Identification of Proteins Associated with Ovarian Cancer Chemotherapy Resistance Using MALDI-MSI
Abstract
1. Introduction
2. Results
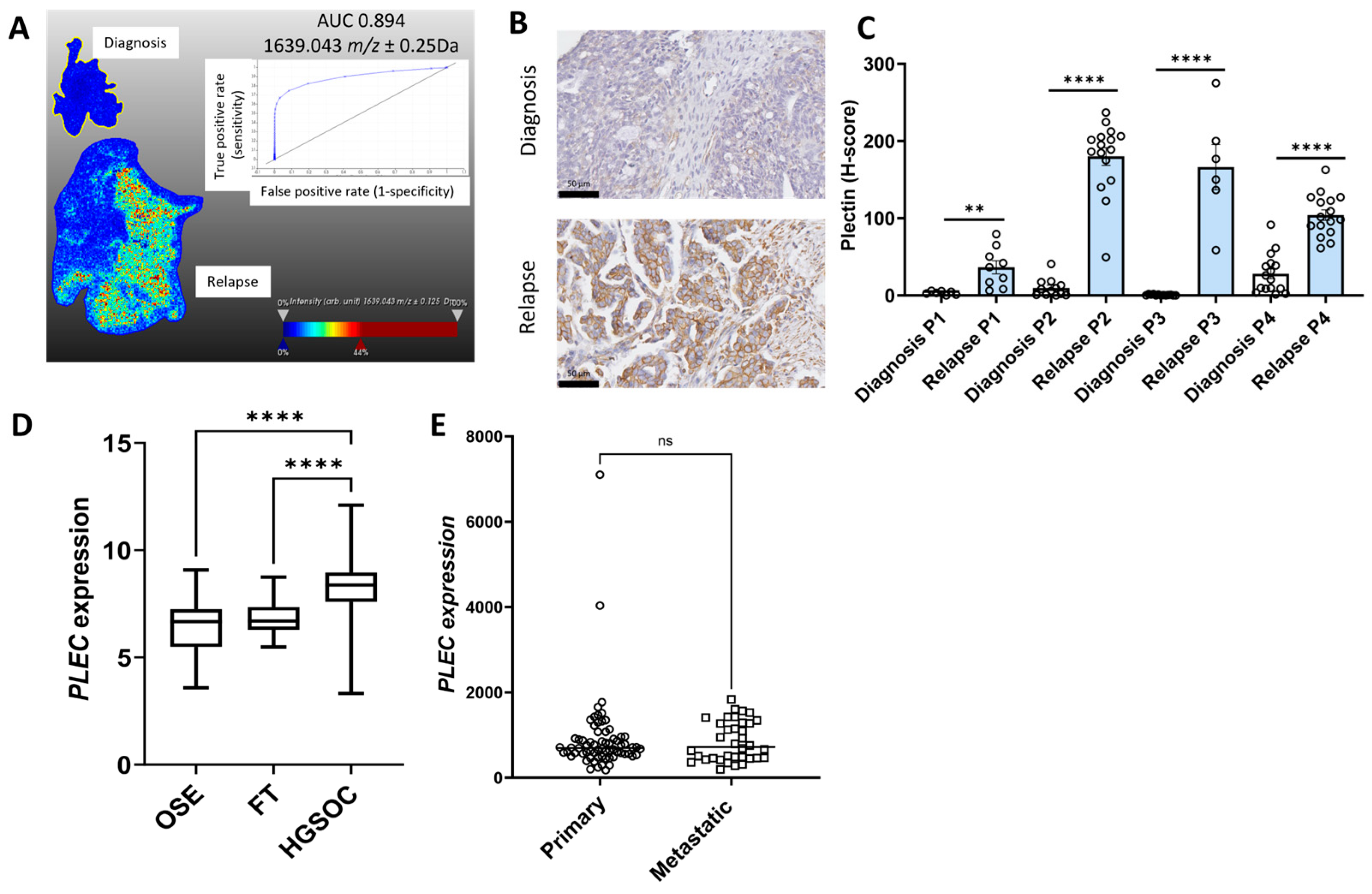
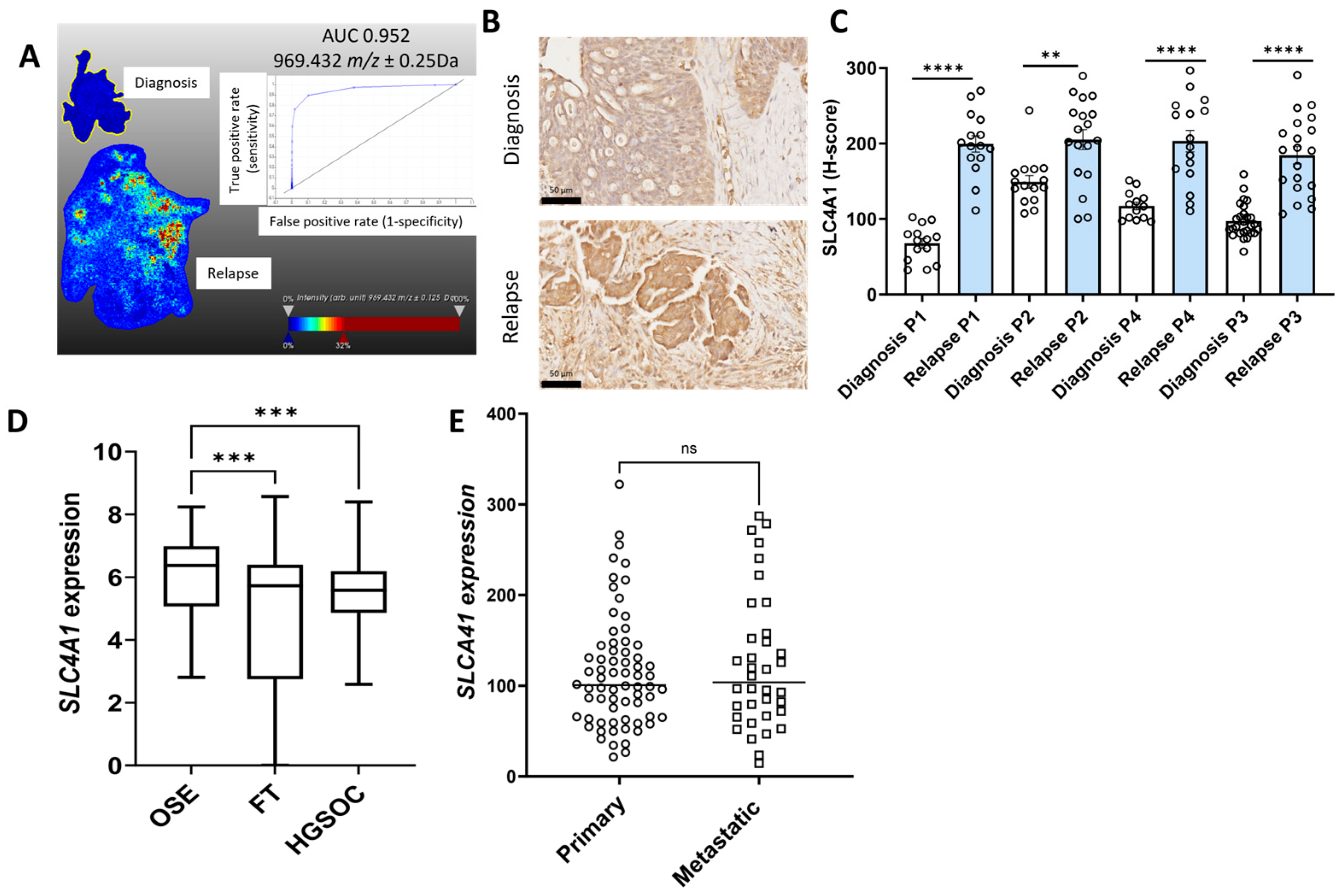
2.1. Identification of Proteins of Interest in Matching HGSOC Tissues at Diagnosis and Relapse Using MALDI-MSI
2.2. Validation of Protein of Interest Increased in Relapse Tissues Compared to Diagnosis Using IHC
2.3. Characterization of the Expression Proteins of Interest in Online Ovarian Cancer Databases
2.4. Relationship Between Proteins of Interest with HGSOC Patient Outcome
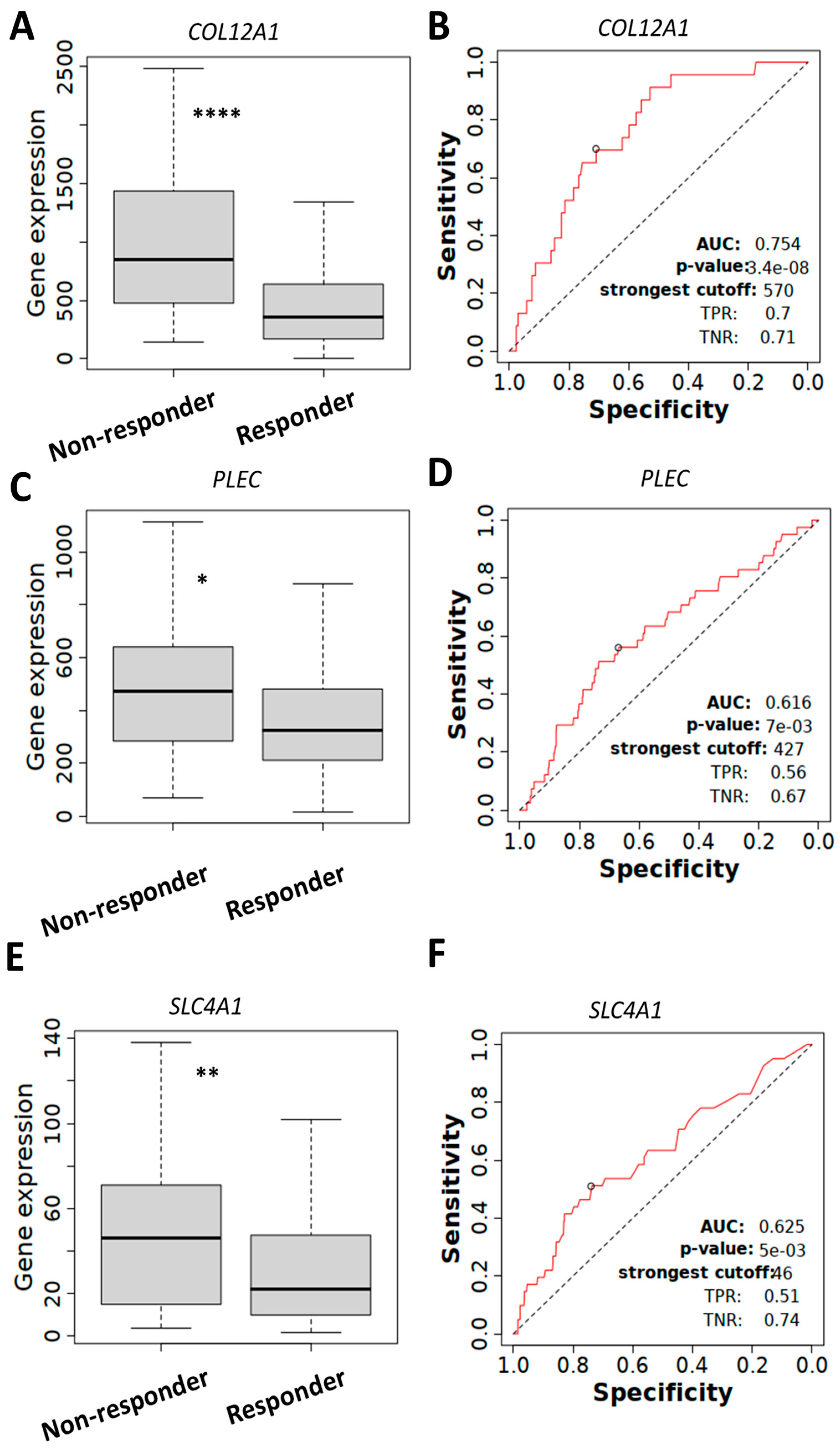
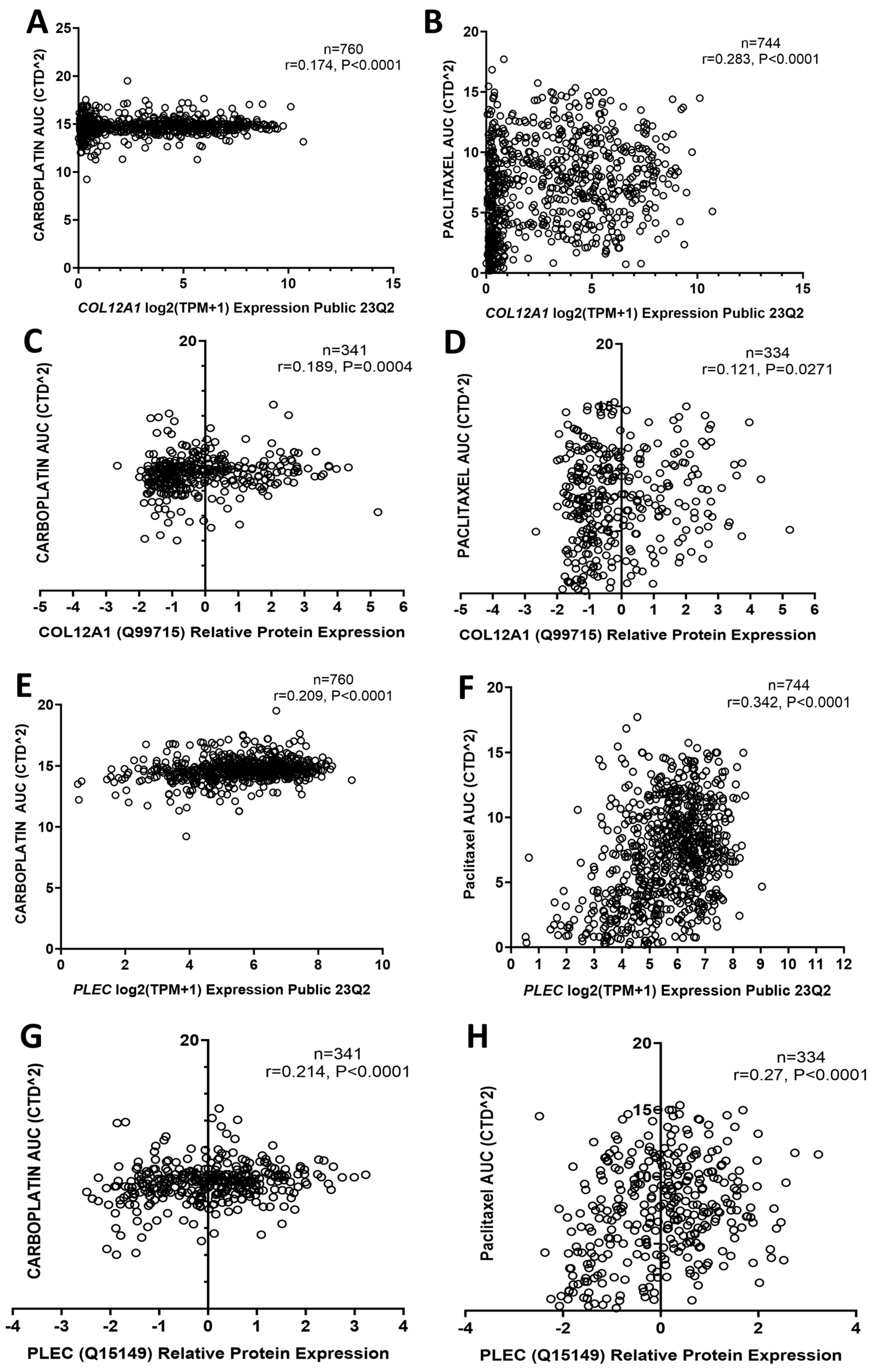
2.5. COL12A1, PLEC, and SLC4A1 Expressions Associated with Chemotherapy Resistance
3. Discussion
4. Materials and Methods
4.1. Patient Cohorts
4.2. MALDI-MSI Preparation and Acquisition
4.3. MALDI-MSI Data Analysis
4.4. Peptide Identification by Nanoflow Liquid Chromatography Tandem Mass Spectrometry (Nano-LC-MS/MS)
4.5. Matching the MALDI-MSI Peak Groups to the Nanoflow Liquid Chromatography Tandem Mass Spectrometry (Nano-LC-MS/MS)
4.6. Ovarian Cancer Online Databases
4.7. IHC
4.8. IHC Assessment
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Colombo, P.E.; Fabbro, M.; Theillet, C.; Bibeau, F.; Rouanet, P.; Ray-Coquard, I. Sensitivity and resistance to treatment in the primary management of epithelial ovarian cancer. Crit. Rev. Oncol. Hematol. 2014, 89, 207–216. [Google Scholar] [CrossRef]
- Havasi, A.; Cainap, S.S.; Havasi, A.T.; Cainap, C. Ovarian Cancer—Insights into Platinum Resistance and Overcoming It. Medicina 2023, 59, 544. [Google Scholar] [CrossRef]
- Salutari, V.; Giudice, E.; Lorusso, D. Maintenance therapy for newly and recurrent epithelial ovarian cancer: Current therapies and future perspectives. Curr. Opin. Obstet. Gynecol. 2024, 36, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Ly, A.; Longuespée, R.; Casadonte, R.; Wandernoth, P.; Schwamborn, K.; Bollwein, C.; Marsching, C.; Kriegsmann, K.; Hopf, C.; Weichert, W.; et al. Site-to-Site Reproducibility and Spatial Resolution in MALDI-MSI of Peptides from Formalin-Fixed Paraffin-Embedded Samples. Proteom. Clin. Appl. 2019, 13, e1800029. [Google Scholar] [CrossRef] [PubMed]
- Meding, S.; Martin, K.; Gustafsson, O.J.; Eddes, J.S.; Hack, S.; Oehler, M.K.; Hoffmann, P. Tryptic peptide reference data sets for MALDI imaging mass spectrometry on formalin-fixed ovarian cancer tissues. J. Proteome Res. 2013, 12, 308–315. [Google Scholar] [CrossRef]
- Aoki, Y.; Toyama, A.; Shimada, T.; Sugita, T.; Aoki, C.; Umino, Y.; Suzuki, A.; Aoki, D.; Daigo, Y.; Nakamura, Y.; et al. A novel method for analyzing formalin-fixed paraffin embedded (FFPE) tissue sections by mass spectrometry imaging. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2007, 83, 205–214. [Google Scholar] [CrossRef]
- Delcourt, V.; Franck, J.; Leblanc, E.; Narducci, F.; Robin, Y.-M.; Gimeno, J.-P.; Quanico, J.; Wisztorski, M.; Kobeissy, F.; Jacques, J.-F.; et al. Combined Mass Spectrometry Imaging and Top-down Microproteomics Reveals Evidence of a Hidden Proteome in Ovarian Cancer. eBioMedicine 2017, 21, 55–64. [Google Scholar] [CrossRef]
- Schwamborn, K.; Weirich, G.; Steiger, K.; Zimmermann, G.; Schmidmayr, M.; Weichert, W.; Caprioli, R.M. Discerning the Primary Carcinoma in Malignant Peritoneal and Pleural Effusions Using Imaging Mass Spectrometry-A Feasibility Study. Proteom. Clin. Appl. 2019, 13, e1800064. [Google Scholar] [CrossRef]
- Longuespée, R.; Gagnon, H.; Boyon, C.; Strupat, K.; Dauly, C.; Kerdraon, O.; Ighodaro, A.; Desmons, A.; Dupuis, J.; Wisztorski, M.; et al. Proteomic analyses of serous and endometrioid epithelial ovarian cancers—Cases studies—Molecular insights of a possible histological etiology of serous ovarian cancer. Proteom. Clin. Appl. 2013, 7, 337–354. [Google Scholar] [CrossRef]
- Klein, O.; Kanter, F.; Kulbe, H.; Jank, P.; Denkert, C.; Nebrich, G.; Schmitt, W.D.; Wu, Z.; Kunze, C.A.; Sehouli, J.; et al. MALDI-Imaging for Classification of Epithelial Ovarian Cancer Histotypes from a Tissue Microarray Using Machine Learning Methods. Proteom. Clin. Appl. 2019, 13, e1700181. [Google Scholar] [CrossRef] [PubMed]
- Kassuhn, W.; Klein, O.; Darb-Esfahani, S.; Lammert, H.; Handzik, S.; Taube, E.T.; Schmitt, W.D.; Keunecke, C.; Horst, D.; Dreher, F.; et al. Classification of Molecular Subtypes of High-Grade Serous Ovarian Cancer by MALDI-Imaging. Cancers 2021, 13, 1512. [Google Scholar] [CrossRef] [PubMed]
- Kulbe, H.; Klein, O.; Wu, Z.; Taube, E.T.; Kassuhn, W.; Horst, D.; Darb-Esfahani, S.; Jank, P.; Abobaker, S.; Ringel, F.; et al. Discovery of Prognostic Markers for Early-Stage High-Grade Serous Ovarian Cancer by Maldi-Imaging. Cancers 2020, 12, 2000. [Google Scholar] [CrossRef]
- Ma, X.; Botros, A.; Yun, S.R.; Park, E.Y.; Kim, O.; Park, S.; Pham, T.H.; Chen, R.; Palaniappan, M.; Matzuk, M.M.; et al. Ultrahigh resolution lipid mass spectrometry imaging of high-grade serous ovarian cancer mouse models. Front. Chem. 2023, 11, 1332816. [Google Scholar] [CrossRef]
- Everest-Dass, A.V.; Briggs, M.T.; Kaur, G.; Oehler, M.K.; Hoffmann, P.; Packer, N.H. N-glycan MALDI Imaging Mass Spectrometry on Formalin-Fixed Paraffin-Embedded Tissue Enables the Delineation of Ovarian Cancer Tissues. Mol. Cell. Proteom. 2016, 15, 3003–3016. [Google Scholar] [CrossRef]
- Briggs, M.T.; Condina, M.R.; Ho, Y.Y.; Everest-Dass, A.V.; Mittal, P.; Kaur, G.; Oehler, M.K.; Packer, N.H.; Hoffmann, P. MALDI Mass Spectrometry Imaging of Early- and Late-Stage Serous Ovarian Cancer Tissue Reveals Stage-Specific N-Glycans. Proteomics 2019, 19, e1800482. [Google Scholar] [CrossRef] [PubMed]
- Mittal, P.; Klingler-Hoffmann, M.; Arentz, G.; Winderbaum, L.; Lokman, N.A.; Zhang, C.; Anderson, L.; Scurry, J.; Leung, Y.; Stewart, C.J.; et al. Lymph node metastasis of primary endometrial cancers: Associated proteins revealed by MALDI imaging. Proteomics 2016, 16, 1793–1801. [Google Scholar] [CrossRef]
- Park, S.J.; Yoon, B.H.; Kim, S.K.; Kim, S.Y. GENT2: An updated gene expression database for normal and tumor tissues. BMC Med. Genom. 2019, 12, 101. [Google Scholar] [CrossRef]
- Ricciardelli, C.; Lokman, N.A.; Cheruvu, S.; Tan, I.A.; Ween, M.P.; Pyragius, C.E.; Ruszkiewicz, A.; Hoffmann, P.; Oehler, M.K. Transketolase is upregulated in metastatic peritoneal implants and promotes ovarian cancer cell proliferation. Clin. Exp. Metastasis 2015, 32, 441–455. [Google Scholar] [CrossRef]
- Januchowski, R.; Świerczewska, M.; Sterzyńska, K.; Wojtowicz, K.; Nowicki, M.; Zabel, M. Increased Expression of Several Collagen Genes is Associated with Drug Resistance in Ovarian Cancer Cell Lines. J. Cancer 2016, 7, 1295–1310. [Google Scholar] [CrossRef]
- Yan, Y.; Liang, Q.; Liu, Y.; Zhou, S.; Xu, Z. COL12A1 as a prognostic biomarker links immunotherapy response in breast cancer. Endocr. Relat. Cancer 2023, 30, e230012. [Google Scholar] [CrossRef] [PubMed]
- Li, S.L.; Ye, F.; Cai, W.J.; Hu, H.D.; Hu, P.; Ren, H.; Zhu, F.F.; Zhang, D.Z. Quantitative proteome analysis of multidrug resistance in human ovarian cancer cell line. J. Cell. Biochem. 2010, 109, 625–633. [Google Scholar] [CrossRef]
- Chudasama, D.; Bo, V.; Hall, M.; Anikin, V.; Jeyaneethi, J.; Gregory, J.; Pados, G.; Tucker, A.; Harvey, A.; Pink, R.; et al. Identification of cancer biomarkers of prognostic value using specific gene regulatory networks (GRN): A novel role of RAD51AP1 for ovarian and lung cancers. Carcinogenesis 2018, 39, 407–417. [Google Scholar] [CrossRef]
- Crespo-Bravo, M.; Hettich, A.; Thorlacius-Ussing, J.; Cox, T.R.; Karsdal, M.A.; Willumsen, N. Type XII collagen is elevated in serum from patients with solid tumors: A non-invasive biomarker of activated fibroblasts. Clin. Exp. Med. 2024, 24, 166. [Google Scholar] [CrossRef] [PubMed]
- Papanicolaou, M.; Parker, A.L.; Yam, M.; Filipe, E.C.; Wu, S.Z.; Chitty, J.L.; Wyllie, K.; Tran, E.; Mok, E.; Nadalini, A.; et al. Temporal profiling of the breast tumour microenvironment reveals collagen XII as a driver of metastasis. Nat. Commun. 2022, 13, 4587. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Z.; Li, J.; Song, S.; Wang, J.; Cai, W.; Hu, W.; Ji, J.; Zhu, Z.; Zang, L.; Yan, R.; et al. A positive feedback between IDO1 metabolite and COL12A1 via MAPK pathway to promote gastric cancer metastasis. J. Exp. Clin. Cancer Res. 2019, 38, 314. [Google Scholar] [CrossRef]
- Xiong, X.; Lai, X.; Zhang, J.; Meng, Q.; Wang, P.; Qin, S.; Liu, W.; Wang, Y.; Yao, Z.; Wang, D.; et al. FBP1 knockdown decreases ovarian cancer formation and cisplatin resistance through EZH2-mediated H3K27me3. Biosci. Rep. 2022, 42, BSR20221002. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, R.; Li, E.; Yan, R.; Ma, B.; Ma, Q. Pan-Cancer Transcriptome and Immune Infiltration Analyses Reveal the Oncogenic Role of Far Upstream Element-Binding Protein 1 (FUBP1). Front. Mol. Biosci. 2022, 9, 794715. [Google Scholar] [CrossRef]
- Xiong, X.; Zhang, J.; Hua, X.; Cao, W.; Qin, S.; Dai, L.; Liu, W.; Zhang, Z.; Li, X.; Liu, Z. FBP1 promotes ovarian cancer development through the acceleration of cell cycle transition and metastasis. Oncol. Lett. 2018, 16, 1682–1688. [Google Scholar] [CrossRef]
- Zhang, J.; Xiong, X.; Hua, X.; Cao, W.; Qin, S.; Dai, L.; Liang, P.; Zhang, H.; Liu, Z. Knockdown of FUSE binding protein 1 enhances the sensitivity of epithelial ovarian cancer cells to carboplatin. Oncol. Lett. 2017, 14, 5819–5824. [Google Scholar] [CrossRef][Green Version]
- Zhang, F.; Xiong, Q.; Wang, M.; Cao, X.; Zhou, C. FUBP1 in human cancer: Characteristics, functions, and potential applications. Transl. Oncol. 2024, 48, 102066. [Google Scholar] [CrossRef] [PubMed]
- Perez, S.M.; Brinton, L.T.; Kelly, K.A. Plectin in Cancer: From Biomarker to Therapeutic Target. Cells 2021, 10, 2246. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, W.; Luo, Q.; Song, G. Plectin: Dual Participation in Tumor Progression. Biomolecules 2024, 14, 1050. [Google Scholar] [CrossRef] [PubMed]
- Wesley, T.; Berzins, S.; Kannourakis, G.; Ahmed, N. The attributes of plakins in cancer and disease: Perspectives on ovarian cancer progression, chemoresistance and recurrence. Cell Commun. Signal. 2021, 19, 55. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Greening, D.; Samardzija, C.; Escalona, R.M.; Chen, M.; Findlay, J.K.; Kannourakis, G. Unique proteome signature of post-chemotherapy ovarian cancer ascites-derived tumor cells. Sci. Rep. 2016, 6, 30061. [Google Scholar] [CrossRef]
- Wesley, T.; Escalona, R.M.; Kannourakis, G.; Ahmed, N. Plakin Expression in Serous Epithelial Ovarian Cancer Has the Potential to Impede Metastatic Spread and Epithelial–Mesenchymal Transition: A Comparative Expression Analysis of Immunohistochemical and In Silico Datasets. Cancers 2024, 16, 4087. [Google Scholar] [CrossRef]
- Perez, S.M.; Dimastromatteo, J.; Landen, C.N., Jr.; Kelly, K.A. A Novel Monoclonal Antibody Targeting Cancer-Specific Plectin Has Potent Antitumor Activity in Ovarian Cancer. Cells 2021, 10, 2218. [Google Scholar] [CrossRef]
- Dasa, S.S.K.; Diakova, G.; Suzuki, R.; Mills, A.M.; Gutknecht, M.F.; Klibanov, A.L.; Slack-Davis, J.K.; Kelly, K.A. Plectin-targeted liposomes enhance the therapeutic efficacy of a PARP inhibitor in the treatment of ovarian cancer. Theranostics 2018, 8, 2782–2798. [Google Scholar] [CrossRef]
- Gorbatenko, A.; Olesen, C.W.; Boedtkjer, E.; Pedersen, S.F. Regulation and roles of bicarbonate transport in cancer. Front. Physiol. 2014, 5, 130. [Google Scholar] [CrossRef]
- Shen, W.W.; Wu, J.; Cai, L.; Liu, B.Y.; Gao, Y.; Chen, G.Q.; Fu, G.H. Expression of anion exchanger 1 sequestrates p16 in the cytoplasm in gastric and colonic adenocarcinoma. Neoplasia 2007, 9, 812–819. [Google Scholar] [CrossRef]
- Schilling, V.; Beyerlein, P.; Chien, J. A Bioinformatics Analysis of Ovarian Cancer Data Using Machine Learning. Algorithms 2023, 16, 330. [Google Scholar] [CrossRef]
- Hao, S.; Meng, Q.; Sun, H.; Li, Y.; Li, Y.; Gu, L.; Liu, B.; Zhang, Y.; Zhou, H.; Xu, Z.; et al. The role of transketolase in human cancer progression and therapy. Biomed. Pharmacother. 2022, 154, 113607. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Ye, M.; Zhou, J.; Zhu, X. Prognostic values of transketolase family genes in ovarian cancer. Oncol. Lett. 2019, 18, 4845–4857. [Google Scholar] [CrossRef]
- Bery, A.; Leung, F.; Smith, C.R.; Diamandis, E.P.; Kulasingam, V. Deciphering the ovarian cancer ascites fluid peptidome. Clin. Proteom. 2014, 11, 13. [Google Scholar] [CrossRef]
- Marcišauskas, S.; Ulfenborg, B.; Kristjansdottir, B.; Waldemarson, S.; Sundfeldt, K. Univariate and classification analysis reveals potential diagnostic biomarkers for early stage ovarian cancer Type 1 and Type 2. J. Proteom. 2019, 196, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Giacomini, I.; Ragazzi, E.; Pasut, G.; Montopoli, M. The Pentose Phosphate Pathway and Its Involvement in Cisplatin Resistance. Int. J. Mol. Sci. 2020, 21, 937. [Google Scholar] [CrossRef]
- Yi, H.; Zheng, X.; Song, J.; Shen, R.; Su, Y.; Lin, D. Exosomes mediated pentose phosphate pathway in ovarian cancer metastasis: A proteomics analysis. Int. J. Clin. Exp. Pathol. 2015, 8, 15719–15728. [Google Scholar]
- Gustafsson, J.O.R.; Eddes, J.S.; Meding, S.; Koudelka, T.; Oehler, M.K.; McColl, S.R.; Hoffmann, P. Internal calibrants allow high accuracy peptide matching between MALDI imaging MS and LC-MS/MS. J. Proteom. 2012, 75, 5093–5105. [Google Scholar] [CrossRef]
- Acland, M.; Mittal, P.; Arentz, G.; Whitehead, F.; Hoffmann, P.; Klingler-Hoffmann, M.; Oehler, M.K. A Protocol for the Acquisition of Comprehensive Proteomics Data from Single Cases Using Formalin-Fixed Paraffin Embedded Sections. Methods Protoc. 2022, 5, 57. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, T.; Zhang, Z.; Payne, S.H.; Zhang, B.; McDermott, J.E.; Zhou, J.Y.; Petyuk, V.A.; Chen, L.; Ray, D.; et al. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell 2016, 166, 755–765. [Google Scholar] [CrossRef]
- Thiele, H.; Heldmann, S.; Trede, D.; Strehlow, J.; Wirtz, S.; Dreher, W.; Berger, J.; Oetjen, J.; Kobarg, J.H.; Fischer, B.; et al. 2D and 3D MALDI-imaging: Conceptual strategies for visualization and data mining. Biochim. Biophys. Acta 2014, 1844, 117–137. [Google Scholar] [CrossRef] [PubMed]
- Price, Z.K.; Lokman, N.A.; Yoshihara, M.; Kajiyama, H.; Oehler, M.K.; Ricciardelli, C. Disabled-2 (DAB2): A Key Regulator of Anti- and Pro-Tumorigenic Pathways. Int. J. Mol. Sci. 2022, 24, 696. [Google Scholar] [CrossRef] [PubMed]
- Fekete, J.T.; Gyorffy, B. ROCplot.org: Validating predictive biomarkers of chemotherapy/hormonal therapy/anti-HER2 therapy using transcriptomic data of 3,104 breast cancer patients. Int. J. Cancer 2019, 145, 3140–3151. [Google Scholar] [CrossRef] [PubMed]
- Győrffy, B. Discovery and ranking of the most robust prognostic biomarkers in serous ovarian cancer. Geroscience 2023, 45, 1889–1898. [Google Scholar] [CrossRef]
- Lokman, N.A.; Elder, A.S.; Ween, M.P.; Pyragius, C.E.; Hoffmann, P.; Oehler, M.K.; Ricciardelli, C. Annexin A2 is regulated by ovarian cancer-peritoneal cell interactions and promotes metastasis. Oncotarget 2013, 4, 1199–1211. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]




| Gene Name | Progression-Free Survival (PFS) | Post-Progression Survival (PPS) | Overall Survival (OS) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | HR | 95% CI | p-Value | |
| COL12A1 | 1.76 | 1.38–2.24 | 3.7 × 10−6 | 1.23 | 0.93–1.63 | 0.14 | 1.38 | 1.05–1.83 | 0.022 |
| FUBP1 | 1.43 | 1.22–1.68 | 1.1 × 10−5 | 1.19 | 0.98–1.45 | 0.082 | 1.16 | 0.97–1.4 | 0.11 |
| PLEC | 1.15 | 0.97–1.36 | 0.1 | 1.3 | 1.07–1.58 | 0.0071 | 1.14 | 0.95–1.37 | 0.15 |
| SLC4A1 | 0.83 | 0.65–1.07 | 0.15 | 0.87 | 0.65–1.15 | 0.32 | 0.84 | 0.63–1.12 | 0.23 |
| TKT | 1.28 | 1.02–1.62 | 0.036 | 1.26 | 0.94–1.68 | 0.12 | 1.4 | 1.06–1.86 | 0.018 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noye, T.M.; Mittal, P.; Price, Z.K.; Fewster, A.; Williams, G.; Pukala, T.L.; Klingler-Hoffmann, M.; Hoffmann, P.; Oehler, M.K.; Lokman, N.A.; et al. Identification of Proteins Associated with Ovarian Cancer Chemotherapy Resistance Using MALDI-MSI. Int. J. Mol. Sci. 2025, 26, 5893. https://doi.org/10.3390/ijms26125893
Noye TM, Mittal P, Price ZK, Fewster A, Williams G, Pukala TL, Klingler-Hoffmann M, Hoffmann P, Oehler MK, Lokman NA, et al. Identification of Proteins Associated with Ovarian Cancer Chemotherapy Resistance Using MALDI-MSI. International Journal of Molecular Sciences. 2025; 26(12):5893. https://doi.org/10.3390/ijms26125893
Chicago/Turabian StyleNoye, Tannith M., Parul Mittal, Zoe K. Price, Annie Fewster, Georgia Williams, Tara L. Pukala, Manuela Klingler-Hoffmann, Peter Hoffmann, Martin K. Oehler, Noor A. Lokman, and et al. 2025. "Identification of Proteins Associated with Ovarian Cancer Chemotherapy Resistance Using MALDI-MSI" International Journal of Molecular Sciences 26, no. 12: 5893. https://doi.org/10.3390/ijms26125893
APA StyleNoye, T. M., Mittal, P., Price, Z. K., Fewster, A., Williams, G., Pukala, T. L., Klingler-Hoffmann, M., Hoffmann, P., Oehler, M. K., Lokman, N. A., & Ricciardelli, C. (2025). Identification of Proteins Associated with Ovarian Cancer Chemotherapy Resistance Using MALDI-MSI. International Journal of Molecular Sciences, 26(12), 5893. https://doi.org/10.3390/ijms26125893







