Biodegradation of Poly(ε-caprolactone): Microorganisms, Enzymes, and Mechanisms
Abstract
1. Introduction
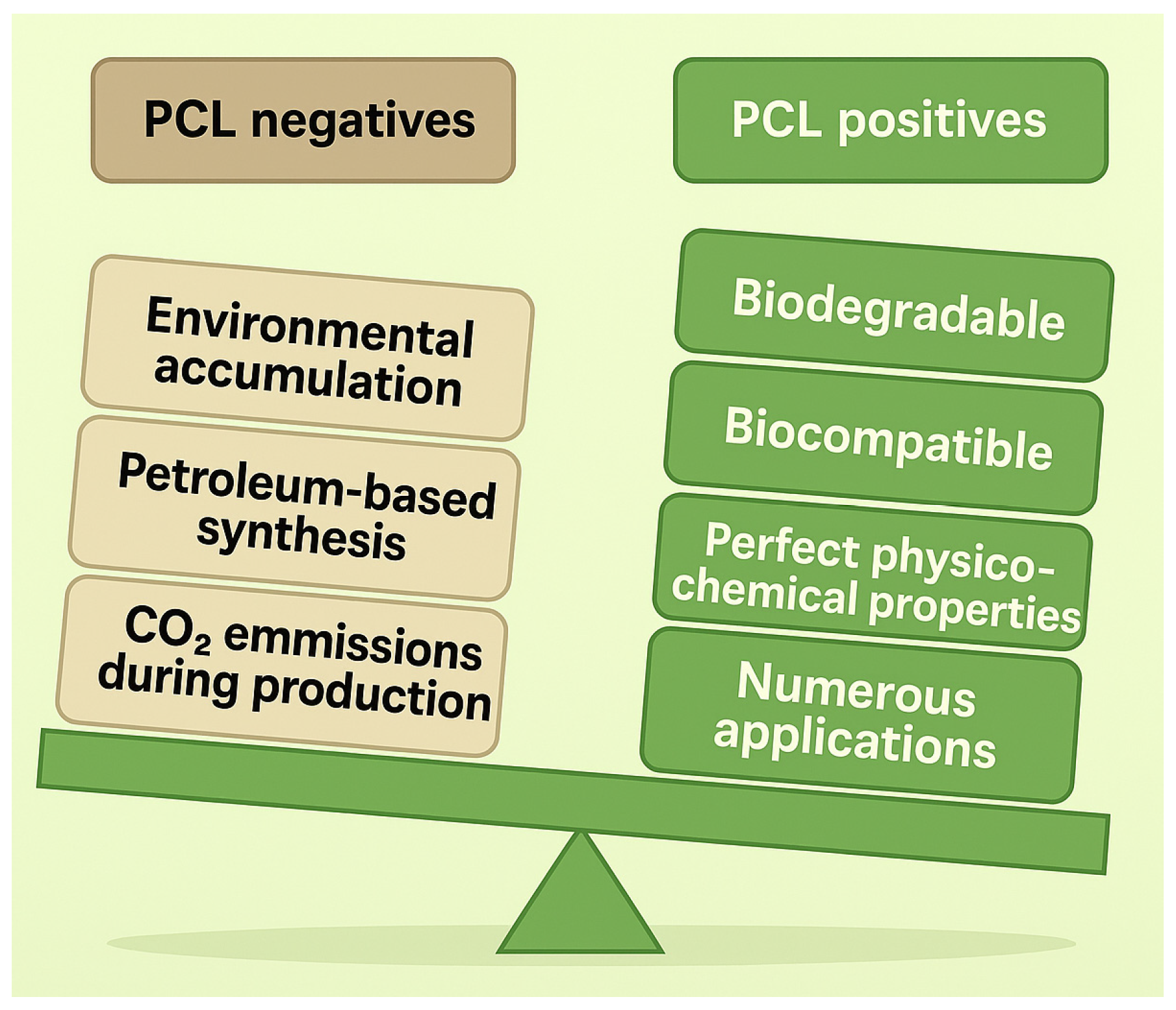
2. PCL Synthesis, Structure, Physicochemical Properties, and Applications
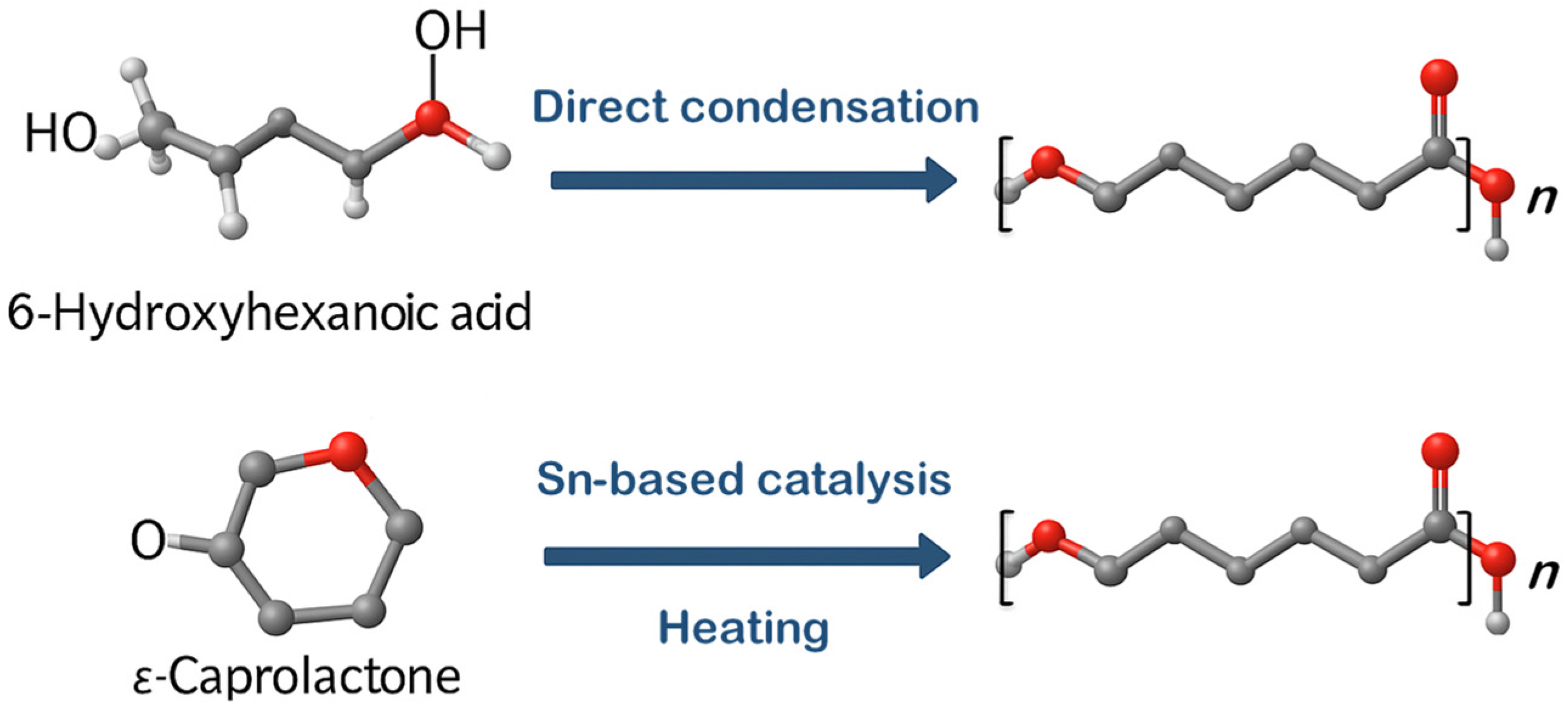
2.1. PCL Synthesis Methods
2.2. PCL Structure and Physicochemical Properties
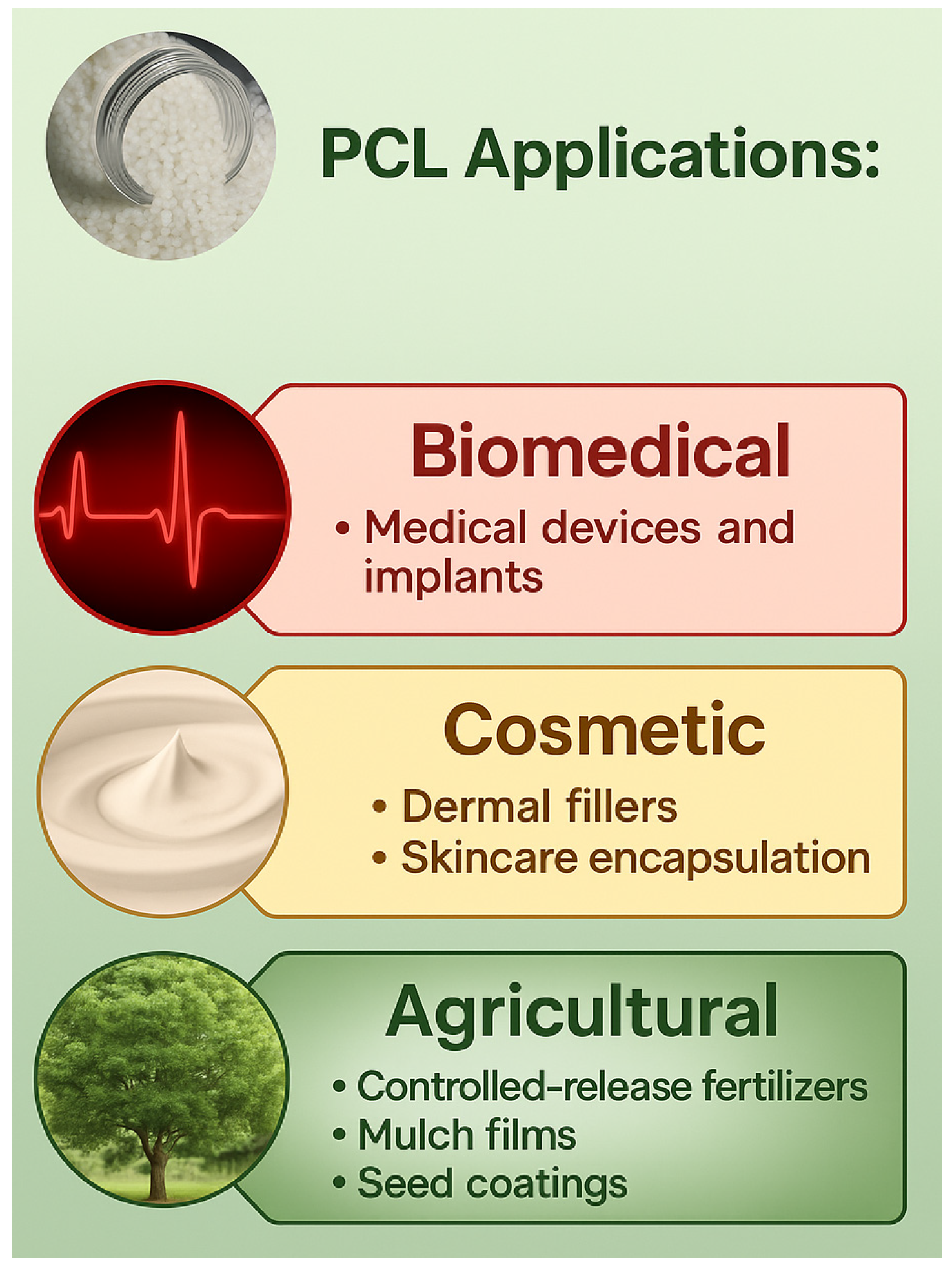
2.3. PCL Applications
2.3.1. Biomedical Applications
2.3.2. Cosmetic, Agricultural, and Environmental Applications
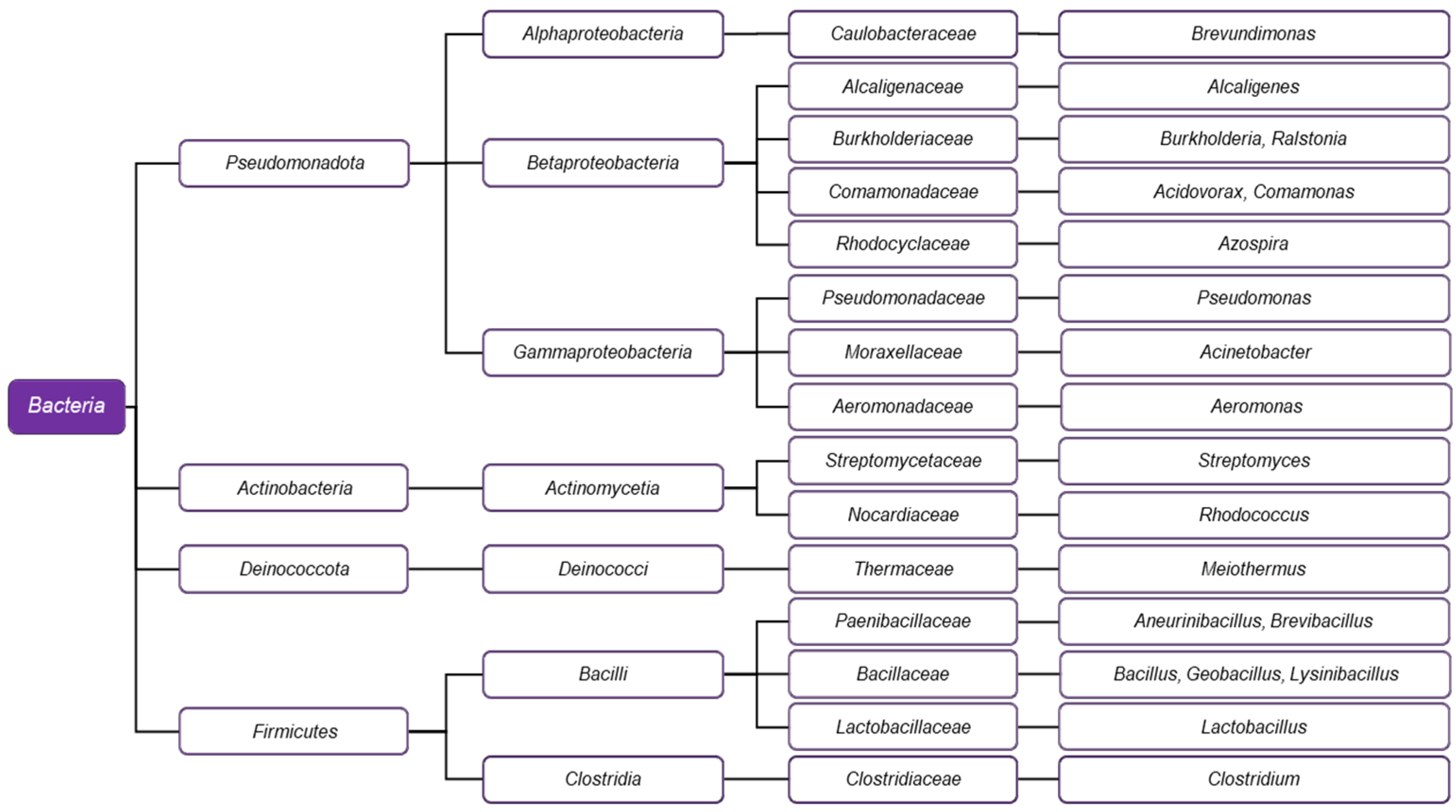
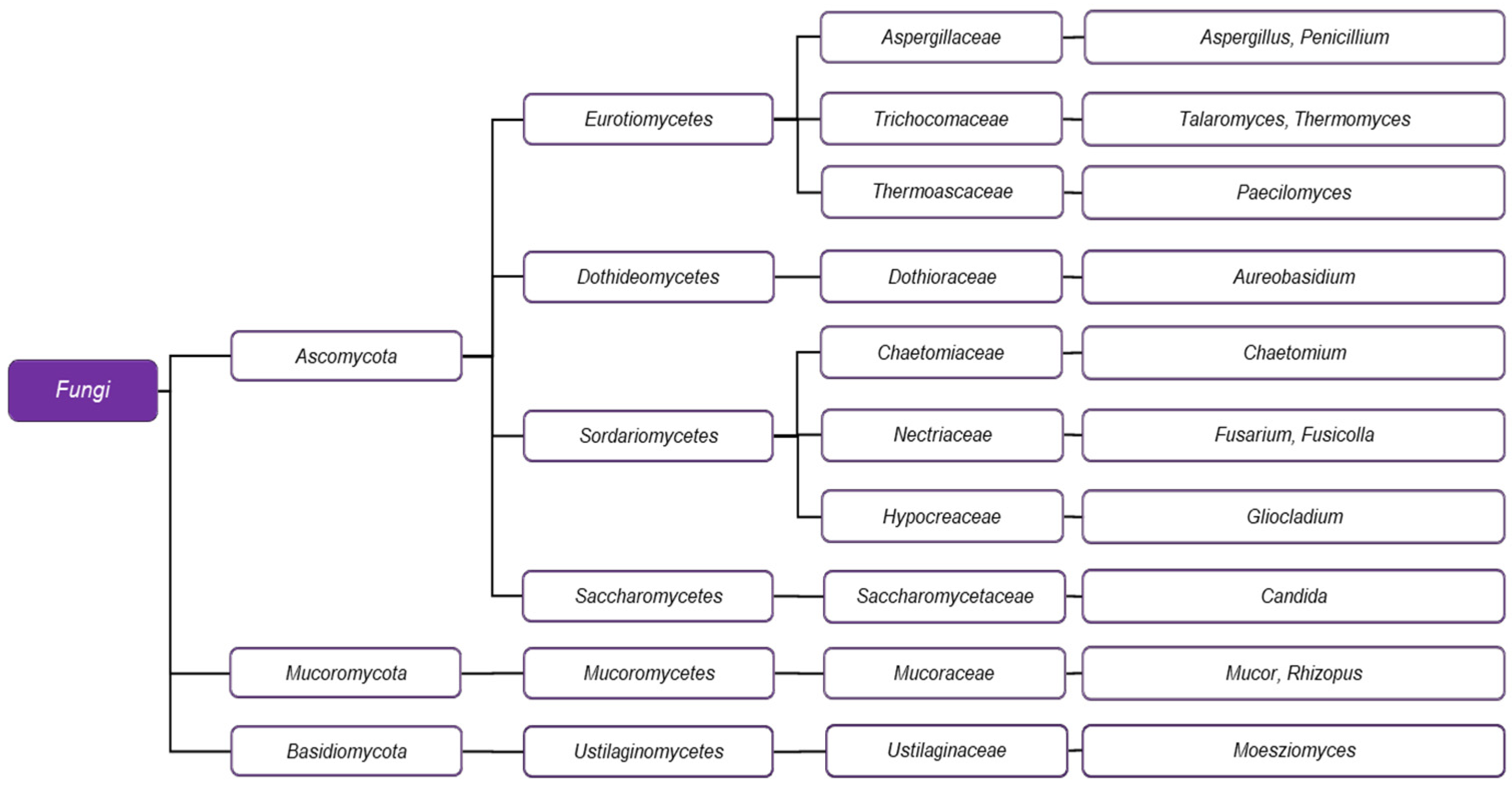
3. Biodiversity of Microorganisms Capable of PCL Biodegradation
3.1. PCL-Degrading Bacteria
3.1.1. Pseudomonadota (Formerly Proteobacteria)
3.1.2. Actinobacteria
3.1.3. Firmicutes and Deinococcota
3.1.4. Putative PCL Degradation by Other Bacterial Species
3.2. PCL-Degrading Fungi
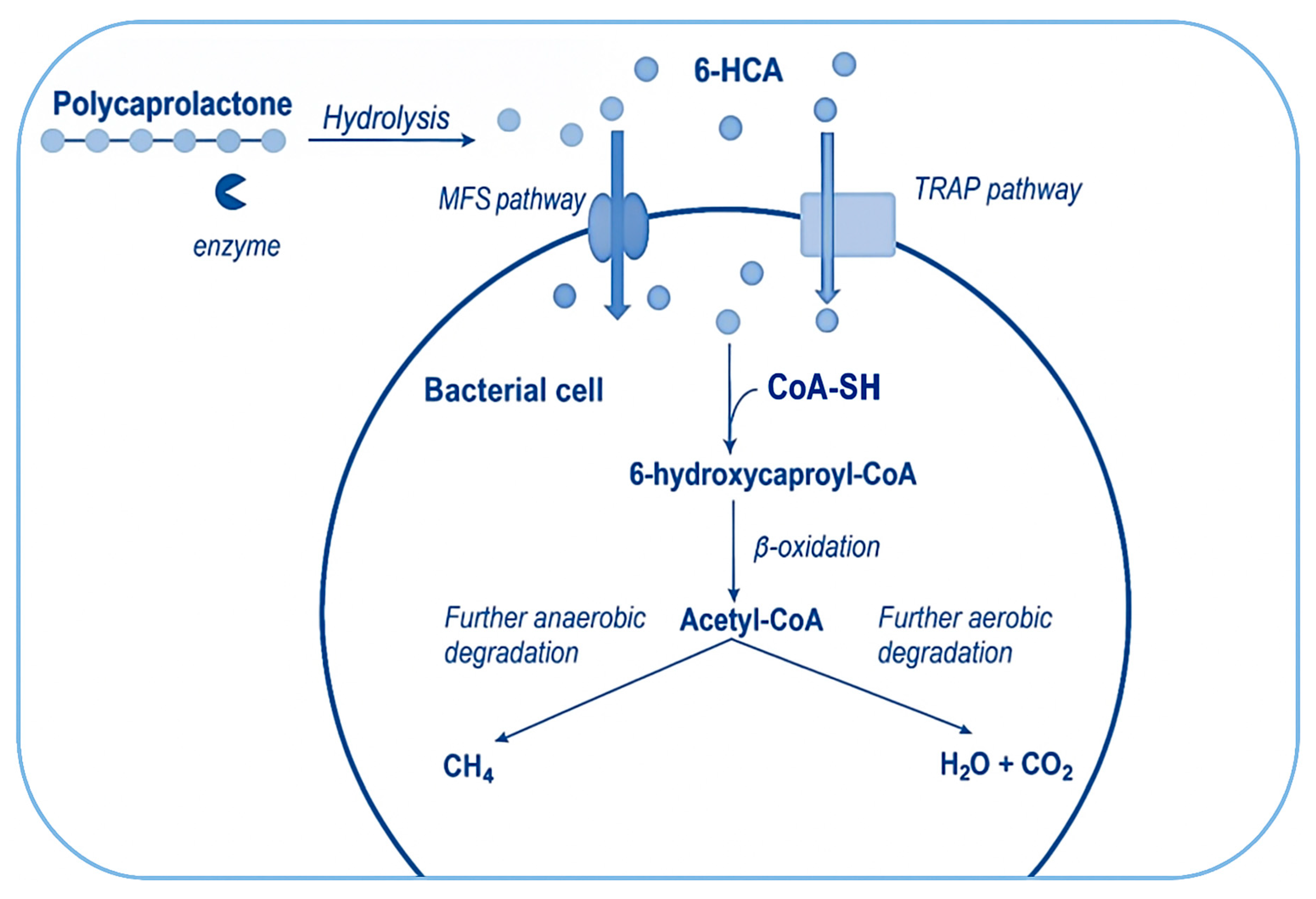
4. Mechanism of Microbial PCL Biodegradation
4.1. Overview of Enzymes Involved in the Process of PCL Degradation
4.2. Enzymes Involved in the Process of PCL Degradation by Bacteria
4.3. Enzymes Involved in the Process of PCL Degradation by Fungi
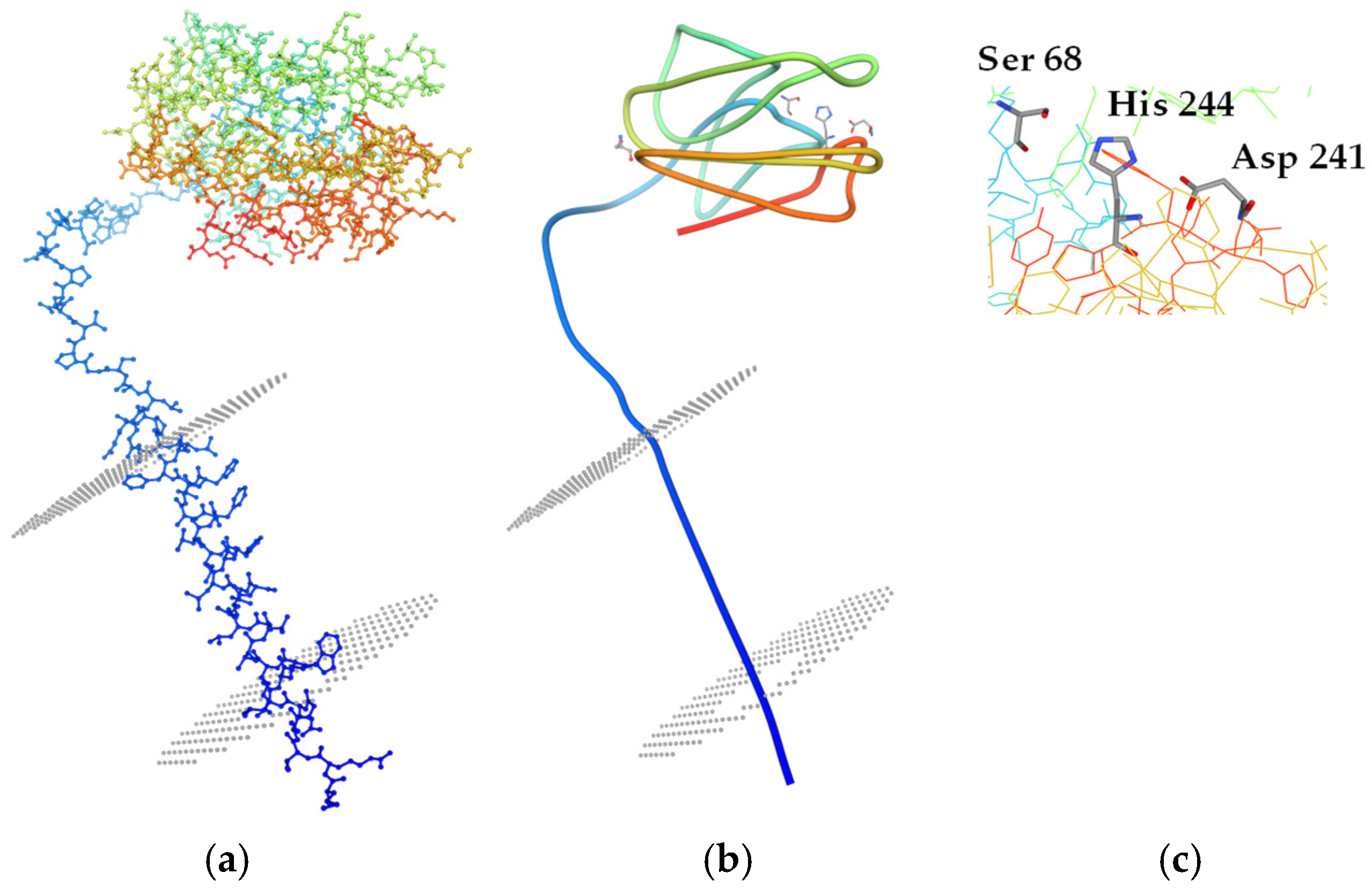
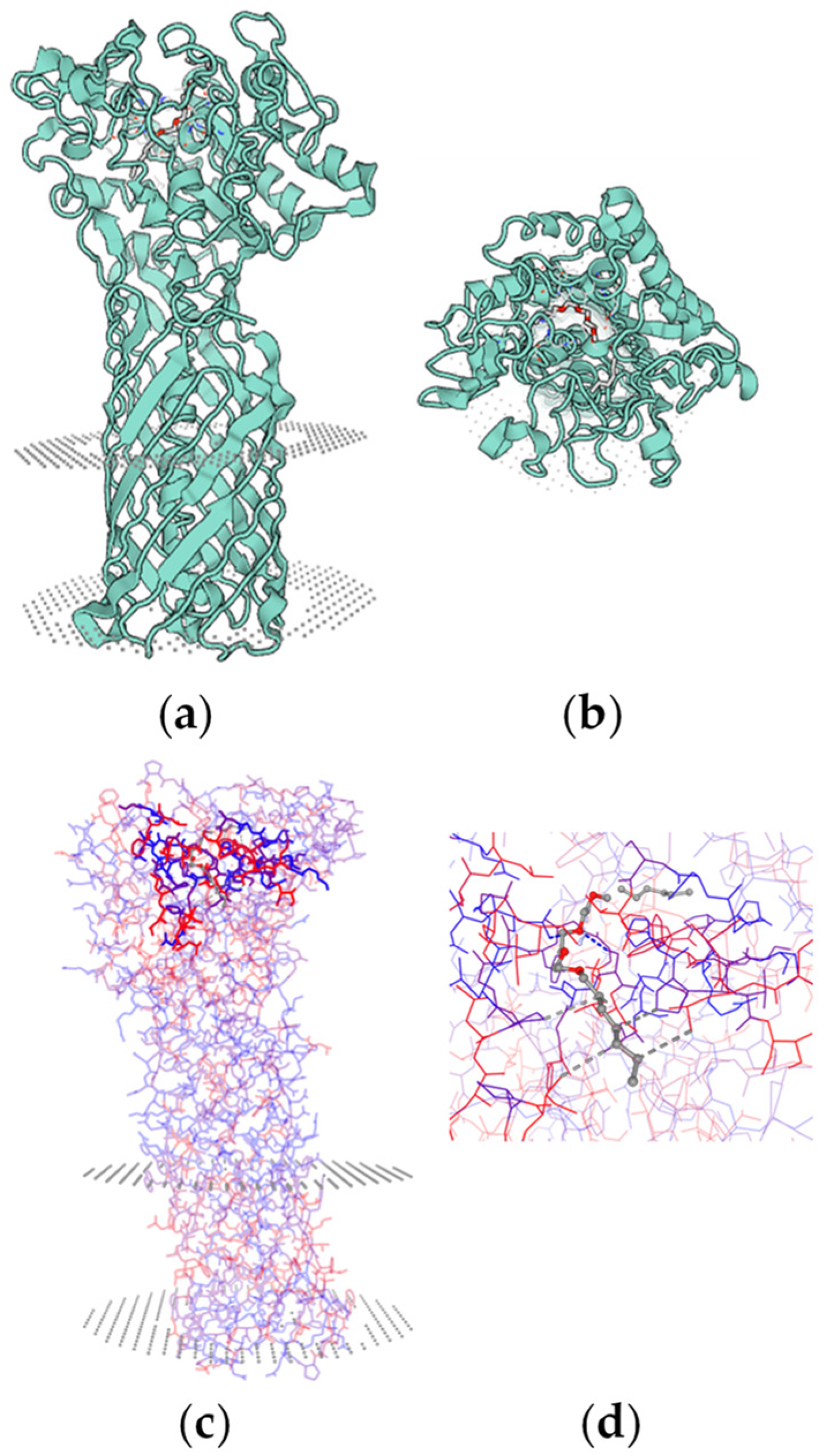
4.4. Catalytic Mechanism of Enzymatic PCL Degradation
5. Conclusions and Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Ramaraju, H.; Verga, A.S.; Steedley, B.J.; Kowblansky, A.P.; Green, G.E.; Hollister, S.J. Investigation of the biodegradation kinetics and associated mechanical properties of 3D-printed polycaprolactone during long-term preclinical testing. Biomaterial 2025, 321, 123257. [Google Scholar] [CrossRef] [PubMed]
- Richert, A.; Kalwasińska, A.; Brzezinska, M.S.; Dąbrowska, G.B. Biodegradability of Novel Polylactide and Polycaprolactone Materials with Bacteriostatic Properties Due to Embedded Birch Tar in Different Environments. Int. J. Mol. Sci. 2021, 22, 10228. [Google Scholar] [CrossRef]
- Injorhor, P.; Trongsatitkul, T.; Wittayakun, J.; Ruksakulpiwat, C.; Ruksakulpiwat, Y. Biodegradable Polylactic Acid-Polyhydroxyalkanoate-Based Nanocomposites with Bio-Hydroxyapatite: Preparation and Characterization. Polymers 2023, 15, 1261. [Google Scholar] [CrossRef]
- Naser, A.Z.; Deiab, I.; Defersha, F.; Yang, S. Expanding Poly(lactic acid) (PLA) and Polyhydroxyalkanoates (PHAs) Applications: A Review on Modifications and Effects. Polymers 2021, 13, 4271. [Google Scholar] [CrossRef]
- Pyo, S.-H.; Park, J.H.; Srebny, V.; Hatti-Kaul, R. A sustainable synthetic route for biobased 6-hydroxyhexanoic acid, adipic acid and ε-caprolactone by integrating bio- and chemical catalysis. Green Chem. 2020, 22, 4450–4455. [Google Scholar] [CrossRef]
- Shekh, M.R.; Kumar, V. Impact of plastic pollution on ecosystems: A review of adverse effects and sustainable solutions. Environ. Monit. Assess. 2025, 197, 264. [Google Scholar] [CrossRef] [PubMed]
- Krobot, Š.; Melčová, V.; Menčík, P.; Kontárová, S.; Rampichová, M.; Hedvičáková, V.; Mojžišová, E.; Baco, A.; Přikryl, R. Poly(3-hydroxybutyrate) (PHB) and Polycaprolactone (PCL) Based Blends for Tissue Engineering and Bone Medical Applications Processed by FDM 3D Printing. Polymers 2023, 15, 2404. [Google Scholar] [CrossRef]
- Hottle, T.A.; Bilec, M.M.; Landis, A.E. Biopolymer production and end of life comparisons using life cycle assessment. Resour. Conserv. Recycl. 2017, 122, 295–306. [Google Scholar] [CrossRef]
- Narancic, T.; Verstichel, S.; Chaganti, S.R.; Morales-Gamez, L.; Kenny, S.T.; De Wilde, B.; Padamati, R.B.; O’Connor, K.E. Biodegradable plastic blends create new possibilities for end-of-life management of plastics but they are not a panacea for plastic pollution. Environ. Sci. Technol. 2018, 52, 10441–10452. [Google Scholar] [CrossRef]
- Mandic, M.; Spasic, J.; Ponjavic, M.; Nikolic, M.; Cosovic, V.R.; O’Connor, K.E.; Nikodinovic-Runic, J.; Djokic, L.; Jeremic, S. Biodegradation of poly(ε-caprolactone) (PCL) and medium chain length polyhydroxyalkanoate (mcl-PHA) using whole cells and cell free protein preparations of Pseudomonas and Streptomyces strains grown on waste cooking oil. Polym. Degrad. Stab. 2019, 162, 160–168. [Google Scholar] [CrossRef]
- Labet, M.; Thielemans, W. Synthesis of PCL: A review. Chem. Soc. Rev. 2009, 38, 3484–3504. [Google Scholar] [CrossRef] [PubMed]
- Martín-González, D.; de la Fuente Tagarro, C.; De Lucas, A.; Bordel, S.; Santos-Beneit, F. Genetic Modifications in Bacteria for the Degradation of Synthetic Polymers: A Review. Int. J. Mol. Sci. 2024, 25, 5536. [Google Scholar] [CrossRef] [PubMed]
- Kayan, G.Ö.; Kayan, A. Polycaprolactone Composites/Blends and Their Applications Especially in Water Treatment. ChemEngineering 2023, 7, 104. [Google Scholar] [CrossRef]
- Woodruff, M.A.; Hutmacher, D.W. The return of a forgotten polymer—Polycaprolactone in the 21st century. Prog. Polym. Sci. 2010, 35, 1217–1256. [Google Scholar] [CrossRef]
- Kayan, A. Recent studies on single site metal alkoxide complexes as catalysts for ring opening polymerization of cyclic compounds. Catal. Surv. Asia 2020, 24, 87–103. [Google Scholar] [CrossRef]
- Grobelny, Z.; Golba, S.; Jurek-Suliga, J. Mechanism of ε-caprolactone polymerization in the presence of alkali metal salts: Investigation of initiation course and determination of polymers structure by MALDI-TOF mass spectrometry. Polym. Bull. 2019, 76, 3501–3515. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, L.; Chen, C.; Huang, P.; Dai, H.; Jiang, W.; Zhou, Y. Living Cationic Polymerization of ε-Caprolactone Catalyzed by a Metal-free Lewis Acid of Trityl Tetrafluoroborate. Macromolecules 2023, 56, 501–509. [Google Scholar] [CrossRef]
- Vidal, J.L.; Yavitt, B.M.; Wheeler, M.D.; Kolwich, J.L.; Donovan, L.N.; Sit, C.S.; Kerton, F.M. Biochar as a sustainable and renewable additive for the production of Poly (ε-caprolactone) composites. Sustain. Chem. Pharm. 2022, 25, 100586. [Google Scholar] [CrossRef]
- Srinivasamurthy, V.S.T.; Böttcher, D.; Engel, J.; Kara, S.; Bornscheuer, U.T. A whole-cell process for the production of ε-caprolactone in aqueous media. Proc. Biochem. 2020, 88, 22–30. [Google Scholar] [CrossRef]
- Schmidt, S.; Scherkus, C.; Muschiol, J.; Menyes, U.; Winkler, T.; Hummel, W.; Gröger, H.; Liese, A.; Herz, H.G.; Bornscheuer, U.T. An enzyme cascade synthesis of ε-caprolactone and its oligomers. Angew. Chem. Int. Ed. Engl. 2015, 54, 2784–2787. [Google Scholar] [CrossRef]
- Dong, H.; Wang, H.; Cao, S.; Shen, J. Lipase-catalyzed polymerization of lactones and linear hydroxyesters. Biotechnol. Lett. 1998, 20, 905–908. [Google Scholar] [CrossRef]
- Yu, W.; Song, X.; Wang, Y.; Zhang, L.; Liu, Y.; Liu, Y. Enhancing the oil/water separation efficiency of polylactic acid fiber membrane via polydimethylsiloxane-PCL copolymer. J. Environ. Chem. Eng. 2024, 12, 114738. [Google Scholar] [CrossRef]
- Thakur, M.; Majid, I.; Hussain, S.; Nanda, V. Poly (ε-caprolactone): A potential polymer for biodegradable food packaging applications. Packag. Technol. Sci. 2021, 34, 449–461. [Google Scholar] [CrossRef]
- Guarino, V.; Gentile, G.; Sorrentino, L.; Ambrosio, L. Polycaprolactone: Synthesis, properties, and applications. In Encyclopedia of Polymer Science and Technology; Wiley: Hoboken, NJ, USA, 2017; pp. 1–36. [Google Scholar] [CrossRef]
- Goel, V.; Luthra, P.; Kapur, G.S.; Ramakumar, S.S.V. Biodegradable/bio-plastics: Myths and realities. J. Polym. Environ. 2021, 29, 3079–3104. [Google Scholar] [CrossRef]
- Radhakrishnan, S.; Nagarajan, S.; Belaid, H.; Farha, C.; Iatsunskyi, I.; Coy, E.; Soussan, L.; Huon, V.; Bares, J.; Belkacemi, K.; et al. Fabrication of 3D printed antimicrobial PCL scaffolds for tissue engineering applications. Mater. Sci. Eng. C 2020, 118, 111525. [Google Scholar] [CrossRef]
- Castro, J.I.; Araujo-Rodríguez, D.G.; Valencia-Llano, C.H.; Tenorio, D.L.; Saavedra, M.; Zapata, P.A.; Grande-Tovar, C.D. Biocompatibility Assessment of PCL/Polylactic Acid/Zinc Oxide Nanoparticle Composites under In Vivo Conditions for Biomedical Applications. Pharmaceutics 2023, 15, 2196. [Google Scholar] [CrossRef]
- Zhou, L.; He, H.; Li, M.-C.; Huang, S.; Mei, C.; Wu, Q. Grafting PCL diol onto cellulose nanocrystals via click chemistry: Enhancing thermal stability and hydrophobic property. Carbohydr. Polym. 2018, 189, 331–341. [Google Scholar] [CrossRef]
- Li, P.; Ruan, L.; Jiang, G.; Sun, Y.; Wang, R.; Gao, X.; Yunusov, K.E.; Aharodnikau, U.E.; Solomevich, S.O. Design of 3D PCL/ε-polylysine-modified chitosan fibrous scaffolds with incorporation of bioactive factors for accelerating wound healing. Acta Biomater. 2022, 152, 197–209. [Google Scholar] [CrossRef]
- Menossi, M.; Salcedo, F.; Rivilli, N.; Nicolini, A.T.; Alvarez, V.A.; Ludueña, L.N. Biodegradable mulch films based on Starch/Poly (Lactic Acid)/Poly (Ε-Caprolactone) ternary blends. J. Polym. Environ. 2022, 31, 2114–2137. [Google Scholar] [CrossRef]
- Salvekar, A.V.; Zhou, Y.; Huang, W.M.; Wong, Y.S.; Venkatraman, S.S.; Shen, Z.; Zhu, G.; Cui, H.P. Shape/temperature memory phenomena in un-crosslinked poly-ε-caprolactone (PCL). Eur. Polym. J. 2015, 72, 282–295. [Google Scholar] [CrossRef]
- Jing, X.; Mi, H.-Y.; Huang, H.-X.; Turng, L.-S. Shape memory thermoplastic polyurethane (TPU)/poly(ε-caprolactone) (PCL) blends as self-knotting sutures. J. Mech. Behav. Biomed. Mater. 2016, 64, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, S.M.; Patil, H.I.; San Martin Martinez, E.; Casañas Pimentel, R.; Ige, P.P. Poly-ε-caprolactone (PCL), a promising polymer for pharmaceutical and biomedical applications: Focus on nanomedicine in cancer. Int. J. Polym. Mater. Polym. Biomater. 2020, 69, 85–126. [Google Scholar] [CrossRef]
- Homaeigohar, S.; Boccaccini, A.R. Nature-Derived and Synthetic Additives to poly (ε-Caprolactone) Nanofibrous Systems for Biomedicine; an Updated Overview. Front. Chem. 2022, 9, 809676. [Google Scholar] [CrossRef]
- Peng, Y.; Lu, Y.; Liu, K.; Liu, S.; Fan, L.; Huang, W. Biodegradable balloon-expandable self-locking PCL stents as buckling explants for the treatment of retinal detachment: An in vitro and in vivo study. J. Biomed. Mater. Res. A 2012, 101, 167–175. [Google Scholar] [CrossRef]
- Silva-López, M.S.; Alcántara-Quintana, L.E. The era of biomaterials: Smart implants? ACS Appl. Bio Mater. 2023, 6, 2982–2994. [Google Scholar] [CrossRef] [PubMed]
- Stafin, K.; Śliwa, P.; Piątkowski, M. Towards Polycaprolactone-Based Scaffolds for Alveolar Bone Tissue Engineering: A Biomimetic Approach in a 3D Printing Technique. Int. J. Mol. Sci. 2023, 24, 16180. [Google Scholar] [CrossRef] [PubMed]
- Bou-Francis, A.; Piercey, M.; Al-Qatami, O.; Mazzanti, G.; Khattab, R.; Ghanem, A. PCL blends for fracture fixation in low load-bearing applications. J. Appl. Polym. Sci. 2020, 137, 48940. [Google Scholar] [CrossRef]
- Raina, N.; Pahwa, R.; Khosla, J.K.; Gupta, P.N.; Gupta, M. PCL-based materials in wound healing applications. Polym. Bull. 2021, 79, 7041–7063. [Google Scholar] [CrossRef]
- Kotcharat, P.; Chuysinuan, P.; Thanyacharoen, T.; Techasakul, S.; Ummartyotin, S. Development of bacterial cellulose and polycaprolactone (PCL) based composite for medical material. Sustain. Chem. Pharm. 2021, 20, 100404. [Google Scholar] [CrossRef]
- Ahmed, M.K.; Zayed, M.A.; El-Dek, S.I.; Hady, M.A.; El Sherbiny, D.H.; Uskoković, V. Nanofibrous ε-polycaprolactone scaffolds containing Ag-doped magnetite nanoparticles: Physicochemical characterization and biological testing for wound dressing applications in vitro and in vivo. Bioact. Mater. 2021, 6, 2070–2088. [Google Scholar] [CrossRef]
- Baptista, C.; Azagury, A.; Shin, H.; Baker, C.M.; Ly, E.; Lee, R.; Mathiowitz, E. The effect of temperature and pressure on PCL morphology. Polymer 2020, 191, 122227. [Google Scholar] [CrossRef]
- Musílková, J.; Beran, M.; Sedlář, A.; Slepička, P.; Bartoš, M.; Kolská, Z.; Havlíčková, Š.; Luňáčková, J.; Svobodová, L.; Froněk, M.; et al. Composite Polylactide/Polycaprolactone Foams with Hierarchical Porous Structure for Pre-Vascularized Tissue Engineering. Int. J. Mol. Sci. 2025, 26, 2974. [Google Scholar] [CrossRef]
- Casanova, F.; Santos, L. Encapsulation of cosmetic active ingredients for topical application—A review. J. Microencapsul. 2015, 33, 1–17. [Google Scholar] [CrossRef]
- Christen, M.-O.; Vercesi, F. PCL: How a Well-Known and Futuristic Polymer Has Become an Innovative Collagen-Stimulator in Esthetics. Clin. Cosm. Investig. Dermatol. 2020, 13, 31–48. [Google Scholar] [CrossRef]
- Qiao, D.; Li, J.; Zhang, S.; Yang, X. Controlled release fertilizer with temperature-responsive behavior coated using polyether polyol (PPG)/PCL (PCL) blend-based polyurethane performs smart nutrient release. Mater. Today Chem. 2022, 26, 101249. [Google Scholar] [CrossRef]
- Yang, N.; Ying, L.; Li, K.; Chen, F.; Zhao, F.; Sun, Z.; Feng, L.; Liu, J. Biodegradable mulching films based on PCL and its porous structure construction. Polymers 2022, 14, 5340. [Google Scholar] [CrossRef]
- Chakkalakkal, N.D.; Thomas, M.; Chittillapilly, P.S.; Sujith, A.; Anjali, P.D. Electrospun polymer nanocomposite membrane as a promising seed coat for controlled release of agrichemicals and improved germination: Towards a better agricultural prospect. J. Clean. Prod. 2022, 377, 134479. [Google Scholar] [CrossRef]
- Hamzah, M.S.A.; Austad, A.; Abd Razak, S.I.; Nayan, N.H.M. Tensile and wettability properties of electrospun polycaprolactone coated with pectin/polyaniline composite for drug delivery application. Int. J. Struct. Integr. 2019, 10, 704–713. [Google Scholar] [CrossRef]
- Almeida, B.C.; Figueiredo, P.; Carvalho, A.T.P. PCL enzymatic hydrolysis: A Mechanistic study. ACS Omega 2019, 4, 6769–6774. [Google Scholar] [CrossRef]
- Nawaz, A.; Hasan, F.; Shah, A.A. Degradation of poly(ε-caprolactone) (PCL) by a newly isolated Brevundimonas sp. strain MRL-AN1 from soil. FEMS Microbiol. Lett. 2015, 362, 1–7. [Google Scholar] [CrossRef]
- Oda, Y.; Oida, N.; Urakami, T.; Tonomura, K. PCL depolymerase produced by the bacterium Alcaligenes faecalis. FEMS Microbiol. Lett. 2006, 152, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Chew, P.L.; Annuar, M.S.M.; Show, P.L.; Ling, T.C. Extractive bioconversion of poly-ϵ-caprolactone by Burkholderia cepacia lipase in an aqueous two-phase system. Biochem. Eng. J. 2015, 101, 9–17. [Google Scholar] [CrossRef]
- Zhang, Y.; Qiu, S. Phylogenomic analysis of the genus Ralstonia based on 686 single-copy genes. Antonie Leeuwenhoek 2015, 109, 71–82. [Google Scholar] [CrossRef]
- Shah, A.A.; Nawaz, A.; Kanwal, L.; Hasan, F.; Khan, S.; Badshah, M. Degradation of poly(ε-caprolactone) by a thermophilic bacterium Ralstonia sp. strain MRL-TL isolated from hot spring. Int. Biodeterior. Biodegrad. 2014, 98, 35–42. [Google Scholar] [CrossRef]
- Luo, G.; Hou, Z.; Gao, J.; Chen, X.; Tan, H.; Fan, L. Performance of PCL-based heterotrophic denitrification for recirculating aquaculture systems with varying hydraulic retention times. Desal. Water Treat. 2018, 116, 103–111. [Google Scholar] [CrossRef]
- Wang, X.; Wang, W.; Zhang, Y.; Sun, Z.; Zhang, J.; Chen, G.; Li, J. Simultaneous nitrification and denitrification by a novel isolated Pseudomonas sp. JQ-H3 using PCL as carbon source. Biores. Technol. 2019, 288, 121506. [Google Scholar] [CrossRef]
- Spearman, S.S.; Irin, F.; Ramesh, S.; Rivero, I.V.; Green, M.J.; Harrysson, O.L.A. Effect of pseudomonas lipase enzyme on the degradation of PCL/PCL-polyglycolide fiber blended nanocomposites. Int. J. Polym. Mater. 2018, 68, 360–367. [Google Scholar] [CrossRef]
- Li, L.; Lin, X.; Bao, J.; Xia, H.; Li, F. Two Extracellular Poly(ε-caprolactone)-Degrading Enzymes From Pseudomonas hydrolytica sp. DSWY01T: Purification, Characterization, and Gene Analysis. Front. Bioeng. Biotechnol. 2022, 10, 835847. [Google Scholar] [CrossRef] [PubMed]
- Inglis, G.D.; Yanke, L.J.; Selinger, L.B. Cutinolytic esterase activity of bacteria isolated from mixed-plant compost and characterization of a cutinase gene from Pseudomonas pseudoalcaligenes. Can. J. Microbiol. 2011, 57, 902–913. [Google Scholar] [CrossRef]
- Benedict, C.V.; Cameron, J.A.; Huang, S.J. PCL degradation by mixed and pure cultures of bacteria and a yeast. J. Appl. Polym. Sci. 1983, 28, 335–342. [Google Scholar] [CrossRef]
- Budkum, J.; Thammasittirong, S.N.-R.; Thammasittirong, A. High poly ε-caprolactone biodegradation activity by a new Acinetobacter seifertii isolate. Folia Microbiol. 2022, 67, 659–669. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.-R.; Jang, Y.-A.; Eom, G.T. Microbial production of adipic acid from 6-hydroxyhexanoic acid for biocatalytic upcycling of PCL. Enz. Microb. Technol. 2024, 181, 110521. [Google Scholar] [CrossRef]
- Brzezinska, M.S.; Walczak, M.; Kalwasińska, A.; Richert, A.; Świątczak, J.; Deja-Sikora, E.; Burkowska-But, A. Biofilm formation during biodegradation of polylactide, poly (3,4 hydroxybutyrate) and poly(ε-caprolactone) in activated sludge. Int. J. Biol. Macromol. 2020, 159, 539–546. [Google Scholar] [CrossRef]
- Chua, T.-K.; Tseng, M.; Yang, M.-K. Degradation of Poly(ε-caprolactone) by thermophilic Streptomyces thermoviolaceus subsp. thermoviolaceus 76T-2. AMB Express 2013, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Atanasova, N.; Stoitsova, S.; Paunova-Krasteva, T.; Kambourova, M. Plastic Degradation by Extremophilic Bacteria. Int. J. Mol. Sci. 2021, 22, 5610. [Google Scholar] [CrossRef] [PubMed]
- Atanasova, N.; Paunova-Krasteva, T.; Stoitsova, S.; Radchenkova, N.; Boyadzhieva, I.; Petrov, K.; Kambourova, M. Degradation of Poly(ε-caprolactone) by a Thermophilic Community and Brevibacillus thermoruber Strain 7 Isolated from Bulgarian Hot Spring. Biomolecules 2021, 11, 1488. [Google Scholar] [CrossRef]
- Shin, N.; Kim, S.H.; Cho, J.Y.; Hwang, J.H.; Kim, H.J.; Oh, S.J.; Park, S.-H.; Park, K.; Bhatia, S.K.; Yang, Y.-H. Fast Degradation of PCL/Poly(butylene adipate-co-terephthalate) Blends by Novel Bacillus Strain NR4 with Broad Degrading Activity. J. Polym. Environ. 2023, 32, 898–912. [Google Scholar] [CrossRef]
- Motiwalla, M.J.; Punyarthi, P.P.; Mehta, M.K.; D’Souza, J.S.; Kelkar-Mane, V. Studies on degradation efficiency of PCL by a naturally-occurring bacterium. J. Environ. Biol. 2013, 34, 43–49. [Google Scholar]
- Wu, K.-J.; Wu, C.-S.; Chang, J.-S. Biodegradability and mechanical properties of PCL composites encapsulating phosphate-solubilizing bacterium Bacillus sp. PG01. Process Biochem. 2007, 42, 669–675. [Google Scholar] [CrossRef]
- Malunavicius, V.; Padaiga, A.; Stankeviciute, J.; Pakalniskis, A.; Gudiukaite, R. Engineered Geobacillus lipolytic enzymes—Attractive polyesterases that degrade PCLs and simultaneously produce esters. Int. J. Biol. Macromol. 2023, 253, 127656. [Google Scholar] [CrossRef]
- Xu, S.; Yamaguchi, T.; Osawa, S.; Suye, S.-I. Biodegradation of Poly (ε-Caprolactone) Film in the Presence of Lysinibacillus sp. 70038 and Characterization of the Degraded Film. Biocontrol Sci. 2007, 12, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Dutta, J.R.; Ganesan, R. Lactobacillus spp. lipase mediated poly (ε-caprolactone) degradation. Int. J. Biol. Macromol. 2016, 95, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Nagarjuna, R.; Dutta, J.R.; Ganesan, R. Enzyme-Embedded Degradation of Poly(ε-caprolactone) using Lipase-Derived from Probiotic Lactobacillus plantarum. ACS Omega 2019, 4, 2844–2852. [Google Scholar] [CrossRef] [PubMed]
- Federle, T.W.; Barlaz, M.A.; Pettigrew, C.A.; Kerr, K.M.; Kemper, J.J.; Nuck, B.A.; Schechtman, L.A. Anaerobic Biodegradation of Aliphatic Polyesters: Poly(3-hydroxybutyrate-co-3-hydroxyoctanoate) and Poly(ε-caprolactone). Biomacromolecules 2002, 3, 813–822. [Google Scholar] [CrossRef]
- Lopardo, C.R.; Urakawa, H. Performance and microbial diversity of bioreactors using PCL and polyhydroxyalkanoate as carbon source and biofilm carrier in a closed recirculating aquaculture system. Aquac. Int. 2019, 27, 1251–1268. [Google Scholar] [CrossRef]
- Rüthi, J.; Rast, B.M.; Qi, W.; Perez-Mon, C.; Pardi-Comensoli, L.; Brunner, I.; Frey, B. The plastisphere microbiome in alpine soils alters the microbial genetic potential for plastic degradation and biogeochemical cycling. J. Hazard. Mater. 2022, 441, 129941. [Google Scholar] [CrossRef]
- Cook, W.J.; Cameron, J.A.; Bell, J.P.; Huang, S.J. Scanning electron microscopic visualization of biodegradation of PCLs by fungi. J. Polym. Sci. Polym. Lett. 1981, 19, 159–165. [Google Scholar] [CrossRef]
- Benedict, C.V.; Cook, W.J.; Jarrett, P.; Cameron, J.A.; Huang, S.J.; Bell, J.P. Fungal degradation of PCLs. J. Appl. Polym. Sci. 1983, 28, 327–334. [Google Scholar] [CrossRef]
- Tilstra, L.; Johnsonbaugh, D. The biodegradation of blends of PCL and polyethylene exposed to a defined consortium of fungi. J. Environ. Polym. Degrad. 1993, 1, 257–267. [Google Scholar] [CrossRef]
- Vivi, V.K.; Martins-Franchetti, S.M.; Attili-Angelis, D. Biodegradation of PCL and PVC: Chaetomium globosum (ATCC 16021) activity. Folia Microbiol. 2018, 64, 1–7. [Google Scholar] [CrossRef]
- Sanchez, J.G.; Tsuchii, A.; Tokiwa, Y. Degradation of PCL at 50 °C by a thermotolerant Aspergillus sp. Biotechnol. Lett. 2000, 22, 849–853. [Google Scholar] [CrossRef]
- Hermanová, S.; Omelková, J.; Voběrková, S.; Bálková, R.; Richtera, L.; Mravcová, L.; Jančář, J. The effect of processing of PCL films on degradation process initiated by Aspergillus oryzae Lipase. Int. J. Polym. Anal. Charact. 2012, 17, 465–475. [Google Scholar] [CrossRef]
- Kim, S.H.; Lee, J.W.; Kim, J.S.; Lee, W.; Park, M.S.; Lim, Y.W. Plastic-inhabiting fungi in marine environments and PCL degradation activity. Antonie Leeuwenhoek 2022, 115, 1379–1392. [Google Scholar] [CrossRef]
- Hosni, A.S.A.; Pittman, J.K.; Robson, G.D. Microbial degradation of four biodegradable polymers in soil and compost demonstrating PCL as an ideal compostable plastic. J. Waste Manag. 2019, 97, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, J.; Jin, Y.; Li, M.; Gu, Z. In vitro enzymatic degradation of the cross-linked poly(ε-caprolactone) implants. Polym. Degrad. Stab. 2014, 112, 10–19. [Google Scholar] [CrossRef]
- Oda, Y.; Asari, H.; Urakami, T.; Tonomura, K. Microbial degradation of poly(3-hydroxybutyrate) and PCL by filamentous fungi. J. Ferment. Bioeng. 1995, 80, 265–269. [Google Scholar] [CrossRef]
- Li, F.; Yu, D.; Lin, X.; Liu, D.; Xia, H.; Chen, S. Biodegradation of poly(ε-caprolactone) (PCL) by a new Penicillium oxalicum strain DSYD05-1. World J. Microbiol. Biotechnol. 2012, 28, 2929–2935. [Google Scholar] [CrossRef]
- Antipova, T.V.; Zhelifonova, V.P.; Zaitsev, K.V.; Nedorezova, P.M.; Aladyshev, A.M.; Klyamkina, A.N.; Kostyuk, S.V.; Danilogorskaya, A.A.; Kozlovsky, A.G. Biodegradation of poly-Ε-caprolactones and poly-L-lactides by fungi. J. Polym. Environ. 2018, 26, 4350–4359. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Ten, L.N.; Das, K.; You, Y.-H.; Jung, H.-Y. Biodegradative Activities of Fungal Strains Isolated from Terrestrial Environments in Korea. Mycobiology 2021, 49, 1–9. [Google Scholar] [CrossRef]
- Urbanek, H. The role of cutinase and cell wall degrading enzymes produced by fusaria in pathogenesis. In Elsevier eBooks Topics in Secondary Metabolism; Elsevier: Amsterdam, The Netherlands, 1989; Volume 2, pp. 243–256. [Google Scholar] [CrossRef]
- Murphy, C.A.; Cameron, J.A.; Huang, S.J.; Vinopal, R.T. Fusarium PCL depolymerase is cutinase. Appl. Environ. Microbiol. 1996, 62, 456–460. [Google Scholar] [CrossRef]
- Murphy, C.A.; Cameron, J.A.; Huang, S.J.; Vinopal, R.T. A second PCL depolymerase from Fusarium, a lipase distinct from cutinase. Appl. Microbiol. Biotechnol. 1998, 50, 692–696. [Google Scholar] [CrossRef]
- Kosiorowska, K.E.; Połomska, X.; Wang, G.; Borodina, I.; Mirończuk, A.M. Efficient Biodegradation of Aliphatic Polyester by Genetically Engineered Strains of the Yeast Yarrowia lipolytica. Int. Biodeterior. Biodegrad. 2021, 161, 105232. [Google Scholar] [CrossRef]
- Murphy, C.A.; Cameron, J.A.; Huang, S.J.; Vinopal, R.T. Inactivation of PCL depolymerase (cutinase) in Fusarium cultures by an extracellular protease. J. Ind. Microbiol. Biotechnol. 1999, 22, 71–77. [Google Scholar] [CrossRef]
- Shi, K.; Jing, J.; Song, L.; Su, T.; Wang, Z. Enzymatic hydrolysis of polyester: Degradation of poly(ε-caprolactone) by Candida antarctica lipase and Fusarium solani cutinase. Int. J. Biol. Macromol. 2019, 144, 183–189. [Google Scholar] [CrossRef]
- Ma, Q.; Shi, K.; Su, T.; Wang, Z. Biodegradation of PCL (PCL) with Different Molecular Weights by Candida antarctica Lipase. J. Polym. Environ. 2020, 28, 2947–2955. [Google Scholar] [CrossRef]
- Arumugasamy, S.K.; Ahmad, Z. Candida antarctica as catalyst for PCL synthesis: Effect of temperature and solvents. Asia-Pac. J. Chem. Eng. 2011, 6, 398–405. [Google Scholar] [CrossRef]
- Pastorino, L.; Pioli, F.; Zilli, M.; Converti, A.; Nicolini, C. Lipase-catalyzed degradation of poly(ε-caprolactone). Enzym. Microb. Technol. 2004, 35, 321–326. [Google Scholar] [CrossRef]
- Arpigny, J.L.; Jaeger, K.-E. Bacterial lipolytic enzymes: Classification and properties. Biochem. J. 1999, 343, 177. [Google Scholar] [CrossRef]
- Holmquist, M. Alpha Beta-Hydrolase Fold enzymes structures, functions and mechanisms. Curr. Prot. Pept. Sci. 2000, 1, 209–235. [Google Scholar] [CrossRef]
- Denessiouk, K.; Denesyuk, A.I.; Permyakov, S.E.; Permyakov, E.A.; Johnson, M.S.; Uversky, V.N. The active site of the SGNH hydrolase-like fold proteins: Nucleophile–oxyanion (Nuc-Oxy) and Acid–Base zones. Curr. Res. Struct. Biol. 2023, 7, 100123. [Google Scholar] [CrossRef]
- Cea-Rama, I.; Coscolín, C.; Gonzalez-Alfonso, J.L.; Raj, J.; Vasiljević, M.; Plou, F.J.; Ferrer, M.; Sanz-Aparicio, J. Crystal structure of a family VIII β-lactamase fold hydrolase reveals the molecular mechanism for its broad substrate scope. FEBS J. 2022, 289, 6714–6730. [Google Scholar] [CrossRef] [PubMed]
- Fojan, P.; Jonson, P.H.; Petersen, M.T.N.; Petersen, S.B. What distinguishes an esterase from a lipase: A novel structural approach. Biochimie 2000, 82, 1033–1041. [Google Scholar] [CrossRef] [PubMed]
- Ali, Y.B.; Verger, R.; Abousalham, A. Lipases or esterases: Does it really matter? Toward a new Bio-Physico-Chemical classification. Methods Mol. Biol. 2012, 861, 31–51. [Google Scholar] [CrossRef]
- Bartnikowski, M.; Dargaville, T.R.; Ivanovski, S.; Hutmacher, D.W. Degradation mechanisms of PCL in the context of chemistry, geometry and environment. Prog. Polym. Sci. 2019, 96, 1–20. [Google Scholar] [CrossRef]
- Badino, S.F.; Bååth, J.A.; Borch, K.; Jensen, K.; Westh, P. Adsorption of enzymes with hydrolytic activity on polyethylene terephthalate. Enz. Microb. Technol. 2021, 152, 109937. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, K.-E.; Ransac, S.; Dijkstra, B.W.; Colson, C.; Heuvel, M.; Misset, O. Bacterial lipases. FEMS Microbiol. Rev. 1994, 15, 29–63. [Google Scholar] [CrossRef]
- Hide, W.; Chan, L.; Li, W. Structure and evolution of the lipase superfamily. J. Lipid Res. 1992, 33, 167–178. [Google Scholar] [CrossRef]
- Drabløs, F.; Petersen, S.B. Identification of conserved residues in family of esterase and lipase sequences. Methods Enzymol. 1997, 286, 28–61. [Google Scholar] [CrossRef]
- Feng, S.; Yue, Y.; Chen, J.; Zhou, J.; Li, Y.; Zhang, Q. Biodegradation mechanism of PCL by a novel esterase MGS0156: A QM/MM approach. Environ. Sci. Process. Impacts 2020, 22, 2332–2344. [Google Scholar] [CrossRef]
- Zampolli, J.; Vezzini, D.; Brocca, S.; Di Gennaro, P. Insights into the biodegradation of polycaprolactone through genomic analysis of two plastic-degrading Rhodococcus bacteria. Front. Microbiol. 2024, 14, 1284956. [Google Scholar] [CrossRef]
- Suzuki, M.; Tachibana, Y.; Oba, K.; Takizawa, R.; Kasuya, K.-I. Microbial degradation of poly(ε-caprolactone) in a coastal environment. Polym. Degr. Stab. 2018, 149, 1–8. [Google Scholar] [CrossRef]
- Drew, D.; North, R.A.; Nagarathinam, K.; Tanabe, M. Structures and general transport mechanisms by the Major Facilitator Superfamily (MFS). Chem. Rev. 2021, 121, 5289–5335. [Google Scholar] [CrossRef] [PubMed]
- Mandrand-Berthelot, M.; Condemine, G.; Hugouvieux-Cotte-Pattat, N. Catabolism of hexuronides, hexuronates, aldonates, and aldarates. EcoSal Plus 2004, 1, 10–1128. [Google Scholar] [CrossRef]
- Mulligan, C.; Fischer, M.; Thomas, G.H. Tripartite ATP-independent periplasmic (TRAP) transporters in bacteria and archaea. FEMS Microbiol. Rev. 2010, 35, 68–86. [Google Scholar] [CrossRef]
- Schulz, H. Beta oxidation of fatty acids. Biochim. Biophys. Acta—Lipids Lipid Metab. 1991, 1081, 109–120. [Google Scholar] [CrossRef]
- Wang, Z.; Hou, X.; Shang, G.; Deng, G.; Luo, K.; Peng, M. Exploring fatty acid Β-Oxidation pathways in bacteria: From general mechanisms to DSF signaling and pathogenicity in Xanthomonas. Curr. Microbiol. 2024, 81, 336. [Google Scholar] [CrossRef]
- Bar–Tana, J.; Rose, G.; Brandes, R.; Shapiro, B. Palmitoyl-coenzyme A synthetase. Mechanism of reaction. Biochem. J. 1973, 131, 199–209. [Google Scholar] [CrossRef]
- Yaneva, N.; Schuster, J.; Schäfer, F.; Lede, V.; Przybylski, D.; Paproth, T.; Harms, H.; Müller, R.H.; Rohwerder, T. Bacterial Acyl-COA mutase specifically catalyzes coenzyme B12-dependent isomerization of 2-Hydroxyisobutyryl-COA and (S)-3-Hydroxybutyryl-COA. J. Biol. Chem. 2012, 287, 15502–15511. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.; Tolar, B.B.; Tosun, B.; Yoshikuni, Y.; Francis, C.A.; Wakatsuki, S.; DeMirci, H. Crystal structure of the 4-hydroxybutyryl-CoA synthetase (ADP-forming) from Nitrosopumilus maritimus. Commun. Biol. 2024, 7, 1364. [Google Scholar] [CrossRef]
- Mahmud, T.; Wenzel, S.C.; Wan, E.; Wen, K.W.; Bode, H.B.; Gaitatzis, N.; Müller, R. A biosynthetic pathway to Isovaleryl-COA in myxobacteria: The involvement of the mevalonate pathway. Chem. BioChem. 2004, 6, 322–330. [Google Scholar] [CrossRef]
- Yang, Q.; Guo, S.; Lu, Q.; Tao, Y.; Zheng, D.; Zhou, Q.; Liu, J. Butyryl/Caproyl-CoA: Acetate CoA-transferase: Cloning, expression and characterization of the key enzyme involved in medium-chain fatty acid biosynthesis. Biosci. Rep. 2021, 41, BSR20211135. [Google Scholar] [CrossRef] [PubMed]

| Property | PCL | PLA | PHA | Reference |
|---|---|---|---|---|
| Chemical structure | Aliphatic polyester | Aliphatic polyester | Polyhydroxyalkanoate | [2,4] |
| Melting temperature (Tm °C) | 58–64 °C | 130–180 °C | 170–180 °C | [6,10] |
| Glass transition temperature (Tg, °C) | −60 °C | 55–60 °C | −10 to 5 °C | [11] |
| Crystallinity (%) | 45–70% | 35–40% | 30–80% | [4,11] |
| Degradation rate (in vivo/in vitro) | Slow (months–years) | Moderate (weeks–months) | Fast (depending on microbial strain) | [14] |
| Biocompatibility | Excellent | Moderate to high | Excellent | [1,2,11] |
| Solubility in organic solvents | High | Poor | Poor | [25,26] |
| Processability | Easy (low Tm, flexible) | Requires high temperature | Brittle, limited solubility | [28,29] |
| Shape memory effect | Present | Absent or weak | Absent | [31,32] |
| Mechanical behavior | Tough, flexible | Stiff, brittle | Variable, often brittle | [14] |
| Class | Type | Substrate Preference | Notable Features |
|---|---|---|---|
| I | Lipases | Long-chain triglycerides | α/β-Hydrolase fold, Interfacial activation, Lid domain |
| II | Esterases | Broad | Divergent, α/β/α sandwich; SGNH conserved sequence, GDSL motif near N-terminus (most) |
| III | Esterases | Short-chain esters | PAF-AH-like, α/β-hydrolase fold |
| IV | Esterases | Short-chain esters | HSL-like, α/β-hydrolase fold |
| V | Esterases | Variable | Dehalogenase and haloperoxidase-like enzymes in extremophiles |
| VI | Cutinases | Esters | Plant cuticle degradation |
| VII | Esterases | Acetylated compounds | Acetylcholinesterase-specific activity |
| VIII | Esterases | Broad | Distinct β-lactamase-like fold, untypical catalytic triad |
| Species/Strain | Source | Enzyme | Properties | Efficiency | Reference |
|---|---|---|---|---|---|
| Brevibacillus thermoruber strain 7 | Marikostinovo Hot Spring, Bulgaria | Lipase | 28 kDa, T range 45–65 °C, Topt 55 °C, pH range 6–9, pHopt 7–8, ↑Ca2+, ↓Mg2+, Co2+, K+, Na2+, Cu2+, Mn2+, Hg2+, Zn2+, Fe3+ | Clear halos on 0.1% PCL-containing agar; deep surface damages on PCL after 1 week of incubation | [67] |
| Brevundimonas sp. MRL-AN1 | Soil sample, Pakistan | PCL-depolymerase | 63.49 kDa, T range 30–37 °C, pH range 6.0–8.0, ↓Fe2+ and Zn2+ | 80% of PCL film degraded in 10 days; prefers C6-C10 p-nitrophenyl acyl esters | [51] |
| Alcaligenes faecalis B273 | Soil and activated sludge | PCL-depolymerase | Topt 40 °C, pHopt 7.0, Km PCL = 0.29 mg/mL | Prefers C10 p-nitrophenyl acyl esters and tributyrin; does not cleave PHB | [52] |
| Burkholderia cepacia ST8 | Soil and water, Malaysia | Lipase | ↑Ca2+ ↓Cu2+ and Co2+ | 179 U/mL in medium with Tween 80 | [53] |
| Ralstonia sp. MRL-TL | Hot spring | Esterase, serine hydrolase family | 50 kDa, Topt 50 °C, pHopt 7.0, preferred substrate p-NP-caproate | 50% of PCL film degraded in 10 days; it also degrades PLA, PES, PHB, and PHBV | [54] |
| Pseudomonas pachastrellae | Coastal seawater, Okinoshima Park, Japan | Cutinase | 30 kDa, optimal at 0.51 M NaCl, operates via surface hydrolysis | Solid-state PCL-hydrolytic activity | [104] |
| Pseudomonas sp. | Sigma-Aldrich (Merck KGaA) | Lipase | Topt 37° C in 0.05 M phosphate buffer solution (PBS) with pH 7 | Visible degradation of the PCL matrix by the end of the first week | [58] |
| Pseudomonas hydrolytica | Lab-maintained strain, originally isolated from forest soil in China | PCLase I and PCLase II | PCLase I: 29,64 kDa; T 50 °C, pH 9; highly stable at pH 12 with 100% activity; ↑Mg2+, Ca2+, Fe3+, and Fe2+; ↓Cu2+ and Co2+ PCLase II: 33,2 kDa; T 40 °C; pH 10; ↑Mg2+ and Ca2+ at 1 Mm, ↑Co2+ and Cu2+, ↓Mg2+ and Ca2+ at 10 mM, ↓Fe2+ | PCL I: degrades PCL (70% weight loss in 3 days), PBS, pNP ester, tributyrin, olive oil, and cutin PCL II: degrades PCL (75% weight loss in 8 days), PHB, PBS, pNP ester, tributyrin, and olive oil | [59] |
| Pseudomonas pseudoalcaligenes | Mixed-plant compost | Cutinase | 32 kDa, consists of 302 amino acids, cutin-induced | Variable PCL degradation; clearing zones up to 65 mm | [61] |
| Acinetobacter seifertii | Soil samples | Esterase with PCL-depolymerase activity | 30–40 °C | - | [62] |
| Streptomyces thermoviolaceus ssp. thermoviolaceus | Soil, Taiwan | Chitinase with PCL-depolymerase activity, PCL depolymerase | Chitinase with PCL-depolymerase activity: 35 kDa PCL depolymerase: 55 kDa | Both form bands in a polyacrylamide gel containing 0.1% PCL after incubation at 45 °C for 30 min | [66] |
| Geobacillus sp. | Lab-maintained strain, originally isolated from a Lithuanian oil well | Lipase/esterase, used for the development of engineered polyesterases: GD-95 RM and GDEst-lip in E. coli | GD-95 RM: LA 1400 U/mg, stable up to 85 °C GDEst-lip: 98 kDa, LA 600 U/mg; active between 5 and 90 °C, pH 6–12, both resistant to organic solvents, Topt 50 °C | GD-95 RM: 264.0 mg and 280.7 mg of PCL45,000 and PCL80,000, 24 h at 30 °C GDEst-lip: 145.5 mg PCL45,000 and 134.0 mg PCL80,000, 24 h | [71] |
| Levilactobacillus brevis | Commercially obtained strain from IMTECH | Lipase | 26 kDa, assayed at 37 °C, pH 7 | 10-day incubation; ~2 wt.% mass loss for 1 mg/mL lipase, increases to ~10 wt.% for 4–5 mg/mL | [73] |
| Lactiplantibacillus plantarum | Commercially obtained strain from IMTECH | Lipase | 66 kDa, assayed at T 37 °C, pH 7; also at pH 8.1 + Tween 20 in an embedded approach | 10-day incubation; ~10 wt % mass loss for 1 mg/mL lipase, increases to ~60 wt.% for 5 mg/mL; 73% mass loss in 8 days for 8% lipase-embedded PCL, crystallinity increases from 39% to 95% | [74] |
| Organism | Source | Enzyme | Properties | Efficiency | Reference |
|---|---|---|---|---|---|
| Aspergillus oryzae | Mucos Pharma 1 | Lipase | Highest activity at Topt 37 °C, pH 7; retains stability at 55 °C | 10% decrease in PCL Mw for 45 days of incubation | [73] |
| Thermomyces lanuginosus | Sigma Aldrich 1 | Lipase | Topt: 37 °C, initial pH: 6.02, decreases during reaction | Mass loss over 5 days: ~54.8% at 0.1 mol% and ~17.3% at 3.0 mol% cross-linking. Corresponding degradation rates: 14.5%/day and 3.5%/day | [75,76] |
| Paecilomyces lilacinus | Soil, activated sludge | PCL depolymerase | T 30 °C; pH 3.5–4.5; expression inhibited by starch, glucose, and lactose | 10% PCL degraded in 10 days | [86] |
| Fusarium moniliforme | University of Connecticut Culture Collection | Cutinase with PCL depolymerase activity | 24 kDa, pH 9–10, induced by 16-hydroxy hexadecanoic acid | Zones of clearing on MM-PCL agar plates after 48 h | [82] |
| Fusarium solani f. sp. pisi 77–102 | Institut fuÈr Genbiologische Forschung | Lipase/Cutinase 2 | Lipase: pH 7.8, assayed at T 30 °C Cutinase: T range 20–70 °C, Topt 30–40 °C, pH range 3–11, pHopt 6–9 | Lipase: clear MM/PCL/agar zones with Tween 80 or tributyrin; lipase/cutinase coexpression: 0.5 g PCL degraded in 144 h | [83,84] |
| Candida rugosa | Sigma-Aldrich 1 | Lipase | T 40 °C, assayed at pH 7.7 | PCL drops from 86,909 g/mol to 80,873 g/mol, and Mw remains at 1.48 | [89] |
| Mucor miehei | Sigma-Aldrich 1 | Lipase | T 40 °C, retains activity up to 60 °C, assayed at pH 7.7 | 74% PCL hydrolyzed 24 h, Mw of PCL drops from 86,909 g/mol to 24,011 g/mol, Mn/Mw increases to 2.39 | [89] |
| Rhizopus delemar | Fluka 1 | Lipase | T 40 °C, assayed at pH 7.7 | PCL drops from 86,909 g/mol to 64,137 g/mol | [89] |
| Moesziomyces antarcticus | Beijing Cliscent Technology; Novozymes China Biotechnology 1 | Lipase | 33 kDa, 317 amino acids, T 45 °C, pH 7.2 | 87% PCL weight loss in 72 h via two-phase degradation: rapid (85%, 0–12 h) and slow (86.9%, 12–20 h), independent of PCL Mw | [86,87] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krumov, N.; Atanasova, N.; Boyadzhieva, I.; Petrov, K.; Petrova, P. Biodegradation of Poly(ε-caprolactone): Microorganisms, Enzymes, and Mechanisms. Int. J. Mol. Sci. 2025, 26, 5826. https://doi.org/10.3390/ijms26125826
Krumov N, Atanasova N, Boyadzhieva I, Petrov K, Petrova P. Biodegradation of Poly(ε-caprolactone): Microorganisms, Enzymes, and Mechanisms. International Journal of Molecular Sciences. 2025; 26(12):5826. https://doi.org/10.3390/ijms26125826
Chicago/Turabian StyleKrumov, Nikolay, Nikolina Atanasova, Ivanka Boyadzhieva, Kaloyan Petrov, and Penka Petrova. 2025. "Biodegradation of Poly(ε-caprolactone): Microorganisms, Enzymes, and Mechanisms" International Journal of Molecular Sciences 26, no. 12: 5826. https://doi.org/10.3390/ijms26125826
APA StyleKrumov, N., Atanasova, N., Boyadzhieva, I., Petrov, K., & Petrova, P. (2025). Biodegradation of Poly(ε-caprolactone): Microorganisms, Enzymes, and Mechanisms. International Journal of Molecular Sciences, 26(12), 5826. https://doi.org/10.3390/ijms26125826








