Identification of SARS-CoV-2 Main Protease Cleavage Sites in Bovine β-Casein
Abstract
1. Introduction
2. Results and Discussion
2.1. In Silico Prediction of SARS-CoV-2 Mpro Cleavage Sites in Casein
2.2. In Vitro Cleavage of Casein Isoforms with SARS-CoV-2 Mpro
2.3. Mass Spectrometry-Based Analysis of SARS-CoV-2 Mpro Cleavage Sites
3. Materials and Methods
3.1. Proteins
3.2. In Silico Cleavage Site Prediction
3.3. Cleavage Reactions Using Casein as Substrate
3.4. Cleavage Site Identification by MALDI-TOF MS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Xu, W.; Liu, Y.; Li, H.; Chen, L. The research progress of SARS-CoV-2 main protease inhibitors from 2020 to 2022. Eur. J. Med. Chem. 2023, 257, 115491. [Google Scholar] [CrossRef]
- Mótyán, J.A.; Mahdi, M.; Hoffka, G.; Tőzsér, J. Potential Resistance of SARS-CoV-2 Main Protease (Mpro) against Protease Inhibitors: Lessons Learned from HIV-1 Protease. Int. J. Mol. Sci. 2022, 23, 3507. [Google Scholar] [CrossRef] [PubMed]
- Denaro, M.; Ferro, E.; Barrano, G.; Meli, S.; Busacca, M.; Corallo, D.; Capici, A.; Zisa, A.; Cucuzza, L.; Gradante, S.; et al. Monitoring of SARS-CoV-2 Infection in Ragusa Area: Next Generation Sequencing and Serological Analysis. Int. J. Mol. Sci. 2023, 24, 4742. [Google Scholar] [CrossRef] [PubMed]
- Mahdi, M.; Kiarie, I.W.; Mótyán, J.A.; Hoffka, G.; Al-Muffti, A.S.; Tóth, A.; Tőzsér, J. Receptor Binding for the Entry Mechanisms of SARS-CoV-2: Insights from the Original Strain and Emerging Variants. Viruses 2025, 17, 691. [Google Scholar] [CrossRef]
- Ihssen, J.; Faccio, G.; Yao, C.; Sirec, T.; Spitz, U. Fluorogenic in vitro activity assay for the main protease Mpro from SARS-CoV-2 and its adaptation to the identification of inhibitors. STAR Protoc. 2021, 2, 100793. [Google Scholar] [CrossRef]
- Dražić, T.; Kühl, N.; Leuthold, M.M.; Behnam, M.A.M.; Klein, C.D. Efficiency Improvements and Discovery of New Substrates for a SARS-CoV-2 Main Protease FRET Assay. SLAS Discov. 2021, 26, 1189–1199. [Google Scholar] [CrossRef]
- Legare, S.; Heide, F.; Bailey-Elkin, B.A.; Stetefeld, J. Improved SARS-CoV-2 main protease high-throughput screening assay using a 5-carboxyfluorescein substrate. J. Biol. Chem. 2022, 298, 101739. [Google Scholar] [CrossRef]
- Suresh, V.; Sheik, D.A.; Detomasi, T.C.; Zhao, T.; Zepeda, T.; Saladi, S.; Rajesh, U.C.; Byers, K.; Craik, C.S.; Davisson, V.J. A Prototype Assay Multiplexing SARS-CoV-2 3CL-Protease and Angiotensin-Converting Enzyme 2 for Saliva-Based Diagnostics in COVID-19. Biosensors 2023, 13, 682. [Google Scholar] [CrossRef]
- Anton, D.B.; Galvez Bulhões Pedreira, J.; Zvirtes, M.L.; Laufer, S.A.; Ducati, R.G.; Goettert, M.; Saraiva Macedo Timmers, L.F. Targeting SARS-CoV-2 Main Protease (MPro) with Kinase Inhibitors: A Promising Approach for Discovering Antiviral and Anti-inflammatory Molecules against SARS-CoV-2. J. Chem. Inf. Model. 2023, 63, 4138–4146. [Google Scholar] [CrossRef]
- Miczi, M.; Golda, M.; Kunkli, B.; Nagy, T.; Tőzsér, J.; Mótyán, J.A. Identification of Host Cellular Protein Substrates of SARS-COV-2 Main Protease. Int. J. Mol. Sci. 2020, 21, 9523. [Google Scholar] [CrossRef] [PubMed]
- Miltner, N.; Kalló, G.; Csősz, É.; Miczi, M.; Nagy, T.; Mahdi, M.; Mótyán, J.A.; Tőzsér, J. Identification of SARS-CoV-2 Main Protease (Mpro) Cleavage Sites Using Two-Dimensional Electrophoresis and In Silico Cleavage Site Prediction. Int. J. Mol. Sci. 2023, 24, 3236. [Google Scholar] [CrossRef] [PubMed]
- Cupp-Enyard, C. Sigma’s Non-specific Protease Activity Assay—Casein as a Substrate. J. Vis. Exp. 2008, 19, 899. [Google Scholar]
- Ong, I.L.H.; Yang, K.L. Recent developments in protease activity assays and sensors. Analyst 2017, 142, 1867–1881. [Google Scholar] [CrossRef]
- Hu, X.; Compton, J.R.; Legler, P.M. Analysis of Group IV Viral SSHHPS Using In Vitro and In Silico Methods. J. Vis. Exp. 2019, 154, e60421. [Google Scholar]
- Morazzani, E.M.; Compton, J.R.; Leary, D.H.; Berry, A.V.; Hu, X.; Marugan, J.J.; Glass, P.J.; Legler, P.M. Proteolytic cleavage of host proteins by the Group IV viral proteases of Venezuelan equine encephalitis virus and Zika virus. Antivir. Res. 2019, 164, 106–122. [Google Scholar] [CrossRef]
- Doctor, K.Z.; Gilmour, E.; Recarte, M.; Beatty, T.R.; Shifa, I.; Stangel, M.; Schwisow, J.; Leary, D.H.; Legler, P.M. Automated SSHHPS Analysis Predicts a Potential Host Protein Target Common to Several Neuroinvasive (+)ssRNA Viruses. Viruses 2023, 15, 542. [Google Scholar] [CrossRef]
- Reynolds, N.D.; Aceves, N.M.; Liu, J.L.; Compton, J.R.; Leary, D.H.; Freitas, B.T.; Pegan, S.D.; Doctor, K.Z.; Wu, F.Y.; Hu, X.; et al. The SARS-CoV-2 SSHHPS Recognized by the Papain-like Protease. ACS Infect. Dis. 2021, 7, 1483–1502. [Google Scholar] [CrossRef]
- Lin, C.W.; Tsai, C.H.; Tsai, F.J.; Chen, P.J.; Lai, C.C.; Wan, L.; Chiu, H.H.; Lin, K.H. Characterization of trans- and cis-cleavage activity of the SARS coronavirus 3CLpro protease: Basis for the in vitro screening of anti-SARS drugs. FEBS Lett. 2004, 574, 131–137. [Google Scholar] [CrossRef]
- Tripathi, P.K.; Upadhyay, S.; Singh, M.; Raghavendhar, S.; Bhardwaj, M.; Sharma, P.; Patel, A.K. Screening and evaluation of approved drugs as inhibitors of main protease of SARS-CoV-2. Int. J. Biol. Macromol. 2020, 164, 2622–2631. [Google Scholar] [CrossRef]
- Gupta, A.; Rani, C.; Pant, P.; Vijayan, V.; Vikram, N.; Kaur, P.; Singh, T.P.; Sharma, S.; Sharma, P. Structure-Based Virtual Screening and Biochemical Validation to Discover a Potential Inhibitor of the SARS-CoV-2 Main Protease. ACS Omega 2020, 5, 33151–33161. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.; Tripathi, P.K.; Singh, M.; Raghavendhar, S.; Bhardwaj, M.; Patel, A.K. Evaluation of medicinal herbs as a potential therapeutic option against SARS-CoV-2 targeting its main protease. Phytother. Res. 2020, 34, 3411–3419. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, A.; Kardam, V.; Inampudi, K.K.; Vrati, S.; Gupta, D.; Singh, A.; Kayampeta, S.R.; Appaiahgari, M.B.; Sehgal, D. Identification of a novel inhibitor of SARS-CoV-2 main protease: An in silico, biochemical, and cell-based approach. FEBS J. 2023, 290, 5496–5513. [Google Scholar] [CrossRef] [PubMed]
- Kiemer, L.; Lund, O.; Brunak, S.; Blom, N. Coronavirus 3CLpro proteinase cleavage sites: Possible relevance to SARS virus pathology. BMC Bioinform. 2004, 5, 72. [Google Scholar] [CrossRef]
- Chen, H.; Zhu, Z.; Qiu, Y.; Ge, X.; Zheng, H.; Peng, Y. Prediction of coronavirus 3C-like protease cleavage sites using machine-learning algorithms. Virol. Sin. 2022, 37, 437–444. [Google Scholar] [CrossRef]
- Scott, B.M.; Lacasse, V.; Blom, D.G.; Tonner, P.D.; Blom, N.S. Predicted coronavirus Nsp5 protease cleavage sites in the human proteome. BMC Genom. Data 2022, 23, 25. [Google Scholar] [CrossRef]
- Goetz, D.H.; Choe, Y.; Hansell, E.; Chen, Y.T.; McDowell, M.; Jonsson, C.B.; Roush, W.R.; McKerrow, J.; Craik, C.S. Substrate specificity profiling and identification of a new class of inhibitor for the major protease of the SARS coronavirus. Biochemistry 2007, 46, 8744–8752. [Google Scholar] [CrossRef]
- Chuck, C.P.; Chow, H.F.; Wan, D.C.; Wong, K.B. Profiling of substrate specificities of 3C-like proteases from group 1, 2a, 2b, and 3 coronaviruses. PLoS ONE 2011, 6, e27228. [Google Scholar] [CrossRef]
- Koudelka, T.; Boger, J.; Henkel, A.; Schönherr, R.; Krantz, S.; Fuchs, S.; Rodríguez, E.; Redecke, L.; Tholey, A. N-Terminomics for the Identification of In Vitro Substrates and Cleavage Site Specificity of the SARS-CoV-2 Main Protease. Proteomics 2021, 21, e2000246. [Google Scholar] [CrossRef]
- Pablos, I.; Machado, Y.; de Jesus, H.C.R.; Mohamud, Y.; Kappelhoff, R.; Lindskog, C.; Vlok, M.; Bell, P.A.; Butler, G.S.; Grin, P.M.; et al. Mechanistic insights into COVID-19 by global analysis of the SARS-CoV-2 3CLpro substrate degradome. Cell Rep. 2021, 37, 109892. [Google Scholar] [CrossRef]
- Taylor, E.W.; Radding, W. Understanding Selenium and Glutathione as Antiviral Factors in COVID-19: Does the Viral Mpro Protease Target Host Selenoproteins and Glutathione Synthesis? Front. Nutr. 2020, 7, 143. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, I.A.; Todd, D.A.; Lima, S.T.; Chekan, J.R.; Chiu, N.H.; Taylor, E.W. SARS-CoV-2 Main Protease Targets Host Selenoproteins and Glutathione Biosynthesis for Knockdown via Proteolysis, Potentially Disrupting the Thioredoxin and Glutaredoxin Redox Cycles. Antioxidants 2023, 12, 559. [Google Scholar] [CrossRef] [PubMed]
- Gazda, L.D.; Joóné Matúz, K.; Nagy, T.; Mótyán, J.A.; Tőzsér, J. Biochemical characterization of Ty1 retrotransposon protease. PLoS ONE 2020, 15, e0227062. [Google Scholar] [CrossRef]
- Kamiński, S.; Cieslińska, A.; Kostyra, E. Polymorphism of bovine beta-casein and its potential effect on human health. J. Appl. Genet. 2007, 48, 189–198. [Google Scholar] [CrossRef]
- Ham, J.S.; Han, G.S.; Jeong, S.G.; Seol, K.H.; Jang, A.R.; Oh, M.H.; Kim, D.H.; Park, Y.W. Determination of molecular weights of caprine milk proteins by matrix-assisted laser desorption/ionization mass spectrometry. J. Dairy Sci. 2012, 95, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Vincent, D.; Elkins, A.; Condina, M.R.; Ezernieks, V.; Rochfort, S. Quantitation and Identification of Intact Major Milk Proteins for High-Throughput LC-ESI-Q-TOF MS Analyses. PLoS ONE 2016, 11, e0163471. [Google Scholar] [CrossRef]
- Meyer, B.; Chiaravalli, J.; Gellenoncourt, S.; Brownridge, P.; Bryne, D.P.; Daly, L.A.; Grauslys, A.; Walter, M.; Agou, F.; Chakrabarti, L.A.; et al. Characterising proteolysis during SARS-CoV-2 infection identifies viral cleavage sites and cellular targets with therapeutic potential. Nat. Commun. 2021, 12, 5553. [Google Scholar] [CrossRef]
- Mahdi, M.; Mótyán, J.A.; Szojka, Z.I.; Golda, M.; Miczi, M.; Tőzsér, J. Analysis of the efficacy of HIV protease inhibitors against SARS-CoV-2′s main protease. Virol. J. 2020, 17, 190. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Breidenbach, J.; Lemke, C.; Pillaiyar, T.; Schäkel, L.; Al Hamwi, G.; Diett, M.; Gedschold, R.; Geiger, N.; Lopez, V.; Mirza, S.; et al. Targeting the Main Protease of SARS-CoV-2: From the Establishment of High Throughput Screening to the Design of Tailored Inhibitors. Angew. Chem. Int. Ed. Engl. 2021, 60, 10423–10429. [Google Scholar] [CrossRef]
- Kuellenberg de Gaudry, D.; Lohner, S.; Bischoff, K.; Schmucker, C.; Hoerrlein, S.; Roeger, C.; Schwingshackl, L.; Meerpohl, J.J. A1- and A2 beta-casein on health-related outcomes: A scoping review of animal studies. Eur. J. Nutr. 2022, 61, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Thiruvengadam, M.; Venkidasamy, B.; Thirupathi, P.; Chung, I.M.; Subramanian, U. β-Casomorphin: A complete health perspective. Food Chem. 2021, 337, 127765. [Google Scholar] [CrossRef] [PubMed]
- Bolat, E.; Eker, F.; Yılmaz, S.; Karav, S.; Oz, E.; Brennan, C.; Proestos, C.; Zeng, M.; Oz, F. BCM-7: Opioid-like Peptide with Potential Role in Disease Mechanisms. Molecules 2024, 29, 2161. [Google Scholar] [CrossRef]
- de Vasconcelos, M.L.; Oliveira, L.M.F.S.; Hill, J.P.; Vidal, A.M.C. Difficulties in Establishing the Adverse Effects of β-Casomorphin-7 Released from β-Casein Variants-A Review. Foods 2023, 12, 3151. [Google Scholar] [CrossRef] [PubMed]
- Yaghi, R.M.; Andrews, C.L.; Wylie, D.C.; Iverson, B.L. High-Resolution Substrate Specificity Profiling of SARS-CoV-2 Mpro; Comparison to SARS-CoV Mpro. ACS Chem. Biol. 2024, 19, 1474–1483. [Google Scholar] [CrossRef]
- van Vliet, V.J.E.; Huynh, N.; Palà, J.; Patel, A.; Singer, A.; Slater, C.; Chung, J.; van Huizen, M.; Teyra, J.; Miersch, S.; et al. Ubiquitin variants potently inhibit SARS-CoV-2 PLpro and viral replication via a novel site distal to the protease active site. PLoS Pathog. 2022, 18, e1011065. [Google Scholar] [CrossRef]
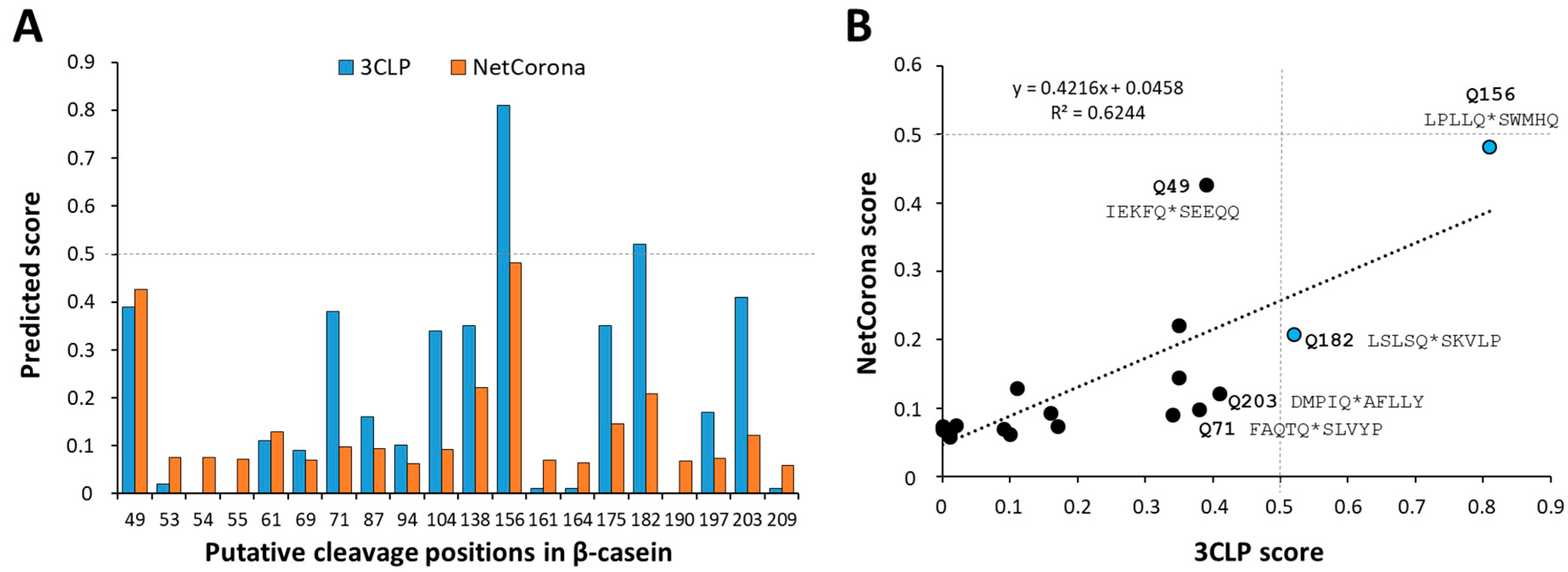
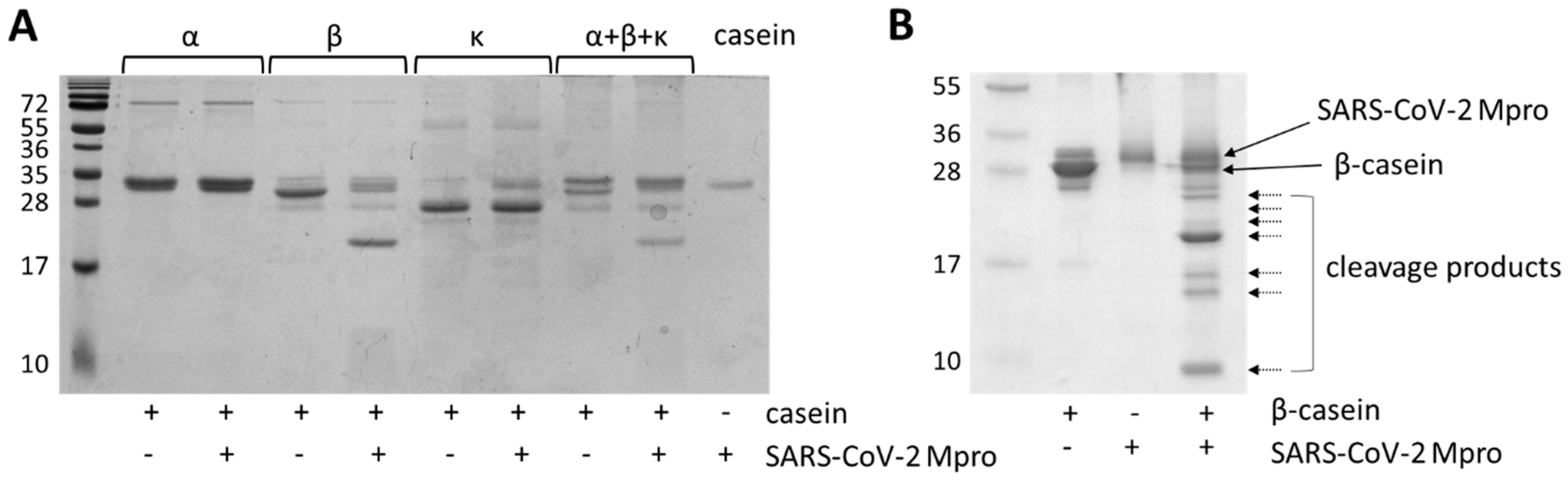
| Cleavage Position(s) | Cleavage Site Sequence | Theoretical | Measured |
|---|---|---|---|
| β-casein * | 24,001 | 24,045 | |
| 71 | FAQTQ*SLVYP | 17,057 | 17,079 |
| 6944 | 6965 | ||
| 87 | NSLPQ*NIPPL | 15,349 | 15,308 |
| 156 | LPLLQ*SWMHQ | 16,349 | 16,373 |
| 7652 | 7658 ** | ||
| 182 | LSLSQ*SKVLP | 4700 | 4687 |
| 71 + 190 | LPVPQ*KAVPY | 13,225 | 13,250 ** |
| 94 + 190 | PPLTQ*TPVVV | 10,753 | 10,770 ** |
| Protein | Manufacturer | Source | UniProt ID |
|---|---|---|---|
| casein sodium salt (α-S1, α-S2, β, κ) * | Sigma-Aldrich, C8654 | bovine milk | - |
| α-casein (α-S1, α-S2) | Sigma-Aldrich, C6780 | bovine milk | P02662 (α-S1) P02663 (α-S2) |
| β-casein | Sigma-Aldrich, C6905 | bovine milk | P02666 |
| κ-casein | Sigma-Aldrich, C0406 | bovine milk | P02668 |
| SARS-CoV-2 Mpro ** | in-house stock | recombinant | P0DTC1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mótyán, J.A.; Nagy, T.; Nagyné Veres, Á.; Golda, M.; Mahdi, M.; Tőzsér, J. Identification of SARS-CoV-2 Main Protease Cleavage Sites in Bovine β-Casein. Int. J. Mol. Sci. 2025, 26, 5829. https://doi.org/10.3390/ijms26125829
Mótyán JA, Nagy T, Nagyné Veres Á, Golda M, Mahdi M, Tőzsér J. Identification of SARS-CoV-2 Main Protease Cleavage Sites in Bovine β-Casein. International Journal of Molecular Sciences. 2025; 26(12):5829. https://doi.org/10.3390/ijms26125829
Chicago/Turabian StyleMótyán, János András, Tibor Nagy, Ágota Nagyné Veres, Mária Golda, Mohamed Mahdi, and József Tőzsér. 2025. "Identification of SARS-CoV-2 Main Protease Cleavage Sites in Bovine β-Casein" International Journal of Molecular Sciences 26, no. 12: 5829. https://doi.org/10.3390/ijms26125829
APA StyleMótyán, J. A., Nagy, T., Nagyné Veres, Á., Golda, M., Mahdi, M., & Tőzsér, J. (2025). Identification of SARS-CoV-2 Main Protease Cleavage Sites in Bovine β-Casein. International Journal of Molecular Sciences, 26(12), 5829. https://doi.org/10.3390/ijms26125829









