RNA Polymerase III-Transcribed RNAs in Health and Disease: Mechanisms, Dysfunction, and Future Directions
Abstract
1. Introduction
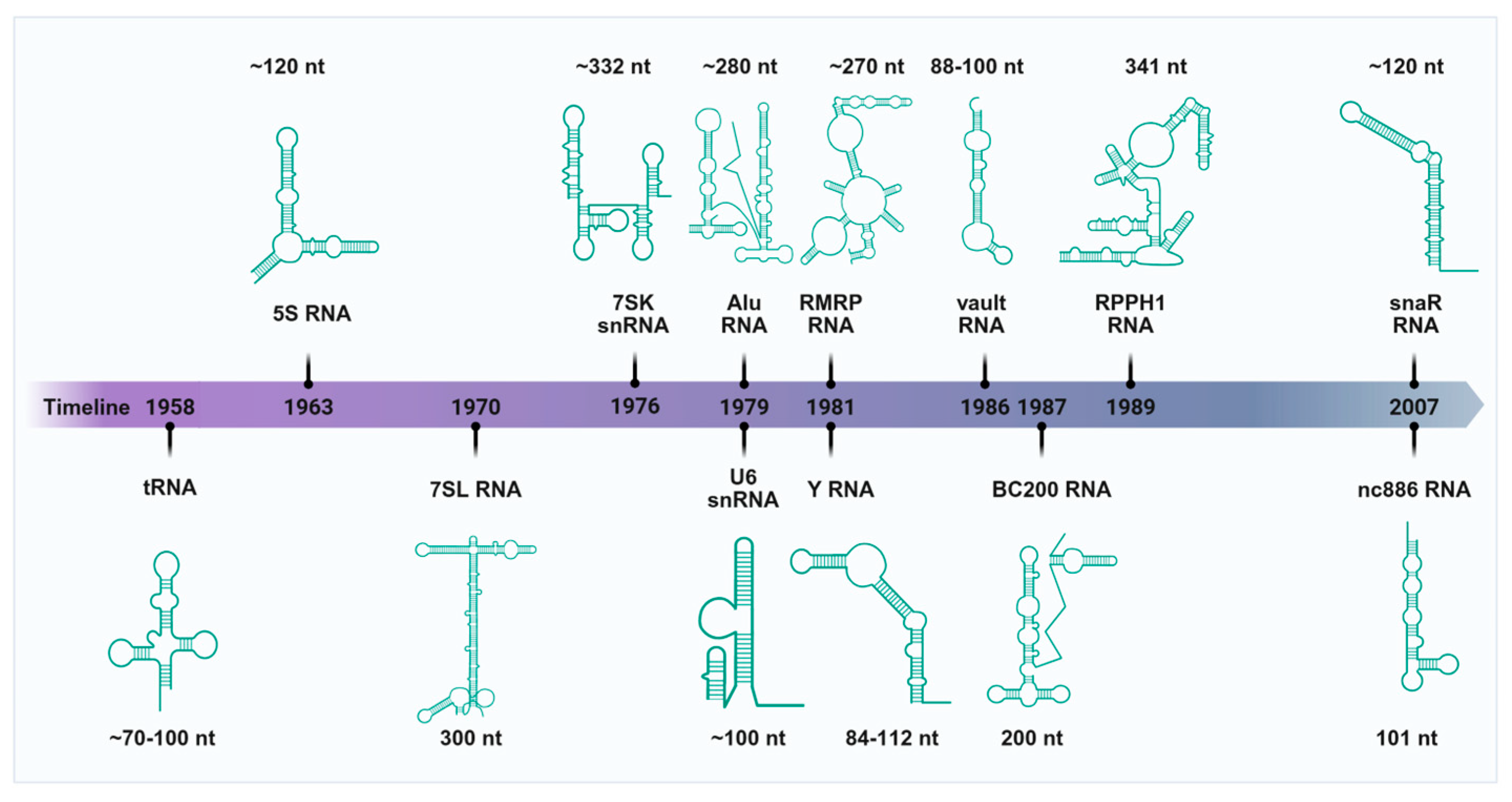
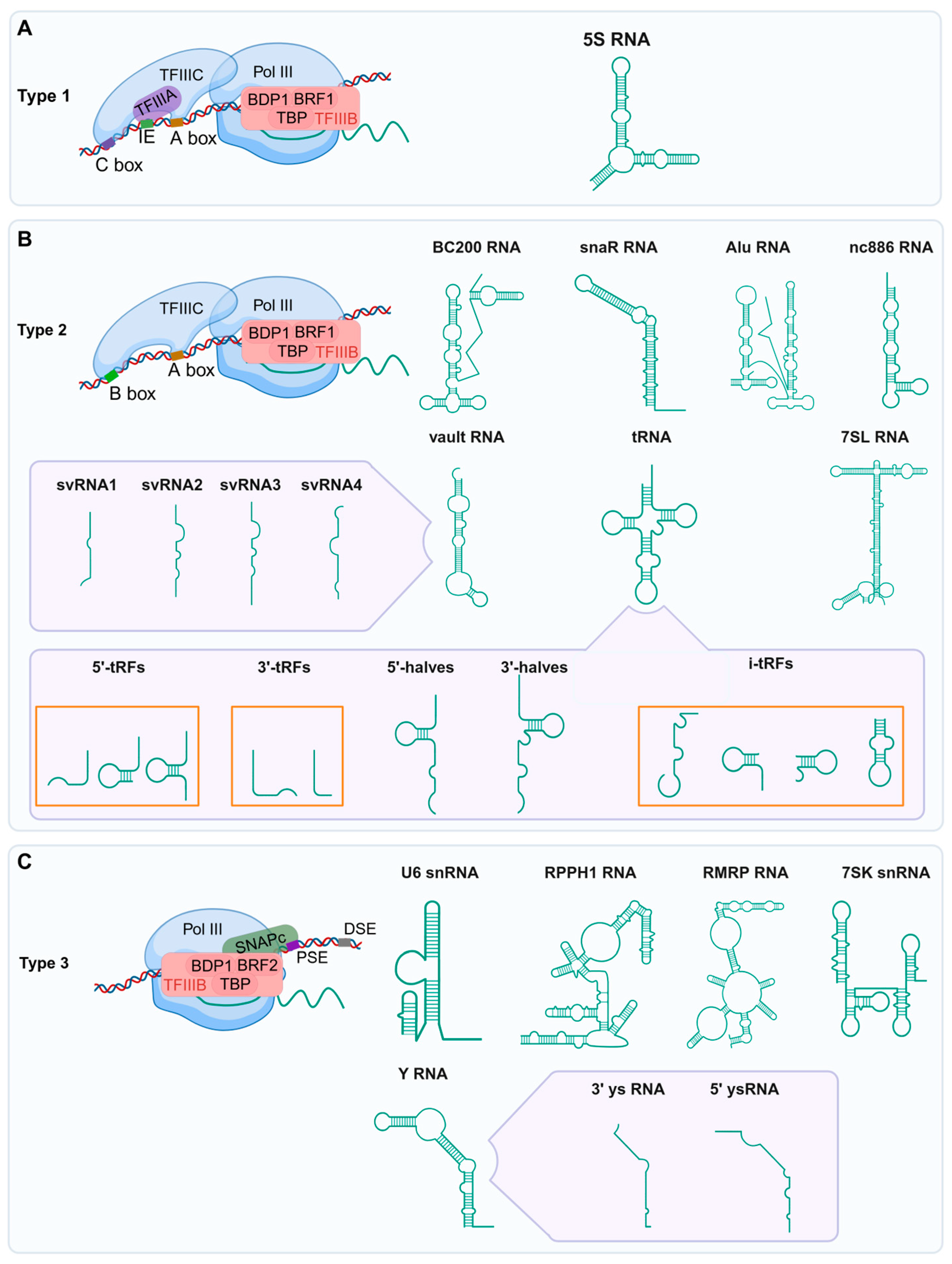
2. RNAs Transcribed by Pol III
2.1. tRNAs and tsRNAs
2.2. 5S rRNA
2.3. U6 snRNA
2.4. 7SK snRNA
2.5. 7SL RNA and Its Evolutionary Derivatives (Alu, BC200, and snaR RNAs)
2.6. RPPH1 and RMRP RNAs
2.7. Y RNA and Y RNA-Derived Small RNAs (ysRNAs)
2.8. Vault RNA (vtRNA) and vtRNA-Associated Small RNAs (vtsRNAs)
2.9. nc886
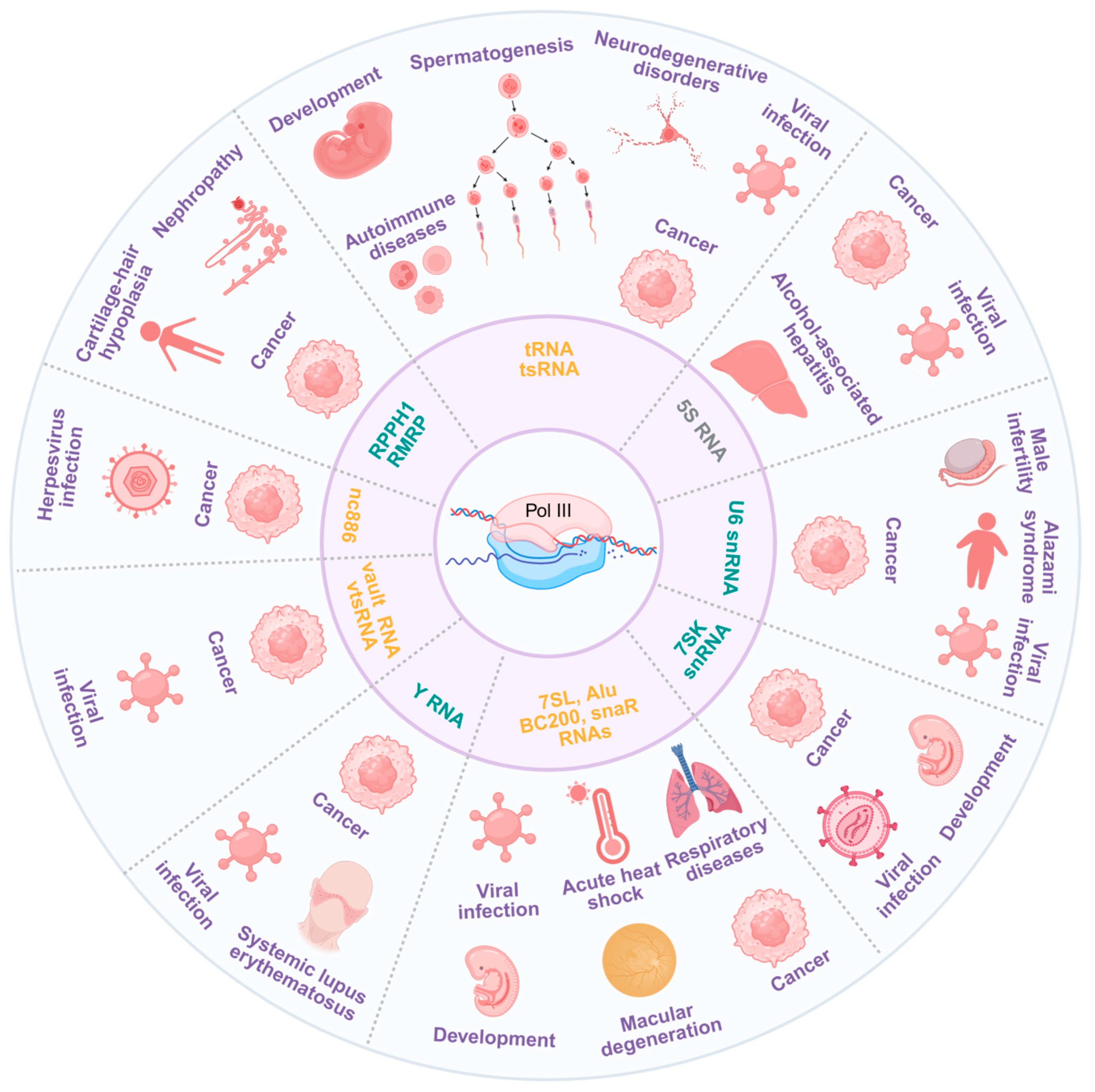
3. Diseases Associated with Pol III Transcription
3.1. Cancer
3.2. Viral Infection
3.3. Other Diseases
| Identity of Pol III Transcriptome | Biological Mechanism | Disease Type | Ref. |
|---|---|---|---|
| tRNA | METTL1-mediated tRNA m7G modification promotes leukaemogenesis of AML via tRNA regulated translational control. | Acute myeloid leukemia | [105] |
| tRNA | METTL1/WDR4-mediated tRNA m7G modification and mRNA translation control promote oncogenesis. | Oncogenesis | [106] |
| tRNA | Amyloid pathology reduces ELP3 expression and tRNA modifications leading to impaired proteostasis. | Alzheimer’s disease | [131] |
| tRNA | tRNA methyltransferase TRMT10A catalyzes N1-methylguanosine (m1G) at position 9 of tRNAs, with its deletion in mice impairing brain function and underscoring tRNA modification’s role in neurodevelopment. | Brain dysfunction | [132] |
| tsRNA | m7G-modified tsRNA-LysTTT catalyzed by METTL1 enhances bladder cancer malignancy. | Bladder cancer malignancy | [109] |
| tsRNA | tRF-23-Q99P9P9NDD promotes gastric cancer progression by modulating lipid metabolism and ferroptosis. | Gastric cancer | [111] |
| tsRNA | HCETSR (tRNA-Glu/TTC-derived) suppresses hepatocellular carcinoma via the SPTBN1/catenin axis. | Hepatocellular carcinoma | [110] |
| tsRNA | tRF-33-P4R8YP9LON4VDP inhibits gastric cancer by regulating STAT3 signaling in an AGO2-dependent manner. | Gastric cancer | [112] |
| tsRNA | 5′tRNA derivative tRF-Tyr competitively binds hnRNPD to modulate the c-Myc/Bcl2/Bax pathway, suppressing gastric cancer. | Gastric cancer | [113] |
| tsRNA | 3′tRF-AlaAGC activates NF-κB signaling via TRADD interaction to drive malignancy and macrophage M2 polarization. | Breast cancer | [114] |
| tsRNA | 3′-pre-tRNA-derived tRF-1-Ser enhances proliferation and stemness by inhibiting MBNL1. | Breast cancer | [115] |
| tsRNA | tsRNA-GlyGCC promotes colorectal cancer progression and 5-FU resistance through SPIB regulation. | Colorectal cancer | [116] |
| tsRNA | tRF-Ala-AGC-3-M8 binds to the EPHA7 3′ UTR region and inhibits EPHA7 translation to attenuate neuroinflammation and neuronal damage in AD patients | Alzheimer’s disease | [135] |
| tsRNA | tsRNAs (rno-tsr007330) modulates myocardial fibrosis via NAT10-mediated EGR3 mRNA acetylation. | Neurodegenerative conditions | [136] |
| tsRNA | tRNA-Cys-5-0007 attenuates ocular angiogenesis and inflammation by targeting VEGFA and TGF-β1. | Neurodegenerative conditions | [137] |
| tsRNA | tRF-Glu-CTC exacerbates neointimal hyperplasia after vascular injury and drives neovascular age-related macular degeneration. | Macular degeneration | [138,139] |
| tsRNA | tRF-His-GTG-1 enhances neutrophil extracellular trap formation and interferon-alpha production via extracellular vesicles in systemic lupus erythematosus. | Systemic lupus erythematosus | [140] |
| tsRNA | tsRNA-Gln-i-0095 suppresses neuroinflammation by downregulating NFIA and TGFBR2 through miRNA-like mechanisms. | Neuroinflammation | [141] |
| tsRNA | Angiogenin-mediated tRNA cleavage generates stress-responsive tsRNAs, while epididymal RNase homologs (RNase 9-12) maintain murine fertility through controlled tRNA processing under physiological conditions. | Infertility | [142] |
| vtRNA | TRIM21 modulates stability of pro-survival, non-coding RNA vtRNA1-1 in human hepatocellular carcinoma cells. | Hepatocellular carcinoma | [117] |
| RPPH1 | RPPH1 enhances breast cancer progression by stabilizing m6A-modified FGFR2 mRNA via IGF2BP2, activating PI3K/AKT signaling. | Breast cancer | [87] |
| RMRP | RMRP accelerates C through the miR-580-3p/ATP13A3 axis. | Esophageal squamous cell carcinoma | [118] |
| RMRP | RMRP promotes ovarian cancer invasion via RAB31-dependent MMP secretion. | Ovarian cancer | [119] |
| RMRP | LncRNA RMRP promotes chondrocyte injury by regulating the FOXC1/RBP4 axis. | Cartilage-hair hypoplasia syndrome | [148] |
| RMRP | RMRP variants inhibit the cell cycle checkpoints pathway in cartilage–hair hypoplasia. | Cartilage-hair hypoplasia syndrome | [149] |
| RMRP | The RMRP gene n.197C>T mutation causes cartilage–hair hypoplasia syndrome. | Cartilage-hair hypoplasia syndrome | [151] |
| RMRP | RMRP accelerates ligamentum flavum hypertrophy by regulating GSDMD-mediated pyroptosis through Gli1 SUMOylation. | Hypertrophy of ligamentum flavum | [154] |
| 7SK snRNA | The m6A-modified 7SK snRNA regulates Pol II transcription in non-small cell lung cancer through P-TEFb. | Lung cancer | [120] |
| Alu RNA | Alu RNA induces epithelial-to-mesenchymal transition in colorectal cancer via NLRP3 inflammasome activation and IL-1β release. | Colorectal cancer | [122] |
| BC200 | BC200 promotes EBV-associated nasopharyngeal carcinoma by sequestering miR-6834-5p to upregulate thymidylate synthase. | EBV-associated nasopharyngeal carcinoma | [123] |
| BC200 | BC200 RNA stabilizes EIF4A3 to modulate viral and host gene expression, suggesting a dual role in maintaining viral latency and cellular homeostasis. | Epstein–Barr virus infection | [129] |
| BC200 | BC200 overexpression in asthma patients mediates inflammatory responses, linking Pol III RNAs to airway pathology. | Asthma | [146] |
| nc886 | nc886 acts as a tumor suppressor by modulating immune responses in prostate cancer. | Prostate cancer | [125] |
| nc886 | TGF-β-induced CpG demethylation reactivates nc886 to drive aggressive ovarian cancer progression. | Ovarian cancer | [126] |
| nc886 | The DUSP11-regulated nc886 represses interferon-stimulated genes to suppress antiviral responses, thereby creating a permissive environment for viral replication. | Kaposi’s sarcoma-associated herpesvirus infection | [130] |
| U6 snRNA | U6 snRNA translocates from the nucleus into extracellular vesicles, facilitating viral dissemination through intercellular communication. | HIV infection | [127] |
| U6 snRNA | THUMPD2-catalyzed N2-methylation of U6 snRNA regulates retinal integrity through pre-mRNA splicing control. | Age-related macular degeneration | [143] |
| 7SL RNA | The ADF-1L protein, derived from the PIF/pioneer transposon, upregulates 7SL RNA expression to bolster host innate immunity against pathogens, illustrating a counteractive host defense mechanism. | Virus infection | [128] |
| 7SL RNA | 7SL RNA and signal recognition particle orchestrate a global cellular response to acute thermal stress. | Acute thermal stress | [145] |
| 5S rRNA | 5S rRNA pseudogene transcripts are associated with interferon production and inflammatory responses in alcohol-associated hepatitis. | Alcohol-associated hepatitis | [147] |
4. Diagnostic and Therapeutic Strategies for Pol III Transcription-Related Diseases
4.1. Therapeutic Interventions
4.2. Diagnostic Biomarkers
| Identity of Pol III Transcriptome | Type | Source | Expression Level | Diagnostic Type of Disease | Ref. |
|---|---|---|---|---|---|
| tsRNA | tsRNA-Gly-CCC-2, tsRNA-Gly-GCC-1, and tsRNA-Lys-CTT-2-M2 | serum | up | tuberculosis | [185] |
| tsRNA | tRF-22-RNLNK88KL, tRF-27-Z3M8ZLSSXUL, and tRF-32-0668K87SERM4P | tissues and plasma | up | colorectal cancer | [186] |
| tsRNA | tRF-24-6VR8K09LE9 | serum | down | gastric cancer | [164] |
| tsRNA | tRF-31-PNR8YP9LON4VD, tRF-30-MIF91SS2P4FI, and tRF-30-IK9NJ4S2I7L7 | serum | up | gastric cancer | [166] |
| tsRNA | tRF-17-18VBY9M | tissues and serum | up | gastric cancer | [167] |
| tsRNA | has-tsr013526 | serum | up | gastric cancer | [168] |
| tsRNA | tiRNA-Gly-GCC-001 | serum | up | major depressive disorder | [173] |
| tsRNA | has-tsr011468 | tissues and serum | Up | lung adenocarcinoma | [187] |
| tsRNA | 5′-tRNA-Glu-TTC-9-1_L30, 5′-tRNA-Val-CAC-3-1_L30, and 5′-M-tRNA-Gln-TTG-3-3_L30 |
serum and semen | up | prostate cancer | [188] |
| tsRNA | tRF-1:28-chrM.Ser-TGA and tiRNA-1:34-Glu-CTC-1-M2 | plasma | up | bladder cancer | [189] |
| tsRNA | tRF-23-R9J89O9N9 | serum | up | hepatocellular carcinoma | [169] |
| tsRNA | tiRNA-Gln-CTG | plasma | down | pre-eclampsia | [174] |
| tsRNA | tRF-33-RZYQHQ9M739P0J | tissues and serum | up | hepatocellular carcinoma | [170] |
| tsRNA | tsRNA-Thr-5-0015 | serum | up | hepatocellular carcinoma | [171] |
| tsRNA | tRF-3a-Pro | serum | up | hepatocellular carcinoma | [172] |
| tsRNA | 5′tiRNA-35-PheGAA-8, tRF3-28-PheGAA-1, tRF3b-PheGAA-6, mt-tRF3-19-ArgTCG, mt-tRF3-20-ArgTCG, and mt-tRF3-21-ArgTCG | peripheral blood mononuclear cells | up (5′tiRNA-35-PheGAA-8, tRF3-28-PheGAA-1, and tRF3b-PheGAA-6)/down (mt-tRF3-19-ArgTCG, mt-tRF3-20-ArgTCG, and mt-tRF3-21-ArgTCG) | nonproliferative diabetic retinopathy | [175] |
| tsRNA | tsRNA-49-73-Glu-CTC | serum | up | non-small cell lung cancer | [190] |
| tsRNA | i-tRF-AsnGTT | serum | down | gastric cancer | [165] |
| vtRNA | vtRNA1-1 | serum | up | hematological disorders | [181] |
| RPPH1 and RMRP RNAs | RPPH1 and RMRP | serum | up | gastric cancer | [176] |
| RPPH1 RNA | RPPH1 | plasma/leukocytes | up (plasma)/down (leukocytes) | pre-eclampsia | [183] |
| RMRP RNA | RMRP | urine | up | bladder cancer | [177] |
| RMRP RNA | RMRP | serum | up | coronary artery disease | [178] |
| RMRP RNA | RMRP | peripheral blood mononuclear cells | up | bipolar disorder | [180] |
| RMRP RNA | RMRP | serum | up | acute coronary syndrome | [179] |
| BC200 | BCYRN1 | serum | up | bladder cancer | [182] |
| BC200 | BCYRN1 | serum | up | multiple sclerosis | [184] |
| Alu RNA | Alu | exosomes in serum | up | colorectal cancer | [122] |
| Y RNA | Y RNA | plasma | up | colorectal cancer | [191] |
5. Conclusions and Further Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Paule, M.R.; White, R.J. Survey and summary: Transcription by RNA polymerases I and III. Nucleic Acids Res. 2000, 28, 1283–1298. [Google Scholar] [CrossRef]
- Girbig, M.; Misiaszek, A.D.; Müller, C.W. Structural insights into nuclear transcription by eukaryotic DNA-dependent RNA polymerases. Nat. Rev. Mol. Cell Biol. 2022, 23, 603–622. [Google Scholar] [CrossRef]
- Hoagland, M.B.; Stephenson, M.L.; Scott, J.F.; Hecht, L.I.; Zamecnik, P.C. A soluble ribonucleic acid intermediate in protein synthesis. J. Biol. Chem. 1958, 231, 241–257. [Google Scholar] [CrossRef] [PubMed]
- Rosset, R.; Monier, R. Apropos of the presence of weak molecular weight RNA in the ribosomes of Escherichia coli. Biochim. Biophys. Acta 1963, 68, 653–656. [Google Scholar] [CrossRef] [PubMed]
- Lerner, M.R.; Steitz, J.A. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc. Natl. Acad. Sci. USA 1979, 76, 5495–5499. [Google Scholar] [CrossRef] [PubMed]
- Zieve, G.; Penman, S. Small RNA species of the HeLa cell: Metabolism and subcellular localization. Cell 1976, 8, 19–31. [Google Scholar] [CrossRef]
- Bishop, J.M.; Levinson, W.E.; Sullivan, D.; Fanshier, L.; Quintrell, N.; Jackson, J. The low molecular weight RNAs of Rous sarcoma virus. II. The 7 S RNA. Virology 1970, 42, 927–937. [Google Scholar] [CrossRef]
- Reddy, R.; Li, W.Y.; Henning, D.; Choi, Y.C.; Nohga, K.; Busch, H. Characterization and subcellular localization of 7-8 S RNAs of Novikoff hepatoma. J. Biol. Chem. 1981, 256, 8452–8457. [Google Scholar] [CrossRef]
- Bartkiewicz, M.; Gold, H.; Altman, S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 1989, 3, 488–499. [Google Scholar] [CrossRef]
- Kedersha, N.L.; Rome, L.H. Isolation and characterization of a novel ribonucleoprotein particle: Large structures contain a single species of small RNA. J. Cell Biol. 1986, 103, 699–709. [Google Scholar] [CrossRef]
- Lerner, M.R.; Boyle, J.A.; Hardin, J.A.; Steitz, J.A. Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science 1981, 211, 400–402. [Google Scholar] [CrossRef] [PubMed]
- Houck, C.M.; Rinehart, F.P.; Schmid, C.W. A ubiquitous family of repeated DNA sequences in the human genome. J. Mol. Biol. 1979, 132, 289–306. [Google Scholar] [CrossRef]
- Watson, J.B.; Sutcliffe, J.G. Primate brain-specific cytoplasmic transcript of the Alu repeat family. Mol. Cell. Biol. 1987, 7, 3324–3327. [Google Scholar] [PubMed]
- Parrott, A.M.; Mathews, M.B. Novel rapidly evolving hominid RNAs bind nuclear factor 90 and display tissue-restricted distribution. Nucleic Acids Res. 2007, 35, 6249–6258. [Google Scholar] [CrossRef] [PubMed]
- Nandy, C.; Mrázek, J.; Stoiber, H.; Grässer, F.A.; Hüttenhofer, A.; Polacek, N. Epstein-barr virus-induced expression of a novel human vault RNA. J. Mol. Biol. 2009, 388, 776–784. [Google Scholar] [CrossRef]
- Lee, K.; Kunkeaw, N.; Jeon, S.H.; Lee, I.; Johnson, B.H.; Kang, G.Y.; Bang, J.Y.; Park, H.S.; Leelayuwat, C.; Lee, Y.S. Precursor miR-886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. RNA 2011, 17, 1076–1089. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Van Bortle, K. The Pol III transcriptome: Basic features, recurrent patterns, and emerging roles in cancer. RNA 2023, 14, e1782. [Google Scholar] [CrossRef]
- Lassar, A.B.; Martin, P.L.; Roeder, R.G. Transcription of class III genes: Formation of preinitiation complexes. Science 1983, 222, 740–748. [Google Scholar] [CrossRef]
- Watt, K.E.; Macintosh, J.; Bernard, G.; Trainor, P.A. RNA Polymerases I and III in development and disease. Semin. Cell Dev. Biol. 2023, 136, 49–63. [Google Scholar] [CrossRef]
- Orellana, E.A.; Siegal, E.; Gregory, R.I. tRNA dysregulation and disease. Nat. Rev. Genet. 2022, 23, 651–664. [Google Scholar] [CrossRef]
- Rappol, T.; Waldl, M.; Chugunova, A.; Hofacker, I.L.; Pauli, A.; Vilardo, E. tRNA expression and modification landscapes, and their dynamics during zebrafish embryo development. Nucleic Acids Res. 2024, 52, 10575–10594. [Google Scholar] [CrossRef] [PubMed]
- Reimão-Pinto, M.M.; Behrens, A.; Forcelloni, S.; Fröhlich, K.; Kaya, S.; Nedialkova, D.D. The dynamics and functional impact of tRNA repertoires during early embryogenesis in zebrafish. EMBO J. 2024, 43, 5747–5779. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Aleksashin, N.A.; Loveland, A.B.; Klepacki, D.; Reier, K.; Kefi, A.; Szal, T.; Remme, J.; Jaeger, L.; Vázquez-Laslop, N.; et al. Ribosome engineering reveals the importance of 5S rRNA autonomy for ribosome assembly. Nat. Commun. 2020, 11, 2900. [Google Scholar] [CrossRef]
- White, R.J. Transcription by RNA polymerase III: More complex than we thought. Nat. Rev. Genet. 2011, 12, 459–463. [Google Scholar] [CrossRef]
- Rajendra, K.C.; Cheng, R.; Zhou, S.; Lizarazo, S.; Smith, D.J.; Van Bortle, K. Evidence of RNA polymerase III recruitment and transcription at protein-coding gene promoters. Mol. Cell 2024, 84, 4111–4124. [Google Scholar]
- Jacobs, R.Q.; Schneider, D.A. Transcription elongation mechanisms of RNA polymerases I, II, and III and their therapeutic implications. J. Biol. Chem. 2024, 300, 105737. [Google Scholar] [CrossRef] [PubMed]
- Yeganeh, M.; Hernandez, N. RNA polymerase III transcription as a disease factor. Genes Dev. 2020, 34, 865–882. [Google Scholar] [CrossRef]
- Tsang, C.K.; Zheng, X.F.S. Role of RNA polymerase III transcription and regulation in ischaemic stroke. RNA Biol. 2024, 21, 1–10. [Google Scholar] [CrossRef]
- Marshall, L.; White, R.J. Non-coding RNA production by RNA polymerase III is implicated in cancer. Nat. Rev. Cancer 2008, 8, 911–914. [Google Scholar] [CrossRef]
- Muthukumar, S.; Li, C.T.; Liu, R.J.; Bellodi, C. Roles and regulation of tRNA-derived small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2024, 25, 359–378. [Google Scholar] [CrossRef]
- Zhou, M.; He, X.; Zhang, J.; Mei, C.; Zhong, B.; Ou, C. tRNA-derived small RNAs in human cancers: Roles, mechanisms, and clinical application. Mol. Cancer 2024, 23, 76. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Gu, J.; Wang, M.; Zhang, J.; Chen, Y.; Jiang, P.; Zhu, T.; Zhang, X. Emerging roles of tRNA-derived fragments in cancer. Mol. Cancer 2023, 22, 30. [Google Scholar] [CrossRef] [PubMed]
- Pliatsika, V.; Loher, P.; Telonis, A.G.; Rigoutsos, I. MINTbase: A framework for the interactive exploration of mitochondrial and nuclear tRNA fragments. Bioinformatics 2016, 32, 2481–2489. [Google Scholar] [CrossRef]
- Hussain, S.; Sajini, A.A.; Blanco, S.; Dietmann, S.; Lombard, P.; Sugimoto, Y.; Paramor, M.; Gleeson, J.G.; Odom, D.T.; Ule, J.; et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013, 4, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Billmeier, M.; Green, D.; Hall, A.E.; Turnbull, C.; Singh, A.; Xu, P.; Moxon, S.; Dalmay, T. Mechanistic insights into non-coding Y RNA processing. RNA Biol. 2022, 19, 468–480. [Google Scholar] [CrossRef]
- Agirrezabala, X.; Frank, J. Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q. Rev. Biophys. 2009, 42, 159–200. [Google Scholar] [CrossRef]
- Schramm, L.; Hernandez, N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002, 16, 2593–2620. [Google Scholar] [CrossRef]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef]
- Kuhle, B.; Chen, Q.; Schimmel, P. tRNA renovatio: Rebirth through fragmentation. Mol. Cell 2023, 83, 3953–3971. [Google Scholar] [CrossRef]
- Megel, C.; Morelle, G.; Lalande, S.; Duchêne, A.M.; Small, I.; Maréchal-Drouard, L. Surveillance and cleavage of eukaryotic tRNAs. Int. J. Mol. Sci. 2015, 16, 1873–1893. [Google Scholar] [CrossRef]
- Wang, J.H.; Chen, W.X.; Mei, S.Q.; Yang, Y.D.; Yang, J.H.; Qu, L.H.; Zheng, L.L. tsRFun: A comprehensive platform for decoding human tsRNA expression, functions and prognostic value by high-throughput small RNA-Seq and CLIP-Seq data. Nucleic Acids Res. 2022, 50, D421–D431. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; He, X.; Liu, C.; Liu, J.; Hu, Q.; Pan, T.; Duan, X.; Liu, B.; Zhang, Y.; Chen, J.; et al. IL-4 Inhibits the Biogenesis of an Epigenetically Suppressive PIWI-Interacting RNA To Upregulate CD1a Molecules on Monocytes/Dendritic Cells. J. Immunol. 2016, 196, 1591–1603. [Google Scholar] [CrossRef] [PubMed]
- Pinzaru, A.M.; Tavazoie, S.F. Transfer RNAs as dynamic and critical regulators of cancer progression. Nat. Rev. Cancer 2023, 23, 746–761. [Google Scholar] [CrossRef]
- Jia, H.; Zhang, L. tRNA-derived small RNAs in disease immunity. Theranostics 2025, 15, 245–257. [Google Scholar] [CrossRef]
- Zacharjasz, J.; Mleczko, A.M.; Bąkowski, P.; Piontek, T.; Bąkowska-Żywicka, K. Small Noncoding RNAs in Knee Osteoarthritis: The Role of MicroRNAs and tRNA-Derived Fragments. Int. J. Mol. Sci. 2021, 22, 5711. [Google Scholar] [CrossRef]
- Wu, D.; Li, X.; Khan, F.A.; Yuan, C.; Pandupuspitasari, N.S.; Huang, C.; Sun, F.; Guan, K. tRNA modifications and tRNA-derived small RNAs: New insights of tRNA in human disease. Cell Biol. Toxicol. 2024, 40, 76. [Google Scholar] [CrossRef] [PubMed]
- Douet, J.; Tourmente, S. Transcription of the 5S rRNA heterochromatic genes is epigenetically controlled in Arabidopsis thaliana and Xenopus laevis. Heredity 2007, 99, 5–13. [Google Scholar] [CrossRef]
- Ciganda, M.; Williams, N. Eukaryotic 5S rRNA biogenesis. RNA 2011, 2, 523–533. [Google Scholar] [CrossRef]
- Engelke, D.R.; Ng, S.Y.; Shastry, B.S.; Roeder, R.G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell 1980, 19, 717–728. [Google Scholar] [CrossRef]
- Lee, B.M.; Xu, J.; Clarkson, B.K.; Martinez-Yamout, M.A.; Dyson, H.J.; Case, D.A.; Gottesfeld, J.M.; Wright, P.E. Induced fit and “lock and key” recognition of 5S RNA by zinc fingers of transcription factor IIIA. J. Mol. Biol. 2006, 357, 275–291. [Google Scholar] [CrossRef]
- Kunkel, G.R.; Pederson, T. Upstream elements required for efficient transcription of a human U6 RNA gene resemble those of U1 and U2 genes even though a different polymerase is used. Genes Dev. 1988, 2, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Shimba, S.; Reddy, R. Purification of human U6 small nuclear RNA capping enzyme. Evidence for a common capping enzyme for gamma-monomethyl-capped small RNAs. J. Biol. Chem. 1994, 269, 12419–12423. [Google Scholar] [CrossRef]
- Trippe, R.; Guschina, E.; Hossbach, M.; Urlaub, H.; Lührmann, R.; Benecke, B.J. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA 2006, 12, 1494–1504. [Google Scholar] [CrossRef] [PubMed]
- Nomura, Y.; Roston, D.; Montemayor, E.J.; Cui, Q.; Butcher, S.E. Structural and mechanistic basis for preferential deadenylation of U6 snRNA by Usb1. Nucleic Acids Res. 2018, 46, 11488–11501. [Google Scholar] [CrossRef]
- Li, X.; Peng, J.; Yi, C. The epitranscriptome of small non-coding RNAs. Noncoding RNA Res. 2021, 6, 167–173. [Google Scholar] [CrossRef]
- Hasler, D.; Meduri, R.; Bąk, M.; Lehmann, G.; Heizinger, L.; Wang, X.; Li, Z.T.; Sement, F.M.; Bruckmann, A.; Dock-Bregeon, A.C.; et al. The Alazami Syndrome-Associated Protein LARP7 Guides U6 Small Nuclear RNA Modification and Contributes to Splicing Robustness. Mol. Cell 2020, 77, 1014–1031. [Google Scholar] [CrossRef]
- Wang, X.; Li, Z.T.; Yan, Y.; Lin, P.; Tang, W.; Hasler, D.; Meduri, R.; Li, Y.; Hua, M.M.; Qi, H.T.; et al. LARP7-Mediated U6 snRNA Modification Ensures Splicing Fidelity and Spermatogenesis in Mice. Mol. Cell 2020, 77, 999–1013. [Google Scholar] [CrossRef]
- Frilander, M.J.; Barborič, M. The Interlocking Lives of LARP7: Fine-Tuning Transcription, RNA Modification, and Splicing through Multiple Non-coding RNAs. Mol. Cell 2020, 78, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ulryck, N.; Herzel, L.; Pythoud, N.; Kleiber, N.; Guérineau, V.; Jactel, V.; Moritz, C.; Bohnsack, M.T.; Carapito, C.; et al. N2-methylguanosine modifications on human tRNAs and snRNA U6 are important for cell proliferation, protein translation and pre-mRNA splicing. Nucleic Acids Res. 2023, 51, 7496–7519. [Google Scholar] [CrossRef]
- Pendleton, K.E.; Chen, B.; Liu, K.; Hunter, O.V.; Xie, Y.; Tu, B.P.; Conrad, N.K. The U6 snRNA m6A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017, 169, 824–835. [Google Scholar] [CrossRef]
- Dietrich, R.C.; Padgett, R.A.; Shukla, G.C. The conserved 3′ end domain of U6atac snRNA can direct U6 snRNA to the minor spliceosome. RNA 2009, 15, 1198–1207. [Google Scholar] [CrossRef] [PubMed]
- Bai, R.; Wan, R.; Wang, L.; Xu, K.; Zhang, Q.; Lei, J.; Shi, Y. Structure of the activated human minor spliceosome. Science 2021, 371, eabg0879. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, S.; Egloff, S.; Eichhorn, C.D.; Hadjian, T.; Zhen, J.; Kiss, T.; Zhou, Z.H.; Feigon, J. Structural basis of RNA conformational switching in the transcriptional regulator 7SK RNP. Mol. Cell 2022, 82, 1724–1736. [Google Scholar] [CrossRef]
- Krueger, B.J.; Jeronimo, C.; Roy, B.B.; Bouchard, A.; Barrandon, C.; Byers, S.A.; Searcey, C.E.; Cooper, J.J.; Bensaude, O.; Cohen, E.A.; et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008, 36, 2219–2229. [Google Scholar] [CrossRef]
- Xue, Y.; Yang, Z.; Chen, R.; Zhou, Q. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res. 2010, 38, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Sedore, S.C.; Byers, S.A.; Biglione, S.; Price, J.P.; Maury, W.J.; Price, D.H. Manipulation of P-TEFb control machinery by HIV: Recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 2007, 35, 4347–4358. [Google Scholar] [CrossRef]
- Li, Z.; Fang, X.; Zhao, B.; Liu, R.; Shen, Y.; Li, T.; Wang, Y.; Guo, Z.; Wang, W.; Zhang, B.; et al. Liquid-liquid phase separation of LARP7 restrains HIV-1 replication. EMBO Rep. 2025, 26, 1935–1956. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Bader, J.; Ramanathan, P.; Hennlein, L.; Meissner, F.; Jablonka, S.; Mann, M.; Fischer, U.; Sendtner, M.; Briese, M. Interaction of 7SK with the Smn complex modulates snRNP production. Nat. Commun. 2021, 12, 1278. [Google Scholar] [CrossRef]
- Ullu, E.; Weiner, A.M. Upstream sequences modulate the internal promoter of the human 7SL RNA gene. Nature 1985, 318, 371–374. [Google Scholar] [CrossRef]
- Sinha, K.M.; Gu, J.; Chen, Y.; Reddy, R. Adenylation of small RNAs in human cells. Development of a cell-free system for accurate adenylation on the 3′-end of human signal recognition particle RNA. J. Biol. Chem. 1998, 273, 6853–6859. [Google Scholar] [CrossRef]
- Ullu, E.; Tschudi, C. Alu sequences are processed 7SL RNA genes. Nature 1984, 312, 171–172. [Google Scholar] [CrossRef] [PubMed]
- Deininger, P. Alu elements: Know the SINEs. Genome Biol. 2011, 12, 236. [Google Scholar] [CrossRef] [PubMed]
- Mariner, P.D.; Walters, R.D.; Espinoza, C.A.; Drullinger, L.F.; Wagner, S.D.; Kugel, J.F.; Goodrich, J.A. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell 2008, 29, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Tlučková, K.; Kaczmarek, B.; Salmazo, A.; Bernecky, C. Mechanism of mammalian transcriptional repression by noncoding RNA. Nat. Struct. Mol. Biol. 2025, 32, 607–612. [Google Scholar] [CrossRef]
- Tiedge, H.; Chen, W.; Brosius, J. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J. Neurosci. 1993, 13, 2382–2390. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, J.; Shin, H.; Jang, S.; Kim, S.C.; Lee, Y. Biosynthesis of brain cytoplasmic 200 RNA. Sci. Rep. 2017, 7, 6884. [Google Scholar] [CrossRef]
- Booy, E.P.; Gussakovsky, D.; Brown, M.; Shwaluk, R.; Nachtigal, M.W.; McKenna, S.A. lncRNA BC200 is processed into a stable Alu monomer. RNA 2024, 30, 1477–1494. [Google Scholar] [CrossRef]
- Mrázek, J.; Kreutmayer, S.B.; Grässer, F.A.; Polacek, N.; Hüttenhofer, A. Subtractive hybridization identifies novel differentially expressed ncRNA species in EBV-infected human B cells. Nucleic Acids Res. 2007, 35, e73. [Google Scholar] [CrossRef]
- Parrott, A.M.; Mathews, M.B. The evolution and consequences of snaR family transposition in primates. Mob. Genet. Elem. 2011, 1, 291–295. [Google Scholar] [CrossRef]
- Van Bortle, K.; Marciano, D.P.; Liu, Q.; Chou, T.; Lipchik, A.M.; Gollapudi, S.; Geller, B.S.; Monte, E.; Kamakaka, R.T.; Snyder, M.P. A cancer-associated RNA polymerase III identity drives robust transcription and expression of snaR-A noncoding RNA. Nat. Commun. 2022, 13, 3007. [Google Scholar] [CrossRef]
- Shaukat, A.N.; Kaliatsi, E.G.; Skeparnias, I.; Stathopoulos, C. The Dynamic Network of RNP RNase P Subunits. Int. J. Mol. Sci. 2021, 22, 10307. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Lin, J.; Zheng, X.; Xu, X. RNase P: Beyond Precursor tRNA Processing. Genom. Proteom. Bioinform. 2024, 22, qzae016. [Google Scholar] [CrossRef]
- Orlovetskie, N.; Mani, D.; Rouvinski, A.; Jarrous, N. Human RNase P exhibits and controls distinct ribonucleolytic activities required for ordered maturation of tRNA. Proc. Natl. Acad. Sci. USA 2023, 120, e2307185120. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Shi, K.; Huang, W.; Zhang, Y.; Chen, Q.; Mou, T.; Wu, Z.; Wei, X. LncRNA RPPH1 acts as a molecular sponge for miR-122 to regulate Wnt1/β-catenin signaling in hepatocellular carcinoma. Int. J. Med. Sci. 2023, 20, 23–34. [Google Scholar] [CrossRef]
- Lin, Y.H.; Chen, C.W.; Cheng, H.C.; Liu, C.J.; Chung, S.T.; Hsieh, M.C.; Tseng, P.L.; Tsai, W.H.; Wu, T.S.; Lai, M.D.; et al. Inhibition of lncRNA RPPH1 activity decreases tumor proliferation and metastasis through down-regulation of inflammation-related oncogenes. Am. J. Transl. Res. 2023, 15, 6701–6717. [Google Scholar] [PubMed]
- Wu, Y.; Cheng, K.; Liang, W.; Wang, X. lncRNA RPPH1 promotes non-small cell lung cancer progression through the miR-326/WNT2B axis. Oncol. Lett. 2020, 20, 105. [Google Scholar] [CrossRef]
- Ning, W.; Yang, J.; Ni, R.; Yin, Q.; Zhang, M.; Zhang, F.; Yang, Y.; Zhang, Y.; Cao, M.; Jin, L.; et al. Hypoxia induced cellular and exosomal RPPH1 promotes breast cancer angiogenesis and metastasis through stabilizing the IGF2BP2/FGFR2 axis. Oncogene 2025, 44, 147–164. [Google Scholar] [CrossRef]
- Perederina, A.; Li, D.; Lee, H.; Bator, C.; Berezin, I.; Hafenstein, S.L.; Krasilnikov, A.S. Cryo-EM structure of catalytic ribonucleoprotein complex RNase MRP. Nat. Commun. 2020, 11, 3474. [Google Scholar] [CrossRef]
- Noh, J.H.; Kim, K.M.; Abdelmohsen, K.; Yoon, J.H.; Panda, A.C.; Munk, R.; Kim, J.; Curtis, J.; Moad, C.A.; Wohler, C.M.; et al. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev. 2016, 30, 1224–1239. [Google Scholar] [CrossRef]
- Yin, H.; Chen, L.; Piao, S.; Wang, Y.; Li, Z.; Lin, Y.; Tang, X.; Zhang, H.; Zhang, H.; Wang, X. M6A RNA methylation-mediated RMRP stability renders proliferation and progression of non-small cell lung cancer through regulating TGFBR1/SMAD2/SMAD3 pathway. Cell Death Differ. 2023, 30, 605–617. [Google Scholar] [CrossRef]
- Wolin, S.L.; Steitz, J.A. Genes for two small cytoplasmic Ro RNAs are adjacent and appear to be single-copy in the human genome. Cell 1983, 32, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, M.P.; Krude, T. Functional roles of non-coding Y RNAs. Int. J. Biochem. Cell Biol. 2015, 66, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Chakrabortty, S.K.; Prakash, A.; Nechooshtan, G.; Hearn, S.; Gingeras, T.R. Extracellular vesicle-mediated transfer of processed and functional RNY5 RNA. RNA 2015, 21, 1966–1979. [Google Scholar] [CrossRef]
- Rutjes, S.A.; van der Heijden, A.; Utz, P.J.; van Venrooij, W.J.; Pruijn, G.J. Rapid nucleolytic degradation of the small cytoplasmic Y RNAs during apoptosis. J. Biol. Chem. 1999, 274, 24799–24807. [Google Scholar] [CrossRef] [PubMed]
- Hahne, J.C.; Lampis, A.; Valeri, N. Vault RNAs: Hidden gems in RNA and protein regulation. Cell. Mol. Life Sci. 2021, 78, 1487–1499. [Google Scholar] [CrossRef]
- Taube, M.; Lisiak, N.; Totoń, E.; Rubiś, B. Human Vault RNAs: Exploring Their Potential Role in Cellular Metabolism. Int. J. Mol. Sci. 2024, 25, 4072. [Google Scholar] [CrossRef]
- Stadler, P.F.; Chen, J.J.; Hackermüller, J.; Hoffmann, S.; Horn, F.; Khaitovich, P.; Kretzschmar, A.K.; Mosig, A.; Prohaska, S.J.; Qi, X.; et al. Evolution of vault RNAs. Mol. Biol. Evol. 2009, 26, 1975–1991. [Google Scholar] [CrossRef]
- Sajini, A.A.; Choudhury, N.R.; Wagner, R.E.; Bornelöv, S.; Selmi, T.; Spanos, C.; Dietmann, S.; Rappsilber, J.; Michlewski, G.; Frye, M. Loss of 5-methylcytosine alters the biogenesis of vault-derived small RNAs to coordinate epidermal differentiation. Nat. Commun. 2019, 10, 2550. [Google Scholar] [CrossRef]
- Prajapat, M.; Sala, L.; Vidigal, J.A. The small noncoding RNA Vaultrc5 is dispensable to mouse development. RNA 2024, 30, 1465–1476. [Google Scholar] [CrossRef]
- Li, F.; Chen, Y.; Zhang, Z.; Ouyang, J.; Wang, Y.; Yan, R.; Huang, S.; Gao, G.F.; Guo, G.; Chen, J.L. Robust expression of vault RNAs induced by influenza A virus plays a critical role in suppression of PKR-mediated innate immunity. Nucleic Acids Res. 2015, 43, 10321–10337. [Google Scholar] [CrossRef]
- Lee, Y.S.; Lee, Y.S. nc886, an RNA Polymerase III-Transcribed Noncoding RNA Whose Expression Is Dynamic and Regulated by Intriguing Mechanisms. Int. J. Mol. Sci. 2023, 24, 8533. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S. Are We Studying Non-Coding RNAs Correctly? Lessons from nc886. Int. J. Mol. Sci. 2022, 23, 4251. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.J.; Kang, D.; Lee, Y.S.; Lee, Y.S. The Versatile Roles of nc886, a Fascinating and Peculiar Regulatory Non-Coding RNA, in Cancer. Int. J. Mol. Sci. 2024, 25, 10825. [Google Scholar] [CrossRef] [PubMed]
- White, R.J. RNA polymerases I and III, growth control and cancer. Nat. Rev. Mol. Cell Biol. 2005, 6, 69–78. [Google Scholar] [CrossRef]
- Zhao, P.; Xia, L.; Chen, D.; Xu, W.; Guo, H.; Xu, Y.; Yan, B.; Wu, X.; Li, Y.; Zhang, Y.; et al. METTL1 mediated tRNA m7G modification promotes leukaemogenesis of AML via tRNA regulated translational control. Exp. Hematol. Oncol. 2024, 13, 8. [Google Scholar] [CrossRef]
- Wang, Z.; Yu, P.; Zou, Y.; Ma, J.; Han, H.; Wei, W.; Yang, C.; Zheng, S.; Guo, S.; Wang, J.; et al. METTL1/WDR4-mediated tRNA m7G modification and mRNA translation control promote oncogenesis and doxorubicin resistance. Oncogene 2023, 42, 1900–1912. [Google Scholar] [CrossRef]
- Han, H.; Yang, C.; Ma, J.; Zhang, S.; Zheng, S.; Ling, R.; Sun, K.; Guo, S.; Huang, B.; Liang, Y.; et al. N7-methylguanosine tRNA modification promotes esophageal squamous cell carcinoma tumorigenesis via the RPTOR/ULK1/autophagy axis. Nat. Commun. 2022, 13, 1478. [Google Scholar] [CrossRef]
- García-Vílchez, R.; Añazco-Guenkova, A.M.; Dietmann, S.; López, J.; Morón-Calvente, V.; D’Ambrosi, S.; Nombela, P.; Zamacola, K.; Mendizabal, I.; García-Longarte, S.; et al. METTL1 promotes tumorigenesis through tRNA-derived fragment biogenesis in prostate cancer. Mol. Cancer 2023, 22, 119. [Google Scholar] [CrossRef]
- Ying, X.; Hu, W.; Huang, Y.; Lv, Y.; Ji, D.; Chen, C.; Yang, B.; Zhang, C.; Liang, Y.; Zhang, H.; et al. A Novel tsRNA, m7G-3′ tiRNA Lys (TTT), Promotes Bladder Cancer Malignancy Via Regulating ANXA2 Phosphorylation. Adv. Sci. 2024, 11, e2400115. [Google Scholar] [CrossRef]
- Rui, T.; Zhu, K.; Mao, Z.; Wu, J.; Pan, Y.; Ye, Q.; Chen, C.; Xiang, A.; Guo, J.; Tang, N.; et al. A Novel tRF, HCETSR, Derived From tRNA-Glu/TTC, Inhibits HCC Malignancy by Regulating the SPBTN1-catenin Complex Axis. Adv. Sci. 2025, 12, e2415229. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, X.; Li, Y.; Li, X.; Huang, Y.; Ju, S. Transfer RNA-derived fragment tRF-23-Q99P9P9NDD promotes progression of gastric cancer by targeting ACADSB. Journal of Zhejiang University. Science 2024, 25, 438–450. [Google Scholar]
- Zhang, S.; Gu, Y.; Ge, J.; Xie, Y.; Yu, X.; Wu, X.; Sun, D.; Zhang, X.; Guo, J.; Guo, J. tRF-33-P4R8YP9LON4VDP inhibits gastric cancer progression via modulating STAT3 signaling pathway in an AGO2-dependent manner. Oncogene 2024, 43, 2160–2171. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Liu, Z.; Peng, L.; Liu, L.; Xie, X.; Zhang, Y.; Gao, Z.; Zhang, C.; Yu, X.; Hu, Y.; et al. A novel 5′tRNA-derived fragment tRF-Tyr inhibits tumor progression by targeting hnRNPD in gastric cancer. Cell Commun. Signal. 2025, 23, 88. [Google Scholar] [CrossRef] [PubMed]
- Mo, D.; Tang, X.; Ma, Y.; Chen, D.; Xu, W.; Jiang, N.; Zheng, J.; Yan, F. tRNA-derived fragment 3′tRF-AlaAGC modulates cell chemoresistance and M2 macrophage polarization via binding to TRADD in breast cancer. J. Transl. Med. 2024, 22, 706. [Google Scholar] [CrossRef]
- Wan, X.; Shi, W.; Ma, L.; Wang, L.; Zheng, R.; He, J.; Wang, Y.; Li, X.; Zha, X.; Wang, J.; et al. A 3′-pre-tRNA-derived small RNA tRF-1-Ser regulated by 25(OH)D promotes proliferation and stemness by inhibiting the function of MBNL1 in breast cancer. Clin. Transl. Med. 2024, 14, e1681. [Google Scholar] [CrossRef]
- Xu, R.; Du, A.; Deng, X.; Du, W.; Zhang, K.; Li, J.; Lu, Y.; Wei, X.; Yang, Q.; Tang, H. tsRNA-GlyGCC promotes colorectal cancer progression and 5-FU resistance by regulating SPIB. J. Exp. Clin. Cancer Res. 2024, 43, 230. [Google Scholar] [CrossRef]
- Kong, E.; Polacek, N. TRIM21 modulates stability of pro-survival non-coding RNA vtRNA1-1 in human hepatocellular carcinoma cells. PLoS Genet. 2025, 21, e1011614. [Google Scholar] [CrossRef]
- Tan, Z.; Luan, S.; Wang, X.; Jiao, W.; Jiang, P. Mechanism study of lncRNA RMRP regulating esophageal squamous cell carcinoma through miR-580-3p/ATP13A3 axis. Discov. Oncol. 2024, 15, 150. [Google Scholar] [CrossRef]
- Lee, K.J.; Ahn, J.H.; Kim, J.H.; Lee, Y.S.; Lee, J.S.; Lee, J.H.; Kim, T.J.; Choi, J.H. Non-coding RNA RMRP governs RAB31-dependent MMP secretion, enhancing ovarian cancer invasion. Biochim. Biophys. Acta Mol. Basis Dis. 2025, 1871, 167781. [Google Scholar] [CrossRef]
- Wang, Y.; Traugot, C.M.; Bubenik, J.L.; Li, T.; Sheng, P.; Hiers, N.M.; Fernandez, P.; Li, L.; Bian, J.; Swanson, M.S.; et al. N6-methyladenosine in 7SK small nuclear RNA underlies RNA polymerase II transcription regulation. Mol. Cell 2023, 83, 3818–3834. [Google Scholar] [CrossRef]
- Abasi, M.; Ranjbari, J.; Ghanbarian, H. 7SK small nuclear RNA (Rn7SK) induces apoptosis through intrinsic and extrinsic pathways in human embryonic kidney cell line. Mol. Biol. Rep. 2024, 51, 96. [Google Scholar] [CrossRef] [PubMed]
- Magliacane Trotta, S.; Adinolfi, A.; D’Orsi, L.; Panico, S.; Mercadante, G.; Mehlen, P.; Ambati, J.; De Falco, S.; Tarallo, V. Cancer-derived exosomal Alu RNA promotes colorectal cancer progression. Exp. Mol. Med. 2024, 56, 700–710. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liu, N.; Cao, P.; Qin, Q.; Li, J.; Yang, L.; Xin, Y.; Jiang, M.; Zhang, S.; Yang, J.; et al. LncRNA BC200 promotes the development of EBV-associated nasopharyngeal carcinoma by competitively binding to miR-6834-5p to upregulate TYMS expression. Int. J. Biol. Macromol. 2024, 278, 134837. [Google Scholar] [CrossRef] [PubMed]
- Raitoharju, E.; Rajić, S.; Marttila, S. Non-coding 886 (nc886/vtRNA2-1), the epigenetic odd duck—Implications for future studies. Epigenetics 2024, 19, 2332819. [Google Scholar] [CrossRef]
- Oliveira-Rizzo, C.; Colantuono, C.L.; Fernández-Alvarez, A.J.; Boccaccio, G.L.; Garat, B.; Sotelo-Silveira, J.R.; Khan, S.; Ignatchenko, V.; Lee, Y.S.; Kislinger, T.; et al. Multi-Omics Study Reveals Nc886/vtRNA2-1 as a Positive Regulator of Prostate Cancer Cell Immunity. J. Proteome Res. 2025, 24, 433–448. [Google Scholar] [CrossRef]
- Ahn, J.H.; Lee, H.S.; Lee, J.S.; Lee, Y.S.; Park, J.L.; Kim, S.Y.; Hwang, J.A.; Kunkeaw, N.; Jung, S.Y.; Kim, T.J.; et al. nc886 is induced by TGF-β and suppresses the microRNA pathway in ovarian cancer. Nat. Commun. 2018, 9, 1166. [Google Scholar] [CrossRef]
- Huang, Y.; Abdelgawad, A.; Gololobova, O.; Liao, Z.; Cong, X.; Batish, M.; Zheng, L.; Witwer, K.W. Enhanced packaging of U6 small nuclear RNA and splicing-related proteins into extracellular vesicles during HIV infection. Sci. Adv. 2025, 11, eadq6557. [Google Scholar] [CrossRef]
- Geng, S.; Lv, X.; Xu, T. PIF/harbinger transposon-derived protein promotes 7SL expression to enhance pathogen resistance. EMBO Rep. 2025, 26, 1196–1211. [Google Scholar] [CrossRef]
- Li, J.; Xin, Y.; Zhang, S.; Li, Y.; Jiang, M.; Zhang, S.; Yang, L.; Yang, J.; Cao, P.; Lu, J. EIF4A3 is stabilized by the long noncoding RNA BC200 to regulate gene expression during Epstein-Barr virus infection. J. Med. Virol. 2024, 96, e29955. [Google Scholar] [CrossRef]
- Jang, J.J.; Lee, M.J.; Lee, M.S.; Myoung, J.; Lee, H.H.; Choi, B.H.; Saruuldalai, E.; Jung, Y.S.; Lee, H.S.; Kim, Y.; et al. The immune sensitivity caused by DUSP11, an RNA 5′-end maturation phosphatase, is adjusted by a human non-coding RNA, nc886. Cell. Mol. Life Sci. 2025, 82, 77. [Google Scholar] [CrossRef]
- Pereira, M.; Ribeiro, D.R.; Berg, M.; Tsai, A.P.; Dong, C.; Nho, K.; Kaiser, S.; Moutinho, M.; Soares, A.R. Amyloid pathology reduces ELP3 expression and tRNA modifications leading to impaired proteostasis. Biochim. Biophys. Acta Mol. Basis Dis. 2024, 1870, 166857. [Google Scholar] [CrossRef] [PubMed]
- Tresky, R.; Miyamoto, Y.; Nagayoshi, Y.; Yabuki, Y.; Araki, K.; Takahashi, Y.; Komohara, Y.; Ge, H.; Nishiguchi, K.; Fukuda, T.; et al. TRMT10A dysfunction perturbs codon translation of initiator methionine and glutamine and impairs brain functions in mice. Nucleic Acids Res. 2024, 52, 9230–9246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, J.; Hou, Y. Classification, function, and advances in tsRNA in non-neoplastic diseases. Cell Death Dis. 2023, 14, 748. [Google Scholar] [CrossRef]
- Zhang, S.; Li, H.; Zheng, L.; Li, H.; Feng, C.; Zhang, W. Identification of functional tRNA-derived fragments in senescence-accelerated mouse prone 8 brain. Aging 2019, 11, 10485–10498. [Google Scholar] [CrossRef]
- Deng, Z.; Li, Y.; Chi, W.; Zhang, W.; Li, F.; Ling, L. tRFAla-AGC-3-M8 attenuates neuroinflammation and neuronal damage in Alzheimer’s disease via the EphA7-ERK1/2-p70S6K signaling pathway. Alzheimers Res. Ther. 2025, 17, 104. [Google Scholar] [CrossRef]
- Hao, Y.; Li, B.; Yin, F.; Liu, W. tRNA-derived small RNA (tsr007330) regulates myocardial fibrosis after myocardial infarction through NAT10-mediated ac4C acetylation of EGR3 mRNA. Biochim. Biophys. Acta Mol. Basis Dis. 2024, 1870, 167267. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, Y.; Zhang, H.Y.; Zhao, Y.; Li, X.M.; Jiang, Y.F.; Yao, M.D.; Jiang, Q.; Yan, B. Dual anti-angiogenic and anti-inflammatory action of tRNA-Cys-5-0007 in ocular vascular disease. J. Transl. Med. 2024, 22, 562. [Google Scholar] [CrossRef]
- Jiang, Q.L.; Xu, J.Y.; Yao, Q.P.; Jiang, R.; Xu, Q.; Zhang, B.T.; Li, T.; Jiang, J. Transfer RNA-derived small RNA tRF-Glu-CTC attenuates neointimal formation via inhibition of fibromodulin. Cell. Mol. Biol. Lett. 2024, 29, 2. [Google Scholar] [CrossRef]
- Liang, Y.; Kong, L.; Zhang, Y.; Zhang, Y.; Shi, M.; Huang, J.; Kong, H.; Qi, S.; Yang, Y.; Hong, J.; et al. Transfer RNA derived fragment, tRF-Glu-CTC, aggravates the development of neovascular age-related macular degeneration. Theranostics 2024, 14, 1500–1516. [Google Scholar] [CrossRef]
- Chen, Y.M.; Tang, K.T.; Liu, H.J.; Huang, S.T.; Liao, T.L. tRF-His-GTG-1 enhances NETs formation and interferon-α production in lupus by extracellular vesicle. Cell Commun. Signal. 2024, 22, 354. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, Y.; Ji, Y.K.; Jiang, Y.F.; Li, D.; Mu, W.; Yao, M.D.; Yao, J.; Yan, B. Co-targeting of glial activation and inflammation by tsRNA-Gln-i-0095 for treating retinal ischemic pathologies. Cell Commun. Signal. 2025, 23, 18. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, J.F.; Gupta, A.; Kharkwal, G.; Linares, E.E.; Holmes, A.D.; Swartz, J.R.; Katzman, S.; Sharma, U. Epididymis-specific RNase A family genes regulate fertility and small RNA processing. J. Biol. Chem. 2024, 300, 107933. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.Q.; Ge, J.Y.; Zhang, X.; Zhu, W.Y.; Lin, L.; Shi, Y.; Xu, B.; Liu, R.J. THUMPD2 catalyzes the N2-methylation of U6 snRNA of the spliceosome catalytic center and regulates pre-mRNA splicing and retinal degeneration. Nucleic Acids Res. 2024, 52, 3291–3309. [Google Scholar] [CrossRef]
- Liu, B.; Wu, T.; Miao, B.A.; Ji, F.; Liu, S.; Wang, P.; Zhao, Y.; Zhong, Y.; Sundaram, A.; Zeng, T.B.; et al. snoRNA-facilitated protein secretion revealed by transcriptome-wide snoRNA target identification. Cell 2025, 188, 465–483. [Google Scholar] [CrossRef]
- Bujisic, B.; Lee, H.G.; Xu, L.; Weissbein, U.; Rivera, C.; Topisirovic, I.; Lee, J.T. 7SL RNA and signal recognition particle orchestrate a global cellular response to acute thermal stress. Nat. Commun. 2025, 16, 1630. [Google Scholar] [CrossRef] [PubMed]
- Shu, M.; Xu, H.; Liang, Y.; Shi, M.; Chen, M.; Guo, J. Long Non-Coding RNA BCYRN1 Drives Cellular Processes to Promote Asthma Development. Int. Arch. Allergy Immunol. 2024, 185, 343–354. [Google Scholar] [CrossRef]
- Wu, J.; Kim, A.; Wu, X.; Ray, S.; Allende, D.S.; Welch, N.; Bellar, A.; Dasarathy, J.; Dasarathy, S.; Nagy, L.E. 5S rRNA pseudogene transcripts are associated with interferon production and inflammatory responses in alcohol-associated hepatitis. Hepatology 2023, 77, 1983–1997. [Google Scholar] [CrossRef]
- Li, J.; Zhou, G.; Chen, T.; Lin, Q.; Yang, Q. LncRNA RMRP promotes chondrocyte injury by regulating the FOXC1/RBP4 axis. Cent. Eur. J. Immunol. 2024, 49, 366–382. [Google Scholar] [CrossRef]
- Gao, J.; Zheng, J.; Chen, S.; Lin, S.; Duan, S. RMRP variants inhibit the cell cycle checkpoints pathway in cartilage-hair hypoplasia. Mol. Med. Rep. 2025, 31, 81. [Google Scholar] [CrossRef]
- Uchida, N.; Ishii, T.; Nishimura, G.; Sato, T.; Kuratsuji, G.; Nagasaki, K.; Hosokawa, Y.; Adachi, E.; Takasawa, K.; Kashimada, K.; et al. RMRP-related short stature: A report of six additional Japanese individuals with cartilage hair hypoplasia and literature review. Am. J. Med. Genet. A 2024, 194, e63562. [Google Scholar] [CrossRef]
- Gomes, M.E.; Kehdy, F.; de Neves-Manta, F.S.; Horovitz, D.D.G.; Sanseverino, M.T.; Leal, G.F.; Felix, T.M.; Cavalcanti, D.P.; Llerena, J.C., Jr.; Gonzalez, S. Identification of a founder effect involving n.197C>T variant in RMRP gene associated to cartilage-hair hypoplasia syndrome in Brazilian patients. Sci. Rep. 2024, 14, 13436. [Google Scholar] [CrossRef]
- Park, J.H.; Im, M.; Kim, Y.J.; Jang, J.H.; Lee, S.M.; Kim, M.S.; Cho, S.Y. Cartilage-hair hypoplasia-anauxetic dysplasia spectrum disorders harboring RMRP mutations in two Korean children: A case report. Medicine 2024, 103, e37247. [Google Scholar] [CrossRef]
- López-Pardo Rico, M.; Moreiras Arias, N.; Buján Bonino, C.; Monteagudo Sánchez, B.; Pérez Feal, P.; Gómez Silva, G.; Suárez Peñaranda, J.M.; Vázquez Osorio, I. Neonatal erythroderma in a patient with cartilage-hair hypoplasia: Identification of a novel RMRP mutation. J. Dtsch. Dermatol. Ges. 2025. ahead of print. [Google Scholar] [CrossRef]
- Yan, X.; Liu, T.; Zhang, R.; Ma, Q.; Sun, C. RMRP accelerates ligamentum flavum hypertrophy by regulating GSDMD-mediated pyroptosis through Gli1 SUMOylation. Front. Immunol. 2024, 15, 1427970. [Google Scholar] [CrossRef] [PubMed]
- Ward, C.; Beharry, A.; Tennakoon, R.; Rozik, P.; Wilhelm, S.D.P.; Heinemann, I.U.; O’Donoghue, P. Mechanisms and Delivery of tRNA Therapeutics. Chem. Rev. 2024, 124, 7976–8008. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Mendonca, C.A.; Yukselen, O.; Muneeruddin, K.; Ren, L.; Liang, J.; Zhou, C.; Xie, J.; Li, J.; et al. AAV-delivered suppressor tRNA overcomes a nonsense mutation in mice. Nature 2022, 604, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Albers, S.; Allen, E.C.; Bharti, N.; Davyt, M.; Joshi, D.; Perez-Garcia, C.G.; Santos, L.; Mukthavaram, R.; Delgado-Toscano, M.A.; Molina, B.; et al. Engineered tRNAs suppress nonsense mutations in cells and in vivo. Nature 2023, 618, 842–848. [Google Scholar] [CrossRef]
- Coller, J.; Ignatova, Z. tRNA therapeutics for genetic diseases. Nat. Rev. Drug Discov. 2024, 23, 108–125. [Google Scholar] [CrossRef] [PubMed]
- Dolgin, E. tRNA therapeutics burst onto startup scene. Nat. Biotechnol. 2022, 40, 283–286. [Google Scholar] [CrossRef]
- Anastassiadis, T.; Köhrer, C. Ushering in the era of tRNA medicines. J. Biol. Chem. 2023, 299, 105246. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, J.; Chen, W.; Gao, L.; He, J.; Xia, Y. Exosomal lncRNA RMRP-shuttled by Olfactory Mucosa-Mesenchymal Stem Cells Suppresses Microglial Pyroptosis to Improve Spinal Cord Injury via EIF4A3/SIRT1. Mol. Neurobiol. 2025. ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Saruuldalai, E.; Lee, H.H.; Lee, Y.S.; Hong, E.K.; Ro, S.; Kim, Y.; Ahn, T.; Park, J.L.; Kim, S.Y.; Shin, S.P.; et al. Adenovirus expressing nc886, an anti-interferon and anti-apoptotic non-coding RNA, is an improved gene delivery vector. Mol. Ther. Nucleic Acids 2024, 35, 102270. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pan, Y.; Li, Z.; Hu, K. tRNA-derived small RNAs: Their role in the mechanisms, biomarkers, and therapeutic strategies of colorectal cancer. J. Transl. Med. 2025, 23, 51. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Ni, K.; Jin, K.; Feng, W.; Ju, S.; Jing, R.; Zong, W. Identification and potential mechanism of a novel gastric cancer suppressor tRF-24-6VR8K09LE9. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2025. ahead of print. [Google Scholar] [CrossRef]
- Jiang, X.; Li, X.; Li, Y.; Zhang, Y.; Gu, X.; Zong, W.; Shen, X.; Ju, S. Systematic assessment of serum i-tRF-AsnGTT in gastric cancer: A potential clinical biomarker. Carcinogenesis 2025, 46, bgae044. [Google Scholar] [CrossRef]
- Yuan, J.; Gu, W.; Xu, T.; Zhang, Y.; Shen, L.; Yan, J.; Guan, X.; Chu, H.; Yuan, R.; Ju, S. Dysregulated transfer RNA-derived small RNAs as potential gastric cancer biomarkers. Exp. Biol. Med. 2024, 249, 10170. [Google Scholar] [CrossRef]
- Mao, C.; Zhang, Z.; Fang, R.; Yuan, W.; Wu, Y.; Cong, H. A novel tRNA-derived fragment tRF-17-18VBY9M works as a potential diagnostic biomarker for gastric cancer. J. Cancer Res. Clin. Oncol. 2024, 150, 263. [Google Scholar] [CrossRef]
- Xu, J.; Wang, Y.; Li, X.; Zheng, M.; Li, Y.; Zhang, W. Clinical value assessment for serum hsa_tsr013526 in the diagnosis of gastric carcinoma. Environ. Toxicol. 2024, 39, 2753–2767. [Google Scholar] [CrossRef]
- Jin, K.; Mao, Z.; Tang, Y.; Feng, W.; Ju, S.; Jing, R.; Chen, J.; Zong, W. tRF-23-R9J89O9N9: A novel liquid biopsy marker for diagnosis of hepatocellular carcinoma. Clin. Chim. Acta 2025, 572, 120261. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Yuan, J.; Zhang, W.; Xu, T.; Jing, R.; Ju, S. Serum tRF-33-RZYQHQ9M739P0J as a novel biomarker for auxiliary diagnosis and disease course monitoring of hepatocellular carcinoma. Heliyon 2024, 10, e30084. [Google Scholar] [CrossRef]
- Jin, K.; Wu, J.; Yang, J.; Chen, B.; Xu, J.; Bao, H.; Zong, W.; Xie, C.; Chen, L.; Wang, F. Identification of serum tsRNA-Thr-5-0015 and combined with AFP and PIVKA-II as novel biomarkers for hepatocellular carcinoma. Sci. Rep. 2024, 14, 28834. [Google Scholar] [CrossRef] [PubMed]
- Si, J.; Zou, Y.; Gao, Y.; Chen, J.; Jiang, W.; Shen, X.; Zhu, C.; Yao, Q. tRF-3a-Pro: A Transfer RNA-Derived Small RNA as a Novel Biomarker for Diagnosis of Hepatitis B Virus-Related Hepatocellular Carcinoma. Cell Prolif. 2025. ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Gao, S.; Xu, M.; Yang, M.; Shen, M.; Liu, J.; Li, G.; Zhuang, D.; Hu, Z.; Wang, C. tiRNA-Gly-GCC-001 in major depressive disorder: Promising diagnostic and therapeutic biomarker. Br. J. Pharmacol. 2024, 181, 1952–1972. [Google Scholar] [CrossRef]
- Wang, Y.; Ji, X.; Shi, H.; Liu, S.; Yu, H. tiRNA-Gln-CTG is Involved in the Regulation of Trophoblast Cell Function in Pre-eclampsia and Serves as a Potent Biomarker. Front. Biosci. (Landmark Ed.) 2025, 30, 26345. [Google Scholar] [CrossRef]
- Ding, C.; Wang, N.; Peng, A.; Wang, Z.; Li, B.; Zhang, X.; Zeng, J.; Zhou, Y. Potential Diagnostic Biomarkers of tRNA-Derived Small RNAs in PBMCs for Nonproliferative Diabetic Retinopathy in Patients with Type 2 Diabetes Mellitus. Transl. Vis. Sci. Technol. 2024, 13, 32. [Google Scholar] [CrossRef]
- Ding, J.; Teng, Y.; Cui, R.; Liu, J.; Xiao, K.; Dong, Z.; Zhang, Y.; Xu, X. LncRNAs in serum-derived extracellular vesicles are potential biomarker and correlated with immune infiltration in gastric cancer. Front. Immunol. 2025, 16, 1533111. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, X.; Wang, X.; Sun, R.; Li, Y.; Li, J.; Quan, W.; Yao, Y.; Hou, Y.; Li, D.; et al. The clinical value of rapidly detecting urinary exosomal lncRNA RMRP in bladder cancer with an RT-RAA-CRISPR/Cas12a method. Clin. Chim. Acta 2024, 562, 119855. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Pu, J.; Jiang, G.; Pan, C.; Xu, J.; Zhang, B.; Bai, M. Analysis of long non-coding RNA RMRP in the diagnosis and prognosis of coronary artery disease. J. Cardiothorac. Surg. 2024, 19, 341. [Google Scholar] [CrossRef]
- Agwa, S.H.A.; Elzahwy, S.S.; Hossam, N.; Yahia, Y.A.; Hamady, S.; Sherif, N.; Elshazly, A.; Darwish, R.M.; Hashim, J.O.; Adly, M.A.; et al. Discriminatory power of a circulating multi-noncoding RNA panel in acute coronary syndrome subtypes: Towards precision detection. Int. J. Biochem. Cell Biol. 2024, 169, 106531. [Google Scholar] [CrossRef]
- Ghamari, M.; Mehrab Mohseni, M.; Taheri, M.; Neishabouri, S.M.; Shirvani-Farsani, Z. Abnormal expression of long non-coding RNAs RMRP, CTC-487M23.5, and DGCR5 in the peripheral blood of patients with bipolar disorder. Metab. Brain Dis. 2024, 39, 313–320. [Google Scholar] [CrossRef]
- Hatayama, Y.; Shimohiro, H.; Hashimoto, Y.; Ichikawa, H.; Kawamura, K.; Motokura, T. Serum vault RNA1-1 levels reflect blood cells and bone marrow. Mol. Cell. Probes 2025, 80, 102018. [Google Scholar] [CrossRef] [PubMed]
- Arima, J.; Yoshino, H.; Fukumoto, W.; Kawahara, I.; Saito, S.; Li, G.; Fukuda, I.; Iizasa, S.; Mitsuke, A.; Sakaguchi, T.; et al. LncRNA BCYRN1 as a Potential Therapeutic Target and Diagnostic Marker in Serum Exosomes in Bladder Cancer. Int. J. Mol. Sci. 2024, 25, 5955. [Google Scholar] [CrossRef] [PubMed]
- Myhrer, D.M.; Frøystad, M.; Paasche Roland, M.C.; Ueland, T.; Lekva, T. The long non-coding RPPH1 is decreased in leukocytes and increased in plasma from women developing pre-eclampsia. Biol. Reprod. 2024, 111, 427–435. [Google Scholar] [CrossRef]
- Tayefeh-Gholami, S.; Akbarzadeh, S.; Rajabi, A.; Najari, P.; Ghasemzadeh, T.; HosseinpourFeizi, M.; Safaralizadeh, R. Investigating SNHG3 and BCYRN1 lncRnas expression in the peripheral blood cells of multiple sclerosis patients. Neurol. Res. 2024, 46, 876–882. [Google Scholar] [CrossRef]
- Huang, Z.; Luo, Q.; Xiong, C.; Zhu, H.; Yu, C.; Xu, J.; Peng, Y.; Li, J.; Le, A. Identification of serum tRNA-derived small RNAs biosignature for diagnosis of tuberculosis. Emerg. Microbes Infect. 2025, 14, 2459132. [Google Scholar] [CrossRef]
- Ye, C.; Cheng, F.; Huang, L.; Wang, K.; Zhong, L.; Lu, Y.; Ouyang, M. New plasma diagnostic markers for colorectal cancer: Transporter fragments of glutamate tRNA origin. J. Cancer 2024, 15, 1299–1313. [Google Scholar] [CrossRef]
- Zhao, P.; Zhu, K.; Xie, C.; Liu, S.; Chen, X. Role and clinical value of serum hsa_tsr011468 in lung adenocarcinoma. Mol. Med. Rep. 2024, 30, 226. [Google Scholar] [CrossRef] [PubMed]
- Ferre-Giraldo, A.; Castells, M.; Sánchez-Herrero, J.F.; López-Rodrigo, O.; de Rocco-Ponce, M.; Bassas, L.; Vigués, F.; Sumoy, L.; Larriba, S. Semen sEV tRF-Based Models Increase Non-Invasive Prediction Accuracy of Clinically Significant Prostate Cancer among Patients with Moderately Altered PSA Levels. Int. J. Mol. Sci. 2024, 25, 10122. [Google Scholar] [CrossRef]
- Xu, X.; Chen, J.; Bai, M.; Liu, T.; Zhan, S.; Li, J.; Ma, Y.; Zhang, Y.; Wu, L.; Zhao, Z.; et al. Plasma tsRNA Signatures Serve as a Novel Biomarker for Bladder Cancer. Cancer Sci. 2025, 116, 1255–1267. [Google Scholar] [CrossRef]
- Li, C.; Zhong, S.; Chen, J.; Mu, X. TsRNA-49-73-Glu-CTC: A promising serum biomarker in non-small cell lung cancer. PLoS ONE 2025, 20, e0320187. [Google Scholar] [CrossRef]
- Tayae, E.; Osman, E.M.; Tawfik, M.R.; Hegazy, N.; Moaaz, M.; Ghazala, R.A. Expression Levels of Plasma YRNAs in Colorectal Cancer as a Potential Noninvasive Biomarker. J. Gastrointest. Cancer 2025, 56, 81. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, L.; Chen, M.; Wang, X. RNA Polymerase III-Transcribed RNAs in Health and Disease: Mechanisms, Dysfunction, and Future Directions. Int. J. Mol. Sci. 2025, 26, 5852. https://doi.org/10.3390/ijms26125852
Sun L, Chen M, Wang X. RNA Polymerase III-Transcribed RNAs in Health and Disease: Mechanisms, Dysfunction, and Future Directions. International Journal of Molecular Sciences. 2025; 26(12):5852. https://doi.org/10.3390/ijms26125852
Chicago/Turabian StyleSun, Longjie, Mingyue Chen, and Xin Wang. 2025. "RNA Polymerase III-Transcribed RNAs in Health and Disease: Mechanisms, Dysfunction, and Future Directions" International Journal of Molecular Sciences 26, no. 12: 5852. https://doi.org/10.3390/ijms26125852
APA StyleSun, L., Chen, M., & Wang, X. (2025). RNA Polymerase III-Transcribed RNAs in Health and Disease: Mechanisms, Dysfunction, and Future Directions. International Journal of Molecular Sciences, 26(12), 5852. https://doi.org/10.3390/ijms26125852






