Bacteria Under Metal Stress—Molecular Mechanisms of Metal Tolerance
Abstract
1. Introduction
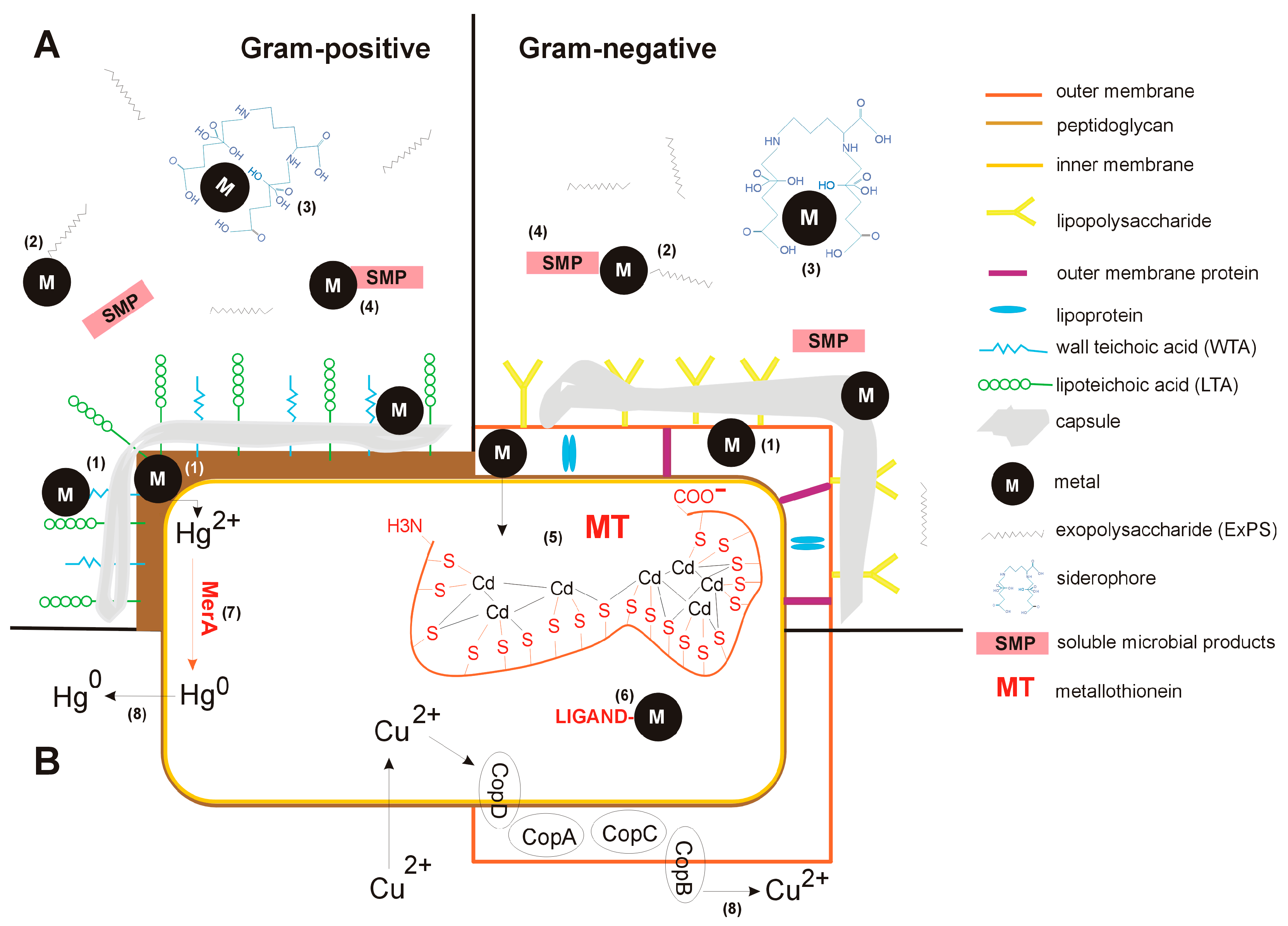
2. Cell Wall Sequestration
3. Extracellular Barriers Immobilizing Metals
3.1. Exopolysaccharides
3.2. Soluble Microbial Products (SMP)
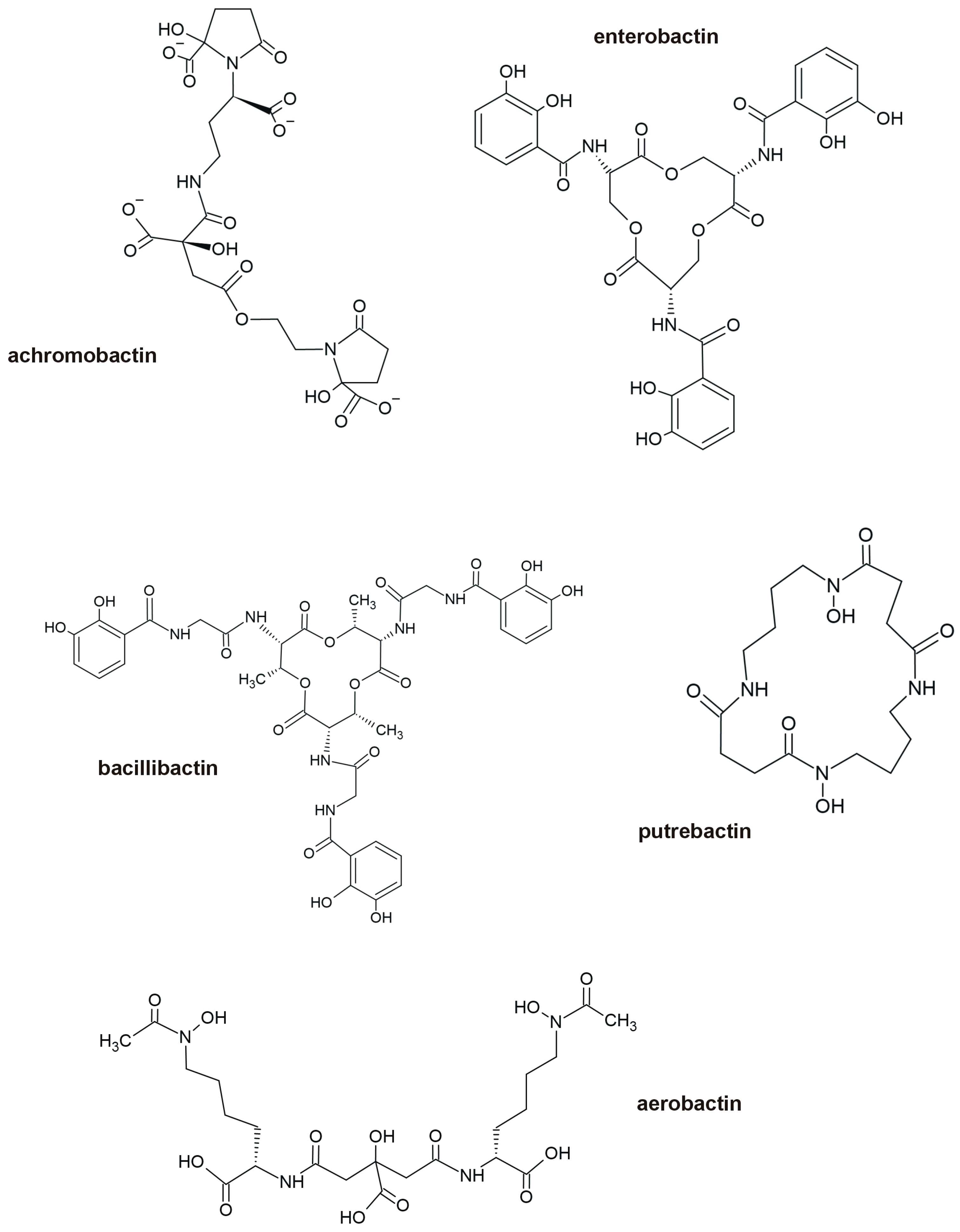
3.3. Siderophores
4. Intracellular Sequestration of Metals
5. Enzymatic Conversion of Metal Ions and/or Their Efflux out of the Cell
6. Regulation of Metal Uptake and Efflux in Bacteria
- (i)
- three families of metal-releasable de-repressors (i.e., ArsR-SmtB, CsoR-RcnR, and CopY), which bind DNA in their apo-form (metal free) and release it upon metal binding;
- (ii)
- three families of metal-inducible co-repressors ((Fur–Zur and Mur), DtxR/MntR, and NikR) which bind DNA and metal simultaneously to repress transcription and dissociate from DNA in the absence of the metal;
- (iii)
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy metal toxicity and the environment. In Molecular, Clinical and Environmental Toxicology; Springer: Basel, Switzerland, 2012; Volume 101, pp. 133–164. [Google Scholar] [CrossRef]
- Hao, X.; Zhu, J.; Rensing, C.; Liu, Y.; Gao, S.; Chen, W.; Huang, Q.; Liu, Y.-R. Recent advances in exploring the heavy metal(loid) resistant microbiome. Comp. Struct. Biotechnol. J. 2021, 19, 94–109. [Google Scholar] [CrossRef] [PubMed]
- Domínguez, D.C. Calcium signaling in Procaryotes. In Calcium and Signal Transduction; Buchholz, J.N., Behringer, E.J., Eds.; IntechOpen: London, UK, 2018; pp. 89–106. [Google Scholar] [CrossRef]
- Wang, T.; Flint, S.; Palmer, J. Magnesium and calcium ions: Roles in bacterial cell attachment and biofilm structure maturation. Biofouling 2019, 35, 959–974. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, J.; Kuan, S.F.; Fu, H.Z.; Lin, H.Y.; Peng, B. Magnesium modulates phospholipid metabolism to promote bacterial phenotypic resistance to antibiotics. eLife 2024, 13, RP100427. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.I.; Sarmento-Ribeiro, A.B.; Gonçalves, A.C. Zinc: From biological functions to therapeutic potential. Int. J. Mol. Sci. 2023, 24, 4822. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Q.; Kobe, B.; Kappler, U. Molybdenum enzymes and how they support virulence in pathogenic bacteria. Front. Microbiol. 2020, 11, 615860. [Google Scholar] [CrossRef]
- Sawers, R.G. Nickel in bacteria and archaea. In Encyclopedia of Metalloproteins; Kretsinger, R.H., Uversky, V.N., Permyakov, E.A., Eds.; Springer: New York, NY, USA, 2013. [Google Scholar] [CrossRef]
- Balali-Mood, M.; Naseri, K.; Tahergorabi, Z.; Khazdair, M.R.; Sadeghi, M. Toxic mechanisms of five heavy metals: Mercury, lead, chromium, cadmium, and arsenic. Front. Pharmacol. 2021, 12, 643972. [Google Scholar] [CrossRef]
- Luo, L.; Wang, B.; Jiang, J.W.; Fitzgerald, M.; Huang, Q.; Yu, Z.; Li, H.; Zhang, J.; Wei, J.; Yang, C.; et al. Heavy metal contaminations in herbal medicines: Determination, comprehensive risk assessments, and solutions. Front. Pharmacol. 2021, 11, 595335. [Google Scholar] [CrossRef]
- Alrashed, M.; Tabassum, H.; Almuhareb, N.; Almutlaq, N.; Alamro, W.; Alenazi, S.T.; Alahmed, L.B.; Al Abudahash, M.M.; Alenzi, N.D. Assessment of DNA damage in relation to heavy metal induced oxidative stress in females with recurrent pregnancy loss (RPL). Saudi J. Biol. Sci. 2021, 28, 5403–5407. [Google Scholar] [CrossRef]
- Thai, T.D.; Lim, W.; Na, D. Synthetic bacteria for the detection and bioremediation of heavy metals. Front. Bioeng. Biotechnol. 2023, 11, 1178680. [Google Scholar] [CrossRef]
- Witter, E.; Gong, P.; Bääth, E.; Marstorp, H. A study of the structure and metal tolerance of the soil microbial community six years after cessation of sewage sludge applications. Environ. Toxicol. Chem. 2000, 19, 1983–1991. [Google Scholar] [CrossRef]
- Abaye, D.A.; Lawlor, K.; Hirsch, P.R.; Brookes, P.C. Changes in the microbial community of an arable soil caused by long-term metal contamination. Europ J. Soil. Sci. 2005, 56, 93–102. [Google Scholar] [CrossRef]
- Carrasco, J.A.; Armario, P.; Pajuelo, E.; Burgos, A.; Caviedes, M.A.; López, R.; Chambera, M.A.; Palomares, A.J. Isolation and characterization of symbiotically effective Rhizobium resistant to arsenic and heavy metals after the toxic spill at the Aznalcóllar pyrite mine. Soil. Biol. Biochem. 2005, 37, 1131–1140. [Google Scholar] [CrossRef]
- Khan, S.; Hesham, A.E.-L.; Qiao, M.; Rehman, S.; He, J.-Z. Effects of Cd and Pb on soil microbial community structure and activities. Environ. Sci. Pollut. Res. 2010, 17, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Oleńska, E.; Imperato, V.; Małek, W.; Włostowski, T.; Wójcik, M.; Swiecicka, I.; Vangronsveld, J.; Thijs, S. Trifolium repens-associated bacteria as a potential tool to facilitate phytostabilization of zinc and lead polluted waste heaps. Plants 2020, 9, 1002. [Google Scholar] [CrossRef]
- Oleńska, E.; Małek, W. Genomic polymorphism of Trifolium repens root nodule symbionts from heavy metal-abundant 100-year-old waste heap in southern Poland. Arch. Microbiol. 2019, 201, 1405–1414. [Google Scholar] [CrossRef]
- Pearson, R.G. Hard and soft acids and bases. J. Am. Chem. Soc. 1963, 85, 3533–3539. [Google Scholar] [CrossRef]
- Minkin, V.I. Glossary of terms used in theoretical organic chemistry. Pure Appl. Chem. 1999, 71, 1919–1981. [Google Scholar] [CrossRef]
- Lippard, S.J.; Berg, J.M. Principles of Bioinorganic Chemistry; University Science Books: Melville, NY, USA, 1994. [Google Scholar]
- Nies, D.H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003, 27, 313–339. [Google Scholar] [CrossRef]
- Valls, M.; de Lorenzo, V. Exploiting the genetic and biochemical capacities of bacteria for the remediation of heavy metal pollution. FEMS Microbiol. Rev. 2002, 26, 327–338. [Google Scholar] [CrossRef]
- Silver, S.; Phung, L.T. Bacterial heavy metal resistance: New surprises. Annu. Rev. Microbiol. 1996, 50, 753–789. [Google Scholar] [CrossRef]
- Oleńska, E.; Małek, W. Mechanisms of heavy metal resistance in bacteria. Adv. Microbiol. 2013, 52, 363–371. [Google Scholar]
- Roy, R.R.; Samanta, S.; Pandit, S.; Naaz, T.; Banerjee, S.; Rawat, J.M.; Chaubey, K.K.; Saha, R.P. An overview of bacteria-mediated heavy metal bioremediation strategies. Appl. Biochem. Biotechnol. 2024, 196, 1712–1751. [Google Scholar] [CrossRef] [PubMed]
- Pande, V.; Pandey, S.C.; Sati, D.; Bhatt, P.; Samant, M. Microbial interventions in bioremediation of heavy metal contaminants in agroecosystem. Front. Microbiol. 2022, 13, 824084. [Google Scholar] [CrossRef]
- Joshi, S.; Gangola, S.; Bhandari, G.; Bhandari, N.S.; Nainwal, D.; Rani, A.; Malik, S.; Slama, P. Rhizospheric bacteria: The key to sustainable heavy metal detoxification strategies. Front. Microbiol. 2023, 14, 1229828. [Google Scholar] [CrossRef]
- Fang, L.; Cai, P.; Chen, W.; Liang, W.; Hong, Z.; Huang, Q. Impact of cell wall structure on the behavior of bacterial cells in the binding of copper and cadmium. Colloids Surf. A Physicochem. Eng. Asp. 2009, 347, 50–55. [Google Scholar] [CrossRef]
- Fein, J.B.; Daughney, C.J.; Yee, N.; Davis, T.A. A chemical equilibrium model for metal adsorption onto bacterial surfaces. Geochim. Cosmochim. Acta 1997, 61, 3319–3328. [Google Scholar] [CrossRef]
- Thomas, K.J., 3rd; Rice, C.V. Revised model of calcium and magnesium binding to the bacterial cell wall. Biometals 2014, 27, 1361–1370. [Google Scholar] [CrossRef] [PubMed]
- Bhavsar, A.P.; Erdman, L.K.; Schertzer, J.W.; Brown, E.D. Teichoic acid is an essential polymer in Bacillus subtilis that is functionally distinct from teichuronic acid. J. Bacteriol. 2004, 186, 7865–7873. [Google Scholar] [CrossRef]
- Cha, J.S.; Cooksey, D.A. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc. Natl. Acad. Sci. USA 1991, 88, 8915–8919. [Google Scholar] [CrossRef]
- Gilotra, U.; Srivastava, S. Plasmid-encoded sequestration of copper by Pseudomonas pickettii strain US321. Curr. Microbiol. 1997, 34, 378–381. [Google Scholar] [CrossRef]
- Slawson, R.M.; Trevors, J.T.; Lee, H. Silver accumulation and resistance in Pseudomonas stutzeri. Arch. Microbiol. 1992, 158, 398–404. [Google Scholar] [CrossRef]
- Klaus, T.; Joerger, R.; Olsson, E.; Granqvist, C.G. Silver-based crystalline nanoparticles, microbially fabricated. Proc. Natl. Acad. Sci. USA 1999, 96, 13611–13614. [Google Scholar] [CrossRef] [PubMed]
- Thelwell, C.; Robinson, N.J.; Turner-Cavet, J.S. An SmtB-like repressor from Synechocystis PCC 6803 regulates a zinc exporter. Proc. Natl. Acad. Sci. USA 1998, 95, 10728–10733. [Google Scholar] [CrossRef]
- Gabr, R.M.; Hassan, S.H.A.; Shoreit, A.A.M. Biosorption of lead and nickel by living and non-living cells of Pseudomonas aeruginosa ASU 6a. Int. Biodeterior. Biodegrad. 2008, 62, 195–203. [Google Scholar] [CrossRef]
- Çabuk, A.; Akar, T.; Tunali, S.; Tabak, Ö. Biosorption characteristics of Bacillus sp. ATS-2 immobilized in silica gel for removal of Pb(II). J. Hazard. Mater. 2006, 136, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Zadeh, P.H.; Fermoso, F.G.; Collins, G.; Serrano, A.; Mills, S.; Abram, F. Impacts of metal stress on extracellular microbial products, and potential for selective metal recovery. Ecotoxicol. Environ. Saf. 2023, 252, 114604. [Google Scholar] [CrossRef]
- Kramer, J.; Özkaya, Ö.; Kümmerli, R. Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 2020, 18, 152–163. [Google Scholar] [CrossRef]
- Wei, X.; Chen, Z.; Liu, A.; Yang, L.; Xu, Y.; Cao, M.; He, N. Advanced strategies for metabolic engineering of Bacillus to produce extracellular polymeric substances. Biotechnol. Adv. 2023, 67, 108199. [Google Scholar] [CrossRef]
- Wu, S.; Huo, H.; Shi, Y.; Zhang, F.; Gu, T.; Li, Z. Extraction and application of extracellular polymeric substances from fungi. Adv. Appl. Microbiol. 2023, 125, 79–106. [Google Scholar] [CrossRef]
- Su, C.-L.; Lau, S.H.; Yeh, H.-Y.; Chang, Y.-T. Biological treatment of benzophenone-type UV filter wastewater in a sequencing batch reactor (SBR). Int. Biodeterior. Biodegrad. 2023, 177, 105534. [Google Scholar] [CrossRef]
- Sankaran, R.; Show, P.L.; Ooi, C.W.; Ling, T.C.; Shu-Jen, C.; Chen, S.Y.; Chang, Y.K. Feasibility assessment of removal of heavy metals and soluble microbial products from aqueous solutions using eggshell wastes. Clean Technol. Environ. Policy 2020, 22, 773–786. [Google Scholar] [CrossRef]
- Thanh, P.M.; Ketheesan, B.; Yan, Z.; Stuckey, D. Trace metal speciation and bioavailability in anaerobic digestion: A review. Biotechnol. Adv. 2016, 34, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Liu, J.; Yang, R.; Wu, L. Distribution, characteristics of extracellular polymeric substances of Phanerochaete chrysosporium under lead ion stress and the influence on Pb removal. Sci. Rep. 2020, 10, 17633. [Google Scholar] [CrossRef]
- Mohite, B.V.; Koli, S.H.; Narkhede, C.P.; Patil, S.N.; Patil, S.V. Prospective of microbial exopolysaccharide for heavy metal exclusion. Appl. Biochem. Biotechnol. 2017, 183, 582–600. [Google Scholar] [CrossRef] [PubMed]
- McSwain, B.S.; Irvine, R.L.; Hausner, M.; Wilderer, P.A. Composition and distribution of extracellular polymeric substances in aerobic flocs and granular sludge. Appl. Environ. Microbiol. 2005, 71, 1051–1057. [Google Scholar] [CrossRef]
- Nwodo, U.U.; Green, E.; Okoh, A.I. Bacterial exopolysaccharides: Functionality and prospects. Int. J. Mol. Sci. 2012, 13, 14002–14015. [Google Scholar] [CrossRef]
- Oleńska, E.; Małek, W.; Kotowska, U.; Wydrych, J.; Polińska, W.; Swiecicka, I.; Thijs, S.; Vangronsveld, J. Exopolysaccharide carbohydrate structure and biofilm formation by Rhizobium leguminosarum bv. trifolii strains inhabiting nodules of Trifolium repens growing on an old Zn–Pb–Cd-polluted waste heap area. Int. J. Mol. Sci. 2021, 22, 2808. [Google Scholar] [CrossRef]
- Meisen, S.; Wingender, J.; Telgheder, U. Analysis of microbial extracellular polysaccharides in biofilms by HPLC. Part I: Development of the analytical method using two complementary stationary phases. Anal. Bioanal. Chem. 2008, 391, 993–1002. [Google Scholar] [CrossRef]
- Kanmani, P.; Lim, S.T. Synthesis and structural characterization of silver nanoparticles using bacterial exopolysaccharide and its antimicrobial activity against food and multidrug resistant pathogens. Process Biochem. 2013, 48, 1099–1106. [Google Scholar] [CrossRef]
- Balíková, K.; Vojtková, H.; Duborská, E.; Kim, H.; Matúš, P.; Urík, M. Role of exopolysaccharides of Pseudomonas in heavy metal removal and other remediation strategies. Polymers 2022, 14, 4253. [Google Scholar] [CrossRef]
- Nouha, K.; Kumar, R.S.; Balasubramanian, S.; Tyagi, R.D. Critical review of EPS production, synthesis and composition for sludge flocculation. J. Environ. Sci. 2018, 66, 225–245. [Google Scholar] [CrossRef] [PubMed]
- Schmid, J.; Sieber, V.; Rehm, B. Bacterial exopolysaccharides: Biosynthesis pathways and engineering strategies. Front. Microbiol. 2015, 6, 496. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.T.; Lam, J.S. Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can. J. Microbiol. 2014, 60, 697–716. [Google Scholar] [CrossRef] [PubMed]
- Rehm, B.H. Bacterial polymers: Biosynthesis, modifications and applications. Nat. Rev. Microbiol. 2010, 8, 578–592. [Google Scholar] [CrossRef]
- Whitney, J.C.; Howell, P.L. Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria. Trends Microbiol. 2013, 21, 63–72. [Google Scholar] [CrossRef]
- Srikanth, R.; Reddy, C.H.S.S.S.; Siddartha, G.; Ramaiah, M.J.; Uppuluri, K.B. Review on production, characterization and applications of microbial levan. Carbohydr. Polym. 2015, 120, 102–114. [Google Scholar] [CrossRef]
- Concórdio-Reis, P.; Freitas, F. Environmental applications: Biopolymer sorbents for heavy metal removal. In Encyclopedia of Polymer Applications; CRC Press: Boca Raton, FL, USA, 2019; pp. 1069–1086. [Google Scholar]
- Biswas, J.K.; Banerjee, A.; Sarkar, B.; Sarkar, D.; Sarkar, S.K.; Rai, M.; Vithanage, M. Exploration of an extracellular polymeric substance from earthworm gut bacterium (Bacillus licheniformis) for bioflocculation and heavy metal removal potential. Appl. Sci. 2020, 10, 349. [Google Scholar] [CrossRef]
- Pérez, J.A.M.; García-Ribera, R.; Quesada, T.; Aguilera, M.; Ramos-Cormenzana, A.; Monteoliva-Sánchez, M. Biosorption of heavy metals by the exopolysaccharide produced by Paenibacillus jamilae. World J. Microbiol. Biotechnol. 2008, 24, 2699–2704. [Google Scholar] [CrossRef]
- Brinza, L.; Dring, M.J.; Gavrilescu, M. Marine micro and macro algal species as biosorbents for heavy metals. Environ. Eng. Manag. J. 2007, 6, 237–251. [Google Scholar] [CrossRef]
- Wang, J.; Chen, C. Biosorbents for heavy metals removal and their future. Biotechnol. Adv. 2009, 27, 195–226. [Google Scholar] [CrossRef]
- Gupta, P.; Diwan, B. Bacterial exopolysaccharide mediated heavy metal removal: A review on biosynthesis, mechanism and remediation strategies. Biotechnol. Rep. 2017, 13, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Concórdio-Reis, P.; Reis, M.A.M.; Freitas, F. Biosorption of heavy metals by the bacterial exopolysaccharide FucoPol. Appl. Sci. 2020, 10, 6708. [Google Scholar] [CrossRef]
- Santamaria, M.; Diaz-Marreto, A.R.; Hernandez, J.; Uutierrez-Navarro, A.M.; Corzo, J. Effect of thorium on the growth and capsule morphology of Bradyrhizobium. Environ. Microbiol. 2003, 5, 916–924. [Google Scholar] [CrossRef]
- Wingender, J.; Strathmann, M.; Rode, A.; Leis, A.; Flemming, H.C. Isolation and biochemical characterization of extracellular polymeric substances from Pseudomonas aeruginosa. Methods Enzymol. 2001, 336, 302–314. [Google Scholar] [CrossRef]
- Jachlewski, S.; Jachlewski, W.D.; Linne, U.; Bräsen, C.; Wingender, J.; Siebers, B. Isolation of extracellular polymeric substances from biofilms of the thermoacidophilic archaeon Sulfolobus acidocaldarius. Front. Bioen Biotechnol. 2015, 3, 123. [Google Scholar] [CrossRef]
- Sutherland, I.W. Biofilm exopolysaccharides: A strong and sticky framework. Microbiology 2001, 147, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Salehizadeh, H.; Shojaosadati, S.A. Removal of metal ions from aqueous solution by polysaccharide produced from Bacillus firmus. Water Res. 2003, 37, 4231–4235. [Google Scholar] [CrossRef]
- Lin, J.; Harichund, C. Production and characterization of heavy-metal removing bacterial bioflocculants. Afr. J. Biotechnol. 2012, 11, 9619–9629. [Google Scholar] [CrossRef]
- Fella-Temzi, S.; Yalaoui-Guellal, D.; Rodriguez-Carvajal, M.A.; Belhadi, D.; Madani, K.; Kaci, Y. Removal of lead by exopolysaccharides from Paenibacillus peoriae strainTS7 isolated from rhizosphere of durum wheat. Biocatal. Agric. Biotechnol. 2018, 16, 425–432. [Google Scholar] [CrossRef]
- Cao, R.; Zhang, Y.; Ju, Y.; Wang, W.; Zhao, Y.; Liu, N.; Zhang, G.; Wang, X.; Xie, X.; Dai, C.; et al. Exopolysaccharide-producing bacteria enhanced Pb immobilization and influenced the microbiome composition in rhizosphere soil of pakchoi (Brassica chinensis L.). Front. Microbiol. 2023, 14, 1117312. [Google Scholar] [CrossRef]
- Zainab, N.; Amna, D.; Din, B.U.; Javed, M.T.; Afridi, M.S.; Mukhtar, T.; Kamran, M.A.; Qurat, U.A.; Khan, A.A.; Ali, J.; et al. Deciphering metal toxicity responses of flax (Linum usitatissimum L.) with exopolysaccharide and ACC-deaminase producing bacteria in industrially contaminated soils. Plant Physiol. Biochem. 2020, 152, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Karthik, C.; Elangovan, N.; Kumar, T.S.; Govindharaju, S.; Barathi, S.; Oves, M.; Arulselvi, P.I. Characterization of multifarious plant growth promoting traits of rhizobacterial strain AR6 under chromium (VI) stress. Microbiol. Res. 2017, 204, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.; Mitra, A.; Roy, M. Halomonas rhizobacteria of Avicennia marina of Indian Sundarbans promote rice growth under saline and heavy metal stresses through exopolysaccharide production. Front. Microbiol. 2019, 10, 1207. [Google Scholar] [CrossRef]
- Rasulov, B.A.; Yili, A.; Aisa, H.A. Biosorption of metal ions by exopolysaccharide produced by Azotobacter chroococcum XU1. J. Environ. Prot. 2013, 4, 989–993. [Google Scholar] [CrossRef]
- Kushwaha, A.; Rani, R.; Kumar, S.; Thomas, T.; David, A.A.; Ahmed, M. A new insight to adsorption and accumulation of high lead concentration by exopolymer and whole cells of lead-resistant bacterium Acinetobacter junii L. Pb1 isolated from coal mine dump. Environ. Sci. Pollut. Res. 2017, 24, 10652–10661. [Google Scholar] [CrossRef]
- Bowman, N.; Patel, D.; Sanchez, A.; Xu, W.; Alsaffar, A.; Tiquia-Arashiro, S.M. Lead-resistant bacteria from Saint Clair River sediments and Pb removal in aqueous solutions. Appl. Microbiol. Biotechnol. 2018, 102, 2391–2398. [Google Scholar] [CrossRef]
- Nadell, C.D.; Drescher, K.; Wingreen, N.S.; Bassler, B.L. Extracellular matrix structure governs invasion resistance in bacterial biofilms. ISME J. 2015, 9, 1700–1709. [Google Scholar] [CrossRef] [PubMed]
- Payne, D.E.; Boles, B.R. Emerging interactions between matrix components during biofilm development. Curr. Genet. 2016, 62, 137–141. [Google Scholar] [CrossRef]
- Molina-Santiago, C.; de Vicente, A.; Romero, D. Bacterial extracellular matrix as a natural source of biotechnologically multivalent materials. Computat Struct. Biotechnol. J. 2021, 19, 2796–2805. [Google Scholar] [CrossRef]
- Limoli, D.H.; Jones, C.J.; Wozniak, D.J. Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol. Spectr. 2015, 3, MB-0011-2014. [Google Scholar] [CrossRef]
- Steinberg, N.; Kolodkin-Gal, I. The matrix reloaded: How sensing the extracellular matrix synchronizes bacterial communities. J. Bacteriol. 2015, 197, 2092–2103. [Google Scholar] [CrossRef]
- Steinberg, N.; Keren-Paz, A.; Hou, O.; Doron, S.; Yanuka-Golub, K.; Olender, T.; Hadar, R.; Rosenberg, G.; Jain, R.; Cámara-Almirón, J.; et al. The extracellular matrix protein TasA is a developmental cue that maintains a motile subpopulation within Bacillus subtilis biofilms. Sci. Signal. 2020, 13, eaaw8905. [Google Scholar] [CrossRef] [PubMed]
- Molina-Santiago, C.; Pearson, J.R.; Navarro, Y.; Berlanga-Clavero, M.V.; Caraballo-Rodriguez, A.M.; Petras, D.; García-Martín, M.L.; Lamon, G.; Haberstein, B.; Cazorla, F.M.; et al. The extracellular matrix protects Bacillus subtilis colonies from Pseudomonas invasion and modulates plant co-colonization. Nat. Commun. 2019, 10, 1919. [Google Scholar] [CrossRef] [PubMed]
- Saba, Y.R.; Ahmed, M.; Sabri, A.N. Potential role of bacterial extracellular polymeric substances as biosorbent material for arsenic bioremediation. Biorem. J. 2019, 23, 72–81. [Google Scholar] [CrossRef]
- Vlamakis, H.; Aguilar, C.; Losick, R.; Kolter, R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008, 22, 945–953. [Google Scholar] [CrossRef]
- Luo, H.W.; Yin, X.; Jubb, A.M.; Chen, H.; Lu, X.; Zhang, W.; Lin, H.; Yu, H.Q.; Liang, L.; Sheng, G.P.; et al. Photochemical reactions between mercury (Hg) and dissolved organic matter decrease Hg bioavailability and methylation. Environ. Pollut. 2017, 220, 1359–1365. [Google Scholar] [CrossRef]
- Xu, H.; Zou, L.; Guan, D.; Li, W.; Jiang, H. Molecular weight-dependent spectral and metal binding properties of sediment dissolved organic matter from different origins. Sci. Total Environ. 2019, 665, 828–835. [Google Scholar] [CrossRef]
- Kostic, I.; Andjelkovic, T.; Nikolic, R.; Cvetkovic, T.; Pavlovic, D.; Bojic, A. Comparative study of binding strengths of heavy metals with humic acid. Hem. Ind. 2013, 67, 773–779. [Google Scholar] [CrossRef]
- Dinh, Q.T.; Li, Z.; Tran, T.A.T.; Wang, D.; Liang, D. Role of organic acids on the bioavailability of selenium in soil: A review. Chemosphere 2017, 184, 618–635. [Google Scholar] [CrossRef]
- Yang, L.; Su, Y.; Xu, Y.; Wang, Z.; Guo, Z.; Weng, S.; Yan, C.; Zhang, S.; Wu, J. Interactions between metal ions and carbohydrates. Coordination behavior of neutral erythritol to Ca(II) and lanthanide ions. Inorg. Chem. 2003, 42, 5844–5856. [Google Scholar] [CrossRef]
- Peng, T.; Liao, W.; Gu, G.; Qiu, G.; Wu, X.; Yang, F.; Zeng, W. Insights into the role of extracellular DNA in heavy metal adsorption. Sci. Total Environ. 2022, 808, 152067. [Google Scholar] [CrossRef] [PubMed]
- Barber-Zucker, S.; Shaanan, B.; Zarivach, R. Transition metal binding selectivity in proteins and its correlation with the phylogenomic classification of the cation diffusion facilitator protein family. Sci. Rep. 2017, 7, 16381. [Google Scholar] [CrossRef]
- Maruyama, T.; Matsushita, H.; Shimada, Y.; Kamata, I.; Hanaki, M.; Sonokawa, S.; Kamiya, N.; Goto, M. Proteins and protein-rich biomass as environmentally friendly adsorbents selective for precious metal ions. Environ. Sci. Technol. 2007, 41, 1359–1364. [Google Scholar] [CrossRef]
- Dertz, E.A.; Xu, J.D.; Stintzi, A.; Raymond, K.N. Bacillibactin-mediated iron transport in Bacillus subtilis. J. Am. Chem. Soc. 2006, 128, 22. [Google Scholar] [CrossRef]
- Albrecht-Gary, A.M.; Blanc, S.; Rochel, N.; Ocacktan, A.Z.; Abdallah, M.A. Bacterial iron transport: Coordination properties of pyoverdin PaA, a peptidic siderophore of Pseudomonas aeruginosa. Inorg. Chem. 1994, 33, 6391–6402. [Google Scholar] [CrossRef]
- Raymond, K.N.; Dertz, E.A.; Kim, S.S. Enterobactin: An archetype for microbial iron transport. Proc. Natl. Acad. Sci. USA 2003, 100, 3584–3588. [Google Scholar] [CrossRef] [PubMed]
- Hider, R.C.; Kong, X. Chemistry and biology of siderophores. Nat. Prod. Rep. 2010, 27, 637–657. [Google Scholar] [CrossRef]
- Virpiranta, H.; Banasik, M.; Taskila, S.; Leiviskä, T.; Halttu, M.; Sotaniemi, V.-H.; Tanskanen, J. Isolation of efficient metal-binding bacteria from boreal peat soils and development of microbial biosorbents for improved nickel scavenging. Water 2020, 12, 2000. [Google Scholar] [CrossRef]
- Brandon, M.S.; Paszczynski, A.J.; Korus, R.; Crawford, R.L. The determination of the stability constant for the iron(II) complex of the biochelator pyridine-2,6-bis(monothiocarboxylic acid). Biodegradation 2003, 14, 73–82. [Google Scholar] [CrossRef]
- Noinaj, N.; Guillier, M.; Barnard, T.J.; Buchanan, S.K. TonB-dependent transporters: Regulation, structure, and function. Annu. Rev. Microbiol. 2010, 64, 43–60. [Google Scholar] [CrossRef]
- Ferguson, A.D.; Deisenhofer, J. TonB-dependent receptors—Structural perspectives. Biochim. Biophys. Acta—Biomembr. 2002, 1565, 318–332. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Singh, P.; Srivastava, A. Synthesis, nature and utility of universal iron chelator—Siderophore: A review. Microbiol. Res. 2018, 212–213, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Pollet, R.M.; Martin, L.M.; Koropatkin, N.M. TonB-dependent transporters in the Bacteroidetes: Unique domain structures and potential functions. Mol. Microbiol. 2021, 115, 490–501. [Google Scholar] [CrossRef] [PubMed]
- Schalk, I.J.; Hannauer, M.; Brand, A. New roles for bacterial siderophores in metal transport and tolerance. Environ. Microbiol. 2011, 13, 2844–2854. [Google Scholar] [CrossRef]
- Rees, D.C.; Johnson, E.; Lewinson, O. ABC transporters: The power to change. Nat. Rev. Mol. Cell Biol. 2009, 10, 218–227. [Google Scholar] [CrossRef]
- Matzanke, B.F.; Anemuller, S.; Schunemann, V.; Trautwein, A.X.; Hantke, K. FhuF, part of a siderophore reductase system. Biochemistry 2004, 43, 1386–1392. [Google Scholar] [CrossRef]
- Fukushima, T.; Allred, B.E.; Sia, A.K.; Nichiporuk, R.; Andersen, U.N.; Raymond, K.N. Gram-positive siderophore-shuttle with iron-exchange from Fe-siderophore to apo-siderophore by Bacillus cereus YxeB. Proc. Natl. Acad. Sci. USA 2013, 110, 13821–13826. [Google Scholar] [CrossRef]
- Chen, Y.; Jurkewitch, E.; Bar-Ness, E.; Hadar, Y. Stability constants of pseudobactin complexes with transition metals. Soil. Sci. Soc. Am. J. 1994, 58, 390–396. [Google Scholar] [CrossRef]
- Hernlem, B.J.; Vane, L.M.; Sayles, G.D. Stability constants for complexes of the siderophore desferrioxamine B with selected heavy metal cations. Inorg. Chim. Acta 1996, 244, 179–184. [Google Scholar] [CrossRef]
- Braud, A.; Hannauer, M.; Mislin, G.L.; Schalk, I.J. The Pseudomonas aeruginosa pyochelin-iron uptake pathway and its metal specificity. J. Bacteriol. 2009, 191, 3517–3525. [Google Scholar] [CrossRef]
- Braud, A.; Hoegy, F.; Jezequel, K.; Lebeau, T.; Schalk, I.J. New insights into the metal specificity of the Pseudomonas aeruginosa pyoverdine-iron uptake pathway. Environ. Microbiol. 2009, 11, 1079–1091. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, M.; Ae, N.; Prasad, M.N.V.; Freitas, H. Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol. 2010, 28, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Braun, V.; Hantke, K. Recent insights into iron import by bacteria. Curr. Opin. Chem. Biol. 2011, 15, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Zloch, M.; Thiem, D.; Gadzala-Kopciuch, R.; Hrynkiewicz, K. Synthesis of siderophores by plant-associated metallotolerant bacteria under exposure to Cd2+. Chemosphere 2016, 156, 312–325. [Google Scholar] [CrossRef]
- Nong, Q.; Yuan, K.; Li, Z.; Chen, P.; Huang, Y.; Hu, L.; Jiang, J.; Luan, T.; Chen, B. Bacterial resistance to lead: Chemical basis and environmental relevance. J. Environ. Sci. 2019, 85, 46–55. [Google Scholar] [CrossRef]
- Lamb, A.L. Breaking a pathogen’s iron will: Inhibiting siderophore production as an antimicrobial strategy. Biochim. Biophys. Acta-Proteins Proteom. 2015, 1854, 1054–1070. [Google Scholar] [CrossRef]
- Xie, B.; Wei, X.; Wan, C.; Zhao, W.; Song, R.; Xin, S.; Song, K. Exploring the biological pathways of siderophores and their multidisciplinary applications: A comprehensive review. Molecules 2024, 29, 2318. [Google Scholar] [CrossRef]
- Lazos, O.; Tosin, M.; Slusarczyk, A.L.; Boakes, S.; Cortés, J.; Sidebottom, P.J.; Leadlay, P.F. Biosynthesis of the putative siderophore erythrochelin requires unprecedented crosstalk between separate nonribosomal peptide gene clusters. Chem. Biol. 2010, 17, 160–173. [Google Scholar] [CrossRef]
- Condurso, H.L.; Bruner, S.D. Structure and noncanonical chemistry of nonribosomal peptide biosynthetic machinery. Nat. Prod. Rep. 2012, 29, 1099–1110. [Google Scholar] [CrossRef]
- Keating, T.A.; Ehmann, D.E.; Kohli, R.M.; Marshall, C.G.; Trauger, J.W.; Walsh, C.T. Chain termination steps in nonribosomal peptide synthetase assembly lines: Directed acyl-S-enzyme breakdown in antibiotic and siderophore biosynthesis. Chem. BioChem 2001, 2, 99–107. [Google Scholar] [CrossRef]
- Keatinge-Clay, A.T. The structures of type I polyketide synthases. Nat. Prod. Rep. 2012, 29, 1050. [Google Scholar] [CrossRef] [PubMed]
- Khosla, C.; Herschlag, D.; Cane, D.E.; Walsh, C.T. Assembly line polyketide synthases: Mechanistic insights and unsolved problems. Biochemistry 2014, 53, 2875–2883. [Google Scholar] [CrossRef]
- Miethke, M.; Bisseret, P.; Beckering, C.L.; Vignard, D.; Eustache, J.; Marahiel, M.A. Inhibition of aryl acid adenylation domains involved in bacterial siderophore synthesis. FEBS J. 2006, 273, 409–419. [Google Scholar] [CrossRef]
- Miethke, M.; Schmidt, S.; Marahiel, M.A. The major facilitator superfamily-type transporter YmfE and the multidrug-efflux activator Mta mediate bacillibactin secretion in Bacillus subtilis. J. Bacteriol. 2008, 190, 5143–5152. [Google Scholar] [CrossRef]
- Nikaido, H.; Pagès, J.-M. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev. 2012, 36, 340–363. [Google Scholar] [CrossRef]
- Lekshmi, M.; Ortiz-Alegria, A.; Kumar, S.; Varela, M.F. Major facilitator superfamily efflux pumps in human pathogens: Role in multidrug resistance and beyond. Curr. Res. Microb. Sci. 2024, 7, 100248. [Google Scholar] [CrossRef] [PubMed]
- Mosa, K.A.; Saadoun, I.; Kumar, K.; Helmy, M.; Dhankher, O.P. Potential biotechnological strategies for the cleanup of heavy metals and metalloids. Front. Plant Sci. 2016, 7, 303. [Google Scholar] [CrossRef]
- Sowmya, M.; Hatha, A.M. Cadmium and lead tolerance mechanisms in bacteria and the role of halotolerant and moderately halophilic bacteria in their remediation. In Handbook of Metal-Microbe Interactions and Bioremediation; CRC Press: Boca Raton, FL, USA, 2017; pp. 557–573. [Google Scholar]
- Braud, A.; Jézéquel, K.; Lebeau, T. Impact of substrates and cell immobilization on siderophore activity by Pseudomonads in a Fe and/or Cr, Hg, Pb containing-medium. J. Hazard. Mater. 2007, 144, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Nnaji, N.D.; Anyanwu, C.U.; Miri, T.; Onyeaka, H. Mechanisms of heavy metal tolerance in bacteria: A review. Sustainability 2024, 16, 11124. [Google Scholar] [CrossRef]
- Chatterjee, S.; Kumari, S.; Rath, S.; Priyadarshanee, M.; Das, S. Diversity, structure and regulation of microbial metallothionein: Metal resistance and possible applications in sequestration of toxic metals. Metallomics 2020, 12, 1637. [Google Scholar] [CrossRef]
- Naik, M.M.; Pandey, A.; Dubey, S.K. Pseudomonas aeruginosa strain WI-1 from Mandovi estuary possesses metallothionein to alleviate lead toxicity and promotes plant growth. Ecotoxicol. Environ. Saf. 2012, 79, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Naik, M.M.; Shamim, K.; Dubey, S.K. Biological characterization of lead-resistant bacteria to explore role of bacterial metallothionein in lead resistance. Curr. Sci. 2012, 103, 426–429. [Google Scholar]
- Blindauer, C.A.; Sadler, P.J. How to hide zinc in a small protein. Acc. Chem. Res. 2005, 38, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Blindauer, C.A. Bacterial metallothioneins: Past, present, and questions for the future. J. Biol. Inorg. Chem. 2011, 16, 1011. [Google Scholar] [CrossRef]
- Morby, A.P.; Turner, J.S.; Huckle, J.W.; Robinson, N.J. SmtB is a metal-dependent repressor of the cyanobacterial metallothionein gene smtA: Identification of a Zn inhibited DNA-protein complex. Nucleic Acids Res. 1993, 21, 921–925. [Google Scholar] [CrossRef]
- Olafson, R.W.; McCubbin, W.D.; Kay, C.M. Primary- and secondary structural analysis of a unique prokaryotic metallothionein from a Synechococcus sp. cyanobaterium. Biochem. J. 1998, 251, 691–699. [Google Scholar] [CrossRef]
- Blindauer, C.A.; Harrison, M.D.; Parkinson, J.A.; Robinson, A.K.; Cavet, J.S.; Robinson, N.J.; Sadler, P.J. A metallothionein containing a zinc finger within a four-metal cluster protects a bacterium from zinc toxicity. Proc. Natl. Acad. Sci. USA 2001, 98, 9593–9598. [Google Scholar] [CrossRef]
- Sharma, J.; Shamim, K.; Dubeya, S.K.; Meena, R.M. Metallothionein assisted periplasmic lead sequestration as lead sulfite by Providencia vermicola strain SJ2A. Sci. Total Environ. 2017, 579, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Helmann, J.D. Metals in motion: Understanding labile metal pools in bacteria. Biochemistry 2025, 64, 329–345. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Terán, I.; Goh, K.G.K.; Ulett, G.C. Resisting death by metal: Metabolism and Cu/Zn homeostasis in bacteria. Emerg. Top. Life Sci. 2024, 8, 45–56. [Google Scholar] [CrossRef]
- Sá, C.; Matos, D.; Pires, A.; Cardoso, P.; Figueira, E. Effects of volatile sulfur compounds on growth and oxidative stress of Rhizobium leguminosarum E20-8 exposed to cadmium. Sci. Total Environ. 2021, 800, 149478. [Google Scholar] [CrossRef] [PubMed]
- Lima, A.I.G.; Corticeiro, S.C.; de Almeida Paula Figueira, E.M. Glutathione-mediated cadmium sequestration in Rhizobium leguminosarum. Enzym. Microb. Technol. 2006, 39, 763–769. [Google Scholar] [CrossRef]
- Stewart, L.J.; Ong, C.-L.Y.; Zhang, M.M.; Brouwer, S.; McIntyre, L.; Davies, M.R.; Walker, M.J.; McEwan, A.G.; Waldron, K.J.; Djoko, K.Y.; et al. Role of glutathione in buffering excess intracellular copper in Streptococcus pyogenes. mBio11 2020, 11, e02804-20. [Google Scholar] [CrossRef] [PubMed]
- Helbig, K.; Bleuel, C.; Krauss, G.J.; Nies, D.H. Glutathione and transition-metal homeostasis in Escherichia coli. J. Bacteriol. 2008, 190, 5431–5438. [Google Scholar] [CrossRef]
- Ma, Z.; Chandrangsu, P.; Helmann, T.C.; Romsang, A.; Gaballa, A.; Helmann, J.D. Bacillithiol is a major buffer of the labile zinc pool in Bacillus subtilis. Mol. Microbiol. 2014, 94, 756–770. [Google Scholar] [CrossRef]
- Kay, K.L.; Hamilton, C.J.; Le Brun, N.E. Mass spectrometric studies of Cu(I)-binding to the N-terminal domains of B. subtilis CopA and influence of bacillithiol. J. Inorg. Biochem. 2019, 190, 24–30. [Google Scholar] [CrossRef]
- Nairn, B.L.; Lonergan, Z.R.; Wang, J.; Braymer, J.J.; Zhang, Y. The response of Acinetobacter baumannii to zinc starvation. Cell Host Microbe 2016, 19, 826–836. [Google Scholar] [CrossRef]
- Strenkert, D.; Schmollinger, S.; Hu, Y.; Hofmann, C.; Holbrook, K.; Liu, H.W.; O Purvine, S.; Nicora, C.D.; Chen, S.; Lipton, M.S.; et al. Zn deficiency disrupts Cu and S homeostasis in Chlamydomonas resulting in over accumulation of Cu and Cysteine. Metallomics 2023, 5, mfad043. [Google Scholar] [CrossRef]
- Thatoi, H.; Das, S.; Mishra, J.; Rath, B.P.; Das, N. Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: A review. J. Environ. Manag. 2014, 146, 383–399. [Google Scholar] [CrossRef]
- Gavrilescu, M. Removal of heavy metals from the environment by biosorption. Eng. Life Sci. 2004, 4, 219–232. [Google Scholar] [CrossRef]
- Chandrangsu, P.; Rensing, C.; Helmann, J.D. Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 2017, 15, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; Chatterjee, S.; Kataki, S.; Rastogi, R.P.; Gupta, D.K. Bacterial tolerance strategies against lead toxicity and their relevance in bioremediation application. Environ. Sci. Pollut. Res. Int. 2021, 28, 14271–14284. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H., Jr. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 2000, 64, 354–411. [Google Scholar] [CrossRef]
- Tseng, T.T.; Gratwick, K.S.; Kollman, J.; Park, D.; Nies, D.H.; Goffeau, A.; Saier, M.H., Jr. The RND permease superfamily: An ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1999, 1, 107–125. [Google Scholar]
- Moreira, M.A.; de Souza, S.C.; de Moraes, C.A. Multidrug efflux systems in Gram-negative bacteria. Braz. J. Microbiol. 2004, 35, 19–28. [Google Scholar] [CrossRef]
- Diels, L.; Dong, Q.; van der Lelie, D.; Baeyens, W.; Mergeay, M. The czc operon of Alcaligenes eutrophus CH34: From resistance mechanism to the removal of heavy metals. J. Ind. Microbiol. 1995, 14, 142–153. [Google Scholar] [CrossRef]
- Grosse, C.; Anton, A.; Hoffmann, T.; Franke, S.; Schleuder, G.; Nies, D.H. Identification of a regulatory pathway that controls the heavy-metal resistance system Czc via promoter czcNp in Ralstonia metallidurans. Arch. Microbiol. 2004, 182, 109–118. [Google Scholar] [CrossRef]
- Macomber, L.; Hausinger, R.P. Mechanisms of nickel toxicity in microorganisms. Metallomics 2011, 3, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Fierros-Romero, G.; Gómez-Ramírez, M.; Sharma, A.; Pless, R.C.; Rojas-Avelizapa, N.G. czcD gene from Bacillus megaterium and Microbacterium liquefaciens as a potential nickel–vanadium soil pollution biomarker. J. Basic Microbiol. 2020, 60, 22–26. [Google Scholar] [CrossRef]
- Haney, C.J.; Grass, G.; Franke, S.; Rensing, C. New developments in the understanding of the cation diffusion facilitator family. J. Ind. Microbiol. Biotechnol. 2005, 32, 215–226. [Google Scholar] [CrossRef]
- Kolaj-Robin, O.; Russell, D.; Hayes, K.A.; Pembroke, J.T.; Soulimane, T. Cation Diffusion Facilitator family: Structure and function. FEBS Lett. 2015, 589, 1283–1295. [Google Scholar] [CrossRef] [PubMed]
- Silver, S.; Phung, L.T. A bacterial view of the periodic table: Genes and proteins for toxic inorganic ions. J. Ind. Microbiol. Biotechnol. 2005, 32, 587–605. [Google Scholar] [CrossRef] [PubMed]
- Oger, C.; Mahillon, J.; Petit, F. Distribution and diversity of a cadmium resistance (cadA) determinant and occurrence of IS257 insertion sequences in Staphylococcal bacteria isolated from a contaminated estuary (Seine, France). FEMS Microbiol. Ecol. 2003, 43, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Prabhakaran, R.; Thamarai, R. Elucidation of the CadA protein 3D Structure and affinity for metals. Bioinform. Biol. Insights. 2024, 8, 11779322241266701. [Google Scholar] [CrossRef]
- Hikal, A.F.; Hasan, S.; Gudeta, D.; Zhao, S.; Foley, S.; Khan, A.A. The acquired pco gene cluster in Salmonella enterica mediates resistance to copper. Front. Microbiol. 2024, 15, 1454763. [Google Scholar] [CrossRef]
- Rensing, C.; Grass, G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 2003, 27, 197–213. [Google Scholar] [CrossRef]
- Giachino, A.; Waldron, K.J. Copper tolerance in bacteria requires the activation of multiple accessory pathways. Mol. Microbiol. 2020, 114, 377–390. [Google Scholar] [CrossRef]
- Festa, R.A.; Thiele, D.J. Copper: An essential metal in biology. Curr. Biol. 2011, 21, R877–R883. [Google Scholar] [CrossRef]
- Chalmers, G.; Rozas, K.M.; Amachawadi, R.G.; Scott, H.M.; Norman, K.N.; Nagaraja, T.G.; Tokach, M.D.; Boerlin, P. Distribution of the pco gene cluster and associated genetic determinants among swine Escherichia coli from a controlled feeding trial. Genes 2018, 18, 504. [Google Scholar] [CrossRef]
- Rosen, B.P. Transport and detoxification systems for transition metals, heavy metals and metalloids in eucaryotic and procaryotic microbes. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2002, 133, 689–693. [Google Scholar] [CrossRef]
- Neubert, M.J.; Dahlmann, E.A.; Ambrose, A.; Johnson, M.D.L. Copper chaperone CupA and zinc control CopY regulation of the Pneumococcal cop operon. ASM J. 2017, 2, e00372-17. [Google Scholar] [CrossRef] [PubMed]
- Mealman, T.D.; Blackburn, N.J.; McEvoy, M.M. Chapter seven—Metal export by CusCFBA, the periplasmic Cu(I)/Ag(I) transport system of Escherichia coli. Curr. Top. Membr. 2012, 69, 163–196. [Google Scholar] [CrossRef]
- Delmar, J.A.; Su, C.C.; Yu, E.W. Heavy metal transport by the CusCFBA efflux system. Protein Sci. 2015, 24, 1720–1736. [Google Scholar] [CrossRef] [PubMed]
- Gudipaty, S.A.; Larsen, A.S.; Rensing, C.; McEvoy, M.M. Regulation of Cu(I)/Ag(I) efflux genes in Escherichia coli by the sensor kinase CusS. FEMS Microbiol. Lett. 2012, 330, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Randall, C.P.; Gupta, A.; Jackson, N.; Busse, D.; O’Neill, A.J. Silver resistance in Gram-negative bacteria: A dissection of endogenous and exogenous mechanisms. J. Antimicrob. Chemother. 2015, 70, 1037–1046. [Google Scholar] [CrossRef]
- Wang, H.; Li, J.; Min, C.; Xia, F.; Tang, M.; Li, J.; Hu, Y.; Zou, M. Characterization of silver resistance and coexistence of sil operon with antibiotic resistance genes among Gram-negative pathogens isolated from wound samples by using whole-genome sequencing. Infect. Drug Resist. 2022, 15, 1425–1437. [Google Scholar] [CrossRef]
- Fang, L.; Li, X.; Li, L.; Li, S.; Liao, X.; Sun, J.; Liu, Y. Co-spread of metal and antibiotic resistance within ST3-IncHI2 plasmids from E. coli isolates of food-producing animals. Sci. Rep. 2016, 6, 25312. [Google Scholar] [CrossRef]
- Gupta, A.; Matsui, K.; Lo, J.F.; Silver, S. Molecular basis for resistance to silver cations in Salmonella. Nat. Med. 1999, 5, 183–188. [Google Scholar] [CrossRef]
- Lee, S.M.; Grass, G.; Rensing, C.; Barrett, S.R.; Yates, C.J.D.; Stoyanov, J.V.; Brown, N.L. The Pco proteins are involved in periplasmic copper handling in Escherichia coli. Biochem. Biophys. Res. Commun. 2002, 295, 616–620. [Google Scholar] [CrossRef]
- Huffman, D.L.; Huyett, J.; Outten, F.W.; Doan, P.E.; Finney, L.A.; Hoffman, B.M.; O’Halloran, T.V. Spectroscopy of Cu(II)-PcoC and the multicopper oxidase function of PcoA, two essential components of Escherichia coli pco copper resistance operon. Biochemistry 2002, 41, 10046–10055. [Google Scholar] [CrossRef]
- Zimmermann, M.; Udagedara, S.R.; Sze, C.M.; Ryan, T.M.; Howlett, G.J.; Xiao, Z.G.; Wedd, A.G. PcoE-A metal sponge expressed to the periplasm of copper resistance Escherichia coli. Implication of its function role in copper resistance. J. Inorg. Biochem. 2012, 115, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Nayeri, N.; Gorecki, K.; Becares, E.R.; Wang, K.; Mahato, D.R.; Andersson, M.; Abeyrathna, S.S.; Lindkvist-Petersson, K.; Meloni, G.; et al. Pco B is a defense outer membrane protein that facilitates cellular uptake of copper. Protein Sci. 2022, 31, 4364. [Google Scholar] [CrossRef]
- Fekih, I.B.; Zhang, C.; Li, Y.P.; Zhao, Y.; Alwathnani, H.A.; Saquib, Q.; Rensing, C.; Cervantes, C. Distribution of arsenic resistance genes in Prokaryotes. Front. Microbiol. 2018, 9, 2473. [Google Scholar] [CrossRef]
- Kabiraj, A.; Biswas, R.; Halder, U.; Bandopadhyay, R. Bacterial arsenic metabolism and its role in arsenic bioremediation. Curr. Microbiol. 2022, 15, 131. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.Y.; Costa, M. Arsenic: A global environmental challenge. Annu. Rev. Pharmacol. Toxicol. 2021, 61, 47–63. [Google Scholar] [CrossRef] [PubMed]
- Busenlehner, L.S.; Pennella, M.A.; Giedroc, D.P. The SmtB/ArsR family of metalloregulatory transcriptional repressors: Structural insights into procaryotic metal resistance. FEMS Microbiol. Rev. 2003, 27, 131–143. [Google Scholar] [CrossRef]
- Ji, G.; Silver, S. Bacterial resistance mechanisms for heavy metals of environmental concern. J. Ind. Microbiol. 1995, 14, 61–75. [Google Scholar] [CrossRef]
- Santini, J.M.; vanden Hoven, R.N. Molybdenum-containing arsenite oxidase of the chemolithoautotrophic arsenite oxidizer NT-26. J. Bacteriol. 2004, 186, 1614–1619. [Google Scholar] [CrossRef]
- vanden Hoven, R.N.; Santini, J.M. Arsenite oxidation by the heterotroph Hydrogenophaga sp. str. NT-14: The arsenite oxidase and its physiological electron acceptor. Biochim. Biophys. Acta 2004, 1656, 148–155. [Google Scholar] [CrossRef]
- Boyd, E.S.; Barkay, T. The mercury resistance operon: From an origin in a geothermal environment to an efficient detoxification machine. Front. Microbiol. 2012, 3, 349. [Google Scholar] [CrossRef]
- Bhat, A.; Sharma, R.; Desigan, K.; Lucas, M.M.; Mishra, A.; Bowers, R.M.; Woyke, T.; Epstein, B.; Tiffin, P.; Pueyo, J.J.; et al. Horizontal gene transfer of the Mer operon is associated with large effects on the transcriptome and increased tolerance to mercury in nitrogen-fixing bacteria. BMC Microbiol. 2024, 24, 247. [Google Scholar] [CrossRef] [PubMed]
- Bruins, M.R.; Kapil, S.; Oehme, F.W. Microbial resistance to metals in the environment. Ecotox. Environ. Safe. 2000, 45, 198–207. [Google Scholar] [CrossRef]
- Dash, H.R.; Das, S. Bioremediation of mercury and the importance of bacterial mer genes. Int. Biodeterior. Biodegrad. 2012, 75, 207–213. [Google Scholar] [CrossRef]
- Tottey, S.; Harvie, D.R.; Robinson, N.J. Understanding how cells allocate metals using metal sensors and metallochaperones. Acc. Chem. Res. 2005, 10, 775–783. [Google Scholar] [CrossRef]
- Chakraborty, U.K.; Park, Y.; Sengupta, K.; Jung, W.; Joshi, C.P.; Francis, D.H.; Chen, P. A ‘through-DNA’ mechanism for co-regulation of metal uptake and efflux. Nat. Commun. 2024, 15, 10555. [Google Scholar] [CrossRef]
- Summers, A.O. Damage control: Regulating defenses against toxic metals and metalloids. Curr. Opin. Microbiol. 2009, 2, 138–144. [Google Scholar] [CrossRef]
- Foster, A.W.; Osman, D.; Robinson, N.J. Metal preferences and metallation. J. Biol. Chem. 2014, 41, 28095–28103. [Google Scholar] [CrossRef] [PubMed]
- Waldron, K.J.; Rutherford, J.C.; Ford, D.; Robinson, N.J. Metalloproteins and metal sensing. Nature 2009, 7257, 823–830. [Google Scholar] [CrossRef]
- Irving, H.; Williams, R.J. Order of stability of metal com plexes. Nature 1948, 162, 746–747. [Google Scholar] [CrossRef]
- Ma, Z.; Jacobsen, F.E.; Giedroc, D.P. Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 2009, 109, 4644–4681. [Google Scholar] [CrossRef]
- Pontel, L.B.; Soncini, F.C. Alternative periplasmic copper-resistance mechanisms in Gram negative bacteria. Mol. Microbiol. 2009, 73, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Ramesh, A.; Ma, Z.; Ward, S.K.; Zhang, L.; George, G.N.; Talaat, A.M.; Sacchettini, J.C.; Giedroc, D.P. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat. Chem. Biol. 2007, 3, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Argüello, J.M.; Eren, E.; González-Guerrero, M. The structure and function of heavy metal transport P1B-ATPases. Biometals 2007, 20, 233–248. [Google Scholar] [CrossRef]
- Liu, T.; Chen, X.; Ma, Z.; Shokes, J.; Hemmingsen, L.; Scott, R.A.; Giedroc, D.P. A Cu(I)-sensing ArsR family metal sensor protein with a relaxed metal selectivity profile. Biochemistry 2008, 47, 10564–10575. [Google Scholar] [CrossRef]
- Bütof, L.; Große, C.; Lilie, H.; Herzberg, M.; Nies, D.H. Interplay between the Zur Regulon Components and Metal Resistance in Cupriavidus metallidurans. J. Bacteriol. 2019, 15, e00192-19. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Renugopalakrishnan, V.; Liepmann, D.; Paulmurugan, R.; Malhotra, B.D. Cell-based biosensors: Recent trends, challenges and future perspectives. Biosens. Bioelectron. 2019, 15, 111435. [Google Scholar] [CrossRef]
- Inda, M.E.; Lu, T.K. Microbes as biosensors. Annu. Rev. Microbiol. 2020, 74, 337–359. [Google Scholar] [CrossRef]
- Velusamy, K.; Periyasamy, S.; Kumar, P.S.; Rangasamy, G.; Pauline, J.M.N.; Ramaraju, P.; Mohanasundaram, S.; Vo, D.-V.N. Biosensor for heavy metals detection in wastewater: A review. Food Chem. Toxicol. 2022, 168, 113307. [Google Scholar] [CrossRef]
- Turner, R.J. The good, the bad, and the ugly of metals as antimicrobials. Biometals 2024, 37, 545–559. [Google Scholar] [CrossRef]
- Wang, C.; Wei, X.; Zhong, L.; Chan, C.L.; Li, H.; Sun, H. Metal-based approaches for the fight against antimicrobial resistance: Mechanisms, opportunities, and challenges. J. Am. Chem. Soc. 2025, 147, 12361–12380. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oleńska, E.; Małek, W.; Swiecicka, I.; Wójcik, M.; Thijs, S.; Vangronsveld, J. Bacteria Under Metal Stress—Molecular Mechanisms of Metal Tolerance. Int. J. Mol. Sci. 2025, 26, 5716. https://doi.org/10.3390/ijms26125716
Oleńska E, Małek W, Swiecicka I, Wójcik M, Thijs S, Vangronsveld J. Bacteria Under Metal Stress—Molecular Mechanisms of Metal Tolerance. International Journal of Molecular Sciences. 2025; 26(12):5716. https://doi.org/10.3390/ijms26125716
Chicago/Turabian StyleOleńska, Ewa, Wanda Małek, Izabela Swiecicka, Małgorzata Wójcik, Sofie Thijs, and Jaco Vangronsveld. 2025. "Bacteria Under Metal Stress—Molecular Mechanisms of Metal Tolerance" International Journal of Molecular Sciences 26, no. 12: 5716. https://doi.org/10.3390/ijms26125716
APA StyleOleńska, E., Małek, W., Swiecicka, I., Wójcik, M., Thijs, S., & Vangronsveld, J. (2025). Bacteria Under Metal Stress—Molecular Mechanisms of Metal Tolerance. International Journal of Molecular Sciences, 26(12), 5716. https://doi.org/10.3390/ijms26125716






