Epigenetic Alterations in Glioblastoma Multiforme as Novel Therapeutic Targets: A Scoping Review
Abstract
1. Introduction
2. Methods
3. Results
4. Discussion
4.1. Glioblastoma Multiforme: An Overview
4.2. DNA Methylation Modulation
4.3. Histone Modification
4.4. Non-Coding RNA Targeting
4.5. Epigenome Editing
4.6. Combination Therapies
4.7. Immunomodulation via Epigenetics
4.8. Emerging Technologies
4.9. Future Directions and Emerging Therapeutic Approaches
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 5hmC | 5-hydroxymethylcytosine |
| 5-HT7R | Serotonin receptor |
| ARL13B | ADP-ribosylation factor-like protein 13B |
| BBB | Blood–brain barrier |
| BET | Bromodomain and extra-terminal tail |
| BETi | BET inhibitor |
| BRD | Bromodomain-containing protein |
| C5aR1 | Complement C5a receptor 1 |
| CAK | CDK-activating kinase |
| CAM | Cell adhesion molecule |
| CASCADES | Cancer stem cell-associated distal enhancer of SOX2 |
| CBX | Chromobox |
| CDK | Cyclin-dependent kinase |
| ceRNA | Competing endogenous RNA |
| Chi3l1 | Chitinase 3-like 1 |
| CIA | Cytosolic iron–sulfur (Fe-S) cluster assembly |
| CK | Creatine kinase |
| CKB | CK, brain-type |
| CNS | Central nervous system |
| COL VI | Collagen VI |
| CpG | Cytosine-phosphate-guanine |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| CTA | Cancer-testis antigen |
| CTCF | CCCTC-binding factor |
| CTD | C-terminal domain |
| CX43 | Connexin 43 |
| d3A | dCas9-DNMT3A catalytic domain fusion protein |
| DLL3 | Delta-like protein 3 |
| DNA-PKcs | DNA-dependent protein kinase catalytic subunit |
| DNMT | DNA methyltransferase |
| DNMTi | DNMT inhibitor |
| DPP-4 | Dipeptidyl peptidase-4 |
| DSB | Double-strand break |
| EGF | Epidermal growth factor |
| EGFR | Epidermal growth factor receptor |
| EMT | Epithelial-mesenchymal transition |
| EpiDoKOL | Epigenetic Domain-specific Knockout Library |
| Erβ | Estrogen receptor β |
| Exo | Biosystems-derived exosome |
| EZH1/2 | Enhancer of Zeste Homologue 1/2 |
| FA | Fanconi anemia |
| GBM | Glioblastoma multiforme, Glioblastoma |
| GCV | Ganciclovir |
| GLUT3 | Glucose transporter 3 |
| GPX4 | Glutathione peroxidase 4 |
| GSC | Glioma stem-like cell |
| H3K27me3 | Histone H3 lysine 27 trimethylation |
| H3K4me3 | Histone H3 lysine 4 trimethylation |
| HAT | Histone acetyltransferase |
| HDAC | Histone deacetylase |
| HDACi | HDAC inhibitor |
| HERV | Human endogenous retrovirus |
| HLA-I | Human leukocyte antigen class I |
| HR | Homologous recombination |
| Hras | Harvey Rat Sarcoma Viral Oncogene Homolog |
| HSV-TK | Herpes simplex virus-thymidine kinase |
| ICL | Interstrand cross-link |
| IDH | Isocitrate dehydrogenase |
| IMPDH2 | lnosine-5′-monophosphate dehydrogenase 2 |
| IκB | Inhibitory κB |
| lncRNA | Long non-coding RNA |
| LSD1 | Lysine-specific histone demethylase 1 |
| m6A | N6-methyladenosine |
| MAO A | Monoamine oxidase A |
| MAZ | Myc-associated zinc finger protein |
| MCM | Mini-chromosome maintenance protein |
| MDH2 | Malate dehydrogenase 2 |
| MDHDH | Malate dehydrogenase degradation helper |
| METTL3/14 | Methyltransferase-like 3/14 |
| MGMT | O6-methylguanine-DNA methyltransferase |
| miRNA, miR | MicroRNA |
| MRI | Magnetic resonance imaging |
| MRN | Mre11-Rad50-NBS1 |
| mRNA | Messenger RNA |
| MTAP | Methylthioadenosine phosphorylase |
| mTORC2 | Mechanistic target of rapamycin complex 2 |
| NA | Not applicable |
| NAMPT | Nicotinamide phosphoribosyl transferase |
| NAMPTi | NAMPT inhibitor |
| NAPRT | Nicotinic acid phosphoribosyl transferase |
| ncRNA | Non-coding RNA |
| NP | Nanoparticle |
| NY-ESO1 | New York esophageal squamous cell carcinoma 1 |
| OGT | O-GlcNAc transferase |
| PARP | Poly-ADP-ribose polymerase |
| PcG | Polycomb group |
| PCr | Phosphocreatine |
| PDGFR | Platelet-derived growth factor receptor |
| PDGFRA | Platelet-derived growth factor receptor A |
| PDIA5 | Protein disulfide isomerase A5 |
| PD-L1 | Programmed death-ligand 1 |
| PEI | Polyethylenimine |
| PHAX | Phosphorylated adaptor for RNA export |
| PKM2 | Pyruvate kinase M2 |
| PLD | Phospholipase D |
| PLEKHA4 | Pleckstrin homology domain containing A4 |
| PLL | Polylysine |
| Pol II | RNA polymerase II |
| PRC1/2 | Polycomb Repressive Complex 1/2 |
| PRISMA-ScR | PRISMA Extension for Scoping Reviews |
| PRMT5/6 | Protein arginine N-methyltransferase 5/6 |
| PTEN | Phosphatase and tensin homolog |
| PTGFRN | Prostaglandin F2 receptor inhibitor |
| RARβ | Retinoic acid receptor β |
| RBBP4 | Retinoblastoma binding protein 4 |
| ROS | Reactive oxygen species |
| Rova-T | Rovalpituzumab tesirine |
| rRNA | Ribosomal RNA |
| RSK4 | Ribosomal S6 kinase 4 |
| RTK | Receptor tyrosine kinase |
| SCD | Stearoyl-CoA desaturase |
| SE | Super-enhancer |
| sgRNA | Single guide RNA |
| SHH | Sonic Hedgehog |
| siRNA | Small interfering RNA |
| snoRNP | Small nucleolar ribonucleoprotein |
| SNX10 | Sorting nexin 10 |
| TAM | Tumor-associated macrophage |
| TE | Transposable element |
| TGF-β | Transforming growth factor β |
| TME | Tumor microenvironment |
| TMZ | Temozolomide |
| TRAIL | Tumor necrosis factor-related apoptosis-inducing ligand |
| TSA | Trichostatin A |
| U3 snoRNA | U3 small nucleolar RNA |
| USP6NL | Ubiquitin-specific peptidase 6 N-terminal-like |
| VEGF | Vascular endothelial growth factor |
| WHO | World Health Organization |
| YY1 | Yin Yang 1 |
Appendix A
| Main Alteration | Pathway Involved | Genes/Proteins Involved | References |
|---|---|---|---|
| DNA Methylation Modulation | Global DNA hypomethylation | ALKBH1 | [177] |
| FGFR3-TACC3 fusions | [178] | ||
| mTORC2 | [45,179] | ||
| NuRD complex | [63,180,181] | ||
| PLEKHA4 | [84] | ||
| Hypomethylation of oncogenes | PPM1D | [182] | |
| Hypermethylation of tumor suppressor genes | DNMT1 | [183,184] | |
| DNMT3A/B | [18,185,186,187] | ||
| MSH2, MSH6 | [18,188] | ||
| Promoter methylation modulation | 5-HT7R | [61] | |
| ANKDD1A | [189] | ||
| ARL13B, IMPDH2 | [53] | ||
| CXCL12, CXCR4 | [190] | ||
| EGFR | [4,60,61,62] | ||
| IDH, 1p/19q co-deletion, G-CIMP status | [20,21] | ||
| LCTL | [191] | ||
| MGMT | [32,50,192,193] | ||
| RARβ | [64] | ||
| REST | [194,195] | ||
| SNX10, PDGFRβ | [55] | ||
| ST6Gal1 | [196] | ||
| TAGLN2 | [197] | ||
| TET, 5mC, 5hmc | [65,66,198,199] | ||
| ZBTB18 | [200] | ||
| Histone Modification | Chromatin remodeling | BRD2, BRD3, BRD4, BRD8 | [104,105,201,202,203] |
| BRG1 | [204] | ||
| Caspase-8 | [107] | ||
| CBX, CBX2, CBX8 | [86,88,205] | ||
| CELF2 | [99] | ||
| Chi3l1 | [106] | ||
| CX43, SHH | [92] | ||
| EGFR, FoxO3a | [90,206,207,208,209] | ||
| HELLS | [210] | ||
| KLHDC8A | [115] | ||
| Lamellipodin, RICTOR | [93] | ||
| Mcl-1 | [211] | ||
| MLL5 | [212] | ||
| NF1 | [18,94] | ||
| OCT4, NANOG, SOX2, CARM1 | [31,33] | ||
| PTEN | [18,85,86] | ||
| PTGFR | [91,129] | ||
| RIT1 | [213] | ||
| SCD | [89] | ||
| SWI-SNF complex | [214] | ||
| TRIM24, TRIM37 | [97,98] | ||
| WNT | [81,83] | ||
| YY1-CDK9 complex | [96] | ||
| Histone methylation | CDK7, CDK9 | [95,215] | |
| EphrinA5 | [216] | ||
| EZH2 | [40,41,42,53,81,82,86,98,217,218,219,220,221,222] | ||
| KDM4B/C, KDM5A, KDM6A/B | [108,109,156,223,224,225] | ||
| KMT2A, WDR5 | [38,226] | ||
| PRMT2, PRMT5, PRMT6, PRMT8 | [110,111,112,113,227,228] | ||
| SLC17A7 | [229] | ||
| WDR82 | [135] | ||
| Histone acetylation | HAT1 | [69] | |
| HDAC1, HDAC2, HDAC6 | [19,70,71,72,73,230] | ||
| Histone acetylation score | [68] | ||
| SIRT1 | [231] | ||
| ncRNA Targeting | miRNA | let-7 family | [232] |
| miR-9 | [220] | ||
| miR-10b | [233] | ||
| miR-26a-5p, miR27a-3p, miR-181a/b, miR-200b/c, miR-498 | [218] | ||
| miR-29s | [186] | ||
| miR-101 | [42] | ||
| miR-124 | [195,234] | ||
| miR-129-2 | [235] | ||
| miR-146a | [121] | ||
| miR-194-3p | [123] | ||
| miR-211 | [236] | ||
| miR-219-1 | [122] | ||
| miR-340 | [237,238] | ||
| miR-486-5p | [239] | ||
| miR-490 | [41] | ||
| miR-524 | [240,241] | ||
| miR-3189 | [40,70] | ||
| Progesterone (P4) | [120] | ||
| lncRNA | CASCADES | [29] | |
| CCND2-AS1 | [242] | ||
| DARS1-AS1 | [128] | ||
| H19 | [243] | ||
| HOTAIR | [203] | ||
| INHEG | [127] | ||
| LINC00511 | [241] | ||
| LINC00945 | [114] | ||
| LINC02283 | [129] | ||
| MDHDH | [130] | ||
| SOX2OT | [244] | ||
| TCONS_00004099 | [43] | ||
| TP53TG1, ENSG000246263 | [245] | ||
| UhyperLncs, UhypoLncs, SNHG | [246] | ||
| XTP6 | [124] | ||
| m6A modifications | GPX4 m6A methylation | [125] | |
| RNA m6A methylation | [126] | ||
| ceRNA | MCM4 | [131] | |
| Epigenome Editing | CRISPR-based targeting | EGFR | [133,247] |
| MGMT | [52] | ||
| CRISPR library screening | ASH2L | [134] | |
| E2F6 | [248] | ||
| ERBIN | [27] | ||
| Immunomodulation | Immune checkpoints and immune evasion | B7-H3 | [249] |
| CTA | [149] | ||
| PD-1, PD-L1 | [155,156,157,250] | ||
| TME and cellular interactions | Integrin β1, DPP-4 | [4,154] | |
| PDIA5 | [157] | ||
| Peripheral immune cells | [152] | ||
| Tumor-associated macrophages | [153] | ||
| Epigenetic and viral elements | CLOCK | [251] | |
| HERV-K | [151] | ||
| Others | Metabolic profile | ASS1 | [252] |
| Autophagy | [11,12,13] | ||
| GABA | [253] | ||
| Gallic acid | [254] | ||
| Glutamine | [30,253] | ||
| GPD1 | [255] | ||
| TME and hypoxia | Gliomagenesis progression | [10,37] | |
| HIF-1α, PAX3/p53 axis | [256] | ||
| Tumor heterogeneity and multiomics approaches | Artificial intelligence algorithm | [36] | |
| Methylome | [6,7,8,9,22,23,24,35,257] | ||
| Multiomics | [258,259,260] | ||
| Tumor heterogeneity | [34,35,261] |
| Drug Class | Molecule/Drug | Target | References |
|---|---|---|---|
| BETi | Birabresib (MK-8628, OTX015) | BRD2, BRD3, BRD4 | [101,102,140] |
| Trotabresib (CC-90010) | BRD4 | [100] | |
| JQ1 | BRD4, HOTAIR | [101,102,158,262] | |
| DNMTi | Decitabine | DNMT1 | [148,263] |
| RG-108 | DNMT | [143,264] | |
| EZHi | DZNep | EZH2 | [143,144,264,265] |
| Tazemetostat (EPZ-6438) | EZH2 | [266,267] | |
| UNC1999 | EZH1, EZH2 | [268] | |
| HDACi | Belinostat (PXD-101) | Class I and II HDACs | [76,77,269] |
| Domatinostat | HDAC1, HDAC2, HDAC3, HDAC11 | [80] | |
| Givinostat | Class I and II HDACs | [46] | |
| LMK235 | HDAC4, HDAC5 | [79] | |
| Panobinostat (LBH589) | Class I, II, and IV HDACs | [144,145,265,269,270,271] | |
| Romidepsin | HDAC1, HDAC2 | [145,269] | |
| Trichostatin A (TSA) | Class I and II HDACs | [78,143,264] | |
| Vorinostat (SAHA) | HDAC1, HDAC2, HDAC3, HDAC6 | [138,139,269,272] | |
| Nanoparticles | NAMPT inhibitors | NAMPT | [39,273] |
| PEG-AuNPs@Hyp | PcG, IDH2 | [162] | |
| RNA interference | SOX2, OLIG2, SALL2, POU3F2 | [274] | |
| siRNA | Cyclophilin A | [275] | |
| Others | ABT-737 | Bcl-2, Bcl-xL | [272] |
| AC1Q3QWB (AQB) | HOTAIR, EZH2 | [141,276] | |
| Aurora kinase A inhibitors | AURKA | [262] | |
| AZD1480 | JAK | [146] | |
| Bevacizumab | VEGF-A | [87] | |
| BIX01294 | G9a, G9a-like protein | [28,143,264] | |
| Dasatinib | Src kinase | [277] | |
| DS-1001b | Mutant IDH1 | [172] | |
| Dual EGFR/BRD4 inhibitors | BRD4, EGFR | [278] | |
| Dual EZH2/HDAC inhibitor | EZH2, HDAC | [279] | |
| EGFR CAR-T cells | EGFR | [158] | |
| GSK-J4 | JMJD3 | [280] | |
| GSK-LSD1 | LSD1 | [141,142] | |
| H3-G34R antibody | H3.3 G34R mutations | [281] | |
| HDAC-MB | HDAC6, MAO A | [159] | |
| Hydrolyzed rutin | Unidentified | [160] | |
| KC7F2 | HIF-1α | [54] | |
| NVP-BEZ235 | PI3K, mTOR | [146] | |
| Olaparib | PARP | [138] | |
| PEI-PLL copolymer | HSV-TK, TRAIL | [161] | |
| Phospho-valproic acid (MDC-1112) | STAT3 | [282] | |
| PRMT5 inhibitors | PRMT5 | [283] | |
| PTC596 | BMI1 | [266,284] | |
| Resveratrol | AMPK, NF-κB, SIRT1 | [285] | |
| Rolipram | DPY30, PDE4B | [117] | |
| Rovalpituzumab tesirine (Rova-T) | DLL3 | [116] | |
| Ruxolitinib | JAK | [146] | |
| Sodium selenite | H3K9m2, HDAC | [286] | |
| SP2509 | KDM1A | [270] | |
| Neural stem cells | HSV-TK, S-TRAIL | [287] |
| Trial Name | Intervention | Number of Participants | Clinical Trial Identifier | Clinical Trial Phase | Current Status | References |
|---|---|---|---|---|---|---|
| Quantitative Magnetic Resonance Spectroscopic Imaging (MRSI) to Predict Early Response to Standard Radiation Therapy (RT)/Temozolomide (TMZ) ± Belinostat Therapy in Newly-Diagnosed Glioblastomas (GBM) | RT/TMZ ± Belinostat | 29 | NCT02137759 | Phase II | Active | [75] |
| A Phase IIa Trial With Dose Optimization of OTX015, a Small Molecule Inhibitor of the Bromodomain and Extra-terminal (BET) Proteins, in Recurrent GBM Patients | Birabresib (OTX015) | 12 | NCT02296476 | Phase II | Terminated | [103] |
| A Pilot Study to Evaluate the Feasibility of the Combined Use of Stereotactic Radiosurgery With Nivolumab and Concurrent Valproate in Patients With Recurrent Glioblastoma | Stereotactic Radiosurgery, Nivolumab, Valproate | 4 | NCT02648633 | Phase I | Terminated | [166] |
| An Open-label, Single-arm, Phase II Study to Evaluate Safety and Efficacy of Doxorubicin in Combination With Radiotherapy, Temozolomide and Valproic Acid in Patients With Glioblastoma Multiforme (GBM) and Diffuse Intrinsic Pontine Glioma (DIPG) | Doxorubicin, RT, TMZ, Valproic acid | 21 | NCT02758366 | Phase II | Terminated | [165] |
| International Cooperative Phase III Trial of the HIT-HGG Study Group for the Treatment of High-Grade Glioma, Diffuse Intrinsic Pontine Glioma, and Gliomatosis Cerebri in Children and Adolescents < 18 Years [HIT-HGG-2013] | Valproic acid, TMZ | 167 | NCT03243461 | Phase III | Active | [164] |
| A Phase 0 First-In-Human Study Using NU-0129: A Spherical Nucleic Acid (SNA) Gold Nanoparticle Targeting BCL2L12 in Recurrent Glioblastoma Multiforme or Gliosarcoma Patients | NU-0129 | 8 | NCT03020017 | Early Phase I | Completed | [173] |
| A Phase 1 Study of DS-1001b in Patients With IDH1 Mutated Gliomas | DS-1001b | 47 | NCT03030066 | Phase I | Active | [171] |
| A Phase 1b/2 Study of FT-2102 in Patients With Advanced Solid Tumors and Gliomas With an IDH1 Mutation | Olutasidenib (FT-2102) | 93 | NCT03684811 | Phase I/II | Completed | [170] |
| A Phase I Trial of Pembrolizumab and Vorinostat Combined With Temozolomide and Radiation Therapy for Newly Diagnosed Glioblastoma | Pembrolizumab, Vorinostat, TMZ, RT | 21 | NCT03426891 | Phase I | Completed | [167] |
| Establishment of a Signature of Circulating microRNA as a Tool to Aid Diagnosis of Primary Brain Tumors in Adults | microRNA analysis [Observational study] | 160 | NCT03630861 | NA | Completed | [174] |
| A Phase 1, Open-label Study to Assess the Pharmacokinetics, Pharmacodynamics and CNS Penetration of CC-90010 in Preoperative Subjects With Progressive or Recurrent Who Grade II Diffuse Astrocytoma, Grade III Anaplastic Astrocytoma and Recurrent Glioblastoma Scheduled for Resection | Trotabresib (CC-90010) | 20 | NCT04047303 | Phase I | Terminated | [168] |
| Phase I Open Label Ascending Dose Study to Assess the Feasibility and Safety of Intermittent Infusions of MTX110 Administered by Convection-Enhanced Delivery (CED) in Patients With Recurrent Glioblastoma (rGBM) (MAGIC-G1) | MTX110 | 36 | NCT05324501 | Phase I | Active | [163] |
| Improving Personalized Glioblastoma Care by Stem Cell Analysis, Omics (Including Immunomics) and Artificial Intelligence Approaches | Biomarker analysis [Observational study] | 120 [Estimated] | NCT05941234 | NA | Active | [175] |
| Monitoring of Patients With Diffuse Gliomas Using Circulating miRNAs | microRNA analysis [Observational study] | 60 [Estimated] | NCT06203496 | NA | Active | [176] |
References
- Verdugo, E.; Puerto, I.; Medina, M.A. An update on the molecular biology of glioblastoma, with clinical implications and progress in its treatment. Cancer Commun. 2022, 42, 1083–1111. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.C.; Ashley, D.M.; Lopez, G.Y.; Malinzak, M.; Friedman, H.S.; Khasraw, M. Management of glioblastoma: State of the art and future directions. CA Cancer J. Clin. 2020, 70, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; Bent, M.J.v.d.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Nayak, C.; Singh, S.K. Integrated Transcriptome Profiling Identifies Prognostic Hub Genes as Therapeutic Targets of Glioblastoma: Evidenced by Bioinformatics Analysis. ACS Omega 2022, 7, 22531–22550. [Google Scholar] [CrossRef]
- Wu, H.; Guo, C.; Wang, C.; Xu, J.; Zheng, S.; Duan, J.; Li, Y.; Bai, H.; Xu, Q.; Ning, F.; et al. Single-cell RNA sequencing reveals tumor heterogeneity, microenvironment, and drug-resistance mechanisms of recurrent glioblastoma. Cancer Sci. 2023, 114, 2609–2621. [Google Scholar] [CrossRef]
- Krabóth, Z.; Tompa, M.; Urbán, P.; Gálik, B.; Kajtár, B.; Gyenesei, A.; Kálmán, B. Glioblastoma epigenomics discloses a complex biology and potential therapeutic targets. Clin. Neurosci. 2024, 77, 27–37. [Google Scholar] [CrossRef]
- Kanwal, R.; Gupta, S. Epigenetic modifications in cancer. Clin. Genet. 2012, 81, 303–311. [Google Scholar] [CrossRef]
- Sadida, H.Q.; Abdulla, A.; Marzooqi, S.A.; Hashem, S.; Macha, M.A.; Akil, A.S.A.-S.; Bhat, A.A. Epigenetic modifications: Key players in cancer heterogeneity and drug resistance. Transl. Oncol. 2024, 39, 101821. [Google Scholar] [CrossRef]
- Lu, Y.; Chan, Y.T.; Tan, H.Y.; Li, S.; Wang, N.; Feng, Y. Epigenetic regulation in human cancer: The potential role of epi-drug in cancer therapy. Mol. Cancer 2020, 19, 79. [Google Scholar] [CrossRef]
- Qiu, Q.; Deng, H.; Song, P.; Liu, Y.; Zhang, M. Lactylation in Glioblastoma: A Novel Epigenetic Modifier Bridging Epigenetic Plasticity and Metabolic Reprogramming. Int. J. Mol. Sci. 2025, 26, 3368. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, J.; Yang, C. Autophagy in brain tumors: Molecular mechanisms, challenges, and therapeutic opportunities. J. Transl. Med. 2025, 23, 52. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.K.; Lee, D.H.; Lee, E.C.; Oh, J.S. Importance of Autophagy Regulation in Glioblastoma with Temozolomide Resistance. Cells 2024, 13, 1332. [Google Scholar] [CrossRef] [PubMed]
- Pizzimenti, C.; Fiorentino, V.; Franchina, M.; Martini, M.; Giuffre, G.; Lentini, M.; Silvestris, N.; Di Pietro, M.; Fadda, G.; Tuccari, G.; et al. Autophagic-Related Proteins in Brain Gliomas: Role, Mechanisms, and Targeting Agents. Cancers 2023, 15, 2622. [Google Scholar] [CrossRef]
- Smith, H.L.; Wadhwani, N.; Horbinski, C. Major Features of the 2021 WHO Classification of CNS Tumors. Neurotherapeutics 2022, 19, 1691–1704. [Google Scholar] [CrossRef]
- Osborn, A.G.; Louis, D.N.; Poussaint, T.Y.; Linscott, L.L.; Salzman, K.L. The 2021 World Health Organization Classification of Tumors of the Central Nervous System: What Neuroradiologists Need to Know. AJNR Am. J. Neuroradiol. 2022, 43, 928–937. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Unruh, D.; Zewde, M.; Buss, A.; Drumm, M.R.; Tran, A.N.; Scholtens, D.M.; Horbinski, C. Methylation and transcription patterns are distinct in IDH mutant gliomas compared to other IDH mutant cancers. Sci. Rep. 2019, 9, 8946. [Google Scholar] [CrossRef]
- Bedics, G.; Szőke, P.; Bátai, B.; Nagy, T.; Papp, G.; Kránitz, N.; Rajnai, H.; Reiniger, L.; Bödör, C.; Scheich, B. Novel, clinically relevant genomic patterns identified by comprehensive genomic profiling in ATRX-deficient IDH-wildtype adult high-grade gliomas. Sci. Rep. 2023, 13, 18436. [Google Scholar] [CrossRef]
- Garrett, M.C.; Albano, R.; Carnwath, T.; Elahi, L.; Behrmann, C.A.; Pemberton, M.; Woo, D.; O’Brien, E.; VanCauwenbergh, B.; Perentesis, J.; et al. HDAC1 and HDAC6 are essential for driving growth in IDH1 mutant glioma. Sci. Rep. 2023, 13, 12433. [Google Scholar] [CrossRef]
- Saaid, A.; Monticelli, M.; Ricci, A.A.; Orlando, G.; Botta, C.; Zeppa, P.; Bianconi, A.; Osella-Abate, S.; Bruno, F.; Pellerino, A.; et al. Prognostic Analysis of the IDH1 G105G (rs11554137) SNP in IDH-Wildtype Glioblastoma. Genes 2022, 13, 1439. [Google Scholar] [CrossRef]
- Zhang, L.; Fritah, S.; Nazarov, P.V.; Kaoma, T.; Van Dyck, E. Impact of IDH Mutations, the 1p/19q Co-Deletion and the G-CIMP Status on Alternative Splicing in Diffuse Gliomas. Int. J. Mol. Sci. 2023, 24, 9825. [Google Scholar] [CrossRef] [PubMed]
- Amirmahani, F.; Kumar, S.; Muthukrishnan, S.D. Epigenetic mechanisms of plasticity and resistance in glioblastoma: Therapeutic targets and implications. Front. Epigenet. Epigenom. 2025, 3, 1519449. [Google Scholar] [CrossRef]
- Shahani, A.; Slika, H.; Elbeltagy, A.; Lee, A.; Peters, C.; Dotson, T.; Raj, D.; Tyler, B. The epigenetic mechanisms involved in the treatment resistance of glioblastoma. Cancer Drug Resist. 2025, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Karbhari, N.; Campian, J.L. Therapeutic Targets in Glioblastoma: Molecular Pathways, Emerging Strategies, and Future Directions. Cells 2025, 14, 494. [Google Scholar] [CrossRef]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.; Horsley, T.; Weeks, L.; et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef]
- Uddin, M.S.; Mamun, A.A.; Alghamdi, B.S.; Tewari, D.; Jeandet, P.; Sarwar, M.S.; Ashraf, G.M. Epigenetics of glioblastoma multiforme: From molecular mechanisms to therapeutic approaches. Semin. Cancer Biol. 2022, 83, 100–120. [Google Scholar] [CrossRef]
- Zhao, J.; Cui, X.; Zhan, Q.; Zhang, K.; Su, D.; Yang, S.; Hong, B.; Wang, Q.; Ju, J.; Cheng, C.; et al. CRISPR-Cas9 library screening combined with an exosome-targeted delivery system addresses tumorigenesis/TMZ resistance in the mesenchymal subtype of glioblastoma. Theranostics 2024, 14, 2835–2855. [Google Scholar] [CrossRef]
- Ciechomska, I.A.; Marciniak, M.P.; Jackl, J.; Kaminska, B. Pre-treatment or Post-treatment of Human Glioma Cells with BIX01294, the Inhibitor of Histone Methyltransferase G9a, Sensitizes Cells to Temozolomide. Front. Pharmacol. 2018, 9, 1271. [Google Scholar] [CrossRef]
- Shahzad, U.; Nikolopoulos, M.; Li, C.; Johnston, M.; Wang, J.J.; Sabha, N.; Varn, F.S.; Riemenschneider, A.; Krumholtz, S.; Krishnamurthy, P.M.; et al. CASCADES, a novel SOX2 super-enhancer-associated long noncoding RNA, regulates cancer stem cell specification and differentiation in glioblastoma. Mol. Oncol. 2024, 19, 764–784. [Google Scholar] [CrossRef]
- Tanaka, K.; Sasayama, T.; Nagashima, H.; Irino, Y.; Takahashi, M.; Izumi, Y.; Uno, T.; Satoh, N.; Kitta, A.; Kyotani, K.; et al. Glioma cells require one-carbon metabolism to survive glutamine starvation. Acta Neuropathol. Commun. 2021, 9, 16. [Google Scholar] [CrossRef]
- Sesé, B.; Iñiguez-Muñoz, S.; Ensenyat-Mendez, M.; Llinàs-Arias, P.; Ramis, G.; Orozco, J.I.J.; de Mattos, S.F.; Villalonga, P.; Marzese, D.M. Glioblastoma Embryonic-like Stem Cells Exhibit Immune-Evasive Phenotype. Cancers 2022, 14, 2070. [Google Scholar] [CrossRef] [PubMed]
- Lyon, J.F.; Vasudevaraja, V.; Mirchia, K.; Walker, J.M.; Corona, R.J.; Chin, L.S.; Tran, I.; Snuderl, M.; Richardson, T.E.; Viapiano, M.S. Spatial progression and molecular heterogeneity of IDH-mutant glioblastoma determined by DNA methylation-based mapping. Acta Neuropathol. Commun. 2021, 9, 120. [Google Scholar] [CrossRef] [PubMed]
- Rios Á, F.L.; Tirapelli, D.; Cirino, M.L.A.; Rodrigues, A.R.; Ramos, E.S.; Carlotti, C.G., Jr. Expression of pluripotency-related genes in human glioblastoma. Neurooncol. Adv. 2022, 4, vdab163. [Google Scholar] [CrossRef]
- Mathur, R.; Wang, Q.; Schupp, P.G.; Nikolic, A.; Hilz, S.; Hong, C.; Grishanina, N.R.; Kwok, D.; Stevers, N.O.; Jin, Q.; et al. Glioblastoma evolution and heterogeneity from a 3D whole-tumor perspective. Cell 2024, 187, 446–463.e416. [Google Scholar] [CrossRef]
- Gempt, J.; Withake, F.; Aftahy, A.K.; Meyer, H.S.; Barz, M.; Delbridge, C.; Liesche-Starnecker, F.; Prokop, G.; Pfarr, N.; Schlegel, J.; et al. Methylation subgroup and molecular heterogeneity is a hallmark of glioblastoma: Implications for biopsy targeting, classification and therapy. ESMO Open 2022, 7, 100566. [Google Scholar] [CrossRef]
- McInerney, C.E.; Lynn, J.A.; Gilmore, A.R.; Flannery, T.; Prise, K.M. Using AI-Based Evolutionary Algorithms to Elucidate Adult Brain Tumor (Glioma) Etiology Associated with IDH1 for Therapeutic Target Identification. Curr. Issues Mol. Biol. 2022, 44, 2982–3000. [Google Scholar] [CrossRef]
- Pine, A.R.; Cirigliano, S.M.; Singhania, R.; Nicholson, J.; da Silva, B.; Leslie, C.S.; Fine, H.A. Microenvironment-Driven Dynamic Chromatin Changes in Glioblastoma Recapitulate Early Neural Development at Single-Cell Resolution. Cancer Res. 2023, 83, 1581–1595. [Google Scholar] [CrossRef]
- Mitchell, K.; Sprowls, S.A.; Arora, S.; Shakya, S.; Silver, D.J.; Goins, C.M.; Wallace, L.; Roversi, G.; Schafer, R.E.; Kay, K.; et al. WDR5 represents a therapeutically exploitable target for cancer stem cells in glioblastoma. Genes Dev. 2023, 37, 86–102. [Google Scholar] [CrossRef]
- Murray, M.A.; Noronha, K.J.; Wang, Y.; Friedman, A.P.; Paradkar, S.; Suh, H.W.; Sundaram, R.K.; Brenner, C.; Saltzman, W.M.; Bindra, R.S. Exploiting Metabolic Defects in Glioma with Nanoparticle-Encapsulated NAMPT Inhibitors. Mol. Cancer Ther. 2024, 23, 1176–1187. [Google Scholar] [CrossRef]
- Sharma, V.; Vinchure, O.S.; Yadav, G.; Sarkar, C.; Kulshreshtha, R. A novel interplay between PRC2 and miR-3189 regulates epithelial–mesenchymal transition (EMT) via modulating COL6A2 in glioblastoma. J. Cell. Physiol. 2024, 239, e31326. [Google Scholar] [CrossRef]
- Vinchure, O.S.; Sharma, V.; Tabasum, S.; Ghosh, S.; Singh, R.P.; Sarkar, C.; Kulshreshtha, R. Polycomb complex mediated epigenetic reprogramming alters TGF-β signaling via a novel EZH2/miR-490/TGIF2 axis thereby inducing migration and EMT potential in glioblastomas. Int. J. Cancer 2019, 145, 1254–1269. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.H.; Li, X.T.; Tao, Z.N.; Deng, Z.T.; Wu, Y.; Zhou, Y.X. Feedback Loop of EZH2/miR-101 Regulates Autophagy and Apoptosis by Targeting mTOR in Glioblastoma Cells. J. Biol. Regul. Homeost. Agents 2024, 38, 1291–1306. [Google Scholar] [CrossRef]
- Wang, Y.; Shan, A.; Zhou, Z.; Li, W.; Xie, L.; Du, B.; Lei, B. LncRNA TCONS_00004099-derived microRNA regulates oncogenesis through PTPRF in gliomas. Ann. Transl. Med. 2021, 9, 1023. [Google Scholar] [CrossRef]
- Klughammer, J.; Kiesel, B.; Roetzer, T.; Fortelny, N.; Nemc, A.; Nenning, K.H.; Furtner, J.; Sheffield, N.C.; Datlinger, P.; Peter, N.; et al. The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space. Nat. Med. 2018, 24, 1611–1624. [Google Scholar] [CrossRef]
- Harachi, M.; Masui, K.; Shimizu, E.; Murakami, K.; Onizuka, H.; Muragaki, Y.; Kawamata, T.; Nakayama, H.; Miyata, M.; Komori, T.; et al. DNA hypomethylator phenotype reprograms glutamatergic network in receptor tyrosine kinase gene-mutated glioblastoma. Acta Neuropathol. Commun. 2024, 12, 40. [Google Scholar] [CrossRef]
- Nakagawa-Saito, Y.; Mitobe, Y.; Togashi, K.; Suzuki, S.; Sugai, A.; Kitanaka, C.; Okada, M. Givinostat Inhibition of Sp1-dependent MGMT Expression Sensitizes Glioma Stem Cells to Temozolomide. Anticancer Res. 2023, 43, 1131–1138. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef]
- Mansouri, A.; Hachem, L.D.; Mansouri, S.; Nassiri, F.; Laperriere, N.J.; Xia, D.; Lindeman, N.I.; Wen, P.Y.; Chakravarti, A.; Mehta, M.P.; et al. MGMT promoter methylation status testing to guide therapy for glioblastoma: Refining the approach based on emerging evidence and current challenges. Neuro Oncol. 2019, 21, 167–178. [Google Scholar] [CrossRef]
- Gilbert, M.R.; Wang, M.; Aldape, K.D.; Stupp, R.; Hegi, M.E.; Jaeckle, K.A.; Armstrong, T.S.; Wefel, J.S.; Won, M.; Blumenthal, D.T.; et al. Dose-dense temozolomide for newly diagnosed glioblastoma: A randomized phase III clinical trial. J. Clin. Oncol. 2013, 31, 4085–4091. [Google Scholar] [CrossRef]
- Lavogina, D.; Laasfeld, T.; Vardja, M.; Lust, H.; Jaal, J. Viability fingerprint of glioblastoma cell lines: Roles of mitotic, proliferative, and epigenetic targets. Sci. Rep. 2021, 11, 20338. [Google Scholar] [CrossRef]
- Wick, W.; Meisner, C.; Hentschel, B.; Platten, M.; Schilling, A.; Wiestler, B.; Sabel, M.C.; Koeppen, S.; Ketter, R.; Weiler, M.; et al. Prognostic or predictive value of MGMT promoter methylation in gliomas depends on IDH1 mutation. Neurology 2013, 81, 1515–1522. [Google Scholar] [CrossRef] [PubMed]
- Zapanta Rinonos, S.; Li, T.; Pianka, S.T.; Prins, T.J.; Eldred, B.S.C.; Kevan, B.M.; Liau, L.M.; Nghiemphu, P.L.; Cloughesy, T.F.; Lai, A. dCas9/CRISPR-based methylation of O-6-methylguanine-DNA methyltransferase enhances chemosensitivity to temozolomide in malignant glioma. J. Neuro-Oncol. 2024, 166, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Shireman, J.M.; Atashi, F.; Lee, G.; Ali, E.S.; Saathoff, M.R.; Park, C.H.; Savchuk, S.; Baisiwala, S.; Miska, J.; Lesniak, M.S.; et al. De novo purine biosynthesis is a major driver of chemoresistance in glioblastoma. Brain 2021, 144, 1230–1246. [Google Scholar] [CrossRef] [PubMed]
- Abbaszade, Z.; Bagca, B.G.; Avci, C.B. Molecular biological investigation of temozolomide and KC7F2 combination in U87MG glioma cell line. Gene 2021, 776, 145445. [Google Scholar] [CrossRef]
- Gimple, R.C.; Zhang, G.; Wang, S.; Huang, T.; Lee, J.; Taori, S.; Lv, D.; Dixit, D.; Halbert, M.E.; Morton, A.R.; et al. Sorting nexin 10 sustains PDGF receptor signaling in glioblastoma stem cells via endosomal protein sorting. JCI Insight 2023, 8, e158077. [Google Scholar] [CrossRef]
- A Phase II Study of PDGFR Kinase Inhibitor in Biomarker-Enriched Recurrent Malignant Gliomas. 2010. Available online: https://clinicaltrials.gov/study/NCT01140568 (accessed on 22 December 2024).
- Kalpathy-Cramer, J.; Chandra, V.; Da, X.; Ou, Y.; Emblem, K.E.; Muzikansky, A.; Cai, X.; Douw, L.; Evans, J.G.; Dietrich, J.; et al. Phase II study of tivozanib, an oral VEGFR inhibitor, in patients with recurrent glioblastoma. J. Neurooncol. 2017, 131, 603–610. [Google Scholar] [CrossRef]
- Dresemann, G.; Weller, M.; Rosenthal, M.A.; Wedding, U.; Wagner, W.; Engel, E.; Heinrich, B.; Mayer-Steinacker, R.; Karup-Hansen, A.; Fluge, O.; et al. Imatinib in combination with hydroxyurea versus hydroxyurea alone as oral therapy in patients with progressive pretreated glioblastoma resistant to standard dose temozolomide. J. Neurooncol. 2010, 96, 393–402. [Google Scholar] [CrossRef]
- Raymond, E.; Brandes, A.A.; Dittrich, C.; Fumoleau, P.; Coudert, B.; Clement, P.M.; Frenay, M.; Rampling, R.; Stupp, R.; Kros, J.M.; et al. Phase II study of imatinib in patients with recurrent gliomas of various histologies: A European Organisation for Research and Treatment of Cancer Brain Tumor Group Study. J. Clin. Oncol. 2008, 26, 4659–4665. [Google Scholar] [CrossRef]
- Wang, Y.; Shu, H.; Liu, J.; Jin, X.; Wang, L.; Qu, Y.; Xia, M.; Peng, P.; Feng, Y.; Wei, M. EGF promotes PKM2 O-GlcNAcylation by stimulating O-GlcNAc transferase phosphorylation at Y976 and their subsequent association. J. Biol. Chem. 2022, 298, 102340. [Google Scholar] [CrossRef]
- Courant, F.; Maravat, M.; Chen, W.; Gosset, D.; Blot, L.; Hervouet-Coste, N.; Sarou-Kanian, V.; Morisset-Lopez, S.; Decoville, M. Expression of the Human Serotonin 5-HT(7) Receptor Rescues Phenotype Profile and Restores Dysregulated Biomarkers in a Drosophila melanogaster Glioma Model. Cells 2022, 11, 1281. [Google Scholar] [CrossRef]
- Su, I.C.; Su, Y.K.; Chuang, H.Y.; Yadav, V.K.; Setiawan, S.A.; Fong, I.H.; Yeh, C.T.; Huang, H.C.; Lin, C.M. Ubiquitin-Specific Protease 6 n-Terminal-like Protein (USP6NL) and the Epidermal Growth Factor Receptor (EGFR) Signaling Axis Regulates Ubiquitin-Mediated DNA Repair and Temozolomide-Resistance in Glioblastoma. Biomedicines 2022, 10, 1531. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Song, C.; Gu, J.; Li, C.; Zang, W.; Shi, L.; Chen, L.; Zhu, L.; Zhou, M.; Wang, T.; et al. RBBP4 regulates the expression of the Mre11-Rad50-NBS1 (MRN) complex and promotes DNA double-strand break repair to mediate glioblastoma chemoradiotherapy resistance. Cancer Lett. 2023, 557, 216078. [Google Scholar] [CrossRef] [PubMed]
- Toprak, C.; Atli, E.I.; Kalkan, R. Methylation of RAR-β is a New Clinical Biomarker for Treatment in Higher-grade Gliomas. Neurolog. Sci. Neurophys. 2023, 40, 152–159. [Google Scholar] [CrossRef]
- Zeng, C.; Song, X.; Zhang, Z.; Cai, Q.Y.; Cai, J.J.; Horbinski, C.; Hu, B.; Cheng, S.Y.; Zhang, W. Dissection of transcriptomic and epigenetic heterogeneity of grade 4 gliomas: Implications for prognosis. Acta Neuropathol. Commun. 2023, 11, 133. [Google Scholar] [CrossRef]
- Lopez-Bertoni, H.; Johnson, A.; Rui, Y.; Lal, B.; Sall, S.; Malloy, M.; Coulter, J.B.; Lugo-Fagundo, M.; Shudir, S.; Khela, H.; et al. Sox2 induces glioblastoma cell stemness and tumor propagation by repressing TET2 and deregulating 5hmC and 5mC DNA modifications. Signal Transduct. Target. Ther. 2022, 7, 37. [Google Scholar] [CrossRef]
- Wang, Z.; Zang, C.; Cui, K.; Schones, D.E.; Barski, A.; Peng, W.; Zhao, K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 2009, 138, 1019–1031. [Google Scholar] [CrossRef]
- Qin, J.J.; Fu, J.; Chen, X.Z. Comprehensive analysis of histone acetylation-related genes in glioblastoma and lower-grade gliomas: Insights into drug sensitivity, molecular subtypes, immune infiltration, and prognosis. J. Gene Med. 2024, 26, e3678. [Google Scholar] [CrossRef]
- Bargiela-Cuevas, S.; Marin, M.; Gabaldon-Ojeda, M.; Klett-Mingo, J.I.; Granado, P.; Sacristan, S.; Esteban-Lasso, A.; Casas, J.G.; Martin, M.E.; González, V.M.M.; et al. Histone Acetyl Transferase 1 Is Overexpressed in Poor Prognosis, High-grade Meningeal and Glial Brain Cancers: Immunohistochemical and Aptahistochemical Study. J. Histochem. Cytochem. 2024, 72, 585–599. [Google Scholar] [CrossRef]
- Kwak, S.; Park, S.H.; Kim, S.H.; Sung, G.J.; Song, J.H.; Jeong, J.H.; Kim, H.; Ha, C.H.; Kim, S.W.; Choi, K.C. miR-3189-targeted GLUT3 repression by HDAC2 knockdown inhibits glioblastoma tumorigenesis through regulating glucose metabolism and proliferation. J. Exp. Clin. Cancer Res. 2022, 41, 87. [Google Scholar] [CrossRef]
- Bahia, R.K.; Hao, X.G.; Hassam, R.; Cseh, O.; Bozek, D.A.; Luchman, H.A.; Weiss, S. Epigenetic and molecular coordination between HDAC2 and SMAD3-SKI regulates essential brain tumour stem cell characteristics. Nat. Commun. 2023, 14, 5051. [Google Scholar] [CrossRef]
- Cascio, C.L.; McNamara, J.B.; Melendez, E.L.; Lewis, E.M.; Dufault, M.E.; Sanai, N.; Plaisier, C.L.; Mehta, S. Nonredundant, isoform-specific roles of HDAC1 in glioma stem cells. JCI Insight 2021, 6, e149232. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Z.; Dong, Y.; Kong, L. Epigenetic targeting of SLC30A3 by HDAC1 is related to the malignant phenotype of glioblastoma. IUBMB Life 2021, 73, 784–799. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Zhao, S.; Yan, F.; Cheng, J.; Huang, L.; Chen, H.; Liu, Q.; Ji, X.; Yuan, Z. HDAC2 selectively regulates FOXO3a-mediated gene transcription during oxidative stress-induced neuronal cell death. J. Neurosci. 2015, 35, 1250–1259. [Google Scholar] [CrossRef]
- Quantitative Magnetic Resonance Spectroscopic Imaging (MRSI) to Predict Early Response to Standard Radiation Therapy (RT)/Temozolomide (TMZ) ± Belinostat Therapy in Newly-Diagnosed Glioblastomas (GBM). 2014. Available online: https://clinicaltrials.gov/study/NCT02137759 (accessed on 22 December 2024).
- Xu, K.; Ramesh, K.; Huang, V.; Gurbani, S.S.; Cordova, J.S.; Schreibmann, E.; Weinberg, B.D.; Sengupta, S.; Voloschin, A.D.; Holdhoff, M.; et al. Final Report on Clinical Outcomes and Tumor Recurrence Patterns of a Pilot Study Assessing Efficacy of Belinostat (PXD-101) with Chemoradiation for Newly Diagnosed Glioblastoma. Tomography 2022, 8, 688–700. [Google Scholar] [CrossRef]
- Chang, C.M.; Ramesh, K.K.; Huang, V.; Gurbani, S.; Kleinberg, L.R.; Weinberg, B.D.; Shim, H.; Shu, H.G. Mutant Isocitrate Dehydrogenase 1 Expression Enhances Response of Gliomas to the Histone Deacetylase Inhibitor Belinostat. Tomography 2023, 9, 942–954. [Google Scholar] [CrossRef]
- Güven, M.; Taşpınar, F.; Denizler-Ebiri, F.N.; Castresana, J.S.; Taşpınar, M. The antagonistic effects of temozolomide and trichostatin a combination on MGMT and DNA mismatch repair pathways in Glioblastoma. Med. Oncol. 2023, 40, 223. [Google Scholar] [CrossRef]
- Chang, H.H.; Chang, Y.Y.; Tsai, B.C.; Chen, L.J.; Chang, A.C.; Chuang, J.Y.; Gean, P.W.; Hsueh, Y.S. A Selective Histone Deacetylase Inhibitor Induces Autophagy and Cell Death via SCNN1A Downregulation in Glioblastoma Cells. Cancers 2022, 14, 4537. [Google Scholar] [CrossRef]
- Nakagawa-Saito, Y.; Saitoh, S.; Mitobe, Y.; Sugai, A.; Togashi, K.; Suzuki, S.; Kitanaka, C.; Okada, M. HDAC Class I Inhibitor Domatinostat Preferentially Targets Glioma Stem Cells over Their Differentiated Progeny. Int. J. Mol. Sci. 2022, 23, 8084. [Google Scholar] [CrossRef]
- Yu, T.; Zhou, F.; Tian, W.; Xu, R.; Wang, B.; Zeng, A.; Zhou, Z.; Li, M.; Wang, Y.; Zhang, J. EZH2 interacts with HP1BP3 to epigenetically activate WNT7B that promotes temozolomide resistance in glioblastoma. Oncogene 2023, 42, 461–470. [Google Scholar] [CrossRef]
- Pang, F.N.; Zhang, L.; Li, M.Y.; Yi, X.C.; Wang, Y.; Yang, P.; Wen, B.; Jiang, J.Q.; Teng, Y.P.; Yang, X.Y.; et al. Ribosomal S6 protein kinase 4 promotes resistance to EZH2 inhibitors in glioblastoma. Cancer Gene Ther. 2023, 30, 1636–1648. [Google Scholar] [CrossRef]
- Tompa, M.; Kajtar, B.; Galik, B.; Gyenesei, A.; Kalman, B. DNA methylation and protein expression of Wnt pathway markers in progressive glioblastoma. Pathol. Res. Pract. 2021, 222, 153429. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Liu, Y.K.; Hong, S.M.; Yang, H.; Guan, B.; Ma, X.D. PLEKHA4 is Associated with Tumour Microenvironment, Stemness, Proliferation and Poor Prognosis of Gliomas. J. Integr. Neurosci. 2023, 22, 135. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Ge, X.; Ding, F.; He, L.; Song, K.; Shi, Z.; Ge, Z.; Zhang, J.; Ji, J.; Wang, X.; et al. Reactivating PTEN to impair glioma stem cells by inhibiting cytosolic iron-sulfur assembly. Sci. Transl. Med. 2024, 16, eadg5553. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, B.; Wang, Y.Z.; Liu, S.H.; Ma, C.K.; Piao, J.M.; Ma, S.Q.; Yu, D.H.; Wu, W. CBX2 enhances the progression and TMZ chemoresistance of glioma via EZH2-mediated epigenetic silencing of PTEN expression. Front. Pharmacol. 2024, 15, 1430891. [Google Scholar] [CrossRef]
- Huang, H.; Song, J.; Liu, Z.; Pan, L.; Xu, G. Autophagy activation promotes bevacizumab resistance in glioblastoma by suppressing Akt/mTOR signaling pathway. Oncol. Lett. 2018, 15, 1487–1494. [Google Scholar] [CrossRef]
- Li, J.; Xu, Z.; Zhou, L.; Hu, K. Expression profile and prognostic values of Chromobox family members in human glioblastoma. Aging (Albany NY) 2022, 14, 1910–1931. [Google Scholar] [CrossRef]
- Oatman, N.; Dasgupta, N.; Arora, P.; Choi, K.; Gawali, M.V.; Gupta, N.; Parameswaran, S.; Salomone, J.; Reisz, J.A.; Lawler, S.; et al. Mechanisms of stearoyl CoA desaturase inhibitor sensitivity and acquired resistance in cancer. Sci. Adv. 2021, 7, eabd7459. [Google Scholar] [CrossRef]
- Yang, C.; Li, Y.S.; Wang, Q.X.; Huang, K.; Wei, J.W.; Wang, Y.F.; Zhou, J.H.; Yi, K.K.; Zhang, K.L.; Zhou, B.C.; et al. EGFR/EGFRvIII remodels the cytoskeleton via epigenetic silencing of AJAP1 in glioma cells. Cancer Lett. 2017, 403, 119–127. [Google Scholar] [CrossRef]
- Mala, U.; Baral, T.K.; Somasundaram, K. Integrative analysis of cell adhesion molecules in glioblastoma identified prostaglandin F2 receptor inhibitor (PTGFRN) as an essential gene. BMC Cancer 2022, 22, 642. [Google Scholar] [CrossRef]
- Torrisi, F.; Alberghina, C.; Lo Furno, D.; Zappalà, A.; Valable, S.; Li Volti, G.; Tibullo, D.; Vicario, N.; Parenti, R. Connexin 43 and Sonic Hedgehog Pathway Interplay in Glioblastoma Cell Proliferation and Migration. Biology 2021, 10, 767. [Google Scholar] [CrossRef]
- Moritz, S.; Krause, M.; Schlatter, J.; Cordes, N.; Vehlow, A. Lamellipodin-RICTOR Signaling Mediates Glioblastoma Cell Invasion and Radiosensitivity Downstream of EGFR. Cancers 2021, 13, 5337. [Google Scholar] [CrossRef] [PubMed]
- Lucas, C.H.G.; Sloan, E.A.; Gupta, R.; Wu, J.; Pratt, D.; Vasudevan, H.N.; Ravindranathan, A.; Barreto, J.; Williams, E.A.; Shai, A.; et al. Multiplatform molecular analyses refine classification of gliomas arising in patients with neurofibromatosis type 1. Acta Neuropathol. 2022, 144, 747–765. [Google Scholar] [CrossRef] [PubMed]
- Bhutada, I.; Khambati, F.; Cheng, S.Y.; Tiek, D.M.; Duckett, D.; Lawrence, H.; Vogelbaum, M.A.; Mo, Q.; Chellappan, S.P.; Padmanabhan, J. CDK7 and CDK9 inhibition interferes with transcription, translation, and stemness, and induces cytotoxicity in GBM irrespective of temozolomide sensitivity. Neuro Oncol. 2024, 26, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.X.; Zhao, L.J.; Shen, J.Z.; Liang, Z.Y.; Wu, Q.L.; Yang, K.L.; Min, L.H.; Gimple, R.C.; Yang, Q.Y.; Bhargava, S.; et al. Transcription Elongation Machinery Is a Druggable Dependency and Potentiates Immunotherapy in Glioblastoma Stem Cells. Cancer Discov. 2022, 12, 502–521. [Google Scholar] [CrossRef]
- Xu, C.X.; Chen, G.Y.; Yu, B.; Sun, B.W.; Zhang, Y.W.; Zhang, M.D.; Yang, Y.; Xiao, Y.C.; Cheng, S.Y.; Li, Y.X.; et al. TRIM24 Cooperates with Ras Mutation to Drive Glioma Progression through snoRNA Recruitment of PHAX and DNA-PKcs. Adv. Sci. 2024, 11, e2400023. [Google Scholar] [CrossRef]
- Cai, L.; Liu, Y.; Li, Y.; Liu, B.; Cao, Y.; Yang, W.; Wang, B.; Sun, T. TRIM37 interacts with EZH2 to epigenetically suppress PTCH1 and regulate stemness in glioma stem cells through sonic hedgehog pathway. J. Neurooncol. 2024, 169, 269–279. [Google Scholar] [CrossRef]
- Turchi, L.; Sakakini, N.; Saviane, G.; Polo, B.; Saurty-Seerunghen, M.S.; Gabut, M.; Gouillou, C.A.; Guerlais, V.; Pasquier, C.; Vignais, M.L.; et al. CELF2 Sustains a Proliferating/OLIG2+ Glioblastoma Cell Phenotype via the Epigenetic Repression of SOX3. Cancers 2023, 15, 5038. [Google Scholar] [CrossRef]
- Tancredi, A.; Gusyatiner, O.; Bady, P.; Buri, M.C.; Lomazzi, R.; Chiesi, D.; Messerer, M.; Hegi, M.E. BET protein inhibition sensitizes glioblastoma cells to temozolomide treatment by attenuating MGMT expression. Cell Death Dis. 2022, 13, 1037. [Google Scholar] [CrossRef]
- Sears, T.K.; Woolard, K.D. R132H IDH1 sensitizes glioma to the antiproliferative and cytotoxic effects of BET inhibition. J. Cancer Res. Clin. Oncol. 2022, 148, 2275–2285. [Google Scholar] [CrossRef]
- Berenguer-Daizé, C.; Astorgues-Xerri, L.; Odore, E.; Cayol, M.; Cvitkovic, E.; Noel, K.; Bekradda, M.; MacKenzie, S.; Rezai, K.; Lokiec, F.; et al. OTX015 (MK-8628), a novel BET inhibitor, displays in vitro and in vivo antitumor effects alone and in combination with conventional therapies in glioblastoma models. Int. J. Cancer 2016, 139, 2047–2055. [Google Scholar] [CrossRef]
- A Phase IIa Trial with Dose Optimization of OTX015, a Small Molecule Inhibitor of the Bromodomain and Extra-Terminal (BET) Proteins, in Recurrent Glioblastoma Multiforme (GBM) Patients. 2014. Available online: https://clinicaltrials.gov/study/NCT02296476 (accessed on 22 December 2024).
- Sun, X.Q.; Klingbeil, O.; Lu, B.; Wu, C.Z.; Ballon, C.; Ouyang, M.; Wu, X.L.; Jin, Y.; Hwangbo, Y.; Huang, Y.H.; et al. BRD8 maintains glioblastoma by epigenetic reprogramming of the p53 network. Nature 2023, 613, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.S.; Qi, Q.H.; Jiang, X.Q.; Wu, J.; Li, Y.Y.; Liu, Z.D.; Cai, Y.; Ran, H.W.; Zhang, S.Y.; Zhang, C.; et al. Phosphocreatine Promotes Epigenetic Reprogramming to Facilitate Glioblastoma Growth Through Stabilizing BRD2. Cancer Discov. 2024, 14, 1547–1565. [Google Scholar] [CrossRef] [PubMed]
- Guetta-Terrier, C.; Karambizi, D.; Akosman, B.; Zepecki, J.P.; Chen, J.S.; Kamle, S.; Fajardo, J.E.; Fiser, A.; Singh, R.; Toms, S.A.; et al. Chi3l1 Is a Modulator of Glioma Stem Cell States and a Therapeutic Target in Glioblastoma. Cancer Res. 2023, 83, 1984–1999. [Google Scholar] [CrossRef]
- Contadini, C.; Ferri, A.; Di Martile, M.; Cirotti, C.; Del Bufalo, D.; De Nicola, F.; Pallocca, M.; Fanciulli, M.; Sacco, F.; Donninelli, G.; et al. Caspase-8 as a novel mediator linking Src kinase signaling to enhanced glioblastoma malignancy. Cell Death Differ. 2023, 30, 417–428. [Google Scholar] [CrossRef]
- Wang, Z.Z.; Cai, H.R.; Li, Z.K.; Sun, W.; Zhao, E.R.; Cui, H.J. Histone demethylase KDM4B accelerates the progression of glioblastoma via the epigenetic regulation of MYC stability. Clin. Epigenet. 2023, 15, 192. [Google Scholar] [CrossRef]
- Lee, D.H.; Kim, G.W.; Yoo, J.; Lee, S.W.; Jeon, Y.H.; Kim, S.Y.; Kang, H.G.; Kim, D.H.; Chun, K.H.; Choi, J.; et al. Histone demethylase KDM4C controls tumorigenesis of glioblastoma by epigenetically regulating p53 and c-Myc. Cell Death Dis. 2021, 12, 89. [Google Scholar] [CrossRef]
- Wang, J.; Xiao, Z.Y.; Li, P.; Wu, C.W.; Li, Y.; Wang, Q.; Chen, Y.M.; Zhou, H.L.; Li, Z.; Wang, Z.T.; et al. PRMT6-CDC20 facilitates glioblastoma progression via the degradation of CDKN1B. Oncogene 2023, 42, 1088–1100. [Google Scholar] [CrossRef]
- Huang, T.; Yang, Y.; Song, X.; Wan, X.; Wu, B.; Sastry, N.; Horbinski, C.M.; Zeng, C.; Tiek, D.; Goenka, A.; et al. PRMT6 methylation of RCC1 regulates mitosis, tumorigenicity, and radiation response of glioblastoma stem cells. Mol. Cell 2021, 81, 1276–1291.e1279. [Google Scholar] [CrossRef]
- Du, C.; Li, S.W.; Singh, S.X.; Roso, K.; Sun, M.A.; Pirozzi, C.J.; Yang, R.; Li, J.L.; He, Y. Epigenetic Regulation of Fanconi Anemia Genes Implicates PRMT5 Blockage as a Strategy for Tumor Chemosensitization. Mol. Cancer Res. 2021, 19, 2046–2056. [Google Scholar] [CrossRef]
- Sachamitr, P.; Ho, J.C.; Ciamponi, F.E.; Ba-Alawi, W.; Coutinho, F.J.; Guilhamon, P.; Kushida, M.M.; Cavalli, F.M.G.; Lee, L.; Rastegar, N.; et al. PRMT5 inhibition disrupts splicing and stemness in glioblastoma. Nat. Commun. 2021, 12, 979. [Google Scholar] [CrossRef]
- Yang, Z.H.; Zheng, Y.F.; Wu, H.Y.; Xie, H.; Zhao, J.J.; Chen, Z.G.; Li, L.X.; Yue, X.Y.; Zhao, B.; Bian, E.R. Integrative analysis of a novel super-enhancer-associated lncRNA prognostic signature and identifying LINC00945 in aggravating glioma progression. Hum. Genom. 2023, 17, 33. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Gimple, R.C.; Wu, X.; Prager, B.C.; Qiu, Z.; Wu, Q.; Daggubati, V.; Mariappan, A.; Gopalakrishnan, J.; Sarkisian, M.R.; et al. Superenhancer activation of KLHDC8A drives glioma ciliation and hedgehog signaling. J. Clin. Investig. 2023, 133, e163592. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, A.S.; Hong, D.S.; Hann, C.L.; Farago, A.F.; Beltran, H.; Waqar, S.N.; Hendifar, A.E.; Anthony, L.B.; Taylor, M.H.; Bryce, A.H.; et al. A phase I/II study of rovalpituzumab tesirine in delta-like 3-expressing advanced solid tumors. NPJ Precis. Oncol. 2021, 5, 74. [Google Scholar] [CrossRef]
- Dixit, D.; Prager, B.C.; Gimple, R.C.; Miller, T.E.; Wu, Q.L.; Yomtoubian, S.; Kidwell, R.L.; Lv, D.G.; Zhao, L.J.; Qiu, Z.X.; et al. Glioblastoma stem cells reprogram chromatin in vivo to generate selective therapeutic dependencies on DPY30 and phosphodiesterases. Sci. Transl. Med. 2022, 14, eabf3917. [Google Scholar] [CrossRef]
- Cescon, M.; Rampazzo, E.; Bresolin, S.; Da Ros, F.; Manfreda, L.; Cani, A.; Della Puppa, A.; Braghetta, P.; Bonaldo, P.; Persano, L. Collagen VI sustains cell stemness and chemotherapy resistance in glioblastoma. Cell. Mol. Life Sci. 2023, 80, 233. [Google Scholar] [CrossRef]
- Lin, H.; Yang, Y.; Hou, C.; Zheng, J.; Lv, G.; Mao, R.; Xu, P.; Chen, S.; Zhou, Y.; Wang, P.; et al. Identification of COL6A1 as the Key Gene Associated with Antivascular Endothelial Growth Factor Therapy in Glioblastoma Multiforme. Genet. Test. Mol. Biomark. 2021, 25, 334–345. [Google Scholar] [CrossRef]
- Velázquez-Vázquez, D.E.; Del Moral-Morales, A.; Cruz-Burgos, J.M.; Martinez-Martinez, E.; Rodriguez-Dorantes, M.; Camacho-Arroyo, I. Expression analysis of progesterone-regulated miRNAs in cells derived from human glioblastoma. Mol. Med. Rep. 2021, 23, 475. [Google Scholar] [CrossRef]
- Cui, T.T.; Bell, E.H.; McElroy, J.; Liu, K.; Sebastian, E.; Johnson, B.; Gulati, P.M.; Becker, A.P.; Gray, A.; Geurts, M.; et al. A Novel miR-146a-POU3F2/SMARCA5 Pathway Regulates Sternness and Therapeutic Response in Glioblastoma. Mol. Cancer Res. 2021, 19, 48–60. [Google Scholar] [CrossRef]
- Ghasemi, A.; Mohammadi, A.; Fallah, S. Epigenetic Modification of MicroRNA-219-1 and Its Association with Glioblastoma Multiforme. Biochemistry 2021, 86, 420–432. [Google Scholar] [CrossRef]
- Jacob, J.R.; Singh, R.; Okamoto, M.; Chakravarti, A.; Palanichamy, K. miRNA-194-3p represses NF-κB in gliomas to attenuate iPSC genes and proneural to mesenchymal transition. iScience 2024, 27, 108650. [Google Scholar] [CrossRef]
- Xiao, F.; Zhu, H.; Xiong, Y.P.; Guo, Y.; Zhang, Z.; Zeng, J.; Xiao, Y.; Liao, B.; Shang, X.S.; Zhao, S.Y.; et al. Positive feedback loop of c-myc/XTP6/NDH2/NF-κB to promote malignant progression in glioblastoma. J. Exp. Clin. Cancer Res. 2024, 43, 187. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.R.; Wang, Z.X.; Yang, Q.Q.; Liu, Y.W.; Gao, Y.S.; Chen, H.F.; Li, A.; Li, R.Q.; Wang, J.; Sun, G. Intracellular C5aR1 inhibits ferroptosis in glioblastoma through METTL3-dependent m6A methylation of GPX4. Cell Death Dis. 2024, 15, 729. [Google Scholar] [CrossRef]
- Tao, M.; Li, X.; He, L.; Rong, X.; Wang, H.; Pan, J.; Lu, Z.; Zhang, X.; Peng, Y. Decreased RNA m(6)A methylation enhances the process of the epithelial mesenchymal transition and vasculogenic mimicry in glioblastoma. Am. J. Cancer Res. 2022, 12, 893–906. [Google Scholar] [PubMed]
- Liu, L.; Liu, Z.; Liu, Q.; Wu, W.; Lin, P.; Liu, X.; Zhang, Y.; Wang, D.; Prager, B.C.; Gimple, R.C.; et al. LncRNA INHEG promotes glioma stem cell maintenance and tumorigenicity through regulating rRNA 2′-O-methylation. Nat. Commun. 2023, 14, 7526. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Wei, Y.; Zhang, Q.; Sun, M.; Wang, Y.; Hou, J.; Zhang, P.; Lv, X.; Su, D.; Jiang, Y.; et al. Multiomics analyses reveal DARS1-AS1/YBX1-controlled posttranscriptional circuits promoting glioblastoma tumorigenesis/radioresistance. Sci. Adv. 2023, 9, eadf3984. [Google Scholar] [CrossRef]
- Goenka, A.; Song, X.; Tiek, D.; Iglesia, R.P.; Lu, M.; Zeng, C.; Horbinski, C.; Zhang, W.; Hu, B.; Cheng, S.Y. Oncogenic long noncoding RNA LINC02283 enhances PDGF receptor A-mediated signaling and drives glioblastoma tumorigenesis. Neuro Oncol. 2023, 25, 1592–1604. [Google Scholar] [CrossRef]
- He, D.; Xin, T.; Pang, B.; Sun, J.; Liu, Z.H.; Qin, Z.; Ji, X.S.; Yang, F.; Wei, Y.B.; Wang, Z.X.; et al. A novel lncRNA MDHDH suppresses glioblastoma multiforme by acting as a scaffold for MDH2 and PSMA1 to regulate NAD+ metabolism and autophagy. J. Exp. Clin. Cancer Res. 2022, 41, 349. [Google Scholar] [CrossRef]
- Yang, S.; Yuan, Y.; Ren, W.; Wang, H.; Zhao, Z.; Zhao, H.; Zhao, Q.; Chen, X.; Jiang, X.; Zhang, L. MCM4 is a novel prognostic biomarker and promotes cancer cell growth in glioma. Front. Oncol. 2022, 12, 1004324. [Google Scholar] [CrossRef]
- Quinn, J.A.; Jiang, S.X.; Reardon, D.A.; Desjardins, A.; Vredenburgh, J.J.; Rich, J.N.; Gururangan, S.; Friedman, A.H.; Bigner, D.D.; Sampson, J.H.; et al. Phase I trial of temozolomide plus O6-benzylguanine 5-day regimen with recurrent malignant glioma. Neuro Oncol. 2009, 11, 556–561. [Google Scholar] [CrossRef]
- Vincent, C.A.; Nissen, I.; Dakhel, S.; Hörnblad, A.; Remeseiro, S. Epigenomic perturbation of novel EGFR enhancers reduces the proliferative and invasive capacity of glioblastoma and increases sensitivity to temozolomide. BMC Cancer 2023, 23, 945. [Google Scholar] [CrossRef]
- Ozyerli-Goknar, E.; Kala, E.Y.; Aksu, A.C.; Bulut, I.; Cingöz, A.; Nizamuddin, S.; Biniossek, M.; Seker-Polat, F.; Morova, T.; Aztekin, C.; et al. Epigenetic-focused CRISPR/Cas9 screen identifies (absent, small, or homeotic)2-like protein (ASH2L) as a regulator of glioblastoma cell survival. Cell Commun. Signal 2023, 21, 328. [Google Scholar] [CrossRef] [PubMed]
- Wadhwani, N.; Nayak, S.; Wang, Y.; Hashizume, R.; Jie, C.; Mania-Farnell, B.; James, C.D.; Xi, G.; Tomita, T. WDR82-Mediated H3K4me3 Is Associated with Tumor Proliferation and Therapeutic Efficacy in Pediatric High-Grade Gliomas. Cancers 2023, 15, 3429. [Google Scholar] [CrossRef] [PubMed]
- Galanis, E.; Jaeckle, K.A.; Maurer, M.J.; Reid, J.M.; Ames, M.M.; Hardwick, J.S.; Reilly, J.F.; Loboda, A.; Nebozhyn, M.; Fantin, V.R.; et al. Phase II trial of vorinostat in recurrent glioblastoma multiforme: A north central cancer treatment group study. J. Clin. Oncol. 2009, 27, 2052–2058. [Google Scholar] [CrossRef]
- A Cancer Research UK Phase I Trial of Olaparib (AZD2281), an Oral PARP Inhibitor, in Combination with Extended Low-Dose Oral Temozolomide in Patients with Relapsed Glioblastoma. 2011. Available online: https://clinicaltrials.gov/study/NCT01390571 (accessed on 22 December 2024).
- Rasmussen, R.D.; Gajjar, M.K.; Jensen, K.E.; Hamerlik, P. Enhanced efficacy of combined HDAC and PARP targeting in glioblastoma. Mol. Oncol. 2016, 10, 751–763. [Google Scholar] [CrossRef]
- Kang, D.W.; Hwang, W.C.; Noh, Y.N.; Kang, Y.; Jang, Y.; Kim, J.A.; Min, D.S. Phospholipase D1 is upregulated by vorinostat and confers resistance to vorinostat in glioblastoma. J. Cell. Physiol. 2021, 236, 549–560. [Google Scholar] [CrossRef]
- Liu, Y.J.; Wang, W.D.; Zhang, D.Y.; Sun, Y.J.; Li, F.Z.; Zheng, M.; Lovejoy, D.B.; Zou, Y.; Shi, B.Y. Brain co-delivery of first-line chemotherapy drug and epigenetic bromodomain inhibitor for multidimensional enhanced synergistic glioblastoma therapy. Exploration 2022, 2, 20210274. [Google Scholar] [CrossRef]
- Zhao, J.X.; Jin, W.L.; Yi, K.K.; Wang, Q.X.; Zhou, J.H.; Tan, Y.L.; Xu, C.; Xiao, M.L.; Hong, B.A.; Xu, F.F.; et al. Combination LSD1 and HOTAIR-EZH2 inhibition disrupts cell cycle processes and induces apoptosis in glioblastoma cells. Pharmacol. Res. 2021, 171, 105764. [Google Scholar] [CrossRef]
- Alabed, S.J.; Zihlif, M.; Taha, M. Discovery of new potent lysine specific histone demythelase-1 inhibitors (LSD-1) using structure based and ligand based molecular modelling and machine learning. RSC Adv. 2022, 12, 35873–35895. [Google Scholar] [CrossRef]
- Alexanian, A.R.; Brannon, A. Unique combinations of epigenetic modifiers synergistically impair the viability of the U87 glioblastoma cell line while exhibiting minor or moderate effects on normal stem cell growth. Med. Oncol. 2022, 39, 86. [Google Scholar] [CrossRef]
- De La Rosa, J.; Urdiciain, A.; Zelaya, M.V.; Zazpe, I.; Meléndez, B.; Rey, J.A.; Idoate, M.A.; Castresana, J.S. APR-246 combined with 3-deazaneplanocin A, panobinostat or temozolomide reduces clonogenicity and induces apoptosis in glioblastoma cells. Int. J. Oncol. 2021, 58, 312–330. [Google Scholar] [CrossRef]
- Pratap, U.P.; Sareddy, G.R.; Liu, Z.X.; Venkata, P.P.; Liu, J.H.; Tang, W.W.; Altwegg, K.A.; Ebrahimi, B.; Li, X.N.; Tekmal, R.R.; et al. Histone deacetylase inhibitors enhance estrogen receptor beta expression and augment agonistmediated tumor suppression in glioblastoma. Neuro-Oncol. Adv. 2021, 3, vdab099. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, N.; Lopez, A.; Cuello, V.; Persans, M.; Schuenzel, E.; Innis-Whitehouse, W.; Keniry, M. NVP-BEZ235 or JAKi Treatment leads to decreased survival of examined GBM and BBC cells. Cancer Treat. Res. Commun. 2021, 27, 100340. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Wang, W.; Li, H.; Jiao, Y.; Huo, R.; Yan, Z.; Wang, J.; Wang, S.; Wang, J.; Chen, D.; et al. Single-Cell Atlas Reveals Complexity of the Immunosuppressive Microenvironment of Initial and Recurrent Glioblastoma. Front. Immunol. 2020, 11, 835. [Google Scholar] [CrossRef] [PubMed]
- Lai, T.J.; Sun, L.; Li, K.; Prins, T.J.; Treger, J.; Li, T.; Sun, M.Z.; Nathanson, D.A.; Liau, L.M.; Lai, A.; et al. Epigenetic Induction of Cancer-Testis Antigens and Endogenous Retroviruses at Single-Cell Level Enhances Immune Recognition and Response in Glioma. Cancer Res. Commun. 2024, 4, 1834–1849. [Google Scholar] [CrossRef]
- Jang, H.J.; Shah, N.M.; Maeng, J.H.; Liang, Y.; Basri, N.L.; Ge, J.; Qu, X.; Mahlokozera, T.; Tzeng, S.C.; Williams, R.B.; et al. Epigenetic therapy potentiates transposable element transcription to create tumor-enriched antigens in glioblastoma cells. Nat. Genet. 2024, 56, 1903–1913. [Google Scholar] [CrossRef]
- Ma, R.; Rei, M.; Woodhouse, I.; Ferris, K.; Kirschner, S.; Chandran, A.; Gileadi, U.; Chen, J.L.; Pereira Pinho, M.; Ariosa-Morejon, Y.; et al. Decitabine increases neoantigen and cancer testis antigen expression to enhance T-cell-mediated toxicity against glioblastoma. Neuro Oncol. 2022, 24, 2093–2106. [Google Scholar] [CrossRef]
- Shah, A.H.; Govindarajan, V.; Doucet-O’Hare, T.T.; Rivas, S.; Ampie, L.; DeMarino, C.; Banasavadi-Siddegowda, Y.K.; Zhang, Y.; Johnson, K.R.; Almsned, F.; et al. Differential expression of an endogenous retroviral element [HERV-K(HML-6)] is associated with reduced survival in glioblastoma patients. Sci. Rep. 2022, 12, 6902. [Google Scholar] [CrossRef]
- Asey, B.; Pantel, T.F.; Mohme, M.; Zghaibeh, Y.; Duhrsen, L.; Silverbush, D.; Schuller, U.; Drexler, R.; Ricklefs, F.L. Peripheral blood-derived immune cell counts as prognostic indicators and their relationship with DNA methylation subclasses in glioblastoma patients. Brain Pathol. 2025, 35, e13334. [Google Scholar] [CrossRef]
- Kloosterman, D.J.; Erbani, J.; Boon, M.; Farber, M.; Handgraaf, S.M.; Ando-Kuri, M.; Sánchez-López, E.; Fontein, B.; Mertz, M.; Nieuwland, M.; et al. Macrophage-mediated myelin recycling fuels brain cancer malignancy. Cell 2024, 187, 5336–5356.e5330. [Google Scholar] [CrossRef]
- Bayik, D.; Bartels, C.F.; Lovrenert, K.; Watson, D.C.; Zhang, D.; Kay, K.; Lee, J.; Lauko, A.; Johnson, S.; Lo, A.; et al. Distinct Cell Adhesion Signature Defines Glioblastoma Myeloid-Derived Suppressor Cell Subsets. Cancer Res. 2022, 82, 4274–4287. [Google Scholar] [CrossRef]
- Hutarew, G.; Hölzl, D.; Schiefer, T.; Langwieder, C.K.; Alinger-Scharinger, B.; Schlicker, H.U.; Schwartz, C.; Sotlar, K.; Kraus, T.F.J. Methylome Profiling of PD-L1-Expressing Glioblastomas Shows Enrichment of Post-Transcriptional and RNA-Associated Gene Regulation. Cancers 2022, 14, 5375. [Google Scholar] [CrossRef] [PubMed]
- Goswami, S.; Raychaudhuri, D.; Singh, P.; Natarajan, S.M.; Chen, Y.; Poon, C.; Hennessey, M.; Tannir, A.J.; Zhang, J.; Anandhan, S.; et al. Myeloid-specific KDM6B inhibition sensitizes glioblastoma to PD1 blockade. Nat. Cancer 2023, 4, 1455–1473. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; He, J.; Chen, R.; Wang, Z.; Dai, Z.; Liang, X.; Wu, W.; Luo, P.; Zhang, J.; Peng, Y.; et al. Pan-Cancer Analysis of the Immunological Role of PDIA5: A Potential Target for Immunotherapy. Front. Immunol. 2022, 13, 881722. [Google Scholar] [CrossRef]
- Xia, L.; Liu, J.Y.; Zheng, Z.Z.; Chen, Y.J.; Ding, J.C.; Hu, Y.H.; Hu, G.S.; Xia, N.S.; Liu, W. BRD4 inhibition boosts the therapeutic effects of epidermal growth factor receptor-targeted chimeric antigen receptor T cells in glioblastoma. Mol. Ther. 2021, 29, 3011–3026. [Google Scholar] [CrossRef]
- Wei, W.Y.; Huang, C.; Zhang, J.; Chen, Q.X.; Liu, Z.Y.; Ren, X.J.; Gan, S.L.; Wu, P.Z.; Wang, D.Q.; Tang, B.Z.; et al. HDAC6-Activatable Multifunctional Near-Infrared Probe for Glioma Cell Detection and Elimination. Anal. Chem. 2024, 96, 2406–2414. [Google Scholar] [CrossRef]
- de Oliveira, C.T.P.; Colenci, R.; Pacheco, C.C.; Mariano, P.M.; do Prado, P.R.; Mamprin, G.P.R.; Santana, M.G.; Gambero, A.; de Oliveira Carvalho, P.; Priolli, D.G. Hydrolyzed Rutin Decreases Worsening of Anaplasia in Glioblastoma Relapse. CNS Neurol. Disord. Drug Targets 2019, 18, 405–412. [Google Scholar] [CrossRef]
- Malik, Y.S.; Sheikh, M.A.; Xing, Z.; Guo, Z.; Zhu, X.; Tian, H.; Chen, X. Polylysine-modified polyethylenimine polymer can generate genetically engineered mesenchymal stem cells for combinational suicidal gene therapy in glioblastoma. Acta Biomater. 2018, 80, 144–153. [Google Scholar] [CrossRef]
- Kaundal, B.; Karmakar, S.; Choudhury, S.R. Mitochondria-targeting nano therapy altering IDH2-mediated EZH2/EZH1 interaction as precise epigenetic regulation in glioblastoma. Biomater. Sci. 2022, 10, 5301–5317. [Google Scholar] [CrossRef]
- Phase I Open Label Ascending Dose Study to Assess the Feasibility and Safety of Intermittent Infusions of MTX110 Administered by Convection-Enhanced Delivery (CED) in Patients with Recurrent Glioblastoma (rGBM) (MAGIC-G1). 2022. Available online: https://clinicaltrials.gov/study/NCT05324501 (accessed on 22 December 2024).
- International Cooperative Phase III Trial of the HIT-HGG Study Group for the Treatment of High Grade Glioma, Diffuse Intrinsic Pontine Glioma, and Gliomatosis Cerebri in Children and Adolescents < 18 Years.(HIT-HGG-2013). 2017. Available online: https://clinicaltrials.gov/study/NCT03243461 (accessed on 22 December 2024).
- An Open-Label, Single-arm, Phase II Study to Evaluate Safety and Efficacy of Doxorubicin in Combination with Radiotherapy, Temozolomide and Valproic Acid in Patients with Glioblastoma Multiforme (GBM) and Diffuse Intrinsic Pontine Glioma (DIPG). 2016. Available online: https://clinicaltrials.gov/study/NCT02758366 (accessed on 22 December 2024).
- A Pilot Study to Evaluate the Feasibility of the Combined Use of Stereotactic Radiosurgery with Nivolumab and Concurrent Valproate in Patients with Recurrent Glioblastoma. 2015. Available online: https://clinicaltrials.gov/study/NCT02648633 (accessed on 22 December 2024).
- A Phase I Trial of Pembrolizumab and Vorinostat Combined with Temozolomide and Radiation Therapy for Newly Diagnosed Glioblastoma. 2018. Available online: https://clinicaltrials.gov/study/NCT03426891 (accessed on 22 December 2024).
- A Phase 1, Open-Label Study to Assess the Pharmacokinetics, Pharmacodynamics and Central Nervous System (CNS) Penetration of CC-90010 in Preoperative Subjects with Progressive or Recurrent Who Grade II Diffuse Astrocytoma, Grade III Anaplastic Astrocytoma and Recurrent Glioblastoma Scheduled for Resection. 2019. Available online: https://clinicaltrials.gov/study/NCT04047303 (accessed on 22 December 2024).
- Vieito, M.; Simonelli, M.; de Vos, F.; Moreno, V.; Geurts, M.; Lorenzi, E.; Macchini, M.; van den Bent, M.J.; Del Conte, G.; de Jonge, M.; et al. Trotabresib (CC-90010) in combination with adjuvant temozolomide or concomitant temozolomide plus radiotherapy in patients with newly diagnosed glioblastoma. Neurooncol. Adv. 2022, 4, vdac146. [Google Scholar] [CrossRef]
- A Phase 1b/2 Study of FT-2102 in Patients with Advanced Solid Tumors and Gliomas with an IDH1 Mutation. 2018. Available online: https://clinicaltrials.gov/study/NCT03684811 (accessed on 22 December 2024).
- A Phase 1 Study of DS-1001b in Patients with IDH1 Mutated Gliomas. 2017. Available online: https://clinicaltrials.gov/study/NCT03030066 (accessed on 22 December 2024).
- Machida, Y.; Nakagawa, M.; Matsunaga, H.; Yamaguchi, M.; Ogawara, Y.; Shima, Y.; Yamagata, K.; Katsumoto, T.; Hattori, A.; Itoh, M.; et al. A Potent Blood-Brain Barrier-Permeable Mutant IDH1 Inhibitor Suppresses the Growth of Glioblastoma with IDH1 Mutation in a Patient-Derived Orthotopic Xenograft Model. Mol. Cancer Ther. 2020, 19, 375–383. [Google Scholar] [CrossRef]
- A Phase 0 First-In-Human Study Using NU-0129: A Spherical Nucleic Acid (SNA) Gold Nanoparticle Targeting BCL2L12 in Recurrent Glioblastoma Multiforme or Gliosarcoma Patients. 2017. Available online: https://clinicaltrials.gov/study/NCT03020017 (accessed on 22 December 2024).
- Establishment of a Signature of Circulating microRNA as a Tool to Aid Diagnosis of Primary Brain Tumors in Adults. 2018. Available online: https://clinicaltrials.gov/study/NCT03630861 (accessed on 22 December 2024).
- Improving Personalised Glioblastoma Care by Stem Cell Analysis, Omics (Including Immunomics) and Artificial Intelligence Approaches. 2023. Available online: https://clinicaltrials.gov/study/NCT05941234 (accessed on 22 December 2024).
- Monitoring of Patients with Diffuse Gliomas Using Circulating miRNAs. 2024. Available online: https://clinicaltrials.gov/study/NCT06203496 (accessed on 22 December 2024).
- Xie, Q.; Wu, T.P.; Gimple, R.C.; Li, Z.; Prager, B.C.; Wu, Q.; Yu, Y.; Wang, P.; Wang, Y.; Gorkin, D.U.; et al. N(6)-methyladenine DNA Modification in Glioblastoma. Cell 2018, 175, 1228–1243.e1220. [Google Scholar] [CrossRef] [PubMed]
- Mata, D.A.; Benhamida, J.K.; Lin, A.L.; Vanderbilt, C.M.; Yang, S.R.; Villafania, L.B.; Ferguson, D.C.; Jonsson, P.; Miller, A.M.; Tabar, V.; et al. Genetic and epigenetic landscape of IDH-wildtype glioblastomas with FGFR3-TACC3 fusions. Acta Neuropathol. Commun. 2020, 8, 186. [Google Scholar] [CrossRef] [PubMed]
- Masui, K.; Harachi, M.; Ikegami, S.; Yang, H.; Onizuka, H.; Yong, W.H.; Cloughesy, T.F.; Muragaki, Y.; Kawamata, T.; Arai, N.; et al. mTORC2 links growth factor signaling with epigenetic regulation of iron metabolism in glioblastoma. J. Biol. Chem. 2019, 294, 19740–19751. [Google Scholar] [CrossRef] [PubMed]
- Moon, B.S.; Cai, M.; Lee, G.; Zhao, T.; Song, X.; Giannotta, S.L.; Attenello, F.J.; Yu, M.; Lu, W. Epigenetic modulator inhibition overcomes temozolomide chemoresistance and antagonizes tumor recurrence of glioblastoma. J. Clin. Investig. 2020, 130, 5782–5799. [Google Scholar] [CrossRef]
- Zhan, X.; Guo, S.; Li, Y.; Ran, H.; Huang, H.; Mi, L.; Wu, J.; Wang, X.; Xiao, D.; Chen, L.; et al. Glioma stem-like cells evade interferon suppression through MBD3/NuRD complex-mediated STAT1 downregulation. J. Exp. Med. 2020, 217, e20191340. [Google Scholar] [CrossRef]
- Zhang, L.W.; Chen, L.H.; Wan, H.; Yang, R.; Wang, Z.H.; Feng, L.; Yang, S.H.; Jones, S.; Wang, S.Z.; Zhou, W.X.; et al. Exome sequencing identifies somatic gain-of-function PPM1D mutations in brainstem gliomas. Nat. Genet. 2014, 46, 726–730. [Google Scholar] [CrossRef]
- Nomura, M.; Saito, K.; Aihara, K.; Nagae, G.; Yamamoto, S.; Tatsuno, K.; Ueda, H.; Fukuda, S.; Umeda, T.; Tanaka, S.; et al. DNA demethylation is associated with malignant progression of lower-grade gliomas. Sci. Rep. 2019, 9, 1903. [Google Scholar] [CrossRef]
- Ning, X.H.; Shi, Z.D.; Liu, X.; Zhang, A.L.; Han, L.; Jiang, K.; Kang, C.S.; Zhang, Q.Y. DNMT1 and EZH2 mediated methylation silences the microRNA-200b/a/429 gene and promotes tumor progression. Cancer Lett. 2015, 359, 198–205. [Google Scholar] [CrossRef]
- Fomchenko, E.I.; Erson-Omay, E.Z.; Zhao, A.; Bindra, R.S.; Huttner, A.; Fulbright, R.K.; Moliterno, J. DNMT3A co-mutation in an IDH1-mutant glioblastoma. Cold Spring Harb. Mol. Case Stud. 2019, 5, a004119. [Google Scholar] [CrossRef]
- Xu, H.; Sun, J.; Shi, C.J.; Sun, C.Y.; Yu, L.; Wen, Y.J.; Zhao, S.J.; Liu, J.; Xu, J.L.; Li, N.N.; et al. miR-29s inhibit the malignant behavior of U87MG glioblastoma cell line by targeting DNMT3A and 3B. Neurosci. Lett. 2015, 590, 40–46. [Google Scholar] [CrossRef]
- Cheray, M.; Pacaud, R.; Nadaradjane, A.; Oliver, L.; Vallette, F.M.; Cartron, P.F. Specific Inhibition of DNMT3A/ISGF3γ Interaction Increases the Temozolomide Efficiency to Reduce Tumor Growth. Theranostics 2016, 6, 1988–1999. [Google Scholar] [CrossRef] [PubMed]
- Indraccolo, S.; Lombardi, G.; Fassan, M.; Pasqualini, L.; Giunco, S.; Marcato, R.; Gasparini, A.; Candiotto, C.; Nalio, S.; Fiduccia, P.; et al. Genetic, Epigenetic, and Immunologic Profiling of MMR-Deficient Relapsed Glioblastoma. Clin. Cancer Res. 2019, 25, 1828–1837. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.B.; Zhang, Y.; She, X.L.; Sun, Y.N.; Fan, L.; Ren, X.; Fu, H.J.; Liu, C.H.; Li, P.Y.; Zhao, C.H.; et al. Hypermethylated gene ANKDD1A is a candidate tumor suppressor that interacts with FIH1 and decreases HIF1α stability to inhibit cell autophagy in the glioblastoma multiforme hypoxia microenvironment. Oncogene 2019, 38, 103–119. [Google Scholar] [CrossRef]
- Wang, S.; Chen, C.; Li, J.; Xu, X.; Chen, W.; Li, F. The CXCL12/CXCR4 axis confers temozolomide resistance to human glioblastoma cells via up-regulation of FOXM1. J. Neurol. Sci. 2020, 414, 116837. [Google Scholar] [CrossRef]
- Su, J.; Ma, Q.Q.; Long, W.Y.; Tang, H.L.; Wu, C.W.; Luo, M.; Wang, X.Y.; Xiao, K.; Li, Y.; Xiao, Q.; et al. LCTL Is a Prognostic Biomarker and Correlates with Stromal and Immune Infiltration in Gliomas. Front. Oncol. 2019, 9, 1083. [Google Scholar] [CrossRef]
- Wang, T.L.; Pickard, A.J.; Gallo, J.M. Histone Methylation by Temozolomide; A Classic DNA Methylating Anticancer Drug. Anticancer Res. 2016, 36, 3289–3299. [Google Scholar]
- O’Regan, C.J.; Kearney, H.; Beausang, A.; Farrell, M.A.; Brett, F.M.; Cryan, J.B.; Loftus, T.E.; Buckley, P.G. Temporal stability of MGMT promoter methylation in glioblastoma patients undergoing STUPP protocol. J. Neuro-Oncol. 2018, 137, 233–240. [Google Scholar] [CrossRef]
- Zhang, D.B.; Li, Y.; Wang, R.; Li, Y.N.; Shi, P.; Kan, Z.M.; Pang, X.N. Inhibition of REST Suppresses Proliferation and Migration in Glioblastoma Cells. Int. J. Mol. Sci. 2016, 17, 664. [Google Scholar] [CrossRef]
- Tivnan, A.; Zhao, J.; Johns, T.G.; Day, B.W.; Stringer, B.W.; Boyd, A.W.; Tiwari, S.; Giles, K.M.; Teo, C.; McDonald, K.L. The tumor suppressor microRNA, miR-124a, is regulated by epigenetic silencing and by the transcriptional factor, REST in glioblastoma. Tumor Biol. 2014, 35, 1459–1465. [Google Scholar] [CrossRef]
- Kroes, R.A.; Moskal, J.R. The role of DNA methylation in ST6Gal1 expression in gliomas. Glycobiology 2016, 26, 1271–1283. [Google Scholar] [CrossRef]
- Beyer, S.J.; Bell, E.H.; McElroy, J.P.; Fleming, J.L.; Cui, T.; Becker, A.; Bassett, E.; Johnson, B.; Gulati, P.; Popp, I.; et al. Oncogenic transgelin-2 is differentially regulated in isocitrate dehydrogenase wild-type vs. mutant gliomas. Oncotarget 2018, 9, 37097–37111. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Alver, B.M.; Li, S.; Hlady, R.A.; Thompson, J.J.; Schroeder, M.A.; Lee, J.H.; Qiu, J.; Schwartz, P.H.; Sarkaria, J.N.; et al. Distinctive epigenomes characterize glioma stem cells and their response to differentiation cues. Genome Biol. 2018, 19, 43. [Google Scholar] [CrossRef] [PubMed]
- Stasik, S.; Juratli, T.A.; Petzold, A.; Richter, S.; Zolal, A.; Schackert, G.; Dahl, A.; Krex, D.; Thiede, C. Exome sequencing identifies frequent genomic loss of TET1 in IDH-wild-type glioblastoma. Neoplasia 2020, 22, 800–808. [Google Scholar] [CrossRef]
- Fedele, V.; Dai, F.; Masilamani, A.P.; Heiland, D.H.; Kling, E.; Gaetjens-Sanchez, A.M.; Ferrarese, R.; Platania, L.; Soroush, D.; Kim, H.; et al. Epigenetic Regulation of ZBTB18 Promotes Glioblastoma Progression. Mol. Cancer Res. 2017, 15, 998–1011. [Google Scholar] [CrossRef]
- Pastori, C.; Daniel, M.; Penas, C.; Volmar, C.H.; Johnstone, A.L.; Brothers, S.P.; Graham, R.M.; Allen, B.; Sarkaria, J.N.; Komotar, R.J.; et al. BET bromodomain proteins are required for glioblastoma cell proliferation. Epigenetics 2014, 9, 611–620. [Google Scholar] [CrossRef]
- Deshmukh, P.; Mathur, S.; Gangadharan, G.; Krishnappa, G.; Nanjaiah, N.D.; Padmanabhan, B. Novel pyrano 1,3 oxazine based ligand inhibits the epigenetic reader hBRD2 in glioblastoma. Biochem. J. 2020, 477, 2263–2279. [Google Scholar] [CrossRef]
- Pastori, C.; Kapranov, P.; Penas, C.; Peschansky, V.; Volmar, C.H.; Sarkaria, J.N.; Bregy, A.; Komotar, R.; St Laurent, G.; Ayad, N.G.; et al. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc. Natl. Acad. Sci. USA 2015, 112, 8326–8331. [Google Scholar] [CrossRef]
- Ganguly, D.; Sims, M.; Cai, C.; Fan, M.Y.; Pfeffer, L.M. Chromatin Remodeling Factor BRG1 Regulates Stemness and Chemosensitivity of Glioma Initiating Cells. Stem Cells 2018, 36, 1804–1815. [Google Scholar] [CrossRef]
- Jia, Y.; Wang, Y.; Zhang, C.; Chen, M.Y. Upregulated CBX8 Promotes Cancer Metastasis via the WNK2/MMP2 Pathway. Mol. Ther.-Oncolytics 2020, 19, 188–196. [Google Scholar] [CrossRef]
- Gimple, R.C.; Kidwell, R.L.; Kim, L.J.Y.; Sun, T.; Gromovsky, A.D.; Wu, Q.; Wolf, M.; Lv, D.; Bhargava, S.; Jiang, L.; et al. Glioma Stem Cell-Specific Superenhancer Promotes Polyunsaturated Fatty-Acid Synthesis to Support EGFR Signaling. Cancer Discov. 2019, 9, 1248–1267. [Google Scholar] [CrossRef]
- Jameson, N.M.; Ma, J.; Benitez, J.; Izurieta, A.; Han, J.Y.; Mendez, R.; Parisian, A.; Furnari, F. Intron 1-Mediated Regulation of EGFR Expression in EGFR-Dependent Malignancies Is Mediated by AP-1 and BET Proteins. Mol. Cancer Res. 2019, 17, 2208–2220. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.R.; Ren, L.; Wu, D.C.; Yang, X.; Zhou, Z.Y.; Nie, Q.M.; Jiang, G.; Xue, S.L.; Weng, W.J.; Qiu, Y.M.; et al. Overexpression of FoxO3a is associated with glioblastoma progression and predicts poor patient prognosis. Int. J. Cancer 2017, 140, 2792–2804. [Google Scholar] [CrossRef] [PubMed]
- Ramis, G.; Villalonga-Planells, R.; Serra-Sitjar, M.; Brell, M.; Fernández de Mattos, S.; Villalonga, P. The tumor suppressor FOXO3a mediates the response to EGFR inhibition in glioblastoma cells. Cell. Oncol. 2019, 42, 521–536. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Dong, Z.; Prager, B.C.; Kim, L.J.K.; Wu, Q.; Gimple, R.C.; Wang, X.; Bao, S.; Hamerlik, P.; Rich, J.N. Chromatin remodeler HELLS maintains glioma stem cells through E2F3 and MYC. JCI Insight 2019, 4, e126140. [Google Scholar] [CrossRef]
- Shang, E.; Nguyen, T.T.T.; Shu, C.; Westhoff, M.A.; Karpel-Massler, G.; Siegelin, M.D. Epigenetic Targeting of Mcl-1 Is Synthetically Lethal with Bcl-xL/Bcl-2 Inhibition in Model Systems of Glioblastoma. Cancers 2020, 12, 2137. [Google Scholar] [CrossRef]
- Gallo, M.; Coutinho, F.J.; Vanner, R.J.; Gayden, T.; Mack, S.C.; Murison, A.; Remke, M.; Li, R.; Takayama, N.; Desai, K.; et al. MLL5 Orchestrates a Cancer Self-Renewal State by Repressing the Histone Variant H3.3 and Globally Reorganizing Chromatin. Cancer Cell 2015, 28, 715–729. [Google Scholar] [CrossRef]
- Khalil, A.; Nemer, G. The potential oncogenic role of the RAS-like GTP-binding gene RIT1 in glioblastoma. Cancer Biomark. 2020, 29, 509–519. [Google Scholar] [CrossRef]
- Hiramatsu, H.; Kobayashi, K.; Kobayashi, K.; Haraguchi, T.; Ino, Y.; Todo, T.; Iba, H. The role of the SWI/SNF chromatin remodeling complex in maintaining the stemness of glioma initiating cells. Sci. Rep. 2017, 7, 889. [Google Scholar] [CrossRef]
- Meng, W.; Wang, J.J.; Wang, B.C.; Liu, F.; Li, M.; Zhao, Y.; Zhang, C.R.; Li, Q.F.; Chen, J.X.; Zhang, L.Y.; et al. CDK7 inhibition is a novel therapeutic strategy against GBM both in vitro and in vivo. Cancer Manag. Res. 2018, 10, 5747–5758. [Google Scholar] [CrossRef]
- Ricci, B.; Millner, T.O.; Pomella, N.; Zhang, X.; Guglielmi, L.; Badodi, S.; Ceric, D.; Gemma, C.; Cognolato, E.; Zhang, Y.; et al. Polycomb-mediated repression of EphrinA5 promotes growth and invasion of glioblastoma. Oncogene 2020, 39, 2523–2538. [Google Scholar] [CrossRef]
- Ma, L.; Lin, K.Y.; Chang, G.D.; Chen, Y.W.; Yue, C.; Guo, Q.; Zhang, S.C.; Jia, Z.L.; Huang, T.T.; Zhou, A.D.; et al. Aberrant Activation of β-Catenin Signaling Drives Glioma Tumorigenesis via USP1-Mediated Stabilization of EZH2. Cancer Res. 2019, 79, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Purkait, S.; Takkar, S.; Malgulwar, P.B.; Kumar, A.; Pathak, P.; Suri, V.; Sharma, M.C.; Suri, A.; Kale, S.S.; et al. Analysis of EZH2: Micro-RNA network in low and high grade astrocytic tumors. Brain Tumor Pathol. 2016, 33, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Harachi, M.; Masui, K.; Honda, H.; Muragaki, Y.; Kawamata, T.; Cavenee, W.K.; Mischel, P.S.; Shibata, N. Dual Regulation of Histone Methylation by mTOR Complexes Controls Glioblastoma Tumor Cell Growth via EZH2 and SAM. Mol. Cancer Res. 2020, 18, 1142–1152. [Google Scholar] [CrossRef] [PubMed]
- Chien, Y.C.; Chen, J.N.; Chen, Y.H.; Chou, R.H.; Lee, H.C.; Yu, Y.L. Epigenetic Silencing of miR-9 Promotes Migration and Invasion by EZH2 in Glioblastoma Cells. Cancers 2020, 12, 1781. [Google Scholar] [CrossRef]
- Yin, Y.; Qiu, S.; Li, X.; Huang, B.; Xu, Y.; Peng, Y. EZH2 suppression in glioblastoma shifts microglia toward M1 phenotype in tumor microenvironment. J. Neuroinflamm. 2017, 14, 220. [Google Scholar] [CrossRef]
- de Vries, N.A.; Hulsman, D.; Akhtar, W.; de Jong, J.; Miles, D.C.; Blom, M.; van Tellingen, O.; Jonkers, J.; van Lohuizen, M. Prolonged Ezh2 Depletion in Glioblastoma Causes a Robust Switch in Cell Fate Resulting in Tumor Progression. Cell Rep. 2015, 10, 383–397. [Google Scholar] [CrossRef]
- Liau, B.B.; Sievers, C.; Donohue, L.K.; Gillespie, S.M.; Flavahan, W.A.; Miller, T.E.; Venteicher, A.S.; Hebert, C.H.; Carey, C.D.; Rodig, S.J.; et al. Adaptive Chromatin Remodeling Drives Glioblastoma Stem Cell Plasticity and Drug Tolerance. Cell Stem Cell 2017, 20, 233–246.e237. [Google Scholar] [CrossRef]
- Romani, M.; Daga, A.; Forlani, A.; Pistillo, M.P.; Banelli, B. Targeting of Histone Demethylases KDM5A and KDM6B Inhibits the Proliferation of Temozolomide-Resistant Glioblastoma Cells. Cancers 2019, 11, 878. [Google Scholar] [CrossRef]
- Chen, Y.H.; Fang, R.P.; Yue, C.; Chang, G.Q.; Li, P.; Guo, Q.; Wang, J.; Zhou, A.D.; Zhang, S.C.; Fuller, G.N.; et al. Wnt-Induced Stabilization of KDM4C Is Required for Wnt/β-Catenin Target Gene Expression and Glioblastoma Tumorigenesis. Cancer Res. 2020, 80, 1049–1063. [Google Scholar] [CrossRef]
- Huang, Y.C.; Lin, S.J.; Shih, H.Y.; Chou, C.H.; Chu, H.H.; Chiu, C.C.; Yuh, C.H.; Yeh, T.H.; Cheng, Y.C. Epigenetic regulation of NOTCH1 and NOTCH3 by KMT2A inhibits glioma proliferation. Oncotarget 2017, 8, 63110–63120. [Google Scholar] [CrossRef][Green Version]
- Dong, F.; Li, Q.; Yang, C.; Huo, D.; Wang, X.; Ai, C.; Kong, Y.; Sun, X.; Wang, W.; Zhou, Y.; et al. PRMT2 links histone H3R8 asymmetric dimethylation to oncogenic activation and tumorigenesis of glioblastoma. Nat. Commun. 2018, 9, 4552. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, S.J.; Dolivo, D.M.; Dominko, T. PRMT8 demonstrates variant-specific expression in cancer cells and correlates with patient survival in breast, ovarian and gastric cancer. Oncol. Lett. 2017, 13, 1983–1989. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.Y.; Lee, H.; Yoon, J.G.; Madan, A.; Wayner, E.; Tonning, S.; Hothi, P.; Schroeder, B.; Ulasov, I.; Foltz, G.; et al. Global analysis of H3K4me3 and H3K27me3 profiles in glioblastoma stem cells and identification of SLC17A7 as a bivalent tumor suppressor gene. Oncotarget 2015, 6, 5369–5381. [Google Scholar] [CrossRef]
- Sferra, R.; Pompili, S.; Festuccia, C.; Marampon, F.; Gravina, G.L.; Ventura, L.; Di Cesare, E.; Cicchinelli, S.; Gaudio, E.; Vetuschi, A. The possible prognostic role of histone deacetylase and transforming growth factor β/Smad signaling in high grade gliomas treated by radio-chemotherapy: A preliminary immunohistochemical study. Eur. J. Histochem. 2017, 61, 2732. [Google Scholar] [CrossRef]
- Laws, M.T.; Bonomi, R.E.; Gelovani, D.J.; Llaniguez, J.; Lu, X.; Mangner, T.; Gelovani, J.G. Noninvasive quantification of SIRT1 expression-activity and pharmacologic inhibition in a rat model of intracerebral glioma using 2-[(18)F]BzAHA PET/CT/MRI. Neuro-Oncol. Adv. 2020, 2, vdaa006. [Google Scholar] [CrossRef]
- Bradley, B.S.; Loftus, J.C.; Mielke, C.J.; Dinu, V. Differential expression of microRNAs as predictors of glioblastoma phenotypes. BMC Bioinform. 2014, 15, 21. [Google Scholar] [CrossRef]
- Teplyuk, N.M.; Uhlmann, E.J.; Wong, A.H.; Karmali, P.; Basu, M.; Gabriely, G.; Jain, A.; Wang, Y.; Chiocca, E.A.; Stephens, R.; et al. MicroRNA-10b inhibition reduces E2F1-mediated transcription and miR-15/16 activity in glioblastoma. Oncotarget 2015, 6, 3770–3783. [Google Scholar] [CrossRef]
- Bhaskaran, V.; Nowicki, M.O.; Idriss, M.; Jimenez, M.A.; Lugli, G.; Hayes, J.L.; Mahmoud, A.B.; Zane, R.E.; Passaro, C.; Ligon, K.L.; et al. The functional synergism of microRNA clustering provides therapeutically relevant epigenetic interference in glioblastoma. Nat. Commun. 2019, 10, 442. [Google Scholar] [CrossRef]
- Tian, X.Y.; Zhang, L.; Sun, L.G.; Li, M. Epigenetic Regulation of miR-129-2 Leads to Overexpression of PDGFRa and FoxP1 in Glioma Cells. Asian Pac. J. Cancer Prev. 2015, 16, 6129–6133. [Google Scholar] [CrossRef]
- Li, W.D.; Miao, X.B.; Liu, L.L.; Zhang, Y.; Jin, X.J.; Luo, X.J.; Gao, H.; Deng, X.B. Methylation-mediated silencing of microRNA-211 promotes cell growth and epithelial to mesenchymal transition through activation of the AKT/β-catenin pathway in GBM. Oncotarget 2017, 8, 25167–25176. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; Zhang, Y.; Wang, H.; Rong, X.; Peng, J.; He, L.; Peng, Y. An miR-340-5p-macrophage feedback loop modulates the progression and tumor microenvironment of glioblastoma multiforme. Oncogene 2019, 38, 7399–7415. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Qiu, S.; Ge, R.; He, L.; Li, M.; Li, Y.; Peng, Y. miR-340 suppresses glioblastoma multiforme. Oncotarget 2015, 6, 9257–9270. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Bertoni, H.; Kotchetkov, I.S.; Mihelson, N.; Lal, B.; Rui, Y.; Ames, H.; Lugo-Fagundo, M.; Guerrero-Cazares, H.; Quiñones-Hinojosa, A.; Green, J.J.; et al. A Sox2:miR-486-5p Axis Regulates Survival of GBM Cells by Inhibiting Tumor Suppressor Networks. Cancer Res. 2020, 80, 1644–1655. [Google Scholar] [CrossRef]
- Zhao, K.; Wang, Q.X.; Wang, Y.F.; Huang, K.; Yang, C.; Li, Y.S.; Yi, K.K.; Kang, C.S. EGFR/c-myc axis regulates TGFβ/Hippo/Notch pathway via epigenetic silencing miR-524 in gliomas. Cancer Lett. 2017, 406, 12–21. [Google Scholar] [CrossRef]
- Du, X.L.; Tu, Y.M.; Liu, S.; Zhao, P.Z.; Bao, Z.Y.; Li, C.; Li, J.H.; Pan, M.H.; Ji, J. LINC00511 contributes to glioblastoma tumorigenesis and epithelial-mesenchymal transition via LINC00511/miR-524-5p/YB1/ZEB1 positive feedback loop. J. Cell. Mol. Med. 2020, 24, 1474–1487. [Google Scholar] [CrossRef]
- Zhang, H.; Wei, D.L.; Wan, L.; Yan, S.F.; Sun, Y.H. Highly expressed lncRNA CCND2-AS1 promotes glioma cell proliferation through Wnt/β-catenin signaling. Biochem. Biophys. Res. Commun. 2017, 482, 1219–1225. [Google Scholar] [CrossRef]
- Jiang, P.F.; Wang, P.; Sun, X.L.; Yuan, Z.S.; Zhan, R.C.; Ma, X.Y.; Li, W.G. Knockdown of long noncoding RNA H19 sensitizes human glioma cells to temozolomide therapy. OncoTargets Ther. 2016, 9, 3501–3509. [Google Scholar] [CrossRef]
- Liu, B.Y.; Zhou, J.; Wang, C.Y.; Chi, Y.; Wei, Q.T.; Fu, Z.; Lian, C.L.; Huang, Q.Z.; Liao, C.X.; Yang, Z.; et al. LncRNA SOX2OT promotes temozolomide resistance by elevating SOX2 expression via ALKBH5-mediated epigenetic regulation in glioblastoma. Cell Death Dis. 2020, 11, 384. [Google Scholar] [CrossRef]
- Fritah, S.; Muller, A.; Jiang, W.; Mitra, R.; Sarmini, M.; Dieterle, M.; Golebiewska, A.; Ye, T.; Van Dyck, E.; Herold-Mende, C.; et al. Temozolomide-Induced RNA Interactome Uncovers Novel LncRNA Regulatory Loops in Glioblastoma. Cancers 2020, 12, 2583. [Google Scholar] [CrossRef]
- Ji, J.H.; Zhao, L.; Zhao, X.X.; Li, Q.P.; An, Y.; Li, L.; Li, D.G. Genome-wide DNA methylation regulation analysis of long non-coding RNAs in glioblastoma. Int. J. Mol. Med. 2020, 46, 224–238. [Google Scholar] [CrossRef]
- Huang, K.; Yang, C.; Wang, Q.X.; Li, Y.S.; Fang, C.; Tan, Y.L.; Wei, J.W.; Wang, Y.F.; Li, X.; Zhou, J.H.; et al. The CRISPR/Cas9 system targeting EGFR exon 17 abrogates NF-κB activation via epigenetic modulation of UBXN1 in EGFRwt/vIII glioma cells. Cancer Lett. 2017, 388, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Liu, X.; Li, Y.; Wang, Q.; Zhou, J.; Wang, Y.; Dong, F.; Yang, C.; Sun, Z.; Fang, C.; et al. Genome-Wide CRISPR-Cas9 Screening Identifies NF-κB/E2F6 Responsible for EGFRvIII-Associated Temozolomide Resistance in Glioblastoma. Adv. Sci. 2019, 6, 1900782. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.W.; Wang, J.H.; Marzese, D.M.; Wang, X.W.; Yang, Z.X.; Li, C.J.; Zhang, H.B.; Zhang, J.S.; Chen, C.C.; Kelly, D.F.; et al. B7H3 regulates differentiation and serves as a potential biomarker and theranostic target for human glioblastoma. Lab. Investig. 2019, 99, 1117–1129. [Google Scholar] [CrossRef] [PubMed]
- Garber, S.T.; Hashimoto, Y.; Weathers, S.P.; Xiu, J.; Gatalica, Z.; Verhaak, R.G.W.; Zhou, S.H.; Fuller, G.N.; Khasraw, M.; de Groot, J.; et al. Immune checkpoint blockade as a potential therapeutic target: Surveying CNS malignancies. Neuro Oncol. 2016, 18, 1357–1366. [Google Scholar] [CrossRef]
- Chen, P.W.; Hsu, W.H.; Chang, A.; Tan, Z.; Lan, Z.D.; Zhou, A.; Spring, D.J.; Lang, F.F.; Wang, Y.A.; DePinho, R.A. Circadian Regulator CLOCK Recruits Immune-Suppressive Microglia into the GBM Tumor Microenvironment. Cancer Discov. 2020, 10, 371–381. [Google Scholar] [CrossRef]
- Mörén, L.; Perryman, R.; Crook, T.; Langer, J.K.; Oneill, K.; Syed, N.; Antti, H. Metabolomic profiling identifies distinct phenotypes for ASS1 positive and negative GBM. BMC Cancer 2018, 18, 167. [Google Scholar] [CrossRef]
- Hujber, Z.; Horváth, G.; Petővári, G.; Krencz, I.; Dankó, T.; Mészáros, K.; Rajnai, H.; Szoboszlai, N.; Leenders, W.P.J.; Jeney, A.; et al. GABA, glutamine, glutamate oxidation and succinic semialdehyde dehydrogenase expression in human gliomas. J. Exp. Clin. Cancer Res. 2018, 37, 271. [Google Scholar] [CrossRef]
- Paolini, A.; Curti, V.; Pasi, F.; Mazzini, G.; Nano, R.; Capelli, E. Gallic acid exerts a protective or an anti-proliferative effect on glioma T98G cells via dose-dependent epigenetic regulation mediated by miRNAs. Int. J. Oncol. 2015, 46, 1491–1497. [Google Scholar] [CrossRef]
- Rusu, P.; Shao, C.; Neuerburg, A.; Acikgöz, A.A.; Wu, Y.; Zou, P.; Phapale, P.; Shankar, T.S.; Döring, K.; Dettling, S.; et al. GPD1 Specifically Marks Dormant Glioma Stem Cells with a Distinct Metabolic Profile. Cell Stem Cell 2019, 25, 241–257.e248. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, H.K.; Huang, Q.F.; Liu, Q.Q.; Guo, Y.B.; Lu, J.J.; Li, X.H.; Xue, C.B.; Han, Q.Q. Transcriptional Repression of p53 by PAX3 Contributes to Gliomagenesis and Differentiation of Glioma Stem Cells. Front. Molec. Neurosci. 2018, 11, 187. [Google Scholar] [CrossRef]
- Yoo, S.; Bieda, M.C. Differences among brain tumor stem cell types and fetal neural stem cells in focal regions of histone modifications and DNA methylation, broad regions of modifications, and bivalent promoters. BMC Genom. 2014, 15, 724. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Du, Z.; Wu, B.; Xia, Q.; Zhao, Y.; Lin, L.; Cai, Z.; Wang, S.; Li, E.; Xu, L.; Li, Y.; et al. LCN2-interacting proteins and their expression patterns in brain tumors. Brain Res. 2019, 1720, 146304. [Google Scholar] [CrossRef] [PubMed]
- Suvà, M.L.; Rheinbay, E.; Gillespie, S.M.; Patel, A.P.; Wakimoto, H.; Rabkin, S.D.; Riggi, N.; Chi, A.S.; Cahill, D.P.; Nahed, B.V.; et al. Reconstructing and Reprogramming the Tumor-Propagating Potential of Glioblastoma Stem-like Cells. Cell 2014, 157, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Frank, M.O.; Koyama, T.; Rhrissorrakrai, K.; Robine, N.; Utro, F.; Emde, A.K.; Chen, B.J.; Arora, K.; Shah, M.; Geiger, H.; et al. Sequencing and curation strategies for identifying candidate glioblastoma treatments. BMC Med. Genom. 2019, 12, 56. [Google Scholar] [CrossRef]
- Parker, N.R.; Hudson, A.L.; Khong, P.; Parkinson, J.F.; Dwight, T.; Ikin, R.J.; Zhu, Y.; Cheng, Z.J.; Vafaee, F.; Chen, J.; et al. Intratumoral heterogeneity identified at the epigenetic, genetic and transcriptional level in glioblastoma. Sci. Rep. 2016, 6, 22477. [Google Scholar] [CrossRef]
- Čančer, M.; Drews, L.F.; Bengtsson, J.; Bolin, S.; Rosén, G.; Westermark, B.; Nelander, S.; Forsberg-Nilsson, K.; Uhrbom, L.; Weishaupt, H.; et al. BET and Aurora Kinase A inhibitors synergize against MYCN-positive human glioblastoma cells. Cell Death Dis. 2019, 10, 881. [Google Scholar] [CrossRef]
- Riccadonna, C.; Yacoub Maroun, C.; Vuillefroy de Silly, R.; Boehler, M.; Calvo Tardón, M.; Jueliger, S.; Taverna, P.; Barba, L.; Marinari, E.; Pellegatta, S.; et al. Decitabine Treatment of Glioma-Initiating Cells Enhances Immune Recognition and Killing. PLoS ONE 2016, 11, e0162105. [Google Scholar] [CrossRef]
- Alexanianl, A.R.; Huang, Y.W. Specific combinations of the chromatin-modifying enzyme modulators significantly attenuate glioblastoma cell proliferation and viability while exerting minimal effect on normal adult stem cells growth. Tumor Biol. 2015, 36, 9067–9072. [Google Scholar] [CrossRef]
- De La Rosa, J.; Urdiciain, A.; Zazpe, I.; Zelaya, M.V.; Meléndez, B.; Rey, J.A.; Idoate, M.A.; Castresana, J.S. The synergistic effect of DZ-NEP, panobinostat and temozolomide reduces clonogenicity and induces apoptosis in glioblastoma cells. Int. J. Oncol. 2020, 56, 283–300. [Google Scholar] [CrossRef]
- Jin, X.; Kim, L.J.Y.; Wu, Q.; Wallace, L.C.; Prager, B.C.; Sanvoranart, T.; Gimple, R.C.; Wang, X.; Mack, S.C.; Miller, T.E.; et al. Targeting glioma stem cells through combined BMI1 and EZH2 inhibition. Nat. Med. 2017, 23, 1352–1361. [Google Scholar] [CrossRef]
- Wiese, M.; Schill, F.; Sturm, D.; Pfister, S.; Hulleman, E.; Johnsen, S.A.; Kramm, C.M. No Significant Cytotoxic Effect of the EZH2 Inhibitor Tazemetostat (EPZ-6438) on Pediatric Glioma Cells with Wildtype Histone 3 or Mutated Histone 3.3. Klin. Padiatr. 2016, 228, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Grinshtein, N.; Rioseco, C.C.; Marcellus, R.; Uehling, D.; Aman, A.; Lun, X.Q.; Muto, O.; Podmore, L.; Lever, J.; Shen, Y.Q.; et al. Small molecule epigenetic screen identifies novel EZH2 and HDAC inhibitors that target glioblastoma brain tumor-initiating cells. Oncotarget 2016, 7, 59360–59376. [Google Scholar] [CrossRef]
- Duncan, R.M.; Reyes, L.; Moats, K.; Robinson, R.M.; Murphy, S.A.; Kaur, B.; Stessman, H.A.F.; Dolloff, N.G. ATF3 Coordinates Antitumor Synergy between Epigenetic Drugs and Protein Disulfide Isomerase Inhibitors. Cancer Res. 2020, 80, 3279–3291. [Google Scholar] [CrossRef]
- Singh, M.M.; Johnson, B.; Venkatarayan, A.; Flores, E.R.; Zhang, J.; Su, X.; Barton, M.; Lang, F.; Chandra, J. Preclinical activity of combined HDAC and KDM1A inhibition in glioblastoma. Neuro Oncol. 2015, 17, 1463–1473. [Google Scholar] [CrossRef]
- Tung, B.; Ma, D.; Wang, S.; Oyinlade, O.; Laterra, J.; Ying, M.; Lv, S.Q.; Wei, S.; Xia, S. Krüppel-like factor 9 and histone deacetylase inhibitors synergistically induce cell death in glioblastoma stem-like cells. BMC Cancer 2018, 18, 1025. [Google Scholar] [CrossRef]
- Foster, K.A.; Jane, E.P.; Premkumar, D.R.; Morales, A.; Pollack, I.F. Co-administration of ABT-737 and SAHA induces apoptosis, mediated by Noxa upregulation, Bax activation and mitochondrial dysfunction in PTEN-intact malignant human glioma cell lines. J. Neuro-Oncol. 2014, 120, 459–472. [Google Scholar] [CrossRef]
- Jung, J.; Kim, L.J.Y.; Wang, X.; Wu, Q.; Sanvoranart, T.; Hubert, C.G.; Prager, B.C.; Wallace, L.C.; Jin, X.; Mack, S.C.; et al. Nicotinamide metabolism regulates glioblastoma stem cell maintenance. JCI Insight 2017, 2, e90019. [Google Scholar] [CrossRef]
- Yu, D.; Khan, O.F.; Suvá, M.L.; Dong, B.Q.; Panek, W.K.; Xiao, T.; Wu, M.J.; Han, Y.; Ahmed, A.U.; Balyasnikova, I.V.; et al. Multiplexed RNAi therapy against brain tumor-initiating cells via lipopolymeric nanoparticle infusion delays glioblastoma progression. Proc. Natl. Acad. Sci. USA 2017, 114, E6147–E6156. [Google Scholar] [CrossRef]
- Saw, P.E.; Zhang, A.; Nie, Y.; Zhang, L.; Xu, Y.; Xu, X. Tumor-Associated Fibronectin Targeted Liposomal Nanoplatform for Cyclophilin A siRNA Delivery and Targeted Malignant Glioblastoma Therapy. Front. Pharmacol. 2018, 9, 1194. [Google Scholar] [CrossRef]
- Li, Y.S.; Ren, Y.; Wang, Y.F.; Tan, Y.L.; Wang, Q.X.; Cai, J.Q.; Zhou, J.H.; Yang, C.; Zhao, K.; Yi, K.K.; et al. A Compound AC1Q3QWB Selectively Disrupts HOTAIR-Mediated Recruitment of PRC2 and Enhances Cancer Therapy of DZNep. Theranostics 2019, 9, 4608–4623. [Google Scholar] [CrossRef]
- Eyler, C.E.; Matsunaga, H.; Hovestadt, V.; Vantine, S.J.; van Galen, P.; Bernstein, B.E. Single-cell lineage analysis reveals genetic and epigenetic interplay in glioblastoma drug resistance. Genome Biol. 2020, 21, 174. [Google Scholar] [CrossRef] [PubMed]
- Allen, B.K.; Mehta, S.; Ember, S.W.J.; Schonbrunn, E.; Ayad, N.; Schürer, S.C. Large-Scale Computational Screening Identifies First in Class Multitarget Inhibitor of EGFR Kinase and BRD4. Sci. Rep. 2015, 5, 16924. [Google Scholar] [CrossRef] [PubMed]
- Romanelli, A.; Stazi, G.; Fioravanti, R.; Zwergel, C.; Di Bello, E.; Pomella, S.; Perrone, C.; Battistelli, C.; Strippoli, R.; Tripodi, M.; et al. Design of First-in-Class Dual EZH2/HDAC Inhibitor: Biochemical Activity and Biological Evaluation in Cancer Cells. ACS Med. Chem. Lett. 2020, 11, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Sui, A.X.; Xu, Y.B.; Li, Y.T.; Hu, Q.L.; Wang, Z.Y.; Zhang, H.T.; Yang, J.J.; Guo, X.Q.; Zhao, W.Q. The pharmacological role of histone demethylase JMJD3 inhibitor GSK-J4 on glioma cells. Oncotarget 2017, 8, 68591–68598. [Google Scholar] [CrossRef]
- Haque, F.; Varlet, P.; Puntonet, J.; Storer, L.; Bountali, A.; Rahman, R.; Grill, J.; Carcaboso, A.M.; Jones, C.; Layfield, R.; et al. Evaluation of a novel antibody to define histone 3.3 G34R mutant brain tumours. Acta Neuropathol. Commun. 2017, 5, 45. [Google Scholar] [CrossRef]
- Luo, D.; Fraga-Lauhirat, M.; Millings, J.; Ho, C.; Villarreal, E.M.; Fletchinger, T.C.; Bonfiglio, J.V.; Mata, L.; Nemesure, M.D.; Bartels, L.E.; et al. Phospho-valproic acid (MDC-1112) suppresses glioblastoma growth in preclinical models through the inhibition of STAT3 phosphorylation. Carcinogenesis 2019, 40, 1480–1491. [Google Scholar] [CrossRef]
- Zhu, K.; Tao, H.; Song, J.L.; Jin, L.; Zhang, Y.; Liu, J.; Chen, Z.; Jiang, C.S.; Luo, C.; Zhang, H. Identification of 5-benzylidene-2-phenylthiazolones as potent PRMT5 inhibitors by virtual screening, structural optimization and biological evaluations. Bioorg. Chem. 2018, 81, 289–298. [Google Scholar] [CrossRef]
- Flamier, A.; Abdouh, M.; Hamam, R.; Barabino, A.; Patel, N.; Gao, A.; Hanna, R.; Bernier, G. Off-target effect of the BMI1 inhibitor PTC596 drives epithelial-mesenchymal transition in glioblastoma multiforme. NPJ Precis. Oncol. 2020, 4, 1. [Google Scholar] [CrossRef]
- Liu, Y.; Song, X.; Wu, M.; Wu, J.; Liu, J. Synergistic Effects of Resveratrol and Temozolomide Against Glioblastoma Cells: Underlying Mechanism and Therapeutic Implications. Cancer Manag. Res. 2020, 12, 8341–8354. [Google Scholar] [CrossRef]
- Berthier, S.; Larrouquère, L.; Champelovier, P.; Col, E.; Lefebvre, C.; Cottet-Rouselle, C.; Arnaud, J.; Garrel, C.; Laporte, F.; Boutonnat, J.; et al. A New Patient-Derived Metastatic Glioblastoma Cell Line: Characterisation and Response to Sodium Selenite Anticancer Agent. Cancers 2018, 11, 12. [Google Scholar] [CrossRef]
- Bhere, D.; Khajuria, R.K.; Hendriks, W.T.; Bandyopadhyay, A.; Bagci-Onder, T.; Shah, K. Stem Cells Engineered During Different Stages of Reprogramming Reveal Varying Therapeutic Efficacies. Stem Cells 2018, 36, 932–942. [Google Scholar] [CrossRef]
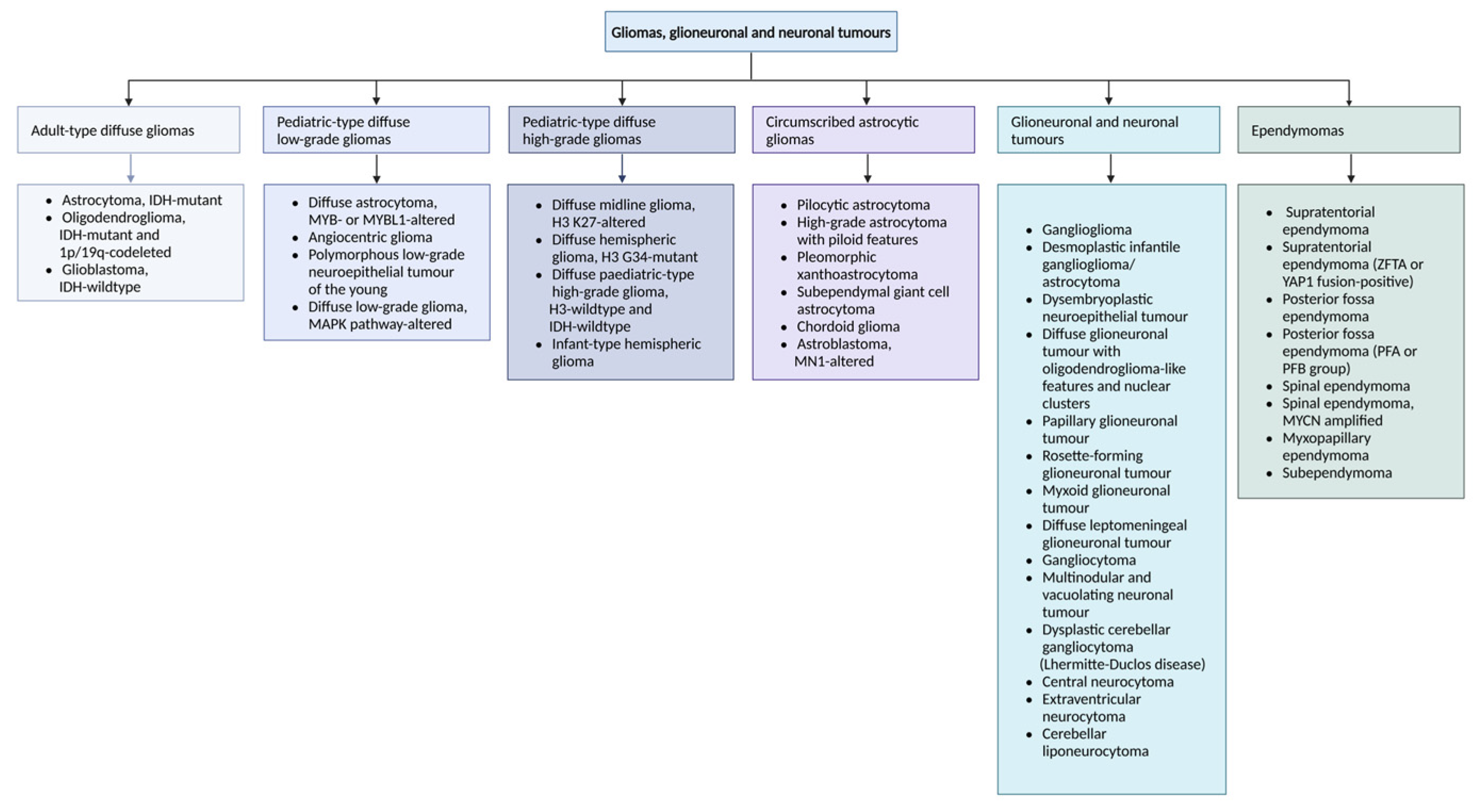
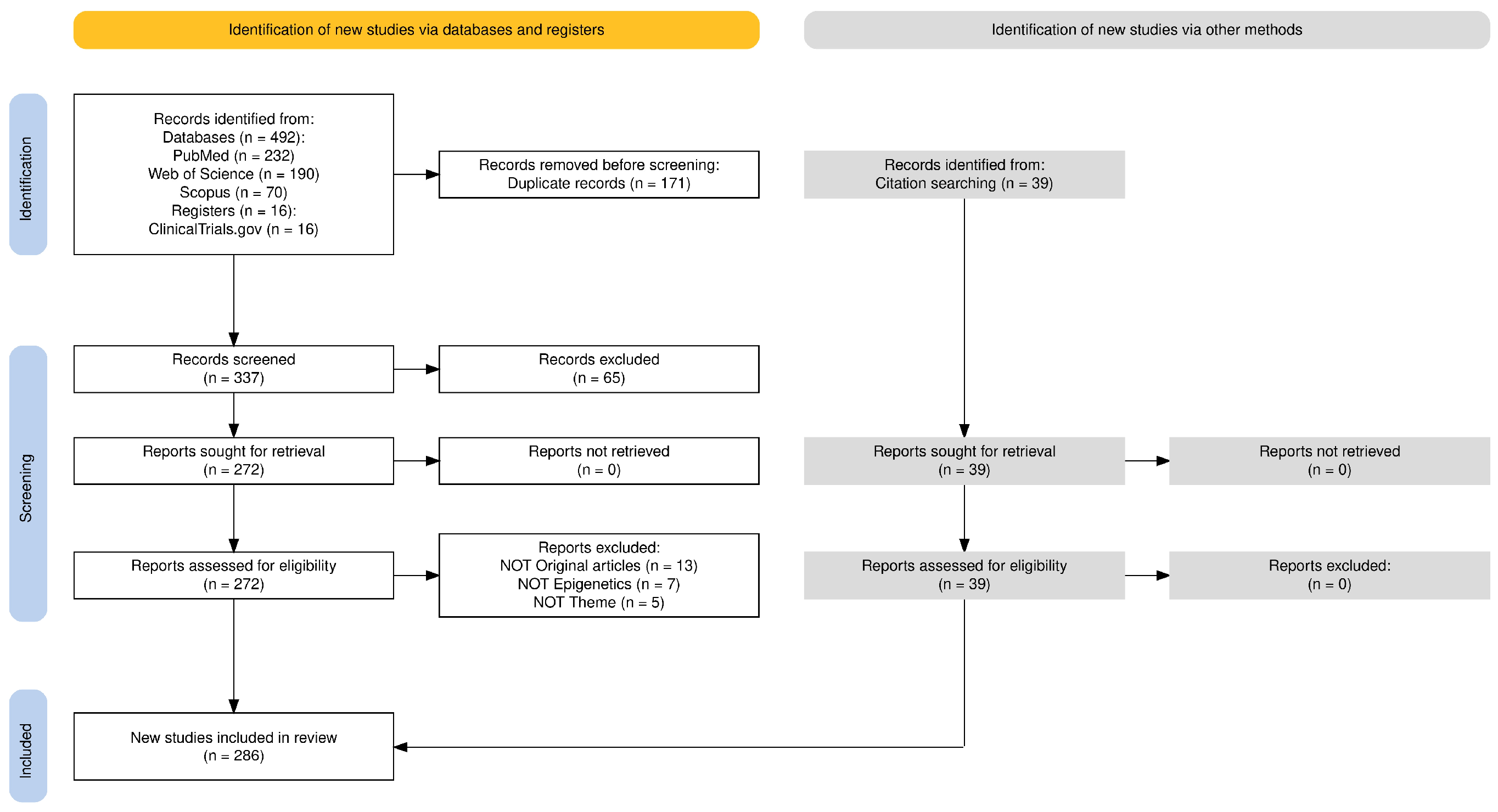
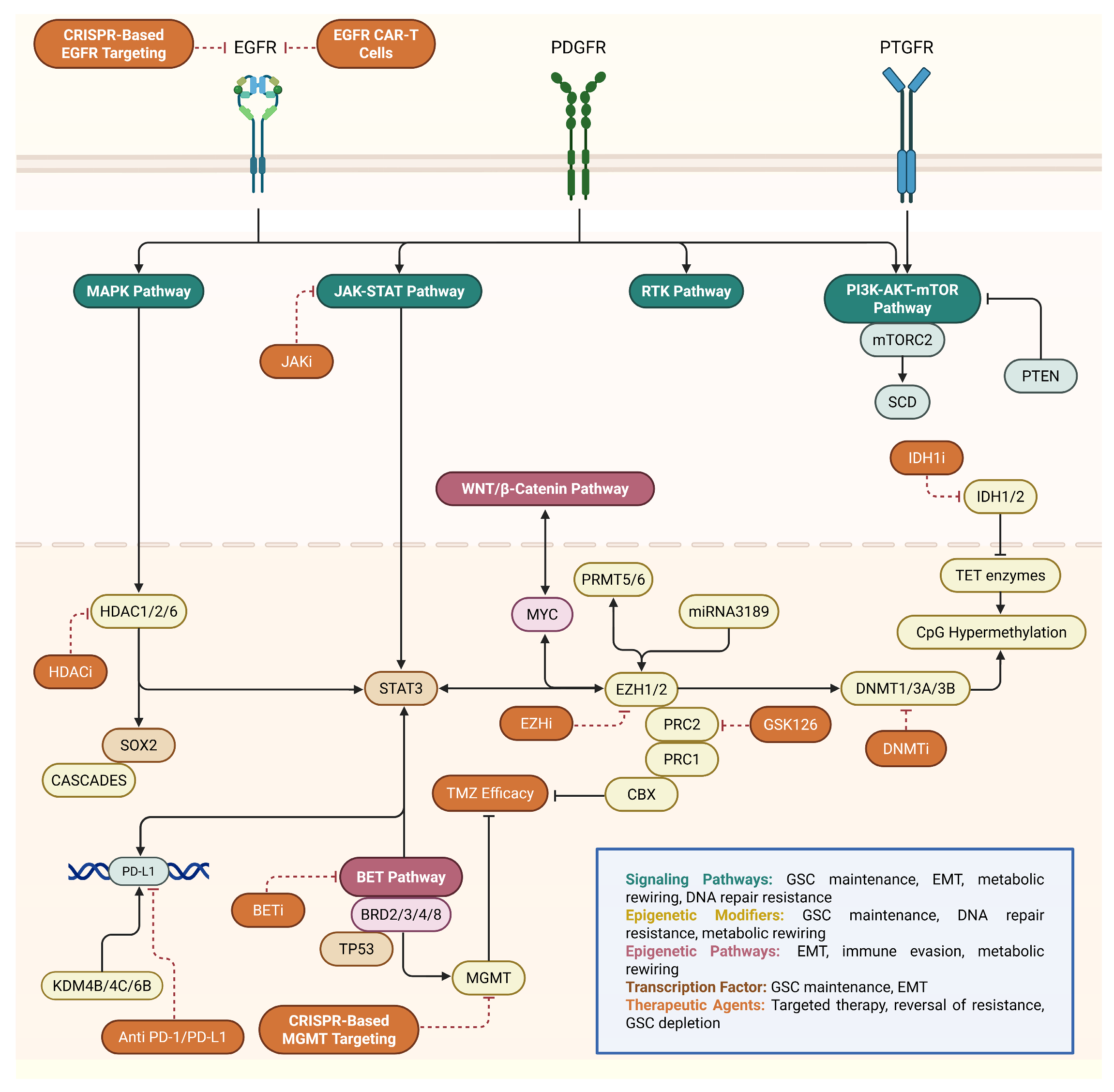
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meleiro, M.; Henrique, R. Epigenetic Alterations in Glioblastoma Multiforme as Novel Therapeutic Targets: A Scoping Review. Int. J. Mol. Sci. 2025, 26, 5634. https://doi.org/10.3390/ijms26125634
Meleiro M, Henrique R. Epigenetic Alterations in Glioblastoma Multiforme as Novel Therapeutic Targets: A Scoping Review. International Journal of Molecular Sciences. 2025; 26(12):5634. https://doi.org/10.3390/ijms26125634
Chicago/Turabian StyleMeleiro, Marco, and Rui Henrique. 2025. "Epigenetic Alterations in Glioblastoma Multiforme as Novel Therapeutic Targets: A Scoping Review" International Journal of Molecular Sciences 26, no. 12: 5634. https://doi.org/10.3390/ijms26125634
APA StyleMeleiro, M., & Henrique, R. (2025). Epigenetic Alterations in Glioblastoma Multiforme as Novel Therapeutic Targets: A Scoping Review. International Journal of Molecular Sciences, 26(12), 5634. https://doi.org/10.3390/ijms26125634









