Genomic Insights into Basal Diptera Phylogeny: The Non-Monophyletic Nature of Blephariceromorpha
Abstract
1. Introduction
2. Results
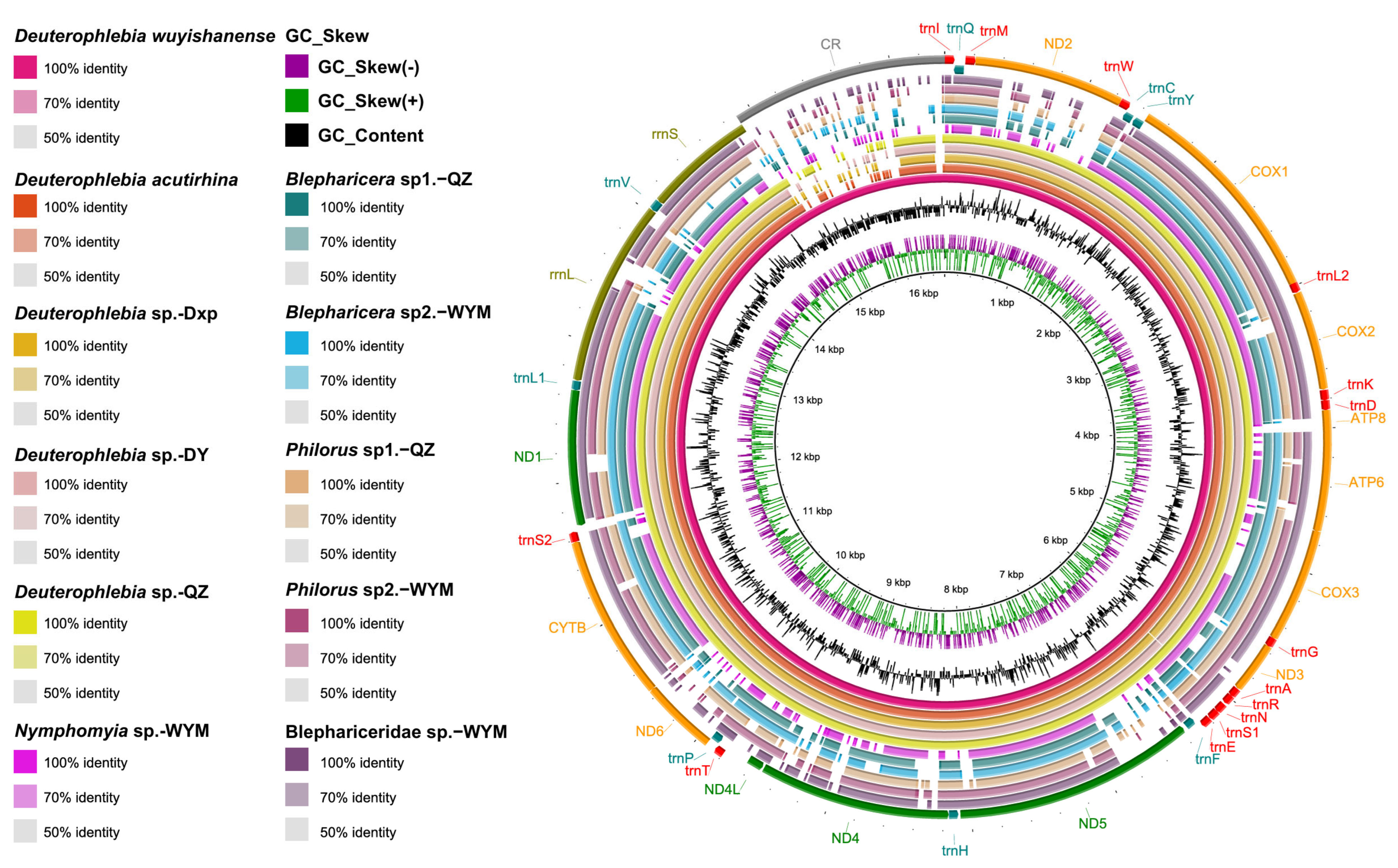
2.1. Mitogenomic and Transcriptomic Data Analysis
2.2. Phylogenetic Analyses Using Nuclear Datasets
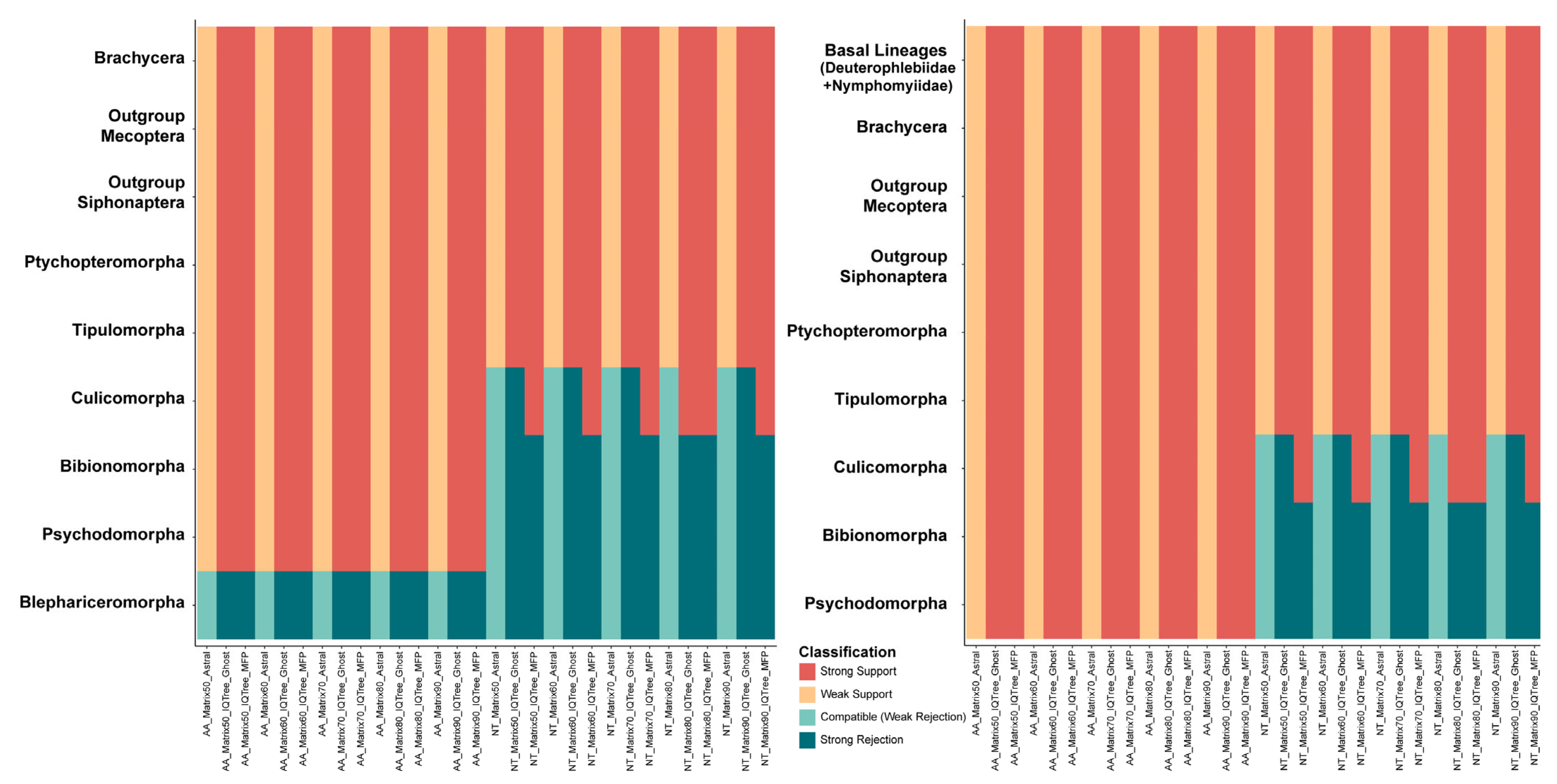
2.3. Phylogenetic Analyses Using Mitochondrial Datasets
3. Discussion
3.1. New Insights into Phylogeny of Nematocera
3.2. Investigation of Basal Lineages in Diptera
3.3. Convergent Evolution of Larval Morphology in Blephariceromorpha
4. Materials and Methods
4.1. Sample Collection
4.2. DNA Extraction, Mitogenomes Sequencing, and Analysis
4.3. RNA Extraction, Sequencing, and Transcriptome Assembly
4.4. Matrix Construction
4.5. Phylogenetic Analysis
4.6. Analyses of Phylogenetic Discordance and Alternative Relationships
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Systema Dipterorum. Available online: http://www.diptera.org/ (accessed on 5 September 2024).
- Wiegmann, B.M.; Trautwein, M.D.; Winkler, I.S.; Barr, N.B.; Kim, J.-W.; Lambkin, C.; Bertone, M.A.; Cassel, B.K.; Bayless, K.M.; Heimberg, A.M.; et al. Episodic Radiations in the Fly Tree of Life. Proc. Natl. Acad. Sci. USA 2011, 108, 5690–5695. [Google Scholar] [CrossRef] [PubMed]
- Yeates, D.; Wiegmann, B. Phylogeny and Evolution of Diptera: Recent Insights and New Perspectivs. In The Evolutionary Biology of Flies; Columbia University Press: New York, NY, USA, 2005. [Google Scholar]
- Yeates, D.K.; Wiegmann, B.M.; Courtney, G.W.; Meier, R.; Lambkin, C.; Pape, T. Phylogeny and Systematics of Diptera: Two Decades of Progress and Prospects. Zootaxa 2007, 1668, 565–590. [Google Scholar] [CrossRef]
- Yeates, D.; Wiegmann, B. Phylogeny of Diptera. In Manual of Afrotropical Diptera; SANBI Graphics & Editing: Pretoria, South Africa, 2017; Volume 4, pp. 253–265. [Google Scholar]
- Borkent, A. The State of Phylogenetic Analysis: Narrow Visions and Simple Answers-Examples from the Diptera (Flies). Zootaxa 2018, 4374, 107–143. [Google Scholar] [CrossRef] [PubMed]
- Savage, J.; Borkent, A.; Brodo, F.; Cumming, J.M.; Curler, G.; Currie, D.C.; de Waard, J.R.; Gibson, J.F.; Hauser, M.; Laplante, L.; et al. Diptera of Canada. ZooKeys 2019, 819, 397–450. [Google Scholar] [CrossRef] [PubMed]
- Gerhardt, R.R.; Hribar, L.J. Chapter 11—Flies (Diptera). In Medical and Veterinary Entomology, 3rd ed.; Mullen, G.R., Durden, L.A., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 171–190. ISBN 978-0-12-814043-7. [Google Scholar]
- Hayon, I.; Mendel, Z.; Dorchin, N. Predatory Gall Midges on Mealybug Pests—Diversity, Life History, and Feeding Behavior in Diverse Agricultural Settings. Biol. Control 2016, 99, 19–27. [Google Scholar] [CrossRef]
- Weaver, S.C.; Reisen, W.K. Present and Future Arboviral Threats. Antivir. Res. 2010, 85, 328–345. [Google Scholar] [CrossRef]
- Wallace, J.B.; Webster, J.R. The Role of Macroinvertebrates in Stream Ecosystem Function. Annu. Rev. Entomol. 1996, 41, 115–139. [Google Scholar] [CrossRef]
- Orford, K.A.; Vaughan, I.P.; Memmott, J. The Forgotten Flies: The Importance of Non-Syrphid Diptera as Pollinators. Proc. R. Soc. B Biol. Sci. 2015, 282, 20142934. [Google Scholar] [CrossRef]
- Oosterbroek, P.; Courtney, G. Phylogeny of the Nematocerous Families of Diptera (Insecta). Zool. J. Linn. Soc. 1995, 115, 267–311. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, D.; Kang, Z. New Data on the Mitochondrial Genome of Nematocera (Lower Diptera): Features, Structures and Phylogenetic Implications. Zool. J. Linn. Soc. 2023, 197, 229–245. [Google Scholar] [CrossRef]
- Gregory, W. Courtney Phylogenetic Analysis of the Blephariceromorpha, with Special Reference to Mountain Midges (Diptera: Deuterophlebiidae. Syst. Entomol. 1991, 16, 137–172. [Google Scholar]
- Wood, D.M. Phylogeny and Classification of the Nenatocera. Man. Nearctic Diptera 1989, 3, 1333–1370. [Google Scholar]
- Bertone, M.A.; Courtney, G.W.; Wiegmann, B.M. Phylogenetics and Temporal Diversification of the Earliest True Flies (Insecta: Diptera) Based on Multiple Nuclear Genes. Syst. Entomol. 2008, 33, 668–687. [Google Scholar] [CrossRef]
- Lambkin, C.L.; Sinclair, B.J.; Pape, T.; Courtney, G.W.; Skevington, J.H.; Meier, R.; Yeates, D.K.; Blagoderov, V.; Wiegmann, B.M. The Phylogenetic Relationships among Infraorders and Superfamilies of Diptera Based on Morphological Evidence. Syst. Entomol. 2013, 38, 164–179. [Google Scholar] [CrossRef]
- Schneeberg, K.; Courtney, G.W.; Beutel, R.G. Adult Head Structures of Deuterophlebiidae (Insecta), a Highly Derived “Ancestral” Dipteran Lineage. Arthropod Struct. Dev. 2011, 40, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Sæther, O.A. Phylogeny of Culicomorpha (Diptera). Syst. Entomol. 2000, 25, 223–234. [Google Scholar] [CrossRef]
- Kapli, P.; Yang, Z.; Telford, M.J. Phylogenetic Tree Building in the Genomic Age. Nat. Rev. Genet. 2020, 21, 428–444. [Google Scholar] [CrossRef]
- Yang, Z.; Bruce, R. Molecular Phylogenetics: Principles and Practice. Nat. Rev. Genet. 2012, 13, 303–314. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect Mitochondrial Genomics: Implications for Evolution and Phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G.; et al. Phylogenomics Resolves the Timing and Pattern of Insect Evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef]
- Ge, X.; Peng, L.; Morse, J.C.; Wang, J.; Zang, H.; Yang, L.; Sun, C.; Wang, B. Phylogenomics Resolves a 100-Year-Old Debate Regarding the Evolutionary History of Caddisflies (Insecta: Trichoptera). Mol. Phylogenet. Evol. 2024, 201, 108196. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Ding, Y.; Tihelka, E.; Cai, C.; Hu, F.; Liu, M.; Zhang, F. Phylogenomics of Elongate-Bodied Springtails Reveals Independent Transitions from Aboveground to Belowground Habitats in Deep Time. Syst. Biol. 2022, 71, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Tihelka, E.; Yu, D.; Chen, W.-J.; Bu, Y.; Cai, C.; Engel, M.S.; Luan, Y.-X.; Zhang, F. Revisiting the Four Hexapoda Classes: Protura as the Sister Group to All Other Hexapods. Proc. Natl. Acad. Sci. USA 2024, 121, e2408775121. [Google Scholar] [CrossRef] [PubMed]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Boore, J.L. The Use of Genome-Level Characters for Phylogenetic Reconstruction. Trends Ecol. Evol. 2006, 21, 439–446. [Google Scholar] [CrossRef]
- Jeffrey, L. Boore Animal Mitochondrial Genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Song, F.; Li, H.; Jiang, P.; Zhou, X.; Liu, J.; Sun, C.; Vogler, A.P.; Cai, W. Capturing the Phylogeny of Holometabola with Mitochondrial Genome Data and Bayesian Site-Heterogeneous Mixture Models. Genome Biol. Evol. 2016, 8, 1411–1426. [Google Scholar] [CrossRef]
- Talavera, G.; Vila, R. What Is the Phylogenetic Signal Limit from Mitogenomes? The Reconciliation between Mitochondrial and Nuclear Data in the Insecta Class Phylogeny. BMC Evol. Biol. 2011, 11, 315. [Google Scholar] [CrossRef]
- Jeffroy, O.; Brinkmann, H.; Delsuc, F.; Philippe, H. Phylogenomics: The Beginning of Incongruence? Trends Genet. 2006, 22, 225–231. [Google Scholar] [CrossRef]
- Bergsten, J. A Review of Long-Branch Attraction. Cladistics 2005, 21, 163–193. [Google Scholar] [CrossRef]
- Feng, S.; Stiller, J.; Deng, Y.; Armstrong, J.; Fang, Q.; Reeve, A.H.; Xie, D.; Chen, G.; Guo, C.; Faircloth, B.C.; et al. Dense Sampling of Bird Diversity Increases Power of Comparative Genomics. Nature 2020, 587, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Stiller, J.; Feng, S.; Chowdhury, A.-A.; Rivas-González, I.; Duchêne, D.A.; Fang, Q.; Deng, Y.; Kozlov, A.; Stamatakis, A.; Claramunt, S.; et al. Complexity of Avian Evolution Revealed by Family-Level Genomes. Nature 2024, 629, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Uribe, J.E.; González, V.L.; Irisarri, I.; Kano, Y.; Herbert, D.G.; Strong, E.E.; Harasewych, M.G. A Phylogenomic Backbone for Gastropod Molluscs. Syst. Biol. 2022, 71, 1271–1280. [Google Scholar] [CrossRef]
- Cox, C.J.; Li, B.; Foster, P.G.; Embley, T.M.; Civáň, P. Conflicting Phylogenies for Early Land Plants Are Caused by Composition Biases among Synonymous Substitutions. Syst. Biol. 2014, 63, 272–279. [Google Scholar] [CrossRef]
- Shin, S.; Clarke, D.J.; Lemmon, A.R.; Moriarty Lemmon, E.; Aitken, A.L.; Haddad, S.; Farrell, B.D.; Marvaldi, A.E.; Oberprieler, R.G.; McKenna, D.D. Phylogenomic Data Yield New and Robust Insights into the Phylogeny and Evolution of Weevils. Mol. Biol. Evol. 2018, 35, 823–836. [Google Scholar] [CrossRef]
- Philippe, H.; Brinkmann, H.; Lavrov, D.V.; Littlewood, D.T.J.; Manuel, M.; Wörheide, G.; Baurain, D. Resolving Difficult Phylogenetic Questions: Why More Sequences Are Not Enough. PLoS Biol. 2011, 9, e1000602. [Google Scholar] [CrossRef]
- Tegenfeldt, F.; Kuznetsov, D.; Manni, M.; Berkeley, M.; Zdobnov, E.M.; Kriventseva, E.V. OrthoDB and BUSCO Update: Annotation of Orthologs with Wider Sampling of Genomes. Nucleic Acids Res. 2025, 53, D516–D522. [Google Scholar] [CrossRef]
- Yeates, D.K.; Wiegmann, B.M. CONGRUENCE AND CONTROVERSY: Toward a Higher-Level Phylogeny of Diptera. Annu. Rev. Entomol. 1999, 44, 397–428. [Google Scholar] [CrossRef] [PubMed]
- Blount, Z.D.; Lenski, R.E.; Losos, J.B. Contingency and Determinism in Evolution: Replaying Life’s Tape. Science 2018, 362, eaam5979. [Google Scholar] [CrossRef]
- Balboa, N. Adaptation and Natural Selection. In Encyclopedia of Evolutionary Psychological Science; Shackelford, T.K., Weekes-Shackelford, V.A., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 58–59. ISBN 978-3-319-19650-3. [Google Scholar]
- Gould, S.J. Wonderful Life: The Burgess Shale and the Nature of History; W.W. Norton & Company: New York, NY, USA, 1989. [Google Scholar]
- Sharon, B.; Emerson, G. A Macroevolutionary Study of Historical Contingency in the Fanged Frogs of Southeast Asia. Biol. J. Linn. Soc. 2001, 73, 139–151. [Google Scholar] [CrossRef]
- Chomicki, G.; Burin, G.; Busta, L.; Gozdzik, J.; Jetter, R.; Mortimer, B.; Bauer, U. Convergence in Carnivorous Pitcher Plants Reveals a Mechanism for Composite Trait Evolution. Science 2024, 383, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Stankowski, S.; Zagrodzka, Z.B.; Garlovsky, M.D.; Pal, A.; Shipilina, D.; Castillo, D.G.; Lifchitz, H.; Moan, A.L.; Leder, E.; Reeve, J.; et al. The Genetic Basis of a Recent Transition to Live-Bearing in Marine Snails. Science 2024, 383, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Varney, R.M.; Speiser, D.I.; Cannon, J.T.; Aguilar, M.A.; Eernisse, D.J.; Oakley, T.H. A Morphological Basis for Path-Dependent Evolution of Visual Systems. Science 2024, 383, 983–987. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; de Pamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A Fast and Versatile Toolkit for Accurate de Novo Assembly of Organelle Genomes. Genome biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Donath, A.; Jühling, F.; Al-Arab, M.; Bernhart, S.H.; Reinhardt, F.; Stadler, P.F.; Middendorf, M.; Bernt, M. Improved Annotation of Protein-Coding Genes Boundaries in Metazoan Mitochondrial Genomes. Nucleic Acids Res. 2019, 47, 10543–10552. [Google Scholar] [CrossRef]
- Allio, R.; Schomaker-Bastos, A.; Romiguier, J.; Prosdocimi, F.; Nabholz, B.; Delsuc, F. MitoFinder: Efficient Automated Large-Scale Extraction of Mitogenomic Data in Target Enrichment Phylogenomics. Mol. Ecol. Resour. 2020, 20, 892–905. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Lin, X.-L.; Liu, Z.; Yan, L.-P.; Duan, X.; Bu, W.-J.; Wang, X.-H.; Zheng, C.-G. Mitogenomes Provide New Insights of Evolutionary History of Boreheptagyiini and Diamesini (Diptera: Chironomidae: Diamesinae). Ecol. Evol. 2022, 12, e8957. [Google Scholar] [CrossRef]
- Ren, L.; Guo, Q.; Yan, W.; Guo, Y.; Ding, Y. The Complete Mitochondria Genome of Calliphora vomitoria (Diptera: Calliphoridae). Mitochondrial DNA Part B 2016, 1, 378–379. [Google Scholar] [CrossRef]
- Qi, Y.; Xu, J.; Tian, X.; Bai, Y.; Gu, X. The Complete Mitochondrial Genome of Hermetia illucens (Diptera: Stratiomyidae). Mitochondrial DNA Part B 2017, 2, 189–190. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, Z.-Q.; Kuermanali, N.; Li, Z.; Chen, S.-J.; Wang, Y.-Z.; Tao, H.; Chen, C.-F. The Complete Mitochondrial Genome of the Parasitic Sheep Ked Melophagus ovinus (Diptera: Hippoboscidae). Mitochondrial DNA Part B 2017, 2, 432–434. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tan, L.; Guan, X.; Zhang, L.; Zhu, F.; Lei, C. The Complete Mitochondrial Genome of the Flea Ceratophyllus wui (Siphonaptera: Ceratophyllidae). Mitochondrial DNA Part B 2018, 3, 401–402. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Han, X.; Liu, Q.; Hou, X. The Mitochondrial Genome of Forcipomyia makanensis (Insecta: Diptera: Ceratopogonidae). Mitochondrial DNA Part B 2019, 4, 344–345. [Google Scholar] [CrossRef]
- Tan, L.; Yao, X.; Liu, J.; Lei, C.; Huang, Q.; Hu, B. The Complete Mitochondrial Genome of the Flea Hystrichopsylla weida qinlingensis (Siphonaptera: Hystrichopsylla). Mitochondrial DNA Part B 2023, 8, 501–503. [Google Scholar] [CrossRef]
- Cameron, S.L.; Lambkin, C.L.; Barker, S.C.; Whiting, M.F. A Mitochondrial Genome Phylogeny of Diptera: Whole Genome Sequence Data Accurately Resolve Relationships over Broad Timescales with High Precision. Syst. Entomol. 2007, 32, 40–59. [Google Scholar] [CrossRef]
- Lin, X.-L.; Zhao, Y.-M.; Yan, L.-P.; Liu, W.-B.; Bu, W.-J.; Wang, X.-H.; Zheng, C.-G. Mitogenomes Provide New Insights into the Evolutionary History of Prodiamesinae (Diptera: Chironomidae). Zool. Scr. 2022, 51, 119–132. [Google Scholar] [CrossRef]
- Milián-García, Y.; Hempel, C.A.; Janke, L.A.A.; Young, R.G.; Furukawa-Stoffer, T.; Ambagala, A.; Steinke, D.; Hanner, R.H. Mitochondrial Genome Sequencing, Mapping, and Assembly Benchmarking for Culicoides Species (Diptera: Ceratopogonidae). BMC Genom. 2022, 23, 584. [Google Scholar] [CrossRef]
- Li Xu-Dong; Chen Bin Sequencing and analysis of the complete mitochondrial genome of Armigeres subalbatus (Diptera: Culicidae). Acta Entomol. Sin. 2018, 61, 114–121. [CrossRef]
- Li, X.; Li, W.; Ding, S.; Cameron, S.L.; Mao, M.; Shi, L.; Yang, D. Mitochondrial Genomes Provide Insights into the Phylogeny of Lauxanioidea (Diptera: Cyclorrhapha). Int. J. Mol. Sci. 2017, 18, 773. [Google Scholar] [CrossRef]
- Li, H.-N.; Pei, W.-Y.; Wang, M.-F.; Chen, B.-Q.; Peng, H.-L.; Cao, R.-J.; Zhao, M.-T.; Yang, J.; Zhang, X.-C.; Zhang, D. Mitochondrial Genomes Provide New Insights into the Phylogeny and Evolution of Anthomyiidae (Insecta: Diptera). Arthropod Syst. Phylogeny 2023, 81, 1051–1062. [Google Scholar] [CrossRef]
- Li, S.-Y.; Chen, M.-H.; Sun, L.; Wang, R.-H.; Li, C.-H.; Gresens, S.; Li, Z.; Lin, X.-L. New Mitogenomes from the Genus Cricotopus (Diptera: Chironomidae, Orthocladiinae): Characterization and Phylogenetic Implications. Arch. Insect Biochem. Physiol. 2024, 115, e22067. [Google Scholar] [CrossRef] [PubMed]
- Beliavskaia, A.; Tan, K.-K.; Sinha, A.; Husin, N.A.; Lim, F.S.; Loong, S.K.; Bell-Sakyi, L.; Carlow, C.K.S.; AbuBakar, S.; Darby, A.C.; et al. Metagenomics of Culture Isolates and Insect Tissue Illuminate the Evolution of Wolbachia, Rickettsia and Bartonella Symbionts in Ctenocephalides spp. Fleas. Microb. Genom. 2023, 9, mgen001045. [Google Scholar] [CrossRef]
- Wong, D.; Norman, H.; Creedy, T.J.; Jordaens, K.; Moran, K.M.; Young, A.; Mengual, X.; Skevington, J.H.; Vogler, A.P. The Phylogeny and Evolutionary Ecology of Hoverflies (Diptera: Syrphidae) Inferred from Mitochondrial Genomes. Mol. Phylogenet. Evol. 2023, 184, 107759. [Google Scholar] [CrossRef]
- Trinca, V.; Uliana, J.V.C.; Ribeiro, G.K.S.; Torres, T.T.; Monesi, N. Characterization of the Mitochondrial Genomes of Bradysia hygida, Phytosciara flavipes and Trichosia splendens (Diptera: Sciaridae) and Novel Insights on the Control Region of Sciarid Mitogenomes. Insect Mol. Biol. 2022, 31, 482–496. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Huang, J.; Wu, H. Mitogenomes Provide Insights into the Phylogeny of Mycetophilidae (Diptera: Sciaroidea). Gene 2021, 783, 145564. [Google Scholar] [CrossRef]
- Li, X.; Ding, S.; Li, X.; Hou, P.; Tang, C.; Yang, D. The Complete Mitochondrial Genome Analysis of Eristalis tenax (Diptera, Syrphidae). Mitochondrial DNA Part B 2017, 2, 654–655. [Google Scholar] [CrossRef]
- Xiang, H.-T.; Wen, F.-Q.; Wang, G.-L. The Complete Nucleotide Sequence of the Mitochondrial Genome of Dorcadia ioffi (Siphonaptera: Vermipsyllidae). Mitochondrial DNA Part B 2017, 2, 389–390. [Google Scholar] [CrossRef]
- Li, N.; Hu, G.-L.; Hua, B.-Z. Complete Mitochondrial Genomes of Bittacus strigosus and Panorpa debilis and Genomic Comparisons of Mecoptera. Int. J. Biol. Macromol. 2019, 140, 672–681. [Google Scholar] [CrossRef]
- Zhang, X.; Kang, Z.; Ding, S.; Wang, Y.; Borkent, C.; Saigusa, T.; Yang, D. Mitochondrial Genomes Provide Insights into the Phylogeny of Culicomorpha (Insecta: Diptera). Int. J. Mol. Sci. 2019, 20, 747. [Google Scholar] [CrossRef]
- Foster, P.G.; de Oliveira, T.M.P.; Bergo, E.S.; Conn, J.E.; Sant’Ana, D.C.; Nagaki, S.S.; Nihei, S.; Lamas, C.E.; González, C.; Moreira, C.C.; et al. Phylogeny of Anophelinae Using Mitochondrial Protein Coding Genes. R. Soc. Open Sci. 2017, 4, 170758. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, V.; Gabrieli, P.; Brandini, S.; Capodiferro, M.R.; Javier, P.A.; Chen, X.-G.; Achilli, A.; Semino, O.; Gomulski, L.M.; Malacrida, A.R.; et al. The Worldwide Spread of the Tiger Mosquito as Revealed by Mitogenome Haplogroup Diversity. Front. Genet. 2016, 7, 208. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Pan, X.; Li, X.; Yu, Y.; Zhang, J.; Jiang, H.; Dou, L.; Zhu, S. The First Complete Mitochondrial Genome of Dacus longicornis (Diptera: Tephritidae) Using next-Generation Sequencing and Mitochondrial Genome Phylogeny of Dacini Tribe. Sci. Rep. 2016, 6, 36426. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, X.; Ding, S.; Wang, N.; Mao, M.; Wang, M.; Yang, D. The Complete Mitochondrial Genome of the Atylotus miser (Diptera: Tabanomorpha: Tabanidae), with Mitochondrial Genome Phylogeny of Lower Brachycera (Orthorrhapha). Gene 2016, 586, 184–196. [Google Scholar] [CrossRef]
- Zhang, D.; Yan, L.; Zhang, M.; Chu, H.; Cao, J.; Li, K.; Hu, D.; Pape, T. Phylogenetic Inference of Calyptrates, with the First Mitogenomes for Gasterophilinae (Diptera: Oestridae) and Paramacronychiinae (Diptera: Sarcophagidae). Int. J. Biol. Sci. 2016, 12, 489–504. [Google Scholar] [CrossRef][Green Version]
- Gao, D.-Z.; Liu, G.-H.; Song, H.-Q.; Wang, G.-L.; Wang, C.-R.; Zhu, X.-Q. The Complete Mitochondrial Genome of Gasterophilus intestinalis, the First Representative of the Family Gasterophilidae. Parasitol. Res. 2016, 115, 2573–2579. [Google Scholar] [CrossRef]
- Deviatiiarov, R.; Kikawada, T.; Gusev, O. The Complete Mitochondrial Genome of an Anhydrobiotic Midge Polypedilum vanderplanki (Chironomidae, Diptera). Mitochondrial DNA Part A 2017, 28, 218–220. [Google Scholar] [CrossRef]
- Briscoe, A.G.; Sivell, D.; Harbach, R.E. The Complete Mitochondrial Genome of Dixella aestivalis (Diptera: Nematocera: Dixidae). Mitochondrial DNA Part A 2017, 28, 83–84. [Google Scholar] [CrossRef]
- Kim, S.; Kim, H.; Shin, S.C. Complete Mitochondrial Genome of the Antarctic Midge Parochlus Steinenii (Diptera: Chironomidae). Mitochondrial DNA Part A 2016, 27, 3475–3476. [Google Scholar] [CrossRef]
- Zhang, N.-X.; Yu, G.; Li, T.-J.; He, Q.-Y.; Zhou, Y.; Si, F.-L.; Ren, S.; Chen, B. The Complete Mitochondrial Genome of Delia Antiqua and Its Implications in Dipteran Phylogenetics. PLoS ONE 2015, 10, e0139736. [Google Scholar] [CrossRef]
- Ye, F.; Liu, T.; King, S.D.; You, P. Mitochondrial Genomes of Two Phlebotomine Sand Flies, Phlebotomus chinensis and Phlebotomus papatasi (Diptera: Nematocera), the First Representatives from the Family Psychodidae. Parasites Vectors 2015, 8, 472. [Google Scholar] [CrossRef] [PubMed]
- Atray, I.; Bentur, J.S.; Nair, S. The Asian Rice Gall Midge (Orseolia oryzae) Mitogenome Has Evolved Novel Gene Boundaries and Tandem Repeats That Distinguish Its Biotypes. PLoS ONE 2015, 10, e0134625. [Google Scholar] [CrossRef][Green Version]
- Hardy, C.M.; Court, L.N.; Morgan, M.J. The Complete Mitochondrial DNA Genome of Aedes Vigilax (Diptera: Culicidae). Mitochondrial DNA Part A 2016, 27, 2552–2553. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Su, S.; Yang, D. The Complete Mitochondrial Genomes of Musca domestica and Scathophaga stercoraria (Diptera: Muscoidea: Muscidae and Scathophagidae). Mitochondrial DNA Part A 2016, 27, 1435–1436. [Google Scholar] [CrossRef]
- Zhong, M.; Wang, X.; Liu, Q.; Luo, B.; Wu, C.; Wen, J. The Complete Mitochondrial Genome of the Scuttle Fly, Megaselia scalaris (Diptera: Phoridae). Mitochondrial DNA Part A 2016, 27, 182–184. [Google Scholar] [CrossRef] [PubMed]
- Zhong, M.; Wang, X.; Liu, Q.; Luo, B.; Wu, C.; Wen, J. The Complete Mitochondrial Genome of the Flesh Fly, Boettcherisca peregrine (Diptera: Sarcophagidae). Mitochondrial DNA Part A 2016, 27, 106–108. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wei, S.; Shi, M.; Chen, X.-X. The Complete Mitochondrial Genome of Neopanorpa pulchra (Mecoptera: Panorpidae). Mitochondrial DNA Part A 2015, 26, 305–306. [Google Scholar] [CrossRef]
- Cameron, S.L. The Complete Mitochondrial Genome of a Flea, Jellisonia amadoi (Siphonaptera: Ceratophyllidae). Mitochondrial DNA Part A 2015, 26, 289–290. [Google Scholar] [CrossRef]
- Chen, S.; Oliveira, M.T.; Sanz, A.; Kemppainen, E.; Fukuoh, A.; Schlicht, B.; Kaguni, L.S.; Jacobs, H.T. A Cytoplasmic Suppressor of a Nuclear Mutation Affecting Mitochondrial Functions in Drosophila. Genetics 2012, 192, 483–493. [Google Scholar] [CrossRef]
- Nelson, L.A.; Cameron, S.L.; Yeates, D.K. The Complete Mitochondrial Genome of the Gall-Forming Fly, Fergusonina taylori Nelson and Yeates (Diptera: Fergusoninidae). Mitochondrial DNA Part A 2011, 22, 197–199. [Google Scholar] [CrossRef]
- Beckenbach, A.T. Mitochondrial Genome Sequences of Nematocera (Lower Diptera): Evidence of Rearrangement Following a Complete Genome Duplication in a Winter Crane Fly. Genome Biol. Evol. 2012, 4, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Behura, S.K.; Lobo, N.F.; Haas, B.; de Bruyn, B.; Lovin, D.D.; Shumway, M.F.; Puiu, D.; Romero-Severson, J.; Nene, V.; Severson, D.W. Complete Sequences of Mitochondria Genomes of Aedes aegypti and Culex quinquefasciatus and Comparative Analysis of Mitochondrial DNA Fragments Inserted in the Nuclear Genomes. Insect Biochem. Mol. Biol. 2011, 41, 770–777. [Google Scholar] [CrossRef]
- Beckenbach, A.T. Mitochondrial Genome Sequences of Representatives of Three Families of Scorpionflies (Order Mecoptera) and Evolution in a Major Duplication of Coding Sequence. Genome 2011, 54, 368–376. [Google Scholar] [CrossRef]
- Wang, S.; Lei, Z.; Wang, H.; Dong, B.; Ren, B. The Complete Mitochondrial Genome of the Leafminer Liriomyza trifolii (Diptera: Agromyzidae). Mol. Biol. Rep. 2011, 38, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Beckenbach, A.T.; Joy, J.B. Evolution of the Mitochondrial Genomes of Gall Midges (Diptera: Cecidomyiidae): Rearrangement and Severe Truncation of tRNA Genes. Genome Biol. Evol. 2009, 1, 278–287. [Google Scholar] [CrossRef]
- Beard, C.B.; Hamm, D.M.; Collins, F.H. The Mitochondrial Genome of the Mosquito Anopheles Gambiae: DNA Sequence, Genome Organization, and Comparisons with Mitochondrial Sequences of Other Insects. Insect Mol. Biol. 1993, 2, 103–124. [Google Scholar] [CrossRef] [PubMed]
- Bingxin, G. Chromosome-Level Genome Assembly of Chironomus striatipennis Kieffer Provides Insights into Benthic Adaptation and Metamorphosis Mechanism. TechRxiv 2022. [Google Scholar]
- Guo, H.; Wang, G.; Zhang, S.T.; Huang, M. Development of SSR Primers for Simulium (Eusimulium) Angustipes (Diptera: Simuliidae) Based on RNA-Seq Dataset. Acta Entomol. Sin. 2018, 61, 815–824. [Google Scholar]
- Driscoll, T.P.; Verhoeve, V.I.; Gillespie, J.J.; Johnston, J.S.; Guillotte, M.L.; Rennoll-Bankert, K.E.; Rahman, M.S.; Hagen, D.; Elsik, C.G.; Macaluso, K.R.; et al. A Chromosome-Level Assembly of the Cat Flea Genome Uncovers Rampant Gene Duplication and Genome Size Plasticity. BMC Biol. 2020, 18, 70. [Google Scholar] [CrossRef]
- Morales-Hojas, R.; Hinsley, M.; Armean, I.M.; Silk, R.; Harrup, L.E.; Gonzalez-Uriarte, A.; Veronesi, E.; Campbell, L.; Nayduch, D.; Saski, C.; et al. The Genome of the Biting Midge Culicoides sonorensis and Gene Expression Analyses of Vector Competence for Bluetongue Virus. BMC Genom. 2018, 19, 624. [Google Scholar] [CrossRef]
- Wang, X.; Xiong, M.; Lei, C.; Zhu, F. The Developmental Transcriptome of the Synanthropic Fly Chrysomya megacephala and Insights into Olfactory Proteins. BMC Genom. 2015, 16, 20. [Google Scholar] [CrossRef] [PubMed]
- Pauli, T.; Burt, T.O.; Meusemann, K.; Bayless, K.; Donath, A.; Podsiadlowski, L.; Mayer, C.; Kozlov, A.; Vasilikopoulos, A.; Liu, S.; et al. New Data, Same Story: Phylogenomics Does Not Support Syrphoidea (Diptera: Syrphidae, Pipunculidae). Syst. Entomol. 2018, 43, 447–459. [Google Scholar] [CrossRef]
- Narayanan Kutty, S.; Meusemann, K.; Bayless, K.M.; Marinho, M.A.T.; Pont, A.C.; Zhou, X.; Misof, B.; Wiegmann, B.M.; Yeates, D.; Cerretti, P.; et al. Phylogenomic Analysis of Calyptratae: Resolving the Phylogenetic Relationships within a Major Radiation of Diptera. Cladistics 2019, 35, 605–622. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Shang, Y.; Yang, L.; Wang, S.; Wang, X.; Chen, S.; Bao, Z.; An, D.; Meng, F.; Cai, J.; et al. Chromosome-Level de Novo Genome Assembly of Sarcophaga peregrina Provides Insights into the Evolutionary Adaptation of Flesh Flies. Mol. Ecol. Resour. 2021, 21, 251–262. [Google Scholar] [CrossRef]
- Husnik, F.; Hypsa, V.; Darby, A. Insect-Symbiont Gene Expression in the Midgut Bacteriocytes of a Blood-Sucking Parasite. Genome Biol. Evol. 2020, 12, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Kingan, S.; Shoue, D.; Nguyen, O.; Froenicke, L.; Galvin, B.; Lambert, C.; Khan, R.; Maheshwari, C.; Weisz, D.; et al. Improved High Quality Sand Fly Assemblies Enabled by Ultra Low Input Long Read Sequencing. Sci. Data 2024, 11, 918. [Google Scholar] [CrossRef]
- Fu, Y.; Fang, X.; Xiao, Y.; Mao, B.; Xu, Z.; Shen, M.; Wang, X. Two Chromosome-Level Genomes of Smittia aterrima and Smittia pratorum (Diptera, Chironomidae). Sci. Data 2024, 11, 165. [Google Scholar] [CrossRef]
- Liu, P.; Yang, W.; Kong, L.; Zhao, S.; Xie, Z.; Zhao, Y.; Wu, Y.; Guo, Y.; Xie, Y.; Liu, T.; et al. A DBHS Family Member Regulates Male Determination in the Filariasis Vector Armigeres Subalbatus. Nat. Commun. 2023, 14, 2292. [Google Scholar] [CrossRef]
- Mahajan, S.; Bachtrog, D. Convergent Evolution of Y Chromosome Gene Content in Flies. Nat. Commun. 2017, 8, 785. [Google Scholar] [CrossRef]
- Lin, M.-D.; Chuang, C.-H.; Kao, C.-H.; Chen, S.-H.; Wang, S.-C.; Hsieh, P.-H.; Chen, G.-Y.; Mao, C.-C.; Li, J.-Y.; Jade Lu, M.-Y.; et al. Decoding the Genome of Bloodsucking Midge Forcipomyia taiwana (Diptera: Ceratopogonidae): Insights into Odorant Receptor Expansion. Insect Biochem. Mol. Biol. 2024, 168, 104115. [Google Scholar] [CrossRef]
- Cohen, C.M.; Cole, T.J.; Brewer, M.S. Pick Your Poison: Molecular Evolution of Venom Proteins in Asilidae (Insecta: Diptera). Toxins 2020, 12, 738. [Google Scholar] [CrossRef] [PubMed]
- Sontowski, R.; Poeschl, Y.; Okamura, Y.; Vogel, H.; Guyomar, C.; Cortesero, A.-M.; van Dam, N.M. A High-Quality Functional Genome Assembly of Delia radicum L. (Diptera: Anthomyiidae) Annotated from Egg to Adult. Mol. Ecol. Resour. 2022, 22, 1954–1971. [Google Scholar] [CrossRef] [PubMed]
- Urban, J.M.; Foulk, M.S.; Bliss, J.E.; Coleman, C.M.; Lu, N.; Mazloom, R.; Brown, S.J.; Spradling, A.C.; Gerbi, S.A. High Contiguity de Novo Genome Assembly and DNA Modification Analyses for the Fungus Fly, Sciara coprophila, Using Single-Molecule Sequencing. BMC Genom. 2021, 22, 643. [Google Scholar] [CrossRef]
- Zhao, C.; Escalante, L.N.; Chen, H.; Benatti, T.R.; Qu, J.; Chellapilla, S.; Waterhouse, R.M.; Wheeler, D.; Andersson, M.N.; Bao, R.; et al. A Massive Expansion of Effector Genes Underlies Gall-Formation in the Wheat Pest Mayetiola destructor. Curr. Biol. 2015, 25, 613–620. [Google Scholar] [CrossRef]
- Shen, X.; Jin, J.; Zhang, G.; Yan, B.; Yu, X.; Wu, H.; Yang, M.; Zhang, F. The Chromosome-Level Genome Assembly of Aphidoletes aphidimyza Rondani (Diptera: Cecidomyiidae). Sci. Data 2024, 11, 785. [Google Scholar] [CrossRef]
- Zhang, B.; Han, H.-B.; Xu, L.-B.; Li, Y.-R.; Song, M.-X.; Liu, A.-P. Transcriptomic Analysis of Diapause-Associated Genes in Exorista civilis Rondani (Diptera:Tachinidae). Arch. Insect Biochem. Physiol. 2021, 107, e21789. [Google Scholar] [CrossRef]
- Schmidt, H.; Hellmann, S.L.; Waldvogel, A.-M.; Feldmeyer, B.; Hankeln, T.; Pfenninger, M. A High-Quality Genome Assembly from Short and Long Reads for the Non-Biting Midge Chironomus riparius (Diptera). G3 2020, 10, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Mahar, J.E.; Shi, M.; Hall, R.N.; Strive, T.; Holmes, E.C. Comparative Analysis of RNA Virome Composition in Rabbits and Associated Ectoparasites. J. Virol. 2020, 94, e02119-19. [Google Scholar] [CrossRef]
- Narayanan Kutty, S.; Wong, W.H.; Meusemann, K.; Meier, R.; Cranston, P.S. A Phylogenomic Analysis of Culicomorpha (Diptera) Resolves the Relationships among the Eight Constituent Families. Syst. Entomol. 2018, 43, 434–446. [Google Scholar] [CrossRef]
- Jia, Z.; Hasi, S.; Vogl, C.; Burger, P.A. Genomic Insights into Evolution and Control of Wohlfahrtia magnifica, a Widely Distributed Myiasis-Causing Fly of Warm-Blooded Vertebrates. Mol. Ecol. Resour. 2022, 22, 2744–2757. [Google Scholar] [CrossRef]
- Amaral, D.T.; Johnson, C.H.; Viviani, V.R. RNA-Seq Analysis of the Blue Light-Emitting Orfelia fultoni (Diptera: Keroplatidae) Suggest Photoecological Adaptations at the Molecular Level. Comp. Biochem. Physiol. Part D: Genom. Proteom. 2021, 39, 100840. [Google Scholar] [CrossRef] [PubMed]
- Melotto, G.; Jones, M.W.; Bosley, K.; Flack, N.; Frank, L.E.; Jacobson, E.; Kipp, E.J.; Nelson, S.; Ramirez, M.; Walls, C.; et al. The Genome of the Soybean Gall Midge (Resseliella maxima). G3 2023, 13, jkad046. [Google Scholar] [CrossRef]
- Anderson, N.; Jaron, K.S.; Hodson, C.N.; Couger, M.B.; Ševčík, J.; Weinstein, B.; Pirro, S.; Ross, L.; Roy, S.W. Gene-Rich X Chromosomes Implicate Intragenomic Conflict in the Evolution of Bizarre Genetic Systems. Proc. Natl. Acad. Sci. USA 2022, 119, e2122580119. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Shaikhutdinov, N.; Kozlova, O.; Itoh, M.; Tagami, M.; Murata, M.; Nishiyori-Sueki, H.; Kojima-Ishiyama, M.; Noma, S.; Cherkasov, A.; et al. High Quality Genome Assembly of the Anhydrobiotic Midge Provides Insights on a Single Chromosome-Based Emergence of Extreme Desiccation Tolerance. NAR Genom. Bioinf. 2022, 4, lqac029. [Google Scholar] [CrossRef]
- Generalovic, T.N.; McCarthy, S.A.; Warren, I.A.; Wood, J.M.D.; Torrance, J.; Sims, Y.; Quail, M.; Howe, K.; Pipan, M.; Durbin, R.; et al. A High-Quality, Chromosome-Level Genome Assembly of the Black Soldier Fly (Hermetia illucens L.). G3 2021, 11, jkab085. [Google Scholar] [CrossRef]
- Sun, X.; Liu, W.; Li, R.; Zhao, C.; Pan, L.; Yan, C. A Chromosome Level Genome Assembly of Propsilocerus akamusi to Understand Its Response to Heavy Metal Exposure. Mol. Ecol. Resour. 2021, 21, 1996–2012. [Google Scholar] [CrossRef]
- Konganti, K.; Guerrero, F.D.; Schilkey, F.; Ngam, P.; Jacobi, J.L.; Umale, P.E.; Perez de Leon, A.A.; Threadgill, D.W. A Whole Genome Assembly of the Horn Fly, Haematobia irritans, and Prediction of Genes with Roles in Metabolism and Sex Determination. G3 2018, 8, 1675–1686. [Google Scholar] [CrossRef]
- Vicoso, B.; Bachtrog, D. Numerous Transitions of Sex Chromosomes in Diptera. PLoS Biol. 2015, 13, e1002078. [Google Scholar] [CrossRef] [PubMed]
- Hoskins, R.A.; Carlson, J.W.; Wan, K.H.; Park, S.; Mendez, I.; Galle, S.E.; Booth, B.W.; Pfeiffer, B.D.; George, R.A.; Svirskas, R.; et al. The Release 6 Reference Sequence of the Drosophila melanogaster Genome. Genome Res. 2015, 25, 445–458. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An Integrated and Scalable Desktop Platform for Streamlined Molecular Sequence Data Management and Evolutionary Phylogenetics Studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation Software for the Fast Assembly of Multi-Gene Datasets with Character Set and Codon Information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Ding, Y.; Zhu, C.D.; Zhou, X.; Orr, M.C.; Scheu, S.; Luan, Y.-X. Phylogenomics from Low-Coverage Whole-Genome Sequencing. Methods Ecol. Evol. 2019, 10, 507–517. [Google Scholar] [CrossRef]
- TransDecoder. Available online: https://github.com/TransDecoder/TransDecoder (accessed on 5 September 2024).
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Kück, P.; Longo, G.C. FASconCAT-G: Extensive Functions for Multiple Sequence Alignment Preparations Concerning Phylogenetic Studies. Front. Zool. 2014, 11, 81. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Crotty, S.M.; Minh, B.Q.; Bean, N.G.; Holland, B.R.; Tuke, J.; Jermiin, L.S.; Haeseler, A.V. GHOST: Recovering Historical Signal from Heterotachously Evolved Sequence Alignments. Syst. Biol. 2019, syz051. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; Von Haeseler, A. Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Calcott, B.; Kainer, D.; Mayer, C.; Stamatakis, A. Selecting Optimal Partitioning Schemes for Phylogenomic Datasets. BMC Evol. Biol. 2014, 14, 82. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Rabiee, M.; Sayyari, E.; Mirarab, S. ASTRAL-III: Polynomial Time Species Tree Reconstruction from Partially Resolved Gene Trees. BMC Bioinf. 2018, 19, 153. [Google Scholar] [CrossRef] [PubMed]
- Lartillot, N.; Lepage, T.; Blanquart, S. PhyloBayes 3: A Bayesian Software Package for Phylogenetic Reconstruction and Molecular Dating. Bioinformatics 2009, 25, 2286–2288. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent Updates to the Phylogenetic Tree Display and Annotation Tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Andrew Rambaut Figtree. Available online: https://tree.bio.ed.ac.uk/software/figtree/ (accessed on 6 September 2024).
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.Y. Ggtree: An r Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Sayyari, E.; Whitfield, J.B.; Mirarab, S. DiscoVista: Interpretable Visualizations of Gene Tree Discordance. Mol. Phylogenet. Evol. 2018, 122, 110–115. [Google Scholar] [CrossRef]
- Strimmer, K.; Von Haeseler, A. Likelihood-Mapping: A Simple Method to Visualize Phylogenetic Content of a Sequence Alignment. Proc. Natl. Acad. Sci. USA 1997, 94, 6815–6819. [Google Scholar] [CrossRef]



| Family | Species | Sampling Localities | mtDNA Accession | Transcriptomes Accession | Sample Sites |
|---|---|---|---|---|---|
| Blephariceridae | Blepharicera sp1.-QZ | Yaowang Mountain, Zhejiang, China | PQ463199 | SRR32672063 | 28°45′25.73″ N, 118°58′34.78″ E |
| Blephariceridae | Blepharicera sp2.-WYM | Wuyi Mountains, Fujian, China | PQ463200 | SRR32672062 | 27°36′15.86″ N, 117°47′03.92″ E |
| Blephariceridae | Blephariceridae sp1.-WYM | Wuyi Mountains, Fujian, China | PQ463201 | SRR32672061 | 27°36′05.30″ N, 117°43′56.06″ E |
| Blephariceridae | Philorus sp1.-QZ | Yaowang Mountain, Zhejiang, China | PQ463208 | SRR32672057 | 28°45′25.73″ N, 118°58′34.78″ E |
| Blephariceridae | Philorus sp2.-WYM | Wuyi Mountains, Fujian, China | PQ463209 | SRR32672056 | 27°36′05.30″ N, 117°43′56.06″ E |
| Deuterophlebiidae | Deuterophlebia acutirhina | Wuyi Mountains, Fujian, China | PQ463202 | SRR32672060 | 27°36′05.30″ N, 117°43′56.06″ E |
| Deuterophlebiidae | Deuterophlebia sp.-Dxp | Linzhi, Xizang, China | PQ463203 | N/A | 29°35′46.15″ N, 94°21′19.52″ E |
| Deuterophlebiidae | Deuterophlebia sp.-DY | Ailao Mountain, Yunnan, China | PQ463204 | N/A | 23°58′13.20″ N, 101°31′37.73″ E |
| Deuterophlebiidae | Deuterophlebia sp.-QZ | Yaowang Mountain, Zhejiang, China | PQ463205 | N/A | 28°45′25.73″ N, 118°58′34.78″ E |
| Deuterophlebiidae | Deuterophlebia wuyiensis | Wuyi Mountains, Fujian, China | PQ463206 | SRR32672059 | 27°44′55.52″ N, 117°40′40.77″ E |
| Nymphomyiidae | Nymphomyia sp.-WYM | Wuyi Mountains, Fujian, China | PQ463207 | SRR32672058 | 27°36′15.86″ N, 117°47′03.92″ E |
| Type 1 | Type 2 | Loci | Site | Description | Abbreviations |
|---|---|---|---|---|---|
| mtDNA | NT | 13 | 7360 | 13PCGs, codon 1 and 2 | 13PCG_NT12 |
| mtDNA | NT | 13 | 11,040 | 13PCGs, codon 1, 2 and 3 | 13PCG_NT123 |
| mtDNA | AA | 13 | 3555 | 13PCGs | 13PCG_AA |
| nDNA | AA | 2931 | 1,344,362 | Taxon occupancy 50% | AA_Matrix50 |
| nDNA | AA | 2648 | 1,167,399 | Taxon occupancy 60% | AA_Matrix60 |
| nDNA | AA | 2204 | 890,185 | Taxon occupancy 70% | AA_Matrix70 |
| nDNA | AA | 1533 | 546,636 | Taxon occupancy 80% | AA_Matrix80 |
| nDNA | AA | 469 | 133,932 | Taxon occupancy 90% | AA_Matrix90 |
| nDNA | NT | 2931 | 4,033,086 | Taxon occupancy 50% | NT_Matrix50 |
| nDNA | NT | 2648 | 3,502,197 | Taxon occupancy 60% | NT_Matrix60 |
| nDNA | NT | 2204 | 2,670,555 | Taxon occupancy 70% | NT_Matrix70 |
| nDNA | NT | 1533 | 1,639,908 | Taxon occupancy 80% | NT_Matrix80 |
| nDNA | NT | 469 | 401,796 | Taxon occupancy 90% | NT_Matrix90 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Ren, J.; Zheng, X.; Cai, L.; Guan, J.; Cai, T.; Xu, X.; Zhen, Y. Genomic Insights into Basal Diptera Phylogeny: The Non-Monophyletic Nature of Blephariceromorpha. Int. J. Mol. Sci. 2025, 26, 5714. https://doi.org/10.3390/ijms26125714
Yang Y, Ren J, Zheng X, Cai L, Guan J, Cai T, Xu X, Zhen Y. Genomic Insights into Basal Diptera Phylogeny: The Non-Monophyletic Nature of Blephariceromorpha. International Journal of Molecular Sciences. 2025; 26(12):5714. https://doi.org/10.3390/ijms26125714
Chicago/Turabian StyleYang, Yaoming, Jiayao Ren, Xuhongyi Zheng, Lingna Cai, Jiayin Guan, Tianlong Cai, Xiaodong Xu, and Ying Zhen. 2025. "Genomic Insights into Basal Diptera Phylogeny: The Non-Monophyletic Nature of Blephariceromorpha" International Journal of Molecular Sciences 26, no. 12: 5714. https://doi.org/10.3390/ijms26125714
APA StyleYang, Y., Ren, J., Zheng, X., Cai, L., Guan, J., Cai, T., Xu, X., & Zhen, Y. (2025). Genomic Insights into Basal Diptera Phylogeny: The Non-Monophyletic Nature of Blephariceromorpha. International Journal of Molecular Sciences, 26(12), 5714. https://doi.org/10.3390/ijms26125714





