; Da Silva, M.A.; Miletti, L.C.; De Araujo, P.S.; Stambuk, B.U. Key Amino Acid Residues of the AGT1 Permease Required for Maltotriose Consumption and Fermentation by Saccharomyces cerevisiae. J. Appl. Microbiol. 2019, 126, 580–594. [Google Scholar] [CrossRef]
- Hatanaka, H.; Toyonaga, H.; Ishida, Y.; Mizohata, E.; Ono, E. Functional Diversity and Plasticity in the Sugar Preferences of Saccharomyces MALT Transporters in Domesticated Yeasts. FEMS Yeast Res. 2022, 22, foac055. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Vásquez, C.I.; García-García, J.H.; Pérez-Ortega, E.R.; Martínez-Segundo, A.G.; Damas-Buenrostro, L.C.; Pereyra-Alférez, B. Expression Patterns of Mal Genes and Association with Differential Maltose and Maltotriose Transport Rate of Two Saccharomyces pastorianus Yeasts. Appl. Environ. Microbiol. 2024, 90, e0039724. [Google Scholar] [CrossRef] [PubMed]
- Faz-Cortez, O.A.; Sánchez-López, A.Y.; Hernández-Vásquez, C.I.; Segura-Ruiz, A.; Pereyra-Alférez, B.; García-García, J.H. Computational Analysis of Polymorphic Residues in Maltose and Maltotriose Transporters of a Wild Saccharomyces cerevisiae Strain. Open Life Sci. 2025, 20, 20251080. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, F.; Vidgren, V.; Ruohonen, L.; Gibson, B. Maltose and Maltotriose Utilisation by Group I Strains of the Hybrid Lager Yeast Saccharomyces pastorianus. FEMS Yeast Res. 2016, 16, fow053. [Google Scholar] [CrossRef]
- Galeote, V.; Novo, M.; Salema-Oom, M.; Brion, C.; Valério, E.; Gonçalves, P.; Dequin, S. FSY1, a Horizontally Transferred Gene in the Saccharomyces cerevisiae EC1118 Wine Yeast Strain, Encodes a High-Affinity Fructose/H+ Symporter. Microbiology 2010, 156, 3754–3761. [Google Scholar] [CrossRef]
- Anjos, J.; Rodrigues De Sousa, H.; Roca, C.; Cássio, F.; Luttik, M.; Pronk, J.T.; Salema-Oom, M.; Gonçalves, P. Fsy1, the Sole Hexose-Proton Transporter Characterized in Saccharomyces Yeasts, Exhibits a Variable Fructose:H+ Stoichiometry. Biochim. Biophys. Acta (BBA) Biomembr. 2013, 1828, 201–207. [Google Scholar] [CrossRef]
- Maier, A.; Völker, B.; Boles, E.; Fuhrmann, G.F. Characterisation of Glucose Transport in Saccharomyces cerevisiae with Plasma Membrane Vesicles (Countertransport) and Intact Cells (Initial Uptake) with Single Hxt1, Hxt2, Hxt3, Hxt4, Hxt6, Hxt7 or Gal2 Transporters. FEMS Yeast Res. 2002, 2, 539–550. [Google Scholar] [CrossRef]
- Postigo, V.; Sánchez, A.; Cabellos, J.M.; Arroyo, T. New Approaches for the Fermentation of Beer: Non-Saccharomyces Yeasts from Wine. Fermentation 2022, 8, 280. [Google Scholar] [CrossRef]
- Verhagen, K.J.A.; Pardijs, I.H.; Van Klaveren, H.M.; Wahl, S.A. A Dive Into Yeast’s Sugar Diet—Comparing the Metabolic Response of Glucose, Fructose, Sucrose, and Maltose Under Dynamic Feast/Famine Conditions. Biotech. Bioeng. 2025, 122, 1035–1050. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, S.; Han, X.; Zhou, Z.; Mao, J. Combined Effects of Fermentation Temperature and Saccharomyces cerevisiae Strains on Free Amino Acids, Flavor Substances, and Undesirable Secondary Metabolites in Huangjiu Fermentation. Food Microbiol. 2022, 108, 104091. [Google Scholar] [CrossRef]
- Simões, J.; Coelho, E.; Magalhães, P.; Brandão, T.; Rodrigues, P.; Teixeira, J.A.; Domingues, L. Exploiting Non-Conventional Yeasts for Low-Alcohol Beer Production. Microorganisms 2023, 11, 316. [Google Scholar] [CrossRef] [PubMed]
- Sahana, G.R.; Balasubramanian, B.; Joseph, K.S.; Pappuswamy, M.; Liu, W.-C.; Meyyazhagan, A.; Kamyab, H.; Chelliapan, S.; Joseph, B.V. A Review on Ethanol Tolerance Mechanisms in Yeast: Current Knowledge in Biotechnological Applications and Future Directions. Process Biochem. 2024, 138, 1–13. [Google Scholar] [CrossRef]
- Li, J.; Yuan, M.; Meng, N.; Li, H.; Sun, J.; Sun, B. Influence of Nitrogen Status on Fermentation Performances of Non-Saccharomyces Yeasts: A Review. Food Sci. Hum. Wellness 2024, 13, 556–567. [Google Scholar] [CrossRef]
- Crépin, L.; Nidelet, T.; Sanchez, I.; Dequin, S.; Camarasa, C. Sequential Use of Nitrogen Compounds by Saccharomyces cerevisiae during Wine Fermentation: A Model Based on Kinetic and Regulation Characteristics of Nitrogen Permeases. Appl. Environ. Microbiol. 2012, 78, 8102–8111. [Google Scholar] [CrossRef]
- Lei, H.; Xu, H.; Feng, L.; Yu, Z.; Zhao, H.; Zhao, M. Fermentation Performance of Lager Yeast in High Gravity Beer Fermentations with Different Sugar Supplementations. J. Biosci. Bioeng. 2016, 122, 583–588. [Google Scholar] [CrossRef]
- Puligundla, P.; Smogrovicova, D.; Mok, C.; Obulam, V.S.R. Recent Developments in High Gravity Beer-Brewing. Innov. Food Sci. Emerg. Technol. 2020, 64, 102399. [Google Scholar] [CrossRef]
- Chotineeranat, S.; Wansuksri, R.; Piyachomkwan, K.; Chatakanonda, P.; Weerathaworn, P.; Sriroth, K. Effect of Calcium Ions on Ethanol Production from Molasses by Saccharomyces cerevisiae. Sugar Tech. 2010, 12, 120–124. [Google Scholar] [CrossRef]
- Nyitrainé Sárdy, D.; Kellner, N.; Magyar, I.; Oláhné Horváth, B. Effects of High Sugar Content on Fermentation Dynamics and Some Metabolites of Wine-Related Yeast Species Saccharomyces cerevisiae, S. uvarum and Starmerella bacillaris. Food Technol. Biotechnol. 2020, 58, 76–83. [Google Scholar] [CrossRef]
- Wang, M.; Sun, Z.; Wang, Y.; Wei, Z.; Chen, B.; Zhang, H.; Guo, X.; Xiao, D. The Effect of Pitching Rate on the Production of Higher Alcohols by Top-Fermenting Yeast in Wheat Beer Fermentation. Ann. Microbiol. 2019, 69, 713–726. [Google Scholar] [CrossRef]
- Kucharczyk, K.; Tuszyński, T. The Effect of Wort Aeration on Fermentation, Maturation and Volatile Components of Beer Produced on an Industrial Scale. J. Inst. Brew. 2017, 123, 31–38. [Google Scholar] [CrossRef]
- Permanasari, A.R.; Syamfitri, A.; Maharani, D.; Setyaningrum, S.; Wibisono, W. The Comparison of Free Cell and Immobilization Cell Fermentation on Bioethanol Production from Sorghum Stem by SSF and SHF Method. J. Tek. Kim. Ling. 2024, 8, 116–126. [Google Scholar] [CrossRef]
- Cheng, Q.; Michels, C.A. MAL11 and MAL61 Encode the Inducible High-Affinity Maltose Transporter of Saccharomyces cerevisiae. J. Bacteriol. 1991, 173, 1817–1820. [Google Scholar] [CrossRef] [PubMed]
- Han, E.; Cotty, F.; Sottas, C.; Jiang, H.; Michels, C.A. Characterization of AGT1 Encoding a General α-glucoside Transporter from Saccharomyces. Mol. Microbiol. 1995, 17, 1093–1107. [Google Scholar] [CrossRef] [PubMed]
- Day, R.E.; Higgins, V.J.; Rogers, P.J.; Dawes, I.W. Characterization of the Putative Maltose Transporters Encoded by YDL247w and YJR160c. Yeast 2002, 19, 1015–1027. [Google Scholar] [CrossRef]
- Dietvorst, J.; Londesborough, J.; Steensma, H.Y. Maltotriose Utilization in Lager Yeast Strains: MTT1 Encodes a Maltotriose Transporter. Yeast 2005, 22, 775–788. [Google Scholar] [CrossRef]
- Salema-Oom, M.; Valadão Pinto, V.; Gonçalves, P.; Spencer-Martins, I. Maltotriose Utilization by Industrial Saccharomyces Strains: Characterization of a New Member of the α-Glucoside Transporter Family. Appl. Environ. Microbiol. 2005, 71, 5044–5049. [Google Scholar] [CrossRef]
- Day, R.E.; Rogers, P.J.; Dawes, I.W.; Higgins, V.J. Molecular Analysis of Maltotriose Transport and Utilization by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2002, 68, 5326–5335. [Google Scholar] [CrossRef]
- Vidgren, V.; Kankainen, M.; Londesborough, J.; Ruohonen, L. Identification of Regulatory Elements in the AGT1 Promoter of Ale and Lager Strains of Brewer’s Yeast. Yeast 2011, 28, 579–594. [Google Scholar] [CrossRef]
- Dietvorst, J.; Walsh, M.C.; Van Heusden, G.P.H.; Steensma, H.Y. Comparison of the MTT1- and MAL31-like Maltose Transporter Genes in Lager Yeast Strains: Maltose Transporter Genes in Lager Yeast Strains. FEMS Microbiol. Lett. 2010, 310, 152–157. [Google Scholar] [CrossRef]
- Qiao, Y.; Li, C.; Lu, X.; Zong, H.; Zhuge, B. Identification of Key Residues for Efficient Glucose Transport by the Hexose Transporter CgHxt4 in High Sugar Fermentation Yeast Candida Glycerinogenes. Appl. Microbiol. Biotechnol. 2021, 105, 7295–7307. [Google Scholar] [CrossRef]
- Yan, N. Structural Biology of the Major Facilitator Superfamily Transporters. Annu. Rev. Biophys. 2015, 44, 257–283. [Google Scholar] [CrossRef] [PubMed]
- Leandro, M.J.; Fonseca, C.; Gonçalves, P. Hexose and Pentose Transport in Ascomycetous Yeasts: An Overview. FEMS Yeast Res. 2009, 9, 511–525. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.S.; Shlykov, M.A.; Castillo, R.; Sun, E.I.; Saier, M.H. The Major Facilitator Superfamily (MFS) Revisited. FEBS J. 2012, 279, 2022–2035. [Google Scholar] [CrossRef] [PubMed]
- Niño-González, M.; Novo-Uzal, E.; Richardson, D.N.; Barros, P.M.; Duque, P. More Transporters, More Substrates: The Arabidopsis Major Facilitator Superfamily Revisited. Mol. Plant 2019, 12, 1182–1202. [Google Scholar] [CrossRef]
- Drew, D.; North, R.A.; Nagarathinam, K.; Tanabe, M. Structures and General Transport Mechanisms by the Major Facilitator Superfamily (MFS). Chem. Rev. 2021, 121, 5289–5335. [Google Scholar] [CrossRef]
- Donzella, L.; Sousa, M.J.; Morrissey, J.P. Evolution and Functional Diversification of Yeast Sugar Transporters. Essays Biochem. 2023, 67, 811–827. [Google Scholar] [CrossRef]
- Serrano, R. Energy Requirements for Maltose Transport in Yeast. Eur. J. Biochem. 1977, 80, 97–102. [Google Scholar] [CrossRef]
- Alves, S.L.; Herberts, R.A.; Hollatz, C.; Trichez, D.; Miletti, L.C.; De Araujo, P.S.; Stambuk, B.U. Molecular Analysis of Maltotriose Active Transport and Fermentation by Saccharomyces cerevisiae Reveals a Determinant Role for the AGT1 Permease. Appl. Environ. Microbiol. 2008, 74, 1494–1501. [Google Scholar] [CrossRef]
- Pellegrini-Calace, M.; Maiwald, T.; Thornton, J.M. PoreWalker: A Novel Tool for the Identification and Characterization of Channels in Transmembrane Proteins from Their Three-Dimensional Structure. PLoS Comput. Biol. 2009, 5, e1000440. [Google Scholar] [CrossRef]
- Busturia, A.; Lagunas, R. Identification of Two Forms of the Maltose Transport System in Saccharomyces cerevisiae and Their Regulation by Catabolite Inactivation. Biochim. Biophys. Acta (BBA)-Biomembr. 1985, 820, 324–326. [Google Scholar] [CrossRef]
- Chang, Y.S.; Dubin, R.A.; Perkins, E.; Michels, C.A.; Needleman, R.B. Identification and Characterization of the Maltose Permease in Genetically Defined Saccharomyces Strain. J. Bacteriol. 1989, 171, 6148–6154. [Google Scholar] [CrossRef] [PubMed]
- Charron, M.J.; Read, E.; Haut, S.R.; Michels, C.A. Molecular Evolution of the Telomere-Associated MAL Loci of Saccharomyces. Genetics 1989, 122, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.A.; Murray, A.W.; Verstrepen, K.J. Rapid Expansion and Functional Divergence of Subtelomeric Gene Families in Yeasts. Curr. Biol. 2010, 20, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Teste, M.-A.; François, J.M.; Parrou, J.-L. Characterization of a New Multigene Family Encoding Isomaltases in the Yeast Saccharomyces cerevisiae, the IMA Family. J. Biol. Chem. 2010, 285, 26815–26824. [Google Scholar] [CrossRef]
- Weller, C.A.; Andreev, I.; Chambers, M.J.; Park, M.; NISC Comparative Sequencing Program; Bloom, J.S.; Sadhu, M.J. Highly Complete Long-Read Genomes Reveal Pangenomic Variation Underlying Yeast Phenotypic Diversity. Genome Res. 2023, 33, 729–740. [Google Scholar] [CrossRef]
- Jespersen, L.; Cesar, L.B.; Meaden, P.G.; Jakobsen, M. Multiple α-Glucoside Transporter Genes in Brewer’s Yeast. Appl. Envrion. Microbiol. 1999, 65, 450–456. [Google Scholar] [CrossRef]
- Vidgren, V.; Ruohonen, L.; Londesborough, J. Characterization and Functional Analysis of the MAL and MPH Loci for Maltose Utilization in Some Ale and Lager Yeast Strains. Appl. Environ. Microbiol. 2005, 71, 7846–7857. [Google Scholar] [CrossRef]
- Needleman, R. Control of Maltase Synthesis in Yeast. Mol. Microbiol. 1991, 5, 2079–2084. [Google Scholar] [CrossRef]
- Stambuk, B.U.; Araujo, P.S. Kinetics of Active α-Glucoside Transport in Saccharomyces cerevisiae. FEMS Yeast Res. 2001, 1, 73–78. [Google Scholar] [CrossRef]
- Stambuk, B.U.; Batista, A.S.; De Araujo, P.S. Kinetics of Active Sucrose Transport in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2000, 89, 212–214. [Google Scholar] [CrossRef]
- Klein, C.J.; Olsson, L.; Rønnow, B.; Mikkelsen, J.D.; Nielsen, J. Alleviation of Glucose Repression of Maltose Metabolism by MIG1 Disruption in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1996, 62, 4441–4449. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bali, M.; Medintz, I.; Michels, C.A. Intracellular Maltose Is Sufficient To Induce MAL Gene Expression in Saccharomyces cerevisiae. Eukaryot. Cell 2002, 1, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Nehlin, J.O.; Ronne, H.; Michels, C.A. MIG1-Dependent and MIG1-Independent Glucose Regulation of MAL Gene Expression in Saccharomyces cerevisiae. Curr. Genet. 1995, 28, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Yue, Y.; Jiang, H.; Zhang, B.; Sherwood, P.W.; Michels, C.A. Analysis of the Mechanism by Which Glucose Inhibits Maltose Induction of MAL Gene Expression in Saccharomyces. Genetics 2000, 154, 121–132. [Google Scholar] [CrossRef]
- Hong, S.H.; Marmur, J. Upstream Regulatory Regions Controlling the Expression of the Yeast Maltase Gene. Mol. Cell. Biol. 1987, 7, 2477–2483. [Google Scholar] [CrossRef]
- Levine, J.; Tanouye, L.; Michels, C.A. The UASMAL Is a Bidirectional Promotor Element Required for the Expression of Both the MAL61 and MAL62 Genes of the Saccharomyces MAL6 Locus. Curr. Genet. 1992, 22, 181–189. [Google Scholar] [CrossRef]
- Bell, P.J.L.; Higgins, V.J.; Dawes, I.W.; Bissinger, P.H. Tandemly Repeated 147 Bp Elements Cause Structural and Functional Variation in Divergent MAL Promoters of Saccharomyces cerevisiae. Yeast 1997, 13, 1135–1144. [Google Scholar] [CrossRef]
- Naumov, G.I.; Naumova, E.S.; Michels, C.A. Genetic Variation of the Repeated MAL Loci in Natural Populations of Saccharomyces cerevisiae and Saccharomyces paradoxus. Genetics 1994, 136, 803–812. [Google Scholar] [CrossRef]
- Baker, E.; Wang, B.; Bellora, N.; Peris, D.; Hulfachor, A.B.; Koshalek, J.A.; Adams, M.; Libkind, D.; Hittinger, C.T. The Genome Sequence of Saccharomyces eubayanus and the Domestication of Lager-Brewing Yeasts. Mol. Biol. Evol. 2015, 32, 2818–2831. [Google Scholar] [CrossRef]
- Brickwedde, A.; Brouwers, N.; Van Den Broek, M.; Gallego Murillo, J.S.; Fraiture, J.L.; Pronk, J.T.; Daran, J.-M.G. Structural, Physiological and Regulatory Analysis of Maltose Transporter Genes in Saccharomyces eubayanus CBS 12357T. Front. Microbiol. 2018, 9, 1786. [Google Scholar] [CrossRef]
- Medintz, I.; Wang, X.; Hradek, T.; Michels, C.A. A PEST-like Sequence in the N-Terminal Cytoplasmic Domain of Saccharomyces Maltose Permease Is Required for Glucose-Induced Proteolysis and Rapid Inactivation of Transport Activity. Biochemistry 2000, 39, 4518–4526. [Google Scholar] [CrossRef] [PubMed]
- Nakao, Y.; Kanamori, T.; Itoh, T.; Kodama, Y.; Rainieri, S.; Nakamura, N.; Shimonaga, T.; Hattori, M.; Ashikari, T. Genome Sequence of the Lager Brewing Yeast, an Interspecies Hybrid. DNA Res. 2009, 16, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Vidgren, V.; Londesborough, J. Characterization of the Saccharomyces bayanus-Type AGT1 Transporter of Lager Yeast: Saccharomyces bayanus-Type AGT1 Transporter of Lager Yeast. J. Inst. Brew. 2012, 118, 148–151. [Google Scholar] [CrossRef]
- Baker, E.P.; Hittinger, C.T. Evolution of a Novel Chimeric Maltotriose Transporter in Saccharomyces eubayanus from Parent Proteins Unable to Perform This Function. PLoS Genet. 2019, 15, e1007786. [Google Scholar] [CrossRef]
- Brouwers, N.; Gorter De Vries, A.R.; Van Den Broek, M.; Weening, S.M.; Elink Schuurman, T.D.; Kuijpers, N.G.A.; Pronk, J.T.; Daran, J.-M.G. In Vivo Recombination of Saccharomyces eubayanus Maltose-Transporter Genes Yields a Chimeric Transporter That Enables Maltotriose Fermentation. PLoS Genet. 2019, 15, e1007853. [Google Scholar] [CrossRef]
- Duval, E.H.; Alves Jr, S.L.; Dunn, B.; Sherlock, G.; Stambuk, B.U. Microarray Karyotyping of Maltose-fermenting Saccharomyces Yeasts with Differing Maltotriose Utilization Profiles Reveals Copy Number Variation in Genes Involved in Maltose and Maltotriose Utilization. J. Appl. Microbiol. 2010, 109, 248–259. [Google Scholar] [CrossRef]
- Vidgren, V.; Multanen, J.-P.; Ruohonen, L.; Londesborough, J. The Temperature Dependence of Maltose Transport in Ale and Lager Strains of Brewer’s Yeast: The Temperature Dependence of Maltose Transport into Yeasts. FEMS Yeast Res. 2010, 10, 402–411. [Google Scholar] [CrossRef]
- Nguyen, H.-V.; Legras, J.-L.; Neuvéglise, C.; Gaillardin, C. Deciphering the Hybridisation History Leading to the Lager Lineage Based on the Mosaic Genomes of Saccharomyces bayanus Strains NBRC1948 and CBS380T. PLoS ONE 2011, 6, e25821. [Google Scholar] [CrossRef]
- Cousseau, F.E.M.; Alves, S.L.; Trichez, D.; Stambuk, B.U. Characterization of Maltotriose Transporters from the Saccharomyces eubayanus Subgenome of the Hybrid Saccharomyces pastorianus Lager Brewing Yeast Strain Weihenstephan 34/70. Lett. Appl. Microbiol. 2013, 56, 21–29. [Google Scholar] [CrossRef]
- Allison, R.M.; Johnson, D.J.; Neale, M.J.; Gray, S. Recombinase-Independent Chromosomal Rearrangements between Dispersed Inverted Repeats in Saccharomyces cerevisiae Meiosis. Nucleic Acids Res. 2023, 51, 9703–9715. [Google Scholar] [CrossRef]
- Crandall, J.G.; Zhou, X.; Rokas, A.; Hittinger, C.T. Specialization Restricts the Evolutionary Paths Available to Yeast Sugar Transporters. Mol. Biol. Evol. 2024, 41, msae228. [Google Scholar] [CrossRef] [PubMed]
- Mefford, H.C. Comparative Sequencing of a Multicopy Subtelomeric Region Containing Olfactory Receptor Genes Reveals Multiple Interactions between Non-Homologous Chromosomes. Hum. Mol. Genet. 2001, 10, 2363–2372. [Google Scholar] [CrossRef] [PubMed]
- Mefford, H.C.; Trask, B.J. The Complex Structure and Dynamic Evolution of Human Subtelomeres. Nat. Rev. Genet. 2002, 3, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Rudd, M.K.; Friedman, C.; Parghi, S.S.; Linardopoulou, E.V.; Hsu, L.; Trask, B.J. Elevated Rates of Sister Chromatid Exchange at Chromosome Ends. PLoS Genet. 2007, 3, e32. [Google Scholar] [CrossRef]
- Linardopoulou, E.V.; Williams, E.M.; Fan, Y.; Friedman, C.; Young, J.M.; Trask, B.J. Human Subtelomeres Are Hot Spots of Interchromosomal Recombination and Segmental Duplication. Nature 2005, 437, 94–100. [Google Scholar] [CrossRef]
- Barton, A.B.; Pekosz, M.R.; Kurvathi, R.S.; Kaback, D.B. Meiotic Recombination at the Ends of Chromosomes in Saccharomyces cerevisiae. Genetics 2008, 179, 1221–1235. [Google Scholar] [CrossRef]
- Dunn, M.J.; Shazib, S.U.A.; Simonton, E.; Slot, J.C.; Anderson, M.Z. Architectural Groups of a Subtelomeric Gene Family Evolve along Distinct Paths in Candida albicans. G3 Genes|Genomes|Genet. 2022, 12, jkac283. [Google Scholar] [CrossRef]
- He, X.; Zhang, J. Rapid Subfunctionalization Accompanied by Prolonged and Substantial Neofunctionalization in Duplicate Gene Evolution. Genetics 2005, 169, 1157–1164. [Google Scholar] [CrossRef]
- Voordeckers, K.; Brown, C.A.; Vanneste, K.; Van Der Zande, E.; Voet, A.; Maere, S.; Verstrepen, K.J. Reconstruction of Ancestral Metabolic Enzymes Reveals Molecular Mechanisms Underlying Evolutionary Innovation through Gene Duplication. PLoS Biol. 2012, 10, e1001446. [Google Scholar] [CrossRef]
- Pougach, K.; Voet, A.; Kondrashov, F.A.; Voordeckers, K.; Christiaens, J.F.; Baying, B.; Benes, V.; Sakai, R.; Aerts, J.; Zhu, B.; et al. Duplication of a Promiscuous Transcription Factor Drives the Emergence of a New Regulatory Network. Nat. Commun. 2014, 5, 4868. [Google Scholar] [CrossRef]
- Henderson, R.; Poolman, B. Proton-Solute Coupling Mechanism of the Maltose Transporter from Saccharomyces cerevisiae. Sci. Rep. 2017, 7, 14375. [Google Scholar] [CrossRef] [PubMed]
- Hatanaka, H.; Omura, F.; Kodama, Y.; Ashikari, T. Gly-46 and His-50 of Yeast Maltose Transporter Mal21p Are Essential for Its Resistance against Glucose-Induced Degradation. J. Biol. Chem. 2009, 284, 15448–15457. [Google Scholar] [CrossRef] [PubMed]
- Brondijk, T.H.C.; Van Der Rest, M.E.; Pluim, D.; De Vries, Y.; Stingl, K.; Poolman, B.; Konings, W.N. Catabolite Inactivation of Wild-Type and Mutant Maltose Transport Proteins in Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 15352–15357. [Google Scholar] [CrossRef]
- Weigle, A.T.; Shukla, D. The Arabidopsis AtSWEET13 Transporter Discriminates Sugars by Selective Facial and Positional Substrate Recognition. Commun. Biol. 2024, 7, 764. [Google Scholar] [CrossRef] [PubMed]
- Chaptal, V.; Kwon, S.; Sawaya, M.R.; Guan, L.; Kaback, H.R.; Abramson, J. Crystal Structure of Lactose Permease in Complex with an Affinity Inactivator Yields Unique Insight into Sugar Recognition. Proc. Natl. Acad. Sci. USA 2011, 108, 9361–9366. [Google Scholar] [CrossRef]
- Guan, L.; Kaback, H.R. Lessons from Lactose Permease. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 67–91. [Google Scholar] [CrossRef]
- Wisedchaisri, G.; Park, M.-S.; Iadanza, M.G.; Zheng, H.; Gonen, T. Proton-Coupled Sugar Transport in the Prototypical Major Facilitator Superfamily Protein XylE. Nat. Commun. 2014, 5, 4521. [Google Scholar] [CrossRef]
- Iancu, C.V.; Zamoon, J.; Woo, S.B.; Aleshin, A.; Choe, J. Crystal Structure of a Glucose/H + Symporter and Its Mechanism of Action. Proc. Natl. Acad. Sci. USA 2013, 110, 17862–17867. [Google Scholar] [CrossRef]
- Smit, A.; Cordero Otero, R.R.; Pretorius, I.S. Differences amongAGT1-Encoded α-Glucoside Transporters and Their Ability to Transport Maltotriose inSaccharomyces Yeasts. Ann. Microbiol. 2007, 57, 77–84. [Google Scholar] [CrossRef]
- Chen, A.; Cheng, Y.; Meng, L.; Chen, J. Key Amino Acid Residues of the Agt1 Transporter for Trehalose Transport by Saccharomyces cerevisiae. J. Fungi 2024, 10, 781. [Google Scholar] [CrossRef]
- Chen, A.; Tapia, H.; Goddard, J.M.; Gibney, P.A. Trehalose and Its Applications in the Food Industry. Comp. Rev. Food Sci. Food Safe 2022, 21, 5004–5037. [Google Scholar] [CrossRef] [PubMed]
- Lucero, P.; Herweijer, M.; Lagunas, R. Catabolite Inactivation of the Yeast Maltose Transporter Is Due to Proteolysis. FEBS Lett. 1993, 333, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Medintz, I.; Jiang, H.; Han, E.K.; Cui, W.; Michels, C.A. Characterization of the Glucose-Induced Inactivation of Maltose Permease in Saccharomyces cerevisiae. J. Bacteriol. 1996, 178, 2245–2254. [Google Scholar] [CrossRef] [PubMed]
- Riballo, E.; Herweijer, M.; Wolf, D.H.; Lagunas, R. Catabolite Inactivation of the Yeast Maltose Transporter Occurs in the Vacuole after Internalization by Endocytosis. J. Bacteriol. 1995, 177, 5622–5627. [Google Scholar] [CrossRef]
- Lucero, P.; Lagunas, R. Catabolite Inactivation of the Yeast Maltose Transporter Requires Ubiquitin-Ligase Npi1/Rsp5 and Ubiquitin-Hydrolase Npi2/Doa4. FEMS Microbiol. Lett. 1997, 147, 273–277. [Google Scholar] [CrossRef]
- Ramos, J.; Cirillo, V.P. Role of Cyclic-AMP-Dependent Protein Kinase in Catabolite Inactivation of the Glucose and Galactose Transporters in Saccharomyces cerevisiae. J. Bacteriol. 1989, 171, 3545–3548. [Google Scholar] [CrossRef]
- Gadura, N.; Michels, C.A. Sequences in the N-Terminal Cytoplasmic Domain of Saccharomyces cerevisiae Maltose Permease Are Required for Vacuolar Degradation but Not Glucose-Induced Internalization. Curr. Genet. 2006, 50, 101–114. [Google Scholar] [CrossRef]
- Clague, M.J.; Urbé, S. Ubiquitin: Same Molecule, Different Degradation Pathways. Cell 2010, 143, 682–685. [Google Scholar] [CrossRef]



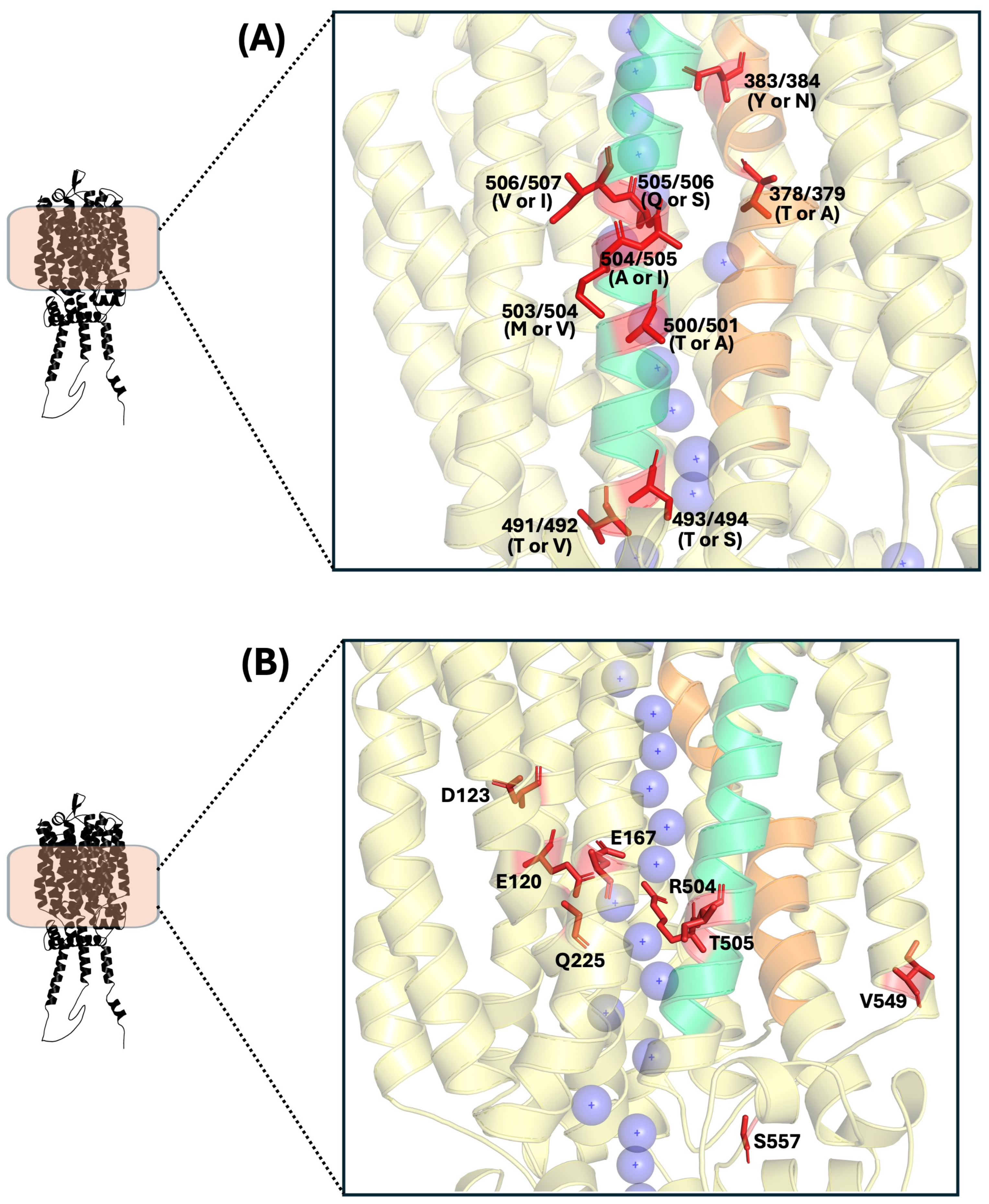


| α-Glucoside Transporter | Key Residue (s) | Region | Importance | Proximity to Transport Channel |
|---|---|---|---|---|
| Agt1p [13] | S128 | TMH1 | Polymorphic residue | High |
| Agt1p [13] | L164 | TMH2 | Polymorphic residue | High |
| Mal31p [14] | V371 | TMH7 | Polymorphic residue | High |
| Mal31p [14] | C374 | TMH7 | Polymorphic residue | High |
| Mal31p [14] | S375 | TMH7 | Polymorphic residue | High |
| Mal31p [14] | A378 | TMH7 | Polymorphic residue | High |
| Mal31p [14] | Y383 | TMH7 | Polymorphic residue | High |
| Mal31p [13] | C154 | TMH2 | Polymorphic residue | High |
| Mal31p [13] | Y157 | TMH2 | Polymorphic residue | High |
| Mal31p [13] | M158 | TMH2 | Polymorphic residue | High |
| Mal31p [13] | M167 | TMH2 | Polymorphic residue | Medium |
| Mal31p [13] | S171 | TMH2 | Polymorphic residue | Medium |
| Mal31p [13] | V503 | TMH11 | Polymorphic residue | High |
| Mal31p [13] | I504 | TMH11 | Polymorphic residue | High |
| Mal31p [13] | Q505 | TMH11 | Polymorphic residue | High |
| Mal31p [13] | V506 | TMH11 | Polymorphic residue | High |
| Mal31p [13] | V508 | TMH11 | Polymorphic residue | High |
| Mal31p [13] | T509 | TMH11 | Polymorphic residue | High |
| Mal31p [13] | M513 | TMH11 | Polymorphic residue | High |
| Mal31p [13] | A366 | TMH7 | Polymorphic residue | High |
| Mal31p [13] | L368 | TMH7 | Polymorphic residue | High |
| Mal31p [13] | G533 | TMH12 | Polymorphic residue | Low |
| Mal31p [13] | F534 | TMH12 | Polymorphic residue | Low |
| Mal31p [13] | L536 | TMH12 | Polymorphic residue | Low |
| Mal31p [13] | A540 | TMH12 | Polymorphic residue | Low |
| Mal31p [13] | V544 | TMH12 | Polymorphic residue | Low |
| Agt1p [11] | R504 | TMH11 | α-glucoside transport | High |
| Agt1p [11] | Q225 | TMH4 | Substrate binding site | High |
| Agt1p [11,92] | E120 | TMH1 | α-glucoside transport, proton translocation | High |
| Agt1p [11,92] | D123 | TMH1 | α-glucoside transport, proton translocation | High |
| Agt1p [11,92] | E167 | TMH2 | α-glucoside transport, proton translocation | High |
| Agt1p [10] | V549 | TMH12 | Maltotriose transport, polymorphic residue | Low |
| Agt1p [10] | T505 | TMH11 | Maltotriose transport, polymorphic residue | High |
| Agt1p [10] | S557 | Cytoplasm | Maltotriose transport, polymorphic residue | Medium |
| Mtt1p [12] | T379 | TMH7 | Maltotriose transport, polymorphic residue | High |
| Mtt1p [12] | N384 | TMH7 | Maltotriose transport, polymorphic residue | High |
| ScMalt#2p [12] | T378 | TMH7 | Maltotriose transport, polymorphic residue | High |
| ScMalt#2p [12] | N383 | TMH7 | Maltotriose transport, polymorphic residue | High |
| ScMalt#5p [12] | T379 | TMH7 | Maltotriose transport, polymorphic residue | High |
| ScMalt#5p [12] | N384 | TMH7 | Maltotriose transport, polymorphic residue | High |
| ScMalt#5p [12] | V492 | TMH11 | Maltose transport, polymorphic residue | High |
| ScMalt#5p [12] | T494 | TMH11 | Maltose transport, polymorphic residue | High |
| ScMalt#5p [12] | T501 | TMH11 | Maltose transport, polymorphic residue | High |
| ScMalt#5p [12] | V504 | TMH11 | Maltose transport, polymorphic residue | High |
| ScMalt#5p [12] | A505 | TMH11 | Maltose transport, polymorphic residue | High |
| ScMalt#5p [12] | S506 | TMH11 | Maltose transport, polymorphic residue | High |
| ScMalt#5p [12] | I507 | TMH11 | Maltose transport, polymorphic residue | High |
| MalT434 [82] | M503 | TMH11 | Maltotriose transport | High |
| MalT434 [82] | C505 | TMH11 | Maltotriose transport | High |
| MalT434 [82] | T508 | TMH11 | Maltotriose transport | High |
| MalT434 [82] | T512 | TMH11 | Maltotriose transport | High |
| MalT434 [82] | F468 | TMH10 | Maltotriose transport | Medium |
| MalT434 [82] | S379 | TMH7 | Maltotriose transport | High |
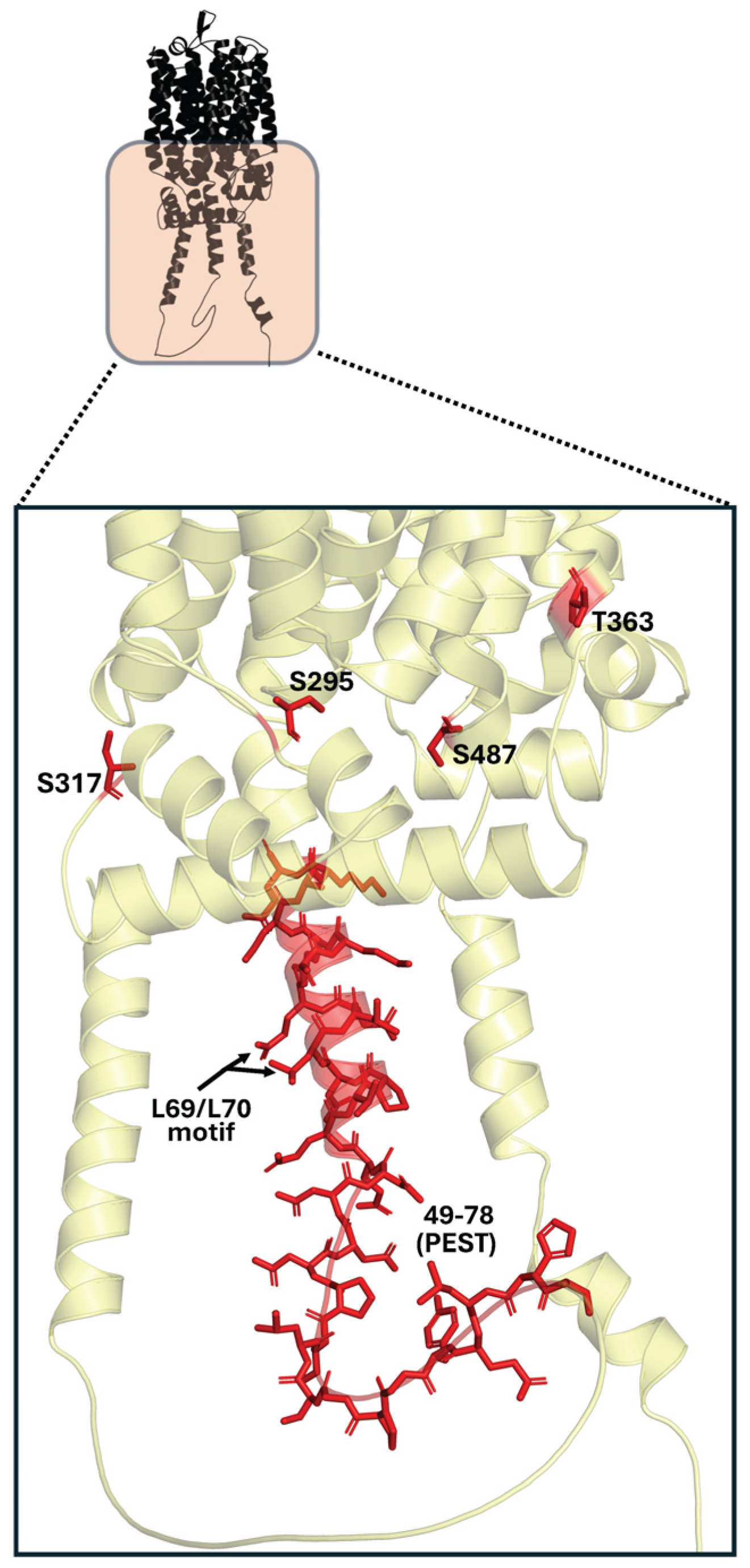
| Mal31p [35,72] | L69 and L70 | Cytoplasm (N-terminal) | Important ubiquitin binding site | Low |
| Mal21p [93] | G46 | Cytoplasm (N-terminal) | Resistance against degradation | Low |
| Mal21p [93] | H50 | Cytoplasm (N-terminal) | Resistance against degradation | Low |
| Mal61p [94] | S295 | Cytoplasm | Protein Kinase C site | Low |
| Mal61p [94] | S317 | Cytoplasm | Protein Kinase C site | Low |
| Mal61p [94] | T363 | Cytoplasm | Protein Kinase A site | Low |
| Mal61p [94] | S487 | Cytoplasm | Protein Kinase C site | Low |
| Mal61p [72] | L69 and L70 | Cytoplasm (N-terminal) | Important ubiquitin binding site | Low |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faz-Cortez, O.A.; García-García, J.H.; Carrizales-Sánchez, A.K.; Fonseca-Peralta, H.M.; Herrera-Gamboa, J.G.; Perez-Ortega, E.R.; Hernández-Vásquez, C.I.; Pereyra-Alférez, B. Maltose and Maltotriose Transporters in Brewer’s Saccharomyces Yeasts: Polymorphic and Key Residues in Their Activity. Int. J. Mol. Sci. 2025, 26, 5943. https://doi.org/10.3390/ijms26135943
Faz-Cortez OA, García-García JH, Carrizales-Sánchez AK, Fonseca-Peralta HM, Herrera-Gamboa JG, Perez-Ortega ER, Hernández-Vásquez CI, Pereyra-Alférez B. Maltose and Maltotriose Transporters in Brewer’s Saccharomyces Yeasts: Polymorphic and Key Residues in Their Activity. International Journal of Molecular Sciences. 2025; 26(13):5943. https://doi.org/10.3390/ijms26135943
Chicago/Turabian StyleFaz-Cortez, Oscar A., Jorge H. García-García, Ana K. Carrizales-Sánchez, Hector M. Fonseca-Peralta, Jessica G. Herrera-Gamboa, Esmeralda R. Perez-Ortega, César I. Hernández-Vásquez, and Benito Pereyra-Alférez. 2025. "Maltose and Maltotriose Transporters in Brewer’s Saccharomyces Yeasts: Polymorphic and Key Residues in Their Activity" International Journal of Molecular Sciences 26, no. 13: 5943. https://doi.org/10.3390/ijms26135943
APA StyleFaz-Cortez, O. A., García-García, J. H., Carrizales-Sánchez, A. K., Fonseca-Peralta, H. M., Herrera-Gamboa, J. G., Perez-Ortega, E. R., Hernández-Vásquez, C. I., & Pereyra-Alférez, B. (2025). Maltose and Maltotriose Transporters in Brewer’s Saccharomyces Yeasts: Polymorphic and Key Residues in Their Activity. International Journal of Molecular Sciences, 26(13), 5943. https://doi.org/10.3390/ijms26135943






