Spectral Flow Cytometry: The Current State and Future of the Technology
Abstract
1. Introduction
2. Design of the Classical and Spectral Flow Cytometer
3. New Fluorescent Molecules
3.1. Fluorescent Proteins (FPs)
3.2. Tandem Dyes
3.3. Small Organic Molecules
3.4. Quantum Dots (Qdots, QD)
3.5. Polymer Dyes
3.6. Multimeric DYES
3.7. DNA-Based Dyes
3.8. Polymer Dots
3.9. Fluorescent Tags of the Future
3.10. Features of Using Dyes in Spectral Flow Cytometry
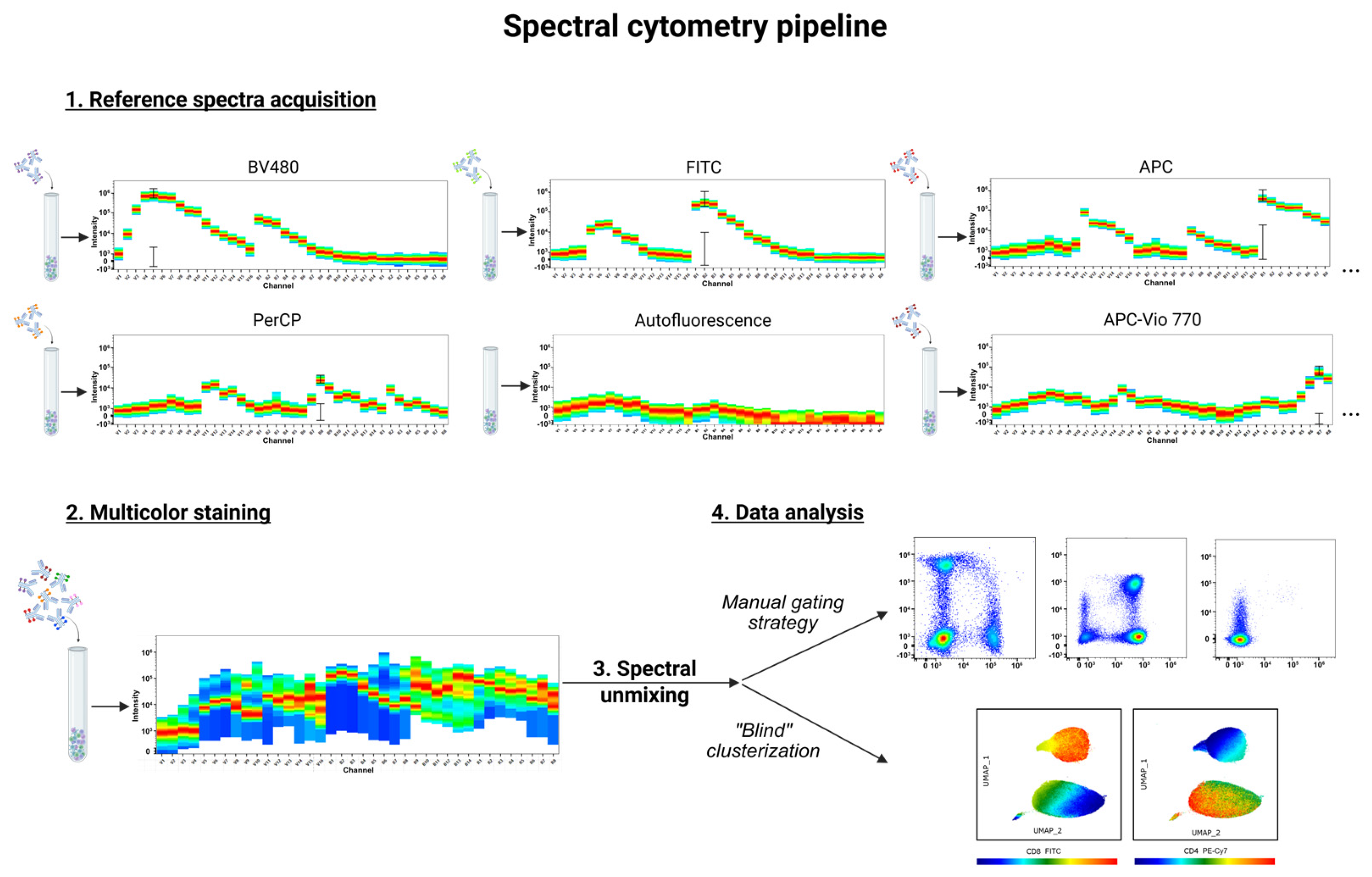
4. The Spectral Cytometry Pipeline
5. Deconvolution of Fluorescence Signal: Compensation and Spectral Unmixing
6. Clustering, Visualization, and Analysis of Multicolor Cytometry Data
7. Deep Immunophenotyping by Spectral Cytometry
8. The Newest Multicolor Panels for Spectral Cytometry
9. Limitations and Challenges of Spectral Cytometry
10. The Future of Spectral Flow Cytometry
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ssRNA-seq | Single-cell mRNA sequencing |
| CITE-Seq | Cellular indexing of transcriptomes and epitopes |
| CyTOF | Mass cytometry method |
| PMT | Photomultiplier tube |
| APD | Avalanche photodiode |
| CMWD | Semiconductor detector array |
| FSC | Forward light scattering |
| SSC | Side light scattering |
| FP | Fluorescent protein |
| FRET | Ferster resonance energy transfer |
| PD-1 | Programmed cell death protein 1 |
| LAG-3 | Lymphocyte activation gene protein 3 |
| MDSCs | Myeloid-derived suppressor cells |
| FOXP3 | Scurfin protein |
| Ki67 | Ki67 proliferation marker |
| CTLA-4 | Class 4 cytotoxic T lymphocyte antigen |
| GITR | Glucocorticoid-induced TNF receptor family protein |
| ICOS | Inducible co-stimulator of T lymphocytes |
| LAP | Leukocyte alkaline phosphatase |
| GARP | Glycoprotein A predominant repeats |
| PD-L1 | Programmed cell death receptor ligand 1 |
| OMIP | Optimized multicolor immunofluorescence panels |
| AF | Autofluorescence |
References
- McKinnon, K.M. Flow Cytometry: An Overview. Curr. Protoc. Immunol. 2018, 120, 5.1.1–5.1.11. [Google Scholar] [CrossRef] [PubMed]
- Khaidukov, S.V.; Zurochka, A.V. Flow Cytometry As a Modern Analytical Tool in Biology and Medicine. Med. Immunol. 2014, 9, 373. [Google Scholar] [CrossRef]
- Fulwyler, M.J. Electronic Separation of Biological Cells by Volume. Science 1965, 150, 910–911. [Google Scholar] [CrossRef]
- Lugli, E.; Roederer, M.; Sottile, R. Multipass High-Dimensional Flow Cytometry. Nat. Biomed. Eng. 2023, 8, 209–211. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.P.; Ostafe, R.; Iyengar, S.N.; Rajwa, B.; Fischer, R. Flow Cytometry: The Next Revolution. Cells 2023, 12, 1875. [Google Scholar] [CrossRef]
- Pisetsky, D.S. Pathogenesis of Autoimmune Disease. Nat. Rev. Nephrol. 2023, 19, 509–524. [Google Scholar] [CrossRef]
- Gonzalez, H.; Hagerling, C.; Werb, Z. Roles of the Immune System in Cancer: From Tumor Initiation to Metastatic Progression. Genes Dev. 2018, 32, 1267–1284. [Google Scholar] [CrossRef]
- Shiravand, Y.; Khodadadi, F.; Kashani, S.M.A.; Hosseini-Fard, S.R.; Hosseini, S.; Sadeghirad, H.; Ladwa, R.; O’Byrne, K.; Kulasinghe, A. Immune Checkpoint Inhibitors in Cancer Therapy. Curr. Oncol. 2022, 29, 3044–3060. [Google Scholar] [CrossRef]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between Differentially Expressed MRNA and MRNA-Protein Correlations in a Xenograft Model System. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef]
- Fernandez, P.; Solenthaler, M.; Spertini, O.; Quarroz, S.; Rovo, A.; Lovey, P.-Y.; Leoncini, L.; Ruault-Jungblut, S.; D’Asaro, M.; Schaad, O.; et al. Using Digital RNA Counting and Flow Cytometry to Compare MRNA with Protein Expression in Acute Leukemias. PLoS ONE 2012, 7, e49010. [Google Scholar] [CrossRef]
- Nettersheim, F.S.; Armstrong, S.S.; Durant, C.; Blanco-Dominguez, R.; Roy, P.; Orecchioni, M.; Suryawanshi, V.; Ley, K. Titration of 124 Antibodies Using CITE-Seq on Human PBMCs. Sci. Rep. 2022, 12, 20817. [Google Scholar] [CrossRef] [PubMed]
- Ornatsky, O.; Bandura, D.; Baranov, V.; Nitz, M.; Winnik, M.A.; Tanner, S. Highly Multiparametric Analysis by Mass Cytometry. J. Immunol. Methods 2010, 361, 1–20. [Google Scholar] [CrossRef]
- Robinson, J.P.; Rajwa, B.; Gregori, G.; Jones, J.; Patsekin, V. Multispectral Cytometry of Single Bio-Particles Using a 32-Channel Detector. In Advanced Biomedical and Clinical Diagnostic Systems III; Vo-Dinh, T., Grundfest, W.S., Benaron, D.A., Cohn, G.E., Eds.; Society of Photo Optical: Bellingham, WA, USA, 2005; p. 359. [Google Scholar]
- Vorobjev, I.A.; Kussanova, A.; Barteneva, N.S. Development of Spectral Imaging Cytometry. In Spectral and Imaging Cytometry. Methods in Molecular Biology; Barteneva, N.S., Vorobjev, I., Eds.; Humana: New York, NY, USA, 2023; pp. 3–22. [Google Scholar]
- Nolan, J.P.; Condello, D. Spectral Flow Cytometry. Curr. Protoc. Cytom. 2013, 63, 1.27.1–1.27.13. [Google Scholar] [CrossRef] [PubMed]
- Cossarizza, A.; Chang, H.; Radbruch, A.; Abrignani, S.; Addo, R.; Akdis, M.; Andrä, I.; Andreata, F.; Annunziato, F.; Arranz, E.; et al. Guidelines for the Use of Flow Cytometry and Cell Sorting in Immunological Studies (Third Edition). Eur. J. Immunol. 2021, 51, 2708–3145. [Google Scholar] [CrossRef] [PubMed]
- Telford, W. Laser Sources for Traditional and Spectral Flow Cytometry. In Flow Cytometry Protocols. Methods in Molecular Biology; Hawley, T.S., Hawley, R., Eds.; Humana: New York, NY, USA, 2024; pp. 33–68. [Google Scholar]
- Siddiqui, S.; Livák, F. Principles of Advanced Flow Cytometry: A Practical Guide. In Methods in Molecular Biology; Bosselut, R., Vacchio, M.S., Eds.; Springer Protocols: New York, NY, USA; Humana: New York, NY, USA, 2023; Volume 2580, pp. 89–114. ISBN 9781071627396. [Google Scholar]
- Robinson, J.P. Spectral Flow Cytometry—Quo Vadimus? Cytom. Part A 2019, 95, 823–824. [Google Scholar] [CrossRef]
- Perfetto, S.P.; Chattopadhyay, P.K.; Roederer, M. Seventeen-Colour Flow Cytometry: Unravelling the Immune System. Nat. Rev. Immunol. 2004, 4, 648–655. [Google Scholar] [CrossRef]
- Robinson, J.P. Flow Cytometry: Past and Future. Biotechniques 2022, 72, 159–169. [Google Scholar] [CrossRef]
- Ortolani, C. Fluorochromes: Overview. In Flow Cytometry Today; Springer: Cham, Switzerland, 2022; pp. 247–258. [Google Scholar]
- Hulspas, R.; Dombkowski, D.; Preffer, F.; Douglas, D.; Kildew-Shah, B.; Gilbert, J. Flow Cytometry and the Stability of Phycoerythrin-tandem Dye Conjugates. Cytom. Part A 2009, 75A, 966–972. [Google Scholar] [CrossRef]
- Sharma, S.; Boyer, J.; Teyton, L. A Practitioner’s View of Spectral Flow Cytometry. Nat. Methods 2024, 21, 740–743. [Google Scholar] [CrossRef]
- Available online: https://www.thermofisher.com/ru/ru/home/life-science/cell-analysis/flow-cytometry/efluor-organic-dyes (accessed on 6 June 2025).
- Fire Dyes. Available online: https://www.biolegend.com/ja-jp/fire-dyes (accessed on 20 November 2024).
- Park, L.M.; Lannigan, J.; Low, Q.; Jaimes, M.C.; Bonilla, D.L. OMIP-069 Version 2: Update to the 40-Color Full Spectrum Flow Cytometry Panel for Deep Immunophenotyping of Major Cell Subsets in Human Peripheral Blood. Cytom. Part A 2024, 105, 791–799. [Google Scholar] [CrossRef]
- Chattopadhyay, P.K.; Perfetto, S.P.; Yu, J.; Roederer, M. The Use of Quantum Dot Nanocrystals in Multicolor Flow Cytometry. WIREs Nanomed. Nanobiotechnol. 2010, 2, 334–348. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, P.K.; Price, D.A.; Harper, T.F.; Betts, M.R.; Yu, J.; Gostick, E.; Perfetto, S.P.; Goepfert, P.; Koup, R.A.; De Rosa, S.C.; et al. Quantum Dot Semiconductor Nanocrystals for Immunophenotyping by Polychromatic Flow Cytometry. Nat. Med. 2006, 12, 972–977. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, P.K.; Gaylord, B.; Palmer, A.; Jiang, N.; Raven, M.A.; Lewis, G.; Reuter, M.A.; Nur-ur Rahman, A.K.M.; Price, D.A.; Betts, M.R.; et al. Brilliant Violet Fluorophores: A New Class of Ultrabright Fluorescent Compounds for Immunofluorescence Experiments. Cytom. Part A 2012, 81A, 456–466. [Google Scholar] [CrossRef]
- Reiber, T.; Zavoiura, O.; Dose, C.; Yushchenko, D.A. Fluorophore Multimerization as an Efficient Approach towards Bright Protein Labels. Eur. J. Org. Chem. 2021, 2021, 2817–2830. [Google Scholar] [CrossRef]
- Dose, C. Bright Fluorochromes Based on Multimerization of Fluorescent Dyes on Branched Polyether Scaffolds. Patent Application No. EP3098269A1, 30 November 2016. [Google Scholar]
- Teo, Y.N.; Kool, E.T. DNA-Multichromophore Systems. Chem. Rev. 2012, 112, 4221–4245. [Google Scholar] [CrossRef]
- Loretan, M.; Domljanovic, I.; Lakatos, M.; Rüegg, C.; Acuna, G.P. DNA Origami as Emerging Technology for the Engineering of Fluorescent and Plasmonic-Based Biosensors. Materials 2020, 13, 2185. [Google Scholar] [CrossRef]
- Schmied, J.J.; Raab, M.; Forthmann, C.; Pibiri, E.; Wünsch, B.; Dammeyer, T.; Tinnefeld, P. DNA Origami–Based Standards for Quantitative Fluorescence Microscopy. Nat. Protoc. 2014, 9, 1367–1391. [Google Scholar] [CrossRef]
- Available online: https://assets.thermofisher.com/TFS-Assets/BID/Handbooks/optimizing-fluorescence-flow-cytometry-handbook.pdf (accessed on 6 June 2025).
- Available online: https://www.thermofisher.com/ru/ru/home/life-science/cell-analysis/flow-cytometry/novafluor-dyes (accessed on 6 June 2025).
- Wu, C.; Chiu, D.T. Highly Fluorescent Semiconducting Polymer Dots for Biology and Medicine. Angew. Chem. Int. Ed. 2013, 52, 3086–3109. [Google Scholar] [CrossRef]
- Li, G.; Liu, Z.; Gao, W.; Tang, B. Recent Advancement in Graphene Quantum Dots Based Fluorescent Sensor: Design, Construction and Bio-Medical Applications. Coord. Chem. Rev. 2023, 478, 214966. [Google Scholar] [CrossRef]
- Shevchenko, Y.; Lurje, I.; Tacke, F.; Hammerich, L. Fluorochrome-Dependent Specific Changes in Spectral Profiles Using Different Compensation Beads or Primary Cells in Full Spectrum Cytometry. Cytom. Part A 2024, 105, 458–463. [Google Scholar] [CrossRef]
- El-Hajjar, L.; Ahmad, F.A.; Nasr, R. A Guide to Flow Cytometry: Components, Basic Principles, Experimental Design, and Cancer Research Applications. Curr. Protoc. 2023, 3, e721. [Google Scholar] [CrossRef] [PubMed]
- Holmberg-thyden, S.; Gr, K.; Ortved, A.; El, D.; Reker, S. A User’ s Guide to Multicolor Flow Cytometry Panels for Comprehensive Immune Profiling. Anal. Biochem. 2021, 627, 114210. [Google Scholar] [CrossRef] [PubMed]
- Monici, M. Cell and Tissue Autofluorescence Research and Diagnostic Applications. Biotechnol. Annu. Rev. 2005, 11, 227–256. [Google Scholar] [PubMed]
- Jameson, V.J.; Luke, T.; Yan, Y.; Hind, A.; Evrard, M.; Man, K.; Mackay, L.K.; Kallies, A.; Villadangos, J.A.; McWilliam, H.E.G.; et al. Unlocking Autofluorescence in the Era of Full Spectrum Analysis: Implications for Immunophenotype Discovery Projects. Cytom. Part A 2022, 101, 922–941. [Google Scholar] [CrossRef]
- Roet, J.E.G.; Mikula, A.M.; de Kok, M.; Chadick, C.H.; Garcia Vallejo, J.J.; Roest, H.P.; van der Laan, L.J.W.; de Winde, C.M.; Mebius, R.E. Unbiased Method for Spectral Analysis of Cells with Great Diversity of Autofluorescence Spectra. Cytom. Part A 2024, 105, 595–606. [Google Scholar] [CrossRef]
- Roederer, M. Spectral Compensation for Flow Cytometry: Visualization Artifacts, Limitations, and Caveats. Cytometry 2001, 45, 194–205. [Google Scholar] [CrossRef]
- Novo, D. A Comparison of Spectral Unmixing to Conventional Compensation for the Calculation of Fluorochrome Abundances from Flow Cytometric Data. Cytom. Part A 2022, 101, 885–891. [Google Scholar] [CrossRef]
- Roederer, M. Compensation in Flow Cytometry. Curr. Protoc. Cytom. 2002, 22, 1.14.1–1.14.20. [Google Scholar] [CrossRef]
- Bhowmick, D.; Lowe, S.K.; Ratliff, M.L. Side-by-Side Comparison of Compensation Beads Used in Polychromatic Flow Cytometry. ImmunoHorizons 2023, 7, 819–833. [Google Scholar] [CrossRef]
- Available online: https://fcsexpressdownloads.s3.amazonaws.com/manual/manual_WIN_RUO/index.html?the_compensation_tool.htm (accessed on 6 June 2025).
- Bendall, S.C.; Nolan, G.P.; Roederer, M.; Chattopadhyay, P.K. A Deep Profiler’s Guide to Cytometry. Trends Immunol. 2012, 33, 323–332. [Google Scholar] [CrossRef]
- Kurita, T. Principal Component Analysis (PCA). In Computer Vision; Springer International Publishing: Cham, Switzerland, 2020; pp. 1013–1016. [Google Scholar]
- Belkina, A.C.; Ciccolella, C.O.; Anno, R.; Halpert, R.; Snyder-cappione, J.E. Automated Optimized Parameters for T-Distributed Stochastic Neighbor Embedding Improve Visualization and Analysis of Large Datasets. Nat. Commun. 2019, 10, 5415. [Google Scholar] [CrossRef] [PubMed]
- Becht, E.; McInnes, L.; Healy, J.; Dutertre, C.A.; Kwok, I.W.H.; Ng, L.G.; Ginhoux, F.; Newell, E.W. Dimensionality Reduction for Visualizing Single-Cell Data Using UMAP. Nat. Biotechnol. 2019, 37, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Liu, S.; Fang, Y.; Xue, X.; Zou, J.; Tseng, G.; Konnikova, L. Recent Advances in Computer-Assisted Algorithms for Cell Subtype Identification of Cytometry Data. Front. Cell Dev. Biol. 2020, 8, 234. [Google Scholar] [CrossRef] [PubMed]
- Pitoiset, F.; Cassard, L.; El Soufi, K.; Boselli, L.; Grivel, J.; Roux, A.; Klatzmann, D.; Chaput, N.; Rosenzwajg, M. Deep Phenotyping of Immune Cell Populations by Optimized and Standardized Flow Cytometry Analyses. Cytom. Part A 2018, 93, 793–802. [Google Scholar] [CrossRef]
- Rincon-Arevalo, H.; Wiedemann, A.; Stefanski, A.-L.; Lettau, M.; Szelinski, F.; Fuchs, S.; Frei, A.P.; Steinberg, M.; Kam-Thong, T.; Hatje, K.; et al. Deep Phenotyping of CD11c+ B Cells in Systemic Autoimmunity and Controls. Front. Immunol. 2021, 12, 635615. [Google Scholar] [CrossRef]
- Niewold, P.; Ashhurst, T.M.; Jonathan, N.; King, C.; Smith, A.L. Evaluating Spectral Cytometry for Immune pro Fi Ling in Viral Disease. Cytom. Part A 2020, 97, 1165–1179. [Google Scholar] [CrossRef]
- Park, L.M.; Lannigan, J.; Jaimes, M.C. OMIP-069: Forty-Color Full Spectrum Flow Cytometry Panel for Deep Immunophenotyping of Major Cell Subsets in Human Peripheral Blood. Cytom. Part A 2020, 97, 1044–1051. [Google Scholar] [CrossRef]
- George, A.P.; Kuzel, T.M.; Zhang, Y.; Zhang, B. The Discovery of Biomarkers in Cancer Immunotherapy. Comput. Struct. Biotechnol. J. 2019, 17, 484–497. [Google Scholar] [CrossRef]
- Saito, A.; Tojo, M.; Kumagai, Y.; Ohzawa, H.; Yamaguchi, H.; Miyato, H.; Sadatomo, A.; Naoi, D.; Ota, G.; Koinuma, K.; et al. Flow Cytometry Detection of Cell Type-Speci Fi c Expression of Programmed Death Receptor Ligand-1 (PD-L1) in Colorectal Cancer Specimens. Heliyon 2021, 7, e05880. [Google Scholar] [CrossRef]
- Huuhtanen, J.; Kreutzman, A.; Mustjoki, S.; Huuhtanen, J.; Kasanen, H.; Peltola, K.; Lönnberg, T.; Glumoff, V.; Brück, O.; Dufva, O.; et al. Single-Cell Characterization of Anti-LAG-3 and Anti-PD-1 Combination Treatment in Patients with Melanoma. J. Clin. Investig. 2023, 133, e164809. [Google Scholar] [CrossRef]
- Yuan, J.; Hegde, P.S.; Clynes, R.; Foukas, P.G.; Harari, A.; Kleen, T.O.; Kvistborg, P.; Maccalli, C.; Maecker, H.T.; Page, D.B.; et al. Novel Technologies and Emerging Biomarkers for Personalized Cancer Immunotherapy. J. Immunother. Cancer 2016, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.C.; Sen, D.R.; Al Abosy, R.; Bi, K.; Virkud, Y.V.; Lafleur, M.W.; Yates, K.B.; Lako, A.; Felt, K.; Naik, G.S.; et al. Subsets of Exhausted CD8+ T Cells Differentially Mediate Tumor Control and Respond to Checkpoint Blockade. Nat. Immunol. 2019, 20, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Dyikanov, D. Comprehensive Peripheral Blood Immunoprofiling Reveals Five Immunotypes with Immunotherapy Response Characteristics in Patients with Cancer. Cancer Cell 2024, 42, 759–779.e12. [Google Scholar] [CrossRef] [PubMed]
- Ohue, Y.; Nishikawa, H. Regulatory T (Treg) Cells in Cancer: Can Treg Cells Be a New Therapeutic Target? Cancer Sci. 2019, 110, 2080–2089. [Google Scholar] [CrossRef]
- Whiteside, T. The Role of Regulatory T Cells in Cancer Immunology. ImmunoTargets Ther. 2015, 4, 159–171. [Google Scholar] [CrossRef]
- Miyara, M.; Yoshioka, Y.; Kitoh, A.; Shima, T.; Wing, K.; Niwa, A.; Heike, T.; Valeyre, D.; Mathian, A.; Nakahata, T.; et al. Functional Delineation and Differentiation Dynamics of Human CD4+ T Cells Expressing the FoxP3 Transcription Factor. Immun. Resour. 2009, 30, 899–911. [Google Scholar] [CrossRef]
- Liston, A.; Aloulou, M. A Fresh Look at a Neglected Regulatory Lineage: CD8+Foxp3+ Regulatory T Cells. Immunol. Lett. 2022, 247, 22–26. [Google Scholar] [CrossRef]
- Vizi, E.S. CD39 and CD73 in Immunity and Inflammation. Trends Mol. Med. 2013, 19, 355–367. [Google Scholar] [CrossRef]
- Zappasodi, R.; Serganova, I.; Cohen, I.J.; Maeda, M.; Shindo, M.; Senbabaoglu, Y.; Watson, M.J.; Leftin, A.; Maniyar, R.; Verma, S.; et al. CTLA-4 Blockade Drives Loss of Treg Stability in Glycolysis-Low Tumours. Nature 2021, 591, 652–658. [Google Scholar] [CrossRef]
- Kornete, M.; Mason, E.S.; Girouard, J.; Lafferty, E.I.; Qureshi, S.; Piccirillo, C.A. Th1-Like ICOS+ Foxp3+ Treg Cells Preferentially Express CXCR3 and Home to β-Islets during Pre-Diabetes in BDC2.5 NOD Mice. PLoS ONE 2015, 10, e0126311. [Google Scholar] [CrossRef]
- Cai, H.; Liu, Y.; Dong, X.; Jiang, F.; Li, H.; Ouyang, S.; Yin, W.; He, T.; Zeng, Q.; Yang, H. Analysis of LAP+ and GARP+ Treg Subsets in Peripheral Blood of Patients with Neuromyelitis Optica Spectrum Disorders. Neurol. Sci. 2023, 44, 1739–1747. [Google Scholar] [CrossRef] [PubMed]
- Diks, A.M.; Overduin, L.A.; Van Leenen, L.D.; Slobbe, L.; Jolink, H.; Visser, L.G.; Van Dongen, J.J.M.; Berkowska, M.A. B-Cell Immunophenotyping to Predict Vaccination Outcome in the Immunocompromised—A Systematic Review. Front. Immunol. 2021, 12, 690328. [Google Scholar] [CrossRef] [PubMed]
- Lushova, A.A.; Zheremyan, E.A.; Astakhova, E.A.; Spiridonova, A.B.; Byazrova, M.G.; Filatov, A.V. B-Lymphocyte Subsets: Functions and Molecular Markers. Immunologiya 2019, 40, 63–76. [Google Scholar] [CrossRef]
- Zabotina, T.N.; Panchuk, I.O.; Tabakov, D.V.; Zakharova, E.N. Myeloid-Derived Suppressor Cells: Origin, Phenotype, Functions, Mechanisms of Interaction with Immune Cells during Tumor Growth. Pract. Oncol. 2020, 21, 249–261. [Google Scholar] [CrossRef]
- Vives-pi, M.; Martinez-caceres, E.M.; Ferna, M.A. Identifying Changes in Peripheral Lymphocyte Subpopulations in Adult Onset Type 1 Diabetes. Front. Immunol. 2021, 12, 784110. [Google Scholar] [CrossRef]
- Na, Y.W.; Liu, Z.H.; Zhao, X.L.; Zong, S.X.; Li, C.M.C.B.T. Circulating Activated Lymphocyte Subsets as Potential Blood Biomarkers of Cancer Progression. Cancer Med. 2020, 9, 5086–5094. [Google Scholar] [CrossRef]
- Georgolopoulos, G.; Iwata, M.; Psatha, N.; Yiangou, M.; Vierstra, J. Unbiased Phenotypic Identification of Functionally Distinct Hematopoietic Progenitors. J. Biol. Res. 2019, 26, 4. [Google Scholar] [CrossRef]
- Mayer, I.M.; Hoelbl-kovacic, A.; Sexl, V. Isolation, Maintenance and Expansion of Adult Hematopoietic Stem / Progenitor Cells and Leukemic Stem Cells. Cancers 2022, 14, 1723. [Google Scholar] [CrossRef]
- Matthes, T. Phenotypic Analysis of Hematopoietic Stem and Progenitor Cell Populations in Acute Myeloid Leukemia Based on Spectral Flow Cytometry, a 20-Color Panel, and Unsupervised Learning Algorithms. Int. J. Mol. Sci. 2024, 25, 2847. [Google Scholar] [CrossRef]
- Garc, G.; Castillo-robleda, A.; Sanz, A. Validation of a Spectral Flow Cytometry Single-Tube Panel for the Clinical Diagnosis and Follow-Up of Children and Adolescents with B-Cell Acute Lymphoblastic Leukemia. Cells 2024, 13, 1891. [Google Scholar] [CrossRef]
- Zhu, Y.; Ma, X.; Hao, Y.; Guan, Y. Myelodysplastic Syndrome Transformed into B-Lineage Acute Lymphoblastic Leukemia: A Case Report. World J. Clin. Cases 2021, 9, 5191–5197. [Google Scholar] [CrossRef] [PubMed]
- Cadinanos-Garai, A.; Flugel, C.L.; Cheung, A.; Jiang, E.; Vaissié, A. High-Dimensional Temporal Mapping of CAR T Cells Reveals Phenotypic and Functional Remodeling during Manufacturing. Mol. Ther. 2025, 33, 2291–2309. [Google Scholar] [CrossRef] [PubMed]
- Wiedeman, A.E.; Linsley, P.S.; Long, S.A.; Wiedeman, A.E.; Muir, V.S.; Rosasco, M.G.; Deberg, H.A.; Presnell, S.; Haas, B.; Dufort, M.J.; et al. Autoreactive CD8+ T Cell Exhaustion Distinguishes Subjects with Slow Type 1 Diabetes Progression Autoreactive CD8+ T Cell Exhaustion Distinguishes Subjects with Slow Type 1 Diabetes Progression. J. Clin. Investig. 2020, 130, 480–490. [Google Scholar] [CrossRef] [PubMed]
- van Dongen, J.J.M.; Lhermitte, L.; Böttcher, S.; Almeida, J.; van der Velden, V.H.J.; Flores-Montero, J.; Rawstron, A.; Asnafi, V.; Lécrevisse, Q.; Lucio, P.; et al. EuroFlow Antibody Panels for Standardized N-Dimensional Flow Cytometric Immunophenotyping of Normal, Reactive and Malignant Leukocytes. Leukemia 2012, 26, 1908–1975. [Google Scholar] [CrossRef]
- Doyle, C.M.; Fewings, N.L.; Ctercteko, G.; Byrne, S.N.; Harman, A.N.; Bertram, K.M. OMIP 082: A 25-Color Phenotyping to Define Human Innate Lymphoid Cells, Natural Killer Cells, Mucosal-Associated Invariant T Cell, and Γδ T Cells from Freshly Isolated Human Intestinal Tissue. Cytom. Part A 2022, 101, 196–202. [Google Scholar] [CrossRef]
- Cuapio, A.; Boulouis, C.; Filipovic, I.; Wullimann, D.; Kammann, T.; Parrot, T.; Chen, P.; Akber, M.; Gao, Y.; Hammer, Q.; et al. NK Cell Frequencies, Function and Correlates to Vaccine Outcome in BNT162b2 MRNA Anti-SARS-CoV-2 Vaccinated Healthy and Immunocompromised Individuals. Mol. Med. 2022, 28, 20. [Google Scholar] [CrossRef]
- Imbratta, C.; Reid, T.D.; Scriba, T.J.; Nemes, E. OMIP-101: 27-Color Flow Cytometry Panel for Immunophenotyping of Major Leukocyte Populations in Fixed Whole Blood. Cytom. Part A 2024, 105, 165–170. [Google Scholar] [CrossRef]
- Backer, R.A.; Probst, H.C.; Clausen, B.E. Multiparameter Flow Cytometric Analysis of the Conventional and Monocyte-Derived DC Compartment in the Murine Spleen. Vaccines 2024, 12, 1294. [Google Scholar] [CrossRef]
- Heubeck, A.; Savage, A.; Hernandez, V.; Reading, J.; Henderson, K.; Torgerson, T.; Bumol, T. Cross-Platform Immunophenotyping of Human Peripheral Blood Mononuclear Cells with Four High-Dimensional Flow Cytometry Panels. Cytom. Part A 2023, 103, 500–517. [Google Scholar] [CrossRef]
- Park, L.M.; Lannigan, J.; Bonilla, D.L.; Low, Q.; Jaimes, M.C. OMIP-109: 45-Color Full Spectrum Flow Cytometry Panel for Deep Immunophenotyping of the Major Lineages Present in Human Peripheral Blood Mononuclear Cells with Emphasis on the T Cell Memory Compartment. Cytometry 2024, 105, 807–815. [Google Scholar] [CrossRef]
- Zacharias, Z.R.; Houtman, J.C.D. OMIP-099: 31-color Spectral Flow Cytometry Panel to Investigate the Steady-state Phenotype of Human T Cells. Cytom. Part A 2024, 105, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Konecny, A.J.; Mage, P.; Tyznik, A.J.; Prlic, M.; Mair, F. 50-Color Phenotyping of the Human Immune System with in-Depth Assessment of T Cells and Dendritic Cells. Cytometry 2025, 105, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Jiwrajka, N.; Tuluc, F.; Jaimes, M.C.; Murray, J.; Anguera, M.C. 30-Color Longitudinal Full-Spectrum Immunophenotyping and Sorting of Human Circulating Immune Cell Subsets Implicated in Systemic Autoimmune Rheumatic Diseases. bioRxiv 2025. [Google Scholar] [CrossRef]
- Spasic, M.; Ogayo, E.R.; Parsons, A.M.; Mittendorf, E.A. Spectral Flow Cytometry Methods and Pipelines for Comprehensive Immunoprofiling of Human Peripheral Blood and Bone Marrow. Cancer Res. Commun. 2024, 4, 895–910. [Google Scholar] [CrossRef]
- Spurgeon, B.E.J.; Iii, A.L.F. OMIP-097: High-Parameter Phenotyping of Human Platelets by Spectral Flow Cytometry. Cytometry 2023, 103, 935–940. [Google Scholar] [CrossRef]
- Dott, T.; Culina, S.; Chemali, R.; Ait, C.; Dubois, F.; Jagla, B.; Marc, J.; Lars, D.; Huetz, F.; Jönsson, F.; et al. Standardized High-Dimensional Spectral Cytometry Protocol and Panels for Whole Blood Immune Phenotyping in Clinical and Translational Studies. Cytometry 2024, 105, 124–138. [Google Scholar] [CrossRef]
- Kare, A.J.; Nichols, L.; Zermeno, R.; Raie, M.N.; Tumbale, S.K.; Ferrara, K.W. OMIP-095: 40-Color Spectral Flow Cytometry Delineates All Major Leukocyte Populations in Murine Lymphoid Tissues. Cytometry 2023, 103, 839–850. [Google Scholar] [CrossRef]
- Victor, A.; Longhini, A.L.F.; Kennedy, M.C.; Wereski, M.G.; Mowla, S.; Xiao, W.; Lowe, S.W.; Levine, R.L.; Gardner, R. Development of a Customizable Mouse Backbone Spectral Fl Ow Cytometry Panel to Delineate Immune Cell Populations in Normal and Tumor Tissues. Front. Immunol. 2024, 15, 1374943. [Google Scholar] [CrossRef]
- Jensen, H.A.; Kim, J. iCoreDrop: A Robust Immune Monitoring Spectral Cytometry Assay with Six Open Channels for Biomarker Flexibility. Cytom. Part A 2023, 103, 405–418. [Google Scholar] [CrossRef]
- Brestoff, J.R. Full Spectrum Flow Cytometry in the Clinical Laboratory. Int. J. Lab. Hematol. 2023, 45, 44–49. [Google Scholar] [CrossRef]

| Characteristics | Sony SA3800 | Sony ID7000 | Cytek Northern Lights | Cytek Aurora | Agilent NovoCyte Opteon | AttuneTM XenithTM | BD FACSymphony™ A5 SE |
|---|---|---|---|---|---|---|---|
| Number of lasers and their wavelengths | up to 4–405/488/561/638 nm | up to 7–320/355/405/488/561/637/808 nm | 3– 405/488/640 nm | 5–355/405/488/561/640 nm | up to 5–349/405/488/561/ 637 nm | 6–349/405/488/561/ 637/781 nm | 5–355/405/488/561/637 nm |
| Detection system | Set of 10 prisms, 32-channel PMT array | 32-channel PMT arrays, individual PMTs | CMWD * | CMWD * | CMWD * | PMT array | Cascade square PMT array |
| Number of detection channels, range of detectable wavelengths | FSC/SSC + 32F ** | FSC/SSC + 184F ** | FSC/2 SSC + 38F ** | FSC/2 SSC + 64F ** | FSC/2 SSC + 73F ** | 3 FSC/3 SSC + 51F ** | FSC/SSC + 48F ** |
| Number of colors in the panel | Not specified | 44 or more | Up to 24 | Up to 40 | Up to 45 | Up to 32 | Up to 40 |
| Software | Sony Spectrum Analyzer | SpectroFlo | Agilent NovoExpress | Attune NxT Cytometric Software | BD FACSDiva | ||
| Type | Fluorochromes | Manufacturer |
|---|---|---|
| Small organic molecules | Spark™, Spark PLUS™ | Biolegend |
| Vio™ | Miltenyi | |
| eFluor™ 450, eFluor™ 660 | Thermofisher | |
| Tandem dyes (fluorescence donor—protein) | Fire™ | Biolegend |
| PerCP-eFluor 710, APC-eFluor 780 | Thermofisher | |
| Astral Leap™ | Biotium | |
| Tandem dyes (fluorescence donor—polymer) | BD Horizon™ BV605, BV650, BV711, BV786 | BD Biosciences |
| Polymer dyes | SuperBright™ | Thermofisher |
| Multimers | VioBright™ | Miltenyi |
| RealBlue™, RealYellow™ | BD Biosciences | |
| KIRAVIA™ | Biolegend | |
| DNA-based dyes | NovaFluor™ | Thermofisher |
| Polymer dots (PDots) | StarBright™ | Bio-Rad |
| Cell Populations | Color Number | Instrument | References |
|---|---|---|---|
| Human PBMC | |||
| B-, T-, γδ-T-, NKT-like, NK-cells, monocytes, basophils, DCs, ILCs | 45 | 5-laser (355, 405, 488, 561, 640 nm) Cytek Aurora | [92] |
| CD4 Treg: naive, memory, TR1-like, activated cells. CD4: TFH, TEMRA, TCM, TEM, naive, TSCM, Th1, Th2. CD8: TEMRA, TCM TEM, naive, TSCM | 31 | 5-laser (355, 405, 488, 561, 640 nm) Cytek Aurora | [93] |
| T-, B-, NK-cells, DCs, ILCs | 50 | 7-laser (320, 355, 405, 488, 561, 637, 808 nm) Sony ID7000 and 5-laser (349, 405, 488, 561, 637 nm) BD FACSDiscover S8 | [94] |
| Subpopulations of T-, B-cells, DCs, plasmacytoid DCs, basophils | 30 | 5-laser (355, 405, 488, 561, 640 nm) Cytek Aurora | [95] |
| 27 T-cell subpopulations, 5 B-cell subpopulations, NK-, NKT-cells | 27 | 4-laser (405, 488, 561, 640 nm) Cytek Aurora | [96] |
| Subpopulations of dendritic cells, NK cells, monocytes, MDSCs | 25 | 4-laser (405, 488, 561, 640 nm) Cytek Aurora | [96] |
| Fresh lysed whole human blood | |||
| CD4-, CD8-, γδ-T-cells, Treg, NKT-like; B-, NK-cells, DCs, monocytes. | 40 | Not stated, probably 5-laser (355, 405, 488, 561, 640 nm) Cytek Aurora | [27] |
| Platelets | 16 | 3-laser (405, 488, 640 nm) Cytek Northern Lights | [97] |
| Subpopulations of dendritic cells, NK cells, monocytes, MDSCs | 35 | 6-laser (320, 355, 405, 488, 561, and 637 nm) Sony ID7000 | [98] |
| T-cell and B-cell subpopulations | 34 | 6-laser (320, 355, 405, 488, 561, and 637 nm) Sony ID7000 | [98] |
| B-cells | 24 | BD FACS Canto II, Cytek Northern Lights (laser config is not stated) | [82] |
| 10 T-cell subpopulations, B-cells, progenitor cells, ILCs | 32 | 4-laser (405, 488, 561, and 640 nm) Cytek Aurora | [96] |
| Mice tissues | |||
| Mouse T-, B-, NK-cells, innate lymphoid and dendritic cells, monocytes, macrophages, basophils, neutrophils, eosinophils | 40 | 5-laser (355, 405, 488, 561, 640 nm) Cytek Aurora | [99] |
| B-cells, subpopulations of T-cells, pDCs, cDCs, macrophages, neutrophils, monocytes | 13 | 5-laser (355 nm, 405 nm, 488 nm, 561 nm, 637 nm) BD FACSymphony S6, 5- laser (355 nm, 405 nm, 488 nm, 561 nm, 637 nm) Sony ID7000, 5-laser (355, 405, 488, 561, 640 nm) Cytek Aurora | [100] |
| Characteristics | Bigfoot™ Spectral Cell Sorter | Cytek Aurora™ CS System | FP7000 Spectral Cell Sorter | BD FACSDiscover™ S8 Cell Sorter |
|---|---|---|---|---|
| Manufacturer | Invitrogen | Cytek Biosciences | Sony | BD Biosciences |
| Number of lasers and their wavelengths | 9 (349/405/445/488/532/561/594/640/785 nm) | 5 (355/405/488/ 561/640 nm) | Up to 6 (320/349/405/488/561/637 nm) | 5 (349/405/488/561/637 nm) |
| Number of colors in the panel | up to 60 | up to 40 | over 44 | up to 38 |
| Sorting possibilities | 1.5, 5, 15 and 50 mL tubes, 10× chips, plates (96-, 384- and 1536-well), PCR strips | 1.5, 5, 15 mL tubes, plates (96- and 384-well) | 1.5, 5, 15 and 50 mL tubes, plates (6-384-well) | 5 mL tubes, plates (96- and 384-well plates) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Astakhova, E.A.; Gubaeva, A.S.; Naumova, D.A.; Egorova, A.E.; Maznina, A.A.; Rybkina, I.G.; Osmanov, I.M.; Tabakov, D.V.; Mityaeva, O.N.; Volchkov, P.Y. Spectral Flow Cytometry: The Current State and Future of the Technology. Int. J. Mol. Sci. 2025, 26, 5911. https://doi.org/10.3390/ijms26125911
Astakhova EA, Gubaeva AS, Naumova DA, Egorova AE, Maznina AA, Rybkina IG, Osmanov IM, Tabakov DV, Mityaeva ON, Volchkov PY. Spectral Flow Cytometry: The Current State and Future of the Technology. International Journal of Molecular Sciences. 2025; 26(12):5911. https://doi.org/10.3390/ijms26125911
Chicago/Turabian StyleAstakhova, E. A., A. S. Gubaeva, D. A. Naumova, A. E. Egorova, A. A. Maznina, I. G. Rybkina, I. M. Osmanov, D. V. Tabakov, O. N. Mityaeva, and P. Yu. Volchkov. 2025. "Spectral Flow Cytometry: The Current State and Future of the Technology" International Journal of Molecular Sciences 26, no. 12: 5911. https://doi.org/10.3390/ijms26125911
APA StyleAstakhova, E. A., Gubaeva, A. S., Naumova, D. A., Egorova, A. E., Maznina, A. A., Rybkina, I. G., Osmanov, I. M., Tabakov, D. V., Mityaeva, O. N., & Volchkov, P. Y. (2025). Spectral Flow Cytometry: The Current State and Future of the Technology. International Journal of Molecular Sciences, 26(12), 5911. https://doi.org/10.3390/ijms26125911





