N6-Methyladenosine Modification of the Three Components “Writers”, “Erasers”, and “Readers” in Relation to Osteogenesis
Abstract
1. Introduction
2. Methods
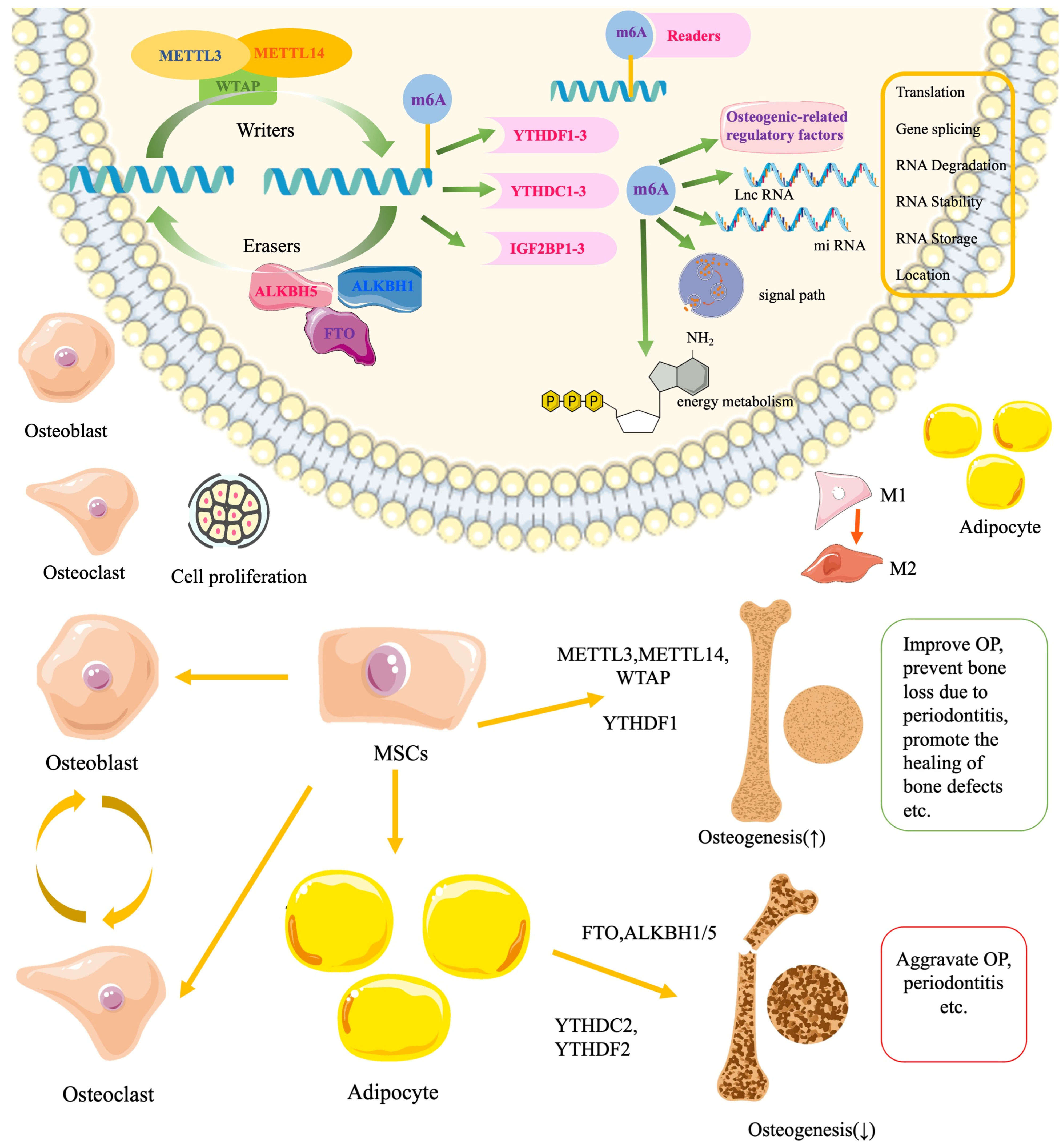
2.1. The Relation of m6A to Osteogenesis and Bone Diseases
2.2. The Role of m6A “Writers” in Osteogenesis
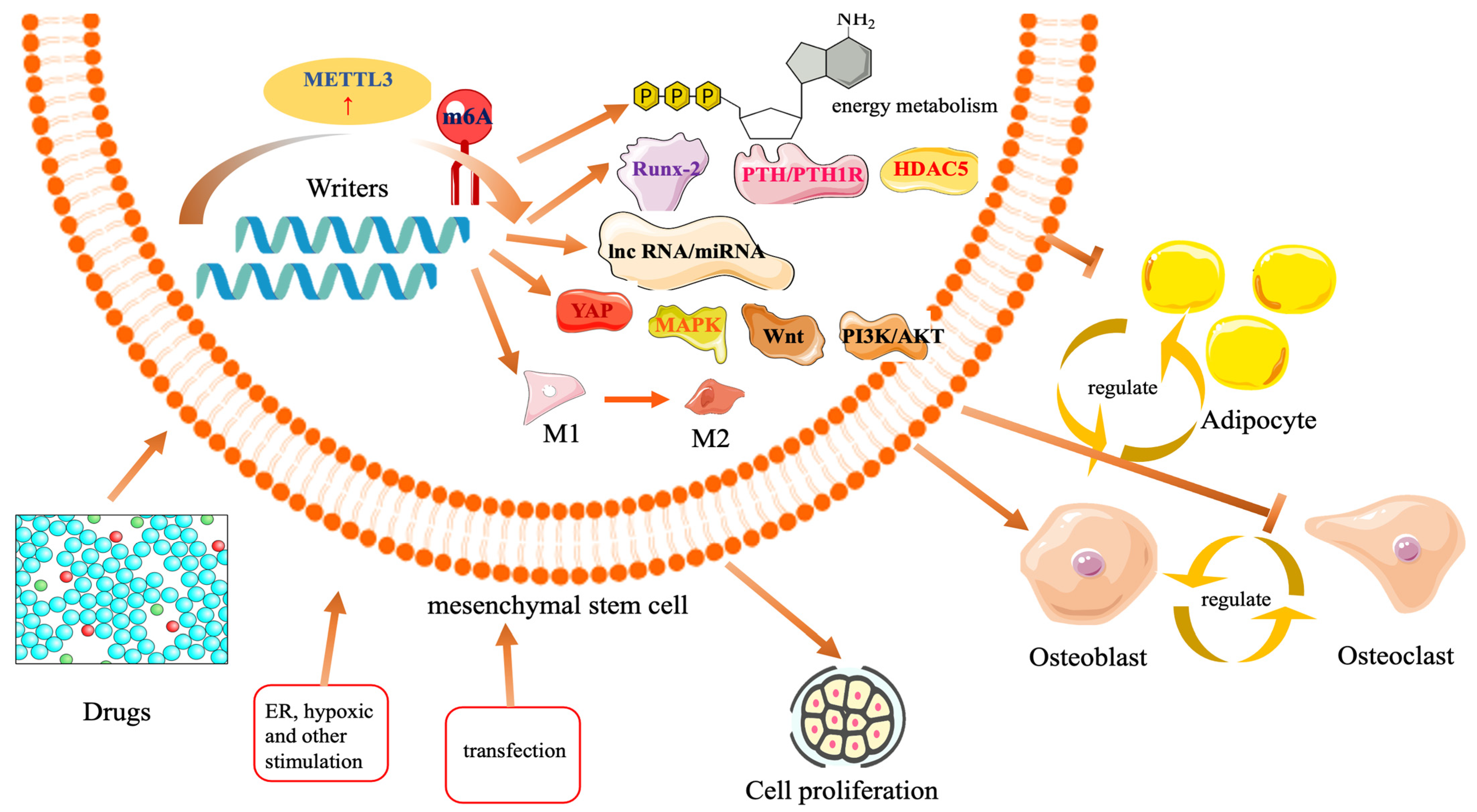
2.2.1. The Role of METTL3 in Osteogenesis
2.2.2. Osteogenic Role of METTL3 in the Microenvironment
2.2.3. Effects of METTL3 Methylation-Related Drugs on Osteogenesis
2.2.4. Negative Role of METTL3 Methylation Modification in Disease
2.3. The Role of METTL14 in Osteogenesis
2.3.1. The Role of WTAP in Osteogenesis
2.3.2. Osteogenic Role of WTAP in the Microenvironment
2.4. Other Methyl Transfer-like Enzymes Related to Osteogenesis Reports
| Regulators | Cells and Species | Research Background | Regulators’ Biological Functions and Involved Signaling Pathways | Ref. |
|---|---|---|---|---|
| METTL3 | 54 to 65 years OP female, C57BL/6 mice BMSCs, OVX C57BL/6 mice OP model | OP | Human OP bone samples: METTL3 (↓), m6A (↓), METTL3 (↑) → m6A methylation of stability (level) (↑) → osteogenesis (↑) → OP (↓); METTL3 (↑) → m6A methylation of pre-miR-320 (↑) → miR-320 (↓) → Runx-2 stability (level) (↑) → osteogenesis (↑) → OP (↓) | [22] |
| METTL3 | SD male rats BMSCs | Skull defects | β-TCP → METTL3 (↑) → m6A methylation of Runx-2 mRNA stability (level) (↑) | [23] |
| METTL3 | BMSCs | Maxillofacial bone defects | METTL3 (↑) → m6A methylation of Runx-2 mRNA stability (level) (↑) | [24] |
| METTL3 | Mouse BMSCs | OP caused by estrogen deficiency | METTL3 (↑) → m6A methylation of PTH/PTH1R (↑) → osteogenesis (↑), lipogenesis (↓) | [25] |
| METTL3 | Mouse Macrophage Cell line RAW 264.7, C57BL/6 mouse BMSCs | Bone fracture healing (migration and differentiation) | Macrophage cell line RAW 264.7: METTL3 (↑) → m6A methylation of HDAC5 (↑); BMSCs: METTL3 (↑) → m6A methylation of Runx-2 mRNA stability (level) (↑) | [26] |
| METTL3 | SD male rats BMSCs | OP | METTL3 (↓) → m6A methylation (↓) → Akt (↓) → VEGF-a, Runx-2, and osterix (↓) | [27] |
| METTL3 | OP-BMSCs, OVX SD rat OP model | OP | METTL3 (↑) → m6A methylation (↑) → Wnt (↑) → osteogenesis (↑) | [28] |
| METTL3 | Human BMSCs | OP | METTL3 (↑) → m6A methylation of LINC00657 (↑) → miR-144-3p (↓) → BMPR1B (↑) → osteogenesis (↑) | [47] |
| METTL3 | Female C57BL/6J mouse BMSCs | OP | METTL3 (↑) → m6A methylation of lncRNA MIR99AHG → lncRNA MIR99AHG (↓) → miR-4660 (↑) → osteogenesis (↑) | [48] |
| METTL3 | Human PDLSCs | Osteogenic differentiation | METTL3 (↑) → m6A methylation (↑) → proliferation, osteogenic differentiation, migration (↑) | [29,30] |
| METTL3 | Human PDLSCs | Osteogenic differentiation | METTL3 (↑) → m6A methylation (↑) → IGF2BP1(↑) → YAP mRNA stability (level) (↑) → osteogenesis (↑) | [31] |
| METTL3 | Human PDLSCs | Osteogenic differentiation | METTL3 (↑) → m6A methylation (↑) → IGF2BP1 (↑) → Runx-2 mRNA stability (level) (↑) → osteogenesis (↑) | [32] |
| METTL3 | Human DPSCs | Osteogenic/odontogenic differentiation | METTL3 (↑) → m6A methylation of lncSNHG7 → Wnt/β-catenin (↑) → osteogenesis (↑) | [33] |
| METTL3 | Human DPSCs | Osteogenic differentiation | METTL3 (↑) → m6A methylation of ACLY and SLC25A1(↑) → osteogenesis and proliferation (↑); IGF2BP2 and IGF2BP2/3 (↑) → ACLY and SLC25A1 stability (↑) | [34] |
| METTL3 | HUC-MSCs | Osteogenic differentiation | METTL3 (↑) → m6A methylation of circCTTN (↑) → osteogenesis (↑) | [35] |
| METTL3 | Human ADSCs | Osteogenic differentiation | METTL3 (↑) → m6A methylation of lncRNA RP11-44 N12.5 (↑) → MAPK (↑) → osteogenesis (↑) | [19] |
| METTL3 | Human PDLSCs | Periodontitis | Periodontitis → METTL3 (↑), METTL3 (↓) → proinflammatory factor, osteogenesis (↓) and PI3K/Akt (↓) | [55] |
| METTL3 | Human PDLSCs | Periodontitis | FOXO1 → METTL3 (↑) → m6A methylation of PI3K/AKT → osteogenesis (↑) | [56] |
| METTL3 | Human PDLSCs | Periodontitis | LPS → METTL3/14 (↑) → m6A methylation of SLC39A9 → SLC39A9 (↓) → zinc (↓) → IL-6 (↑) | [57] |
| METTL3 | Human PDLSCs | Periodontitis | METTL3 (↑) → m6A methylation of lncRNA CUTALP (↑) → miR-30b-3p (↓) → Runx2 (↑) | [58] |
| METTL3 | Human PDLSCs | Periodontitis | METTL3 (↑) → m6A methylation of IncRNA 4114 (↑) → osteogenesis (↑) | [59] |
| METTL3 | MC3T3-E1 | LPS-induced inflammation | METTL3 (↓) → Smad7 and Smurf1 (↑) → osteogenesis (↓), METTL3 (↑) → MAPK (↑) → ERK, p38, JNK, and p65 phosphorylation | [85] |
| METTL3 | MC3T3-E1, Male C57BL/6 mice | Periodontitis | METTL3 (↑) → Wnt/β-catenin/c-Myc → Ribosome and mitochondrial function (↑), METTL3 (↓) → periodontitis was aggravated in mice | [60] |
| METTL3 | MC3T3-E1 | ER Stress | METTL3 (↓) → YTHDF2-mediated Grp78 mRNA degradation → apoptosis and differentiation of MC3T3-E1 cells were impaired | [87] |
| METTL3 | Human BMSCs | Osteomyelitis | METTL3 (↑) → m6A methylation of pri-miR-320a → miR-320a (↓) → PIK3CA (↑), regulates osteogenesis, oxidative stress, inflammation | [64] |
| METTL14 | Human BMSCs | OP | METTL3 (↑) → β-catenin (↑) → TCF1 (↑) → Runx-2 (↑) → osteogenesis (↑) | [49] |
| METTL14 | Human BMSCs, OVX-C57BL/6 mice OP model | OP | METTL14 (↓) → Smad1 protein level (↓) → osteogenesis (↓); METTL14 (↑) → C57BL/6 mice having increased bone mass | [50] |
| METTL14 | Human BMSCs, OVX SD rat OP model | OP | ICA → METTL14 (↑) → P4HB mRNA stability (level) (↑) → osteogenesis (↑), restores bone mass in OP rats | [51] |
| METTL14 | C57BL/6 mice-BMSCs, OVX C57BL/6 mice OP model | OP | METTL3 (↑) → m6A methylation of beclin-1, IGF2BPs maintain beclin-1 stability → osteogenesis (↑), prevent the development of OP, and inhibit osteoclast differentiation | [52] |
| METTL14 | Mouse BMSCs, BMMs, OVX C57BL/6 mice OP model | OP | METTL14 (↑) → m6A methylation of SIRT1 → SIRT1 mRNA stability (level) (↑) → regulate the osteogenesis of mouse BMSCs and inhibit osteoclast differentiation of BMMs, alleviating the progression of OP in mice | [53] |
| METTL14 | Human patients BMSCs | Steroid-associated osteonecrosis of the femoral head | METTL14 (↑) → m6A methylation of PTPN6 → PTPN6 mRNA stability (level) (↑) → PTPN6 combine with GSK-3β → Wnt activation → BMSC proliferation and osteogenesis (↑) | [65] |
| METTL14 | Human PDLSCs | Biomechanics | METTL14 → m6A methylation of IGF1 → IGF1 (level) (↑) → osteogenesis (↑) | [98] |
| METTL14 | Human healthy and patients BMSCs | Ankylosing spondylitis | TNF-α → METTL14 → m6A methylation of ELMO1 → BMSC migration (↑), exacerbating the progression of the disease | [67] |
| WTAP | Mice BMSCs | OP | WTAP → m6A methylation of pri-miR-181a and pri-miR-181c → miR-181a and miR-181c (↑) → SFRP1 (↓) → osteogenesis (↑) | [44] |
| WTAP | Mice BMSCs | OP | WTAP → m6A methylation of miR-29b-3p → HDAC4 (↓) → osteogenesis (↑), adipogenesis (↓) | [45] |
| WTAP | Ligamentum Flavum | Ossification of the ligamentum flavum | WTAP → m6A methylation of circCDK14 → IGF2BP3 regulates circCDK14 stability → miR-93-5p (↓) → AFF4 (↑) → osteogenesis (↑) | [68] |
| WTAP | Human SCAPs | Osteogenic differentiation | WTAP → Runx-2 stability → osteogenesis (↑) | [101] |
| WTAP | Mice BMSCs and BMMs C57BL/6 mice | Periodontitis | LPS → WTAP (↑) WTAP (↓) → promotes M1 transformation to M2, osteogenesis (↑); WTAP (↓) → decreased bone loss in mice with periodontitis | [46] |
| METTL5 | SuMSCs | Osteogenic differentiation | METTL5 → Wnt → promotes skull and suture development | [102] |
| METTL7a | Human BMSCs | Bone damage from bisphosphonates | METTL7a → m6A methylation → saves bone damage from bisphosphonates | [66] |
| METTL3 | Human BMSCs | Osteogenic differentiation | METTL3 → m6A methylation of BMP2 → BMP2 mRNA degradation | [103] |
| METTL3 | MC3TCE-1 | Osteogenic differentiation and fracture healing | METTL3 → m6A methylation of miR-7212-5p → osteogenesis (↓) | [62] |
| METTL3 | Menstrual blood MSCs | Osteogenic differentiation | METTL3 → m6A methylation of MyD88 → MyD88 (↑) → NF-κB activate osteogenesis (↓) | [104] |
| METTL14 | Human BMSCs | Osteogenic differentiation | METTL14 → m6A methylation of miR-873 → osteogenesis and proliferation (↓) | [105] |
| m6A | Human ADSCs | Osteogenic differentiation | The total level of m6A decreased with the increase in osteogenic differentiation of ADSCs | [106] |
| METTL3 | Human healthy and peri-implantitis patients | Peri-implantitis | High METTL3 m6A methylation levels were detected in peri-implantitis | [107] |
| METTL3 | Human gingival fibroblasts, male C57BL/6J mice | Periodontitis | METTL3 (↑) → m6A methylation of TNFAIP3 → TNFAIP3 (↓) → decreases the ubiquitination of NEK7 → NLRP3 (↑) | [61] |
2.5. The Role of m6A “Erasers” in Osteogenesis
2.5.1. The Role of FTO in Osteogenesis and the Microenvironment
2.5.2. The Role of ALKBH1 and ALKBH5 in Osteogenesis and Microenvironment
| Regulators | Cells and Species | Research Background | Regulators’ Biological Functions and Involved Signaling Pathways | Ref. |
|---|---|---|---|---|
| FTO | Human BMSCs | Osteogenic differentiation | FTO (↑) → m6A methylation of Runx-2 (↓) → Runx-2 (↓) → osteogenesis (↓) | [39] |
| FTO | Human BMSCs | OP | FTO (↑) → YTHDF1 → PPARG (↓) → osteogenesis (↑) | [113] |
| FTO | MC3T3-E1 | OP | FTO (↑) → m6A methylation of PDIA3 (↓) → PDIA3 (↑) → osteogenesis (↑), PDIA3 → USP phosphorylation (↑) | [114] |
| FTO | Human DPSCs | Osteogenic differentiation | FTO (↑) → miR-7974 stability (↓) → FKBP15 (↑) → osteogenesis (↓) | [40] |
| FTO | Osteoclast precursor cells, C57BL/6 mice | Periodontitis | FTO (↑) → YTHDF2 dependency mode → CDK2 stability (↑) → promotes the proliferation of osteoclasts and inhibits their apoptosis; FTO (↓) → inhibits bone loss in periodontitis | [115] |
| FTO | C57BL/6 mice BMSCs, C57BL/6 mice | Diabetes mellitus combined with periodontitis | AGEs damage bone formation in an FTO demethylation-dependent manner | [41] |
| ALKBH5 | Mice BMSCs | Osteogenic differentiation | ALKBH5 → m6A methylation of PRMT6 (↓) → PRMT6 (↓) → PI3K/AKT (↓) → osteogenesis (↓) | [119] |
| ALKBH5 | Human BMSCs | OP | ALKBH5 → m6A methylation of VDAC3 (↓) → osteogenesis (↓) and aggravates OP | [42] |
| ALKBH5 | Male embryonic rat-osteoblasts | Osteogenic differentiation | ALKBH5 (↓) → Runx-2 stability (↓) → osteogenesis (↓) | [120] |
| ALKBH5 | Human ADSCs, nude mice mandibular defect model | Osteogenic differentiation | ALKBH5 (↓) → m6A methylation of lnc-AK311120 (↑) → lnc-AK311120 (↓) → osteogenesis (↓) | [121] |
| ALKBH5 | Male C57BL/6J mice | Type II diabetes and peri-implantitis | ALKBH5 (↑) in type II diabetes and peri-implantitis | [122] |
| ALKBH1 | Mice BMSCs, mice | OP | ALKBH1 (↓) → adipogenesis (↑), osteogenesis (↓), loses bone mass and increases fat content | [123] |
| ALKBH1 | Vascular Smooth Muscle Cell | Chronic nephrosis | ALKBH1 → promotes Oct4 combination with BMP2 → angiostenosis | [69] |
2.6. The Role of “Readers” in Osteogenesis
2.6.1. The Role of YTHDF in Osteogenesis
2.6.2. Osteogenic Role of IGF2BP2 in the Microenvironment
| Regulators | Cells and Species | Research Background | Regulators’ Biological Functions and Involved Signaling Pathways | Ref. |
|---|---|---|---|---|
| YTHDF1 | Human BMSCs | Osteogenic differentiation | YTHDF1 → m6A methylation of ZNF839 → ZNF839 interacts with Runx-2 → osteogenesis (↑) | [36] |
| YTHDF1 | Male SD rat BMSCs | Osteogenic differentiation | YTHDF1 → autophagy and β-catenin → proliferation and osteogenesis (↑) | [37] |
| YTHDF1 | MC3T3-E1 | Osteogenesis under hypoxic conditions | YTHDF1 → m6A methylation of THBS1 → THBS stability (↑) → osteogenesis (↑) | [38] |
| YTHDC1 | Mice osteoclasts | OP | YTHDC1 → PTPN6 mRNA stability (↑) → osteoclasis (↓), regulates the process of OP | [54] |
| YTHDC2 | Rat BMSCs | Osteogenic differentiation | YTHDC2 → Runx-2 (↓) → osteogenesis (↓) | [134] |
| YTHDF2 | HUC-MSCs | Osteogenic differentiation | miR-615-3p → YTHDF2 → m6A methylation of FBLN1 (↓) → FBLN1 (↓) → osteogenesis (↓) | [43] |
| YTHDF2 | Human BMSCs | SONFH | miR-27a → YTHDF2 (↓) → alleviates SONFH | [135] |
| YTHDF3 | Human BMSCs | OP | YTHDF3 level (↓) in OP | [136] |
| YTHDC/YTHDF | Orofacial MSCs | Periodontal bone defects | Quercetin → YTHDC/YTHDF → Period1 (↓) → osteogenesis (↑) | [137] |
| IGF2BP1 | HGFs | Periodontitis | IGF2BP, IL-17 (↑) in periodontitis | [138] |
| IGF2BP2 | Mice BMSCs Mice | Periodontitis | IGF2BP2 controls early periodontitis development, exacerbates advanced periodontitis | [139] |
| IGF2BP2 | Ortica valve interstitial cells | Calcific aortic valve disease | IGF2BP2 interact with circHIPK3 → Kremen1 (↑) → Wnt (↓) | [70] |
| IGF2BP3 | MC3T3-E1 | Fracture healing | IGF2BP3 → m6A methylation of miR-23a-3p → Smad5 (↓) → osteogenesis (↓) → delayed fracture healing | [63] |
3. Limitations and Perspectives
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| m6A | N6-methyl adenosine |
| m1A | N1-methyladenosine |
| m5C | 5-methylcytosine |
| m7G | 7-methylguanine |
| METTL3 | methyltransferase-like protein 3 |
| METTL14 | methyltransferase-like protein 14 |
| WTAP | Wilms’ tumor 1-associated protein |
| FTO | fat mass and obesity-associated protein |
| ALKBH5 | ALKB homolog 5 |
| YTH | YT521-B homology |
| YTHDF1–3 | YTH domain family 1–3 |
| YTHDC1–2 | YTH domain-containing proteins 1–2 |
| IGF2BPs | insulin-like growth factor 2 mRNA-binding proteins |
| METTL16 | methyltransferase-like protein 16 |
| BMSCs | bone marrow mesenchymal stem cells |
| Runx-2 | runt-related transcription factor-2 |
| OP | osteoporosis |
| OVX | ovariectomy |
| PTH | parathyroid hormone |
| PTH1R | parathyroid hormone 1 receptor |
| HDAC5 | histone deacetylase 5 |
| SD | Sprague Dawley |
| VEGF | vascular endothelial growth factor |
| ALP | alkaline phosphatase |
| AKT | protein kinase B |
| LncRNAs | long non-coding RNAs |
| PDLSCs | periodontal ligament stem cells |
| YAP | Yes-associated protein |
| eIF3a | eukaryotic translation initiation factor 3a |
| DPSCs | dental pulp stem cells |
| ACLY | ATP citrate lyase |
| NOP2 | nucleolar protein 2 |
| HUC-MSCs | human umbilical cord mesenchymal stem cells |
| MAPK | mitogen-activated protein kinase |
| ADSCs | adipose-derived mesenchymal stem cells |
| PI3K | phosphatidylinositol 3-kinase |
| FOXO1 | forkhead box protein O1 |
| LPS | lipopolysaccharide |
| SLC39A9 | solute carrier family 39 member 9 |
| JNK | c-Jun N-terminal kinase |
| MC3T3-E1 | mouse embryo osteoblast precursor cells |
| ERK | extracellular regulated protein kinase |
| ER | endoplasmic reticulum |
| Grp78 | glucose-regulated protein 78 |
| UPR | unfolded protein response |
| PIK3CA | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| OM | osteomyelitis |
| MTX | methotrexate |
| SAM | S-adenosylmethionine |
| HIF-1α | hypoxia-inducible factor-1α |
| SLC25A1 | mitochondrial citrate transporter |
| HOXD8 | homeobox D8 |
| ITGA5 | integrin alpha 5 |
| AVMSCs | aortic valve mesenchymal cells |
| TWIST1 | twist family bHLH transcription factor 1 |
| USP8 | ubiquitin carboxyl-terminal hydrolase 8 |
| ICA | icariin |
| TCF1 | transcription factor 1 |
| P4HB | prolyl 4-hydroxylase subunit beta |
| SIRT1 | sirtuin 1 |
| BMMs | bone marrow mononuclear macrophages |
| PTPN6 | protein tyrosine phosphatase non-receptor type 6 |
| SONFH | steroid-associated osteonecrosis of the femoral head |
| IGF1 | insulin-like growth factor 1 |
| TNF-α | tumor necrosis factor-α |
| AS | ankylosing spondylitis |
| HDAC4 | histone deacetylase 4 |
| AFF4 | ALF transcription elongation factor 4 |
| SCAPs | stem cells from apical papilla |
| SuMSCs | suture mesenchyme stem cells |
| ALKBH9 | ALKB homolog 9 |
| PVT1 | plasmacytoma variant translocation 1 |
| FKBP15 | FK506-binding protein 15 |
| PPARG | peroxisome proliferative-activated receptor |
| PDIA3 | protein disulfide isomerase family A member 3 |
| USP20 | ubiquitin-specific peptidase 20 |
| OPN | osteopontin |
| DM | diabetes mellitus |
| AGEs | advanced glycation end products |
| PRMT6 | protein arginine N-methyltransferase 6 |
| Oct4 | octamer binding transcription factor 4 |
| BMP2 | bone morphogenetic protein 2 |
| VSMCs | vascular smooth muscle cells |
| hnRNPs | heterogeneous nuclear ribonucleoproteins |
| FMRP | fragile X mental retardation protein |
| Prrc2a | proline-rich coiled-coil 2A |
| LRPPRC | leucine-rich pentatricopeptide repeat-containing protein |
| FBLN1 | fibulin-1 |
| PMOP | postmenopausal osteoporosis |
| OMSCs | orofacial mesenchymal stem cells |
| HGF | human gingival fibroblasts |
| AVICs | aortic valve interstitial cells |
| CAVD | calcific aortic valve disease |
| MenSCs | mesenchymal stem cells |
| TREM-1 | triggering receptor expressed on myeloid cells-1 |
| IL | interleukin |
| NLRP3 | NLR family pyrin domain-containing protein 3 |
References
- Zaidi, M. Skeletal remodeling in health and disease. Nat. Med. 2007, 13, 791–801. [Google Scholar] [CrossRef]
- Huang, M.; Guo, J.; Liu, L.; Jin, H.; Chen, X.; Zou, J. m6A demethylase FTO and osteoporosis: Potential therapeutic interventions. Front. Cell Dev. Biol. 2023, 11, 1275475. [Google Scholar] [CrossRef] [PubMed]
- Siris, E.S.; Adler, R.; Bilezikian, J.; Bolognese, M.; Dawson-Hughes, B.; Favus, M.J.; Harris, S.T.; Jan de Beur, S.M.; Khosla, S.; Lane, N.E.; et al. The clinical diagnosis of osteoporosis: A position statement from the National Bone Health Alliance Working Group. Osteoporos. Int. 2014, 25, 1439–1443. [Google Scholar] [CrossRef] [PubMed]
- De Martinis, M.; Sirufo, M.M.; Polsinelli, M.; Placidi, G.; Di Silvestre, D.; Ginaldi, L. Gender Differences in Osteoporosis: A Single-Center Observational Study. World J. Men’s Health 2021, 39, 750–759. [Google Scholar] [CrossRef]
- Eastell, R.; O’Neill, T.W.; Hofbauer, L.C.; Langdahl, B.; Reid, I.R.; Gold, D.T.; Cummings, S.R. Postmenopausal osteoporosis. Nat. Rev. Dis. Primers 2016, 2, 16069. [Google Scholar] [CrossRef]
- Ebeling, P.R.; Nguyen, H.H.; Aleksova, J.; Vincent, A.J.; Wong, P.; Milat, F. Secondary Osteoporosis. Endocr. Rev. 2022, 43, 240–313. [Google Scholar] [CrossRef]
- Dai, Z.; Ramesh, V.; Locasale, J.W. The evolving metabolic landscape of chromatin biology and epigenetics. Nat. Rev. Genet. 2020, 21, 737–753. [Google Scholar] [CrossRef]
- Luo, M. Chemical and Biochemical Perspectives of Protein Lysine Methylation. Chem. Rev. 2018, 118, 6656–6705. [Google Scholar] [CrossRef]
- Zhou, H.; Rauch, S.; Dai, Q.; Cui, X.; Zhang, Z.; Nachtergaele, S.; Sepich, C.; He, C.; Dickinson, B.C. Evolution of a reverse transcriptase to map N1-methyladenosine in human messenger RNA. Nat. Methods 2019, 16, 1281–1288. [Google Scholar] [CrossRef]
- Blanco, S.; Bandiera, R.; Popis, M.; Hussain, S.; Lombard, P.; Aleksic, J.; Sajini, A.; Tanna, H.; Cortés-Garrido, R.; Gkatza, N.; et al. Stem cell function and stress response are controlled by protein synthesis. Nature 2016, 534, 335–340. [Google Scholar] [CrossRef]
- Zhang, L.S.; Liu, C.; Ma, H.; Dai, Q.; Sun, H.L.; Luo, G.; Zhang, Z.; Zhang, L.; Hu, L.; Dong, X.; et al. Transcriptome-wide Mapping of Internal N7-Methylguanosine Methylome in Mammalian mRNA. Mol. Cell 2019, 74, 1304–1316.e8. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, Q.; Kaboli, P.J.; Shen, J.; Li, M.; Wu, X.; Yin, J.; Zhang, H.; Wu, Y.; Lin, L.; et al. m1A Regulated Genes Modulate PI3K/AKT/mTOR and ErbB Pathways in Gastrointestinal Cancer. Transl. Oncol. 2019, 12, 1323–1333. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Song, J.; Yuan, W.; Zhang, W.; Sun, Z. Roles of RNA Methylation on Tumor Immunity and Clinical Implications. Front. Immunol. 2021, 12, 641507. [Google Scholar] [CrossRef]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef]
- Liu, N.; Pan, T. N6-methyladenosine–encoded epitranscriptomics. Nat. Struct. Mol. Biol. 2016, 23, 98–102. [Google Scholar] [CrossRef]
- Shi, H.; Wei, J.; He, C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef]
- Bi, Z.; Liu, Y.; Zhao, Y.; Yao, Y.; Wu, R.; Liu, Q.; Wang, Y.; Wang, X. A dynamic reversible RNA N6—Methyladenosine modification: Current status and perspectives. J. Cell. Physiol. 2019, 234, 7948–7956. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.S.; Yang, Y.G. Dynamic transcriptomic m6A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef]
- Song, Y.; Pan, Y.; Wu, M.; Sun, W.; Luo, L.; Zhao, Z.; Liu, J. METTL3-Mediated lncRNA m6A Modification in the Osteogenic Differentiation of Human Adipose-Derived Stem Cells Induced by NEL-Like 1 Protein. Stem Cell Rev. Rep. 2021, 17, 2276–2290. [Google Scholar] [CrossRef]
- Gu, J.; Zhan, Y.; Zhuo, L.; Zhang, Q.; Li, G.; Li, Q.; Qi, S.; Zhu, J.; Lv, Q.; Shen, Y.; et al. Biological functions of m6A methyltransferases. Cell Biosci. 2021, 11, 15. [Google Scholar] [CrossRef]
- Chen, X.; Hua, W.; Huang, X.; Chen, Y.; Zhang, J.; Li, G. Regulatory Role of RNA N6-Methyladenosine Modification in Bone Biology and Osteoporosis. Front. Endocrinol. 2019, 10, 911. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Yuan, Y.; He, M.; Gong, R.; Lei, H.; Zhou, H.; Wang, W.; Du, W.; Ma, T.; Liu, S.; et al. m6A Methylation of Precursor-miR-320/RUNX2 Controls Osteogenic Potential of Bone Marrow-Derived Mesenchymal Stem Cells. Mol. Ther. Nucleic Acids 2020, 19, 421–436. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.; Sun, X.; Li, W.; Chu, W.; Zhang, Y.; Li, Y.; Wang, Z.; Zhou, X.; Ma, J.; Xu, C.; et al. 3D-Printed beta-Tricalcium Phosphate Scaffolds Promote Osteogenic Differentiation of Bone Marrow-Deprived Mesenchymal Stem Cells in an N6-methyladenosine-Dependent Manner. Int. J. Bioprint. 2022, 8, 544. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Zhang, G.; Wang, K.; Yang, Z.; Tan, Y. METTL3 potentiates osteogenic differentiation of bone marrow mesenchymal stem cells via IGF2BP1/m6A/RUNX2. Oral. Dis. 2024, 30, 1313–1321. [Google Scholar] [CrossRef]
- Wu, Y.; Xie, L.; Wang, M.; Xiong, Q.; Guo, Y.; Liang, Y.; Li, J.; Sheng, R.; Deng, P.; Wang, Y.; et al. Mettl3-mediated m6A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat. Commun. 2018, 9, 4772. [Google Scholar] [CrossRef]
- Lei, H.; He, M.; He, X.; Li, G.; Wang, Y.; Gao, Y.; Yan, G.; Wang, Q.; Li, T.; Liu, G.; et al. METTL3 induces bone marrow mesenchymal stem cells osteogenic differentiation and migration through facilitating M1 macrophage differentiation. Am. J. Transl. Res. 2021, 13, 4376–4388. [Google Scholar]
- Tian, C.; Huang, Y.; Li, Q.; Feng, Z.; Xu, Q. Mettl3 Regulates Osteogenic Differentiation and Alternative Splicing of Vegfa in Bone Marrow Mesenchymal Stem Cells. Int. J. Mol. Sci. 2019, 20, 551. [Google Scholar] [CrossRef]
- Wu, T.; Tang, H.; Yang, J.; Yao, Z.; Bai, L.; Xie, Y.; Li, Q.; Xiao, J. METTL3-m6 A methylase regulates the osteogenic potential of bone marrow mesenchymal stem cells in osteoporotic rats via the Wnt signalling pathway. Cell Prolif. 2022, 55, e13234. [Google Scholar] [CrossRef]
- Yao, C.; Xin, H.; Liu, S.; She, P.; Ma, Y.; Hu, M.; Jin, Y. Role of METTL3-mediated m6A modification in osteogenic differentiation of periodontal ligament stem cells extracted from adult periodontal ligaments ex-vivo. Cell. Mol. Biol. 2023, 69, 23–28. [Google Scholar]
- Huang, J.; Guo, C.; Wang, Y.; Zhou, Y. Role of N6-adenosine-methyltransferase subunits METTL3 and METTL14 in the biological properties of periodontal ligament cells. Tissue Cell 2023, 82, 102081. [Google Scholar] [CrossRef]
- Sun, X.; Meng, X.; Piao, Y.; Dong, S.; Dong, Q. METTL3 promotes the osteogenic differentiation of human periodontal ligament cells by increasing YAP activity via IGF2BP1 and YTHDF1-mediated m6A modification. J. Periodontal Res. 2024, 59, 1017–1030. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Meng, X.; Piao, Y.; Dong, S.; Dong, Q. METTL3 Promotes Osteogenic Differentiation of Human Periodontal Ligament Stem Cells through IGF2BP1-Mediated Regulation of Runx2 Stability. Int. J. Med. Sci. 2024, 21, 664–673. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zeng, J.; Jiang, C.; Chen, J.; Song, C.; Chen, M.; Wu, B. METTL3-Mediated lncSNHG7 m6A Modification in the Osteogenic/Odontogenic Differentiation of Human Dental Stem Cells. J. Clin. Med. 2022, 12, 113. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Ji, Y.; Han, L.; Zhang, J.; Ni, Y.; Cheng, Y.; Zhang, Y. METTL3-Dependent Glycolysis Regulates Dental Pulp Stem Cell Differentiation. J. Dent. Res. 2022, 101, 580–589. [Google Scholar] [CrossRef]
- Chen, S.; Duan, X.; He, Y.; Chen, W. METTL3 promotes osteogenic differentiation of human umbilical cord mesenchymal stem cells by up-regulating m6A modification of circCTTN. Biosci. Rep. 2024, 44, BSR20231186. [Google Scholar] [CrossRef]
- Liu, T.; Zheng, X.; Wang, C.; Wang, C.; Jiang, S.; Li, B.; Chen, P.; Xu, W.; Zheng, H.; Yang, R.; et al. The m6A “reader” YTHDF1 promotes osteogenesis of bone marrow mesenchymal stem cells through translational control of ZNF839. Cell Death Dis. 2021, 12, 1078. [Google Scholar] [CrossRef]
- Gao, X.; Wang, J.; Wang, Y.; Li, W.; Pan, Z. The m6A Reader YTHDF1 Accelerates the Osteogenesis of Bone Marrow Mesenchymal Stem Cells Partly via Activation of the Autophagy Signaling Pathway. Stem Cells Int. 2023, 2023, 5563568. [Google Scholar] [CrossRef]
- Shi, D.; Liu, X.; Li, X.; Li, T.; Liu, J.; Wu, L. Yth m6A RNA-Binding Protein 1 Regulates Osteogenesis of MC3T3-E1 Cells under Hypoxia via Translational Control of Thrombospondin-1. Int. J. Mol. Sci. 2023, 24, 1741. [Google Scholar] [CrossRef]
- Wang, J.; Fu, Q.; Yang, J.; Liu, J.L.; Hou, S.M.; Huang, X.; Cao, J.S.; Liu, T.L.; Wang, K.Z. RNA N6-methyladenosine demethylase FTO promotes osteoporosis through demethylating Runx2 mRNA and inhibiting osteogenic differentiation. Aging 2021, 13, 21134–21141. [Google Scholar] [CrossRef]
- Zheng, L.; Li, Z.; Wang, B.; Sun, R.; Sun, Y.; Ren, J.; Zhao, J. M6A Demethylase Inhibits Osteogenesis of Dental Follicle Stem Cells via Regulating miR-7974/FKBP15 Pathway. Int. J. Mol. Sci. 2023, 24, 16121. [Google Scholar] [CrossRef]
- Zhou, J.; Zhu, Y.; Ai, D.; Zhou, M.; Li, H.; Li, G.; Zheng, L.; Song, J. Advanced glycation end products impair bone marrow mesenchymal stem cells osteogenesis in periodontitis with diabetes via FTO-mediated N6-methyladenosine modification of sclerostin. J. Transl. Med. 2023, 21, 781. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, S.; Hu, D.; Zhang, L.; Shi, S. ALKBH5 regulates etoposide-induced cellular senescence and osteogenic differentiation in osteoporosis through mediating the m6A modification of VDAC3. Sci. Rep. 2024, 14, 23461. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, W.; Liu, H.; Zhang, C.; Cao, Y.; Long, L.; Han, X.; Wang, Y.; Yan, F.; Li, G.; et al. miR615–3p inhibited FBLN1 and osteogenic differentiation of umbilical cord mesenchymal stem cells by associated with YTHDF2 in a m6A-miRNA interaction manner. Cell Prolif. 2024, 57, e13607. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Liu, J.; Zhang, L.; Li, X.; Sun, Z.; Dai, Z.; Ma, J.; Jiao, G.; Chen, Y. WTAP-mediated m6A modification modulates bone marrow mesenchymal stem cells differentiation potential and osteoporosis. Cell Death Dis. 2023, 14, 33. [Google Scholar] [CrossRef]
- Liu, J.; You, Y.; Sun, Z.; Zhang, L.; Li, X.; Dai, Z.; Ma, J.; Chen, Y.; Jiao, G. WTAP-Mediated m6A RNA Methylation Regulates the Differentiation of Bone Marrow Mesenchymal Stem Cells via the miR-29b-3p/HDAC4 Axis. Stem Cells Transl. Med. 2023, 12, 307–321. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Y.; Niu, Y.; Li, Y.; Hu, Z.; Sun, S.; Chen, Y.; Hu, B.; Huang, Y.; Deng, X. The role of WTAP in regulating macrophage-mediated osteoimmune responses and tissue regeneration in periodontitis. Front. Immunol. 2024, 15, 1423378. [Google Scholar] [CrossRef]
- Peng, J.; Zhan, Y.; Zong, Y. METTL3-mediated LINC00657 promotes osteogenic differentiation of mesenchymal stem cells via miR-144-3p/BMPR1B axis. Cell Tissue Res. 2022, 388, 301–312. [Google Scholar] [CrossRef]
- Li, L.; Wang, B.; Zhou, X.; Ding, H.; Sun, C.; Wang, Y.; Zhang, F.; Zhao, J. METTL3-mediated long non-coding RNA MIR99AHG methylation targets miR-4660 to promote bone marrow mesenchymal stem cell osteogenic differentiation. Cell Cycle 2023, 22, 476–493. [Google Scholar] [CrossRef]
- Wang, X.; Zou, C.; Li, M.; Hou, C.; Jiang, W.; Bian, Z.; Zhu, L. METTL14 upregulates TCF1 through m6A mRNA methylation to stimulate osteogenic activity in osteoporosis. Hum. Cell 2023, 36, 178–194. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y. Downregulation of METTL14 improves postmenopausal osteoporosis via IGF2BP1 dependent posttranscriptional silencing of SMAD1. Cell Death Dis. 2022, 13, 919. [Google Scholar] [CrossRef]
- Jin, Y.; Wu, A.; Bian, S.; Teng, J. Icariin upregulates methyltransferase-like 14-mediated prolyl 4-hydroxylase beta subunit m6A modification to promote osteogenic differentiation of bone marrow stem cells. Exp. Cell Res. 2024, 440, 114138. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Lei, H.; He, X.; Liu, Y.; Wang, A.; Ren, Z.; Liu, X.; Yan, G.; Wang, W.; Wang, Y.; et al. METTL14 Regulates Osteogenesis of Bone Marrow Mesenchymal Stem Cells via Inducing Autophagy Through m6A/IGF2BPs/Beclin-1 Signal Axis. Stem Cells Transl. Med. 2022, 11, 987–1001. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, R.; Zhu, X.; Zhang, X.; Lian, N. METTL14 alleviates the development of osteoporosis in ovariectomized mice by upregulating m6A level of SIRT1 mRNA. Bone 2023, 168, 116652. [Google Scholar] [CrossRef]
- Zhang, M.; Guan, J.; Yu, S.; Zhang, Y.; Cheng, L.; Zhang, Y. YTHDC1 inhibits osteoclast differentiation to alleviate osteoporosis by enhancing PTPN6 messenger RNA stability in an m6A-hUR-dependent manner. J. Leukoc. Biol. 2024, 115, 1154–1164. [Google Scholar] [CrossRef]
- Chen, H.; Peng, L.; Wang, Z.; He, Y.; Zhang, X. Influence of METTL3 knockdown on PDLSC osteogenesis in E. coli LPS-induced inflammation. Oral. Dis. 2024, 30, 3225–3238. [Google Scholar] [CrossRef]
- Wang, Q.; Shi, W.; Lin, S.; Wang, H. FOXO1 regulates osteogenic differentiation of periodontal ligament stem cells through the METTL3 signaling pathway. J. Orthop. Surg. Res. 2023, 18, 637. [Google Scholar] [CrossRef]
- Huang, J.; Wang, Y.; Zhou, Y. METTL3 and METTL14 regulate IL-6 expression via RNA m6A modification of zinc transporter SLC39A9 and DNA methylation of IL-6 in periodontal ligament cells. Biochim. Biophys. Acta Mol. Cell Res. 2024, 1871, 119605. [Google Scholar] [CrossRef]
- Chen, X.; Qin, Y.; Wang, X.; Lei, H.; Zhang, X.; Luo, H.; Guo, C.; Sun, W.; Fang, S.; Qin, W.; et al. METTL3-Mediated m6A Modification Regulates the Osteogenic Differentiation through LncRNA CUTALP in Periodontal Mesenchymal Stem Cells of Periodontitis Patients. Stem Cells Int. 2024, 2024, 3361794. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, J.; Gao, J.; Sun, W.; Chen, X.; Wang, X.; Qin, W.; Jin, Z. N6-methyladenosine promotes osteogenic differentiation of PDLSCs from periodontitis patients. Oral. Dis. 2024, 30, 1322–1336. [Google Scholar] [CrossRef]
- Zhang, Y.; Kong, Y.; Zhang, W.; He, J.; Zhang, Z.; Cai, Y.; Zhao, Y.; Xu, Q. METTL3 promotes osteoblast ribosome biogenesis and alleviates periodontitis. Clin. Epigenetics 2024, 16, 18. [Google Scholar] [CrossRef]
- Zhou, X.; Yang, X.; Huang, S.; Lin, G.; Lei, K.; Wang, Q.; Lin, W.; Li, H.; Qi, X.; Seriwatanachai, D.; et al. Inhibition of METTL3 Alleviates NLRP3 Inflammasome Activation via Increasing Ubiquitination of NEK7. Adv. Sci. 2024, 11, e2308786. [Google Scholar] [CrossRef] [PubMed]
- Mi, B.; Xiong, Y.; Yan, C.; Chen, L.; Xue, H.; Panayi, A.C.; Hu, L.; Hu, Y.; Zhou, W.; Cao, F.; et al. Methyltransferase-like 3-mediated N6-methyladenosine modification of miR-7212-5p drives osteoblast differentiation and fracture healing. J. Cell Mol. Med. 2020, 24, 6385–6396. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.; Sun, J.; Dai, Z.; Guo, L.; Tao, D.; Li, H.; Chen, B.; Zhou, R. Unraveling IGFBP3-mediated m6A modification in fracture healing. Pathol. Res. Pract. 2024, 255, 155220. [Google Scholar] [CrossRef]
- Gao, D.; Shi, J.; Lu, S.; Li, J.; Lv, K.; Xu, Y.; Song, M. METTL3 accelerates staphylococcal protein A (SpA)-induced osteomyelitis progression by regulating m6A methylation-modified miR-320a. J. Orthop. Surg. Res. 2024, 19, 729. [Google Scholar] [CrossRef]
- Cheng, C.; Zhang, H.; Zheng, J.; Jin, Y.; Wang, D.; Dai, Z. METTL14 benefits the mesenchymal stem cells in patients with steroid-associated osteonecrosis of the femoral head by regulating the m6A level of PTPN6. Aging 2021, 13, 25903–25919. [Google Scholar] [CrossRef]
- Jin, Y.; Han, X.; Wang, Y.; Fan, Z. METTL7A-mediated m6A modification of corin reverses bisphosphonates-impaired osteogenic differentiation of orofacial BMSCs. Int. J. Oral. Sci. 2024, 16, 42. [Google Scholar] [CrossRef]
- Xie, Z.; Yu, W.; Zheng, G.; Li, J.; Cen, S.; Ye, G.; Li, Z.; Liu, W.; Li, M.; Lin, J.; et al. TNF-α-mediated m6A modification of ELMO1 triggers directional migration of mesenchymal stem cell in ankylosing spondylitis. Nat. Commun. 2021, 12, 5373. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, L.; Xiang, Q.; Lin, J.; Jiang, S.; Li, W. N6-methyladenosine-modified circCDK14 promotes ossification of the ligamentum flavum via epigenetic modulation by targeting AFF4. Cell Mol. Life Sci. 2024, 81, 436. [Google Scholar] [CrossRef]
- Ouyang, L.; Su, X.; Li, W.; Tang, L.; Zhang, M.; Zhu, Y.; Xie, C.; Zhang, P.; Chen, J.; Huang, H. ALKBH1-demethylated DNA N6-methyladenine modification triggers vascular calcification via osteogenic reprogramming in chronic kidney disease. J. Clin. Investig. 2021, 131, e146985. [Google Scholar] [CrossRef]
- Xian, G.; Huang, R.; Xu, M.; Zhao, H.; Xu, X.; Chen, Y.; Ren, H.; Xu, D.; Zeng, Q. Noncoding RNA regulates the expression of Krm1 and Dkk2 to synergistically affect aortic valve lesions. Exp. Mol. Med. 2024, 56, 1560–1573. [Google Scholar] [CrossRef]
- Reichel, M.; Köster, T.; Staiger, D. Marking RNA: m6A writers, readers, and functions in Arabidopsis. J. Mol. Cell Biol. 2019, 11, 899–910. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Li, H.B.; Yin, Z.; Flavell, R.A. Recent advances in dynamic m6A RNA modification. Open Biol. 2016, 6, 160003. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m6A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- Zhang, L.; Xue, Z.; Yan, J.; Wang, J.; Liu, Q.; Jiang, H. LncRNA Riken-201 and Riken-203 modulates neural development by regulating the Sox6 through sequestering miRNAs. Cell Prolif. 2019, 52, e12573. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, Y.; Shu, Y.; He, J.; Gao, W. Interaction between N6-methyladenosine (m6A) modification and noncoding RNAs in cancer. Mol. Cancer 2020, 19, 94. [Google Scholar] [CrossRef]
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef]
- Lan, Y.; Liu, B.; Guo, H. The role of M6A modification in the regulation of tumor-related lncRNAs. Mol. Ther. Nucleic Acids 2021, 24, 768–779. [Google Scholar] [CrossRef]
- Yan, C.P.; Wang, X.K.; Jiang, K.; Yin, C.; Xiang, C.; Wang, Y.; Pu, C.; Chen, L.; Li, Y.L. β-Ecdysterone Enhanced Bone Regeneration Through the BMP-2/SMAD/RUNX2/Osterix Signaling Pathway. Front. Cell Dev. Biol. 2022, 10, 883228. [Google Scholar] [CrossRef]
- Jathar, S.; Kumar, V.; Srivastava, J.; Tripathi, V. Technological Developments in lncRNA Biology. Adv. Exp. Med. Biol. 2017, 1008, 283–323. [Google Scholar]
- Zhang, X.; Zhang, S.; Yan, X.; Shan, Y.; Liu, L.; Zhou, J.; Kuang, Q.; Li, M.; Long, H.; Lai, W. m6A regulator-mediated RNA methylation modification patterns are involved in immune microenvironment regulation of periodontitis. J. Cell Mol. Med. 2021, 25, 3634–3645. [Google Scholar] [CrossRef] [PubMed]
- Pajarinen, J.; Lin, T.; Gibon, E.; Kohno, Y.; Maruyama, M.; Nathan, K.; Lu, L.; Yao, Z.; Goodman, S.B. Mesenchymal stem cell-macrophage crosstalk and bone healing. Biomaterials 2019, 196, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Gao, J.; Jia, M.; Ma, X.; Lei, Z.; Da, F.; Yan, F.; Zhang, H.; Zhou, Y.; Li, M.; et al. Exendin-4 Induces Bone Marrow Stromal Cells Migration Through Bone Marrow-Derived Macrophages Polarization via PKA-STAT3 Signaling Pathway. Cell Physiol. Biochem. 2017, 44, 1696–1714. [Google Scholar] [CrossRef] [PubMed]
- Vi, L.; Baht, G.S.; Whetstone, H.; Ng, A.; Wei, Q.; Poon, R.; Mylvaganam, S.; Grynpas, M.; Alman, B.A. Macrophages promote osteoblastic differentiation in-vivo: Implications in fracture repair and bone homeostasis. J. Bone Miner. Res. 2015, 30, 1090–1102. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, X.; Li, D.; Cai, L.; Xu, Q. METTL3 Regulates Osteoblast Differentiation and Inflammatory Response via Smad Signaling and MAPK Signaling. Int. J. Mol. Sci. 2019, 21, 199. [Google Scholar] [CrossRef]
- Sun, Z.; Brodsky, J.L. Protein quality control in the secretory pathway. J. Cell Biol. 2019, 218, 3171–3187. [Google Scholar] [CrossRef]
- Kong, Y.; Zhang, Y.; Cai, Y.; Li, D.; Yi, B.; Xu, Q. METTL3 mediates osteoblast apoptosis by regulating endoplasmic reticulum stress during LPS-induced inflammation. Cell Signal. 2022, 95, 110335. [Google Scholar] [CrossRef]
- Hetz, C.; Zhang, K.; Kaufman, R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020, 21, 421–438. [Google Scholar] [CrossRef]
- Chotikatum, S.; Naim, H.Y.; El-Najjar, N. Inflammation induced ER stress affects absorptive intestinal epithelial cells function and integrity. Int. Immunopharmacol. 2018, 55, 336–344. [Google Scholar] [CrossRef]
- Wang, M.; Kaufman, R.J. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 2016, 529, 326–335. [Google Scholar] [CrossRef]
- Zhang, W.; Bai, Y.; Hao, L.; Zhao, Y.; Zhang, L.; Ding, W.; Qi, Y.; Xu, Q. One-carbon metabolism supports S-adenosylmethionine and m6A methylation to control the osteogenesis of bone marrow stem cells and bone formation. J. Bone Miner. Res. 2024, 39, 1356–1370. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, W.; E, Y.; Rui, L.; Jia, C.; Zhu, W. Qianggu Decoction Alleviated Osteoporosis by Promoting Osteogenesis of BMSCs through Mettl3-Mediated m6A Methylation. Adv. Biol. 2024, 8, e2400341. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Li, Y.L.; Wang, J.; Dong, R.C.; Wei, J.; Ma, Y.; Liu, Y.Q. Chinese Ecliptae herba (Eclipta prostrata (L.) L.) extract and its component wedelolactone enhances osteoblastogenesis of bone marrow mesenchymal stem cells via targeting METTL3-mediated m6A RNA methylation. J. Ethnopharmacol. 2023, 312, 116433. [Google Scholar] [CrossRef]
- Dirckx, N.; Tower, R.J.; Mercken, E.M.; Vangoitsenhoven, R.; Moreau-Triby, C.; Breugelmans, T.; Nefyodova, E.; Cardoen, R.; Mathieu, C.; Van der Schueren, B.; et al. Vhl deletion in osteoblasts boosts cellular glycolysis and improves global glucose metabolism. J. Clin. Investig. 2018, 128, 1087–1105. [Google Scholar] [CrossRef]
- Ye, W.; Liu, Z.; Liu, Y.; Xiao, H.; Tan, Q.; Yan, A.; Zhu, G. METTL3 promotes the osteogenic differentiation of periosteum-derived MSCs via regulation of the HOXD8/ITGA5 axis in congenital pseudarthrosis. Regen. Ther. 2024, 26, 42–49. [Google Scholar] [CrossRef]
- Zhou, T.; Han, D.; Liu, J.; Shi, J.; Zhu, P.; Wang, Y.; Dong, N. Factors influencing osteogenic differentiation of human aortic valve interstitial cells. J. Thorac. Cardiovasc. Surg. 2021, 161, e163–e185. [Google Scholar] [CrossRef]
- Yuan, X.; Shi, L.; Guo, Y.; Sun, J.; Miao, J.; Shi, J.; Chen, Y. METTL3 Regulates Ossification of the Posterior Longitudinal Ligament via the lncRNA XIST/miR-302a-3p/USP8 Axis. Front. Cell Dev. Biol. 2021, 9, 629895. [Google Scholar] [CrossRef]
- Wu, Z. Compression Promotes the Osteogenic Differentiation of Human Periodontal Ligament Stem Cells by Regulating METTL14-mediated IGF1. Curr. Stem Cell Res. Ther. 2024, 19, 1120–1128. [Google Scholar] [CrossRef]
- Liu, C.X.; Chen, L.L. Circular RNAs: Characterization, cellular roles, and applications. Cell 2022, 185, 2016–2034. [Google Scholar] [CrossRef]
- Xiang, Q.; Kang, L.; Wang, J.; Liao, Z.; Song, Y.; Zhao, K.; Wang, K.; Yang, C.; Zhang, Y. CircRNA-CIDN mitigated compression loading-induced damage in human nucleus pulposus cells via miR-34a-5p/SIRT1 axis. EBioMedicine 2020, 53, 102679. [Google Scholar] [CrossRef]
- Gao, W.; Miao, X.; Xu, T. Wilms tumor 1-associated protein mediated m6A modification promotes osteogenic differentiation of stem cells from human exfoliated deciduous teeth. J. Dent. Sci. 2024, 19, 2305–2314. [Google Scholar] [CrossRef] [PubMed]
- Lei, K.; Xu, R.; Wang, Q.; Xiong, Q.; Zhou, X.; Li, Q.; Seriwatanachai, D.; Lin, S.; Zhou, C.; Yuan, Q. METTL5 regulates cranial suture fusion via Wnt signaling. Fundam. Res. 2023, 3, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, M.; Ma, L.; Dang, X.; Du, G. piRNA-36741 regulates BMP2-mediated osteoblast differentiation via METTL3 controlled m6A modification. Aging 2021, 13, 23361–23375. [Google Scholar] [CrossRef]
- Yu, J.; Shen, L.; Liu, Y.; Ming, H.; Zhu, X.; Chu, M.; Lin, J. The m6A methyltransferase METTL3 cooperates with demethylase ALKBH5 to regulate osteogenic differentiation through NF-kappaB signaling. Mol. Cell Biochem. 2020, 463, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Liao, B.; Zhao, J.; Li, X.; Yan, K.; Ren, K.; Zhang, X.; Bao, X.; Guo, W. METTL14 mediates m6a modification on osteogenic proliferation and differentiation of bone marrow mesenchymal stem cells by regulating the processing of pri-miR-873. Mol. Med. Rep. 2023, 28, 166. [Google Scholar] [CrossRef]
- Sun, W.; Song, Y.; Xia, K.; Yu, L.; Huang, X.; Zhao, Z.; Liu, J. Transcriptome-wide m6A methylome during osteogenic differentiation of human adipose-derived stem cells. Stem Cell Res. Ther. 2021, 12, 489. [Google Scholar] [CrossRef]
- Krishnamoorthy, H.S.; Kannan, B.; Ganapathy, D.; Jayaseelan, V.P.; Arumugam, P. Dysregulated m6A methylation modification is associated with human peri-implantitis—A pilot study. J. Stomatol. Oral. Maxillofac. Surg. 2023, 124, 101550. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Yang, S.; Wei, J.; Cui, Y.H.; Park, G.; Shah, P.; Deng, Y.; Aplin, A.E.; Lu, Z.; Hwang, S.; He, C.; et al. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 2019, 10, 2782. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, L.; Wang, Y. ALKBH5-mediated m6A demethylation of lncRNA PVT1 plays an oncogenic role in osteosarcoma. Cancer Cell Int. 2020, 20, 34. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Yang, Y.; Kang, M.; Wang, Y.; Wang, Y.; Bi, Y.; He, S.; Shimamoto, F. m6A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol. Cancer 2020, 19, 3. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.S.; Zhang, M.; Chen, P.; Xiong, X.F.; Liu, P.Q.; Wang, H.B.; Wang, J.J.; Shen, J. The m6A demethylase FTO promotes the osteogenesis of mesenchymal stem cells by downregulating PPARG. Acta Pharmacol. Sin. 2022, 43, 1311–1323. [Google Scholar] [CrossRef]
- Zhang, F.; Liu, C.; Chen, Z.; Zhao, C. A novel PDIA3/FTO/USP20 positive feedback regulatory loop induces osteogenic differentiation of preosteoblast in osteoporosis. Cell Biol. Int. 2024, 48, 541–550. [Google Scholar] [CrossRef]
- He, J.; Zhao, Y.; Zhang, Y.; Zhang, Z.; Li, D.; Xu, Q. FTO regulates osteoclast development by modulating the proliferation and apoptosis of osteoclast precursors in inflammatory conditions. Cell Signal 2024, 117, 111098. [Google Scholar] [CrossRef]
- Shanbhogue, V.V.; Hansen, S.; Frost, M.; Brixen, K.; Hermann, A.P. Bone disease in diabetes: Another manifestation of microvascular disease? Lancet Diabetes Endocrinol. 2017, 5, 827–838. [Google Scholar] [CrossRef]
- Lalla, E.; Papapanou, P.N. Diabetes mellitus and periodontitis: A tale of two common interrelated diseases. Nat. Rev. Endocrinol. 2011, 7, 738–748. [Google Scholar] [CrossRef]
- Graves, D.T.; Ding, Z.; Yang, Y. The impact of diabetes on periodontal diseases. Periodontol 2000 2020, 82, 214–224. [Google Scholar] [CrossRef]
- Li, Z.; Wang, P.; Li, J.; Xie, Z.; Cen, S.; Li, M.; Liu, W.; Ye, G.; Zheng, G.; Ma, M.; et al. The N6-methyladenosine demethylase ALKBH5 negatively regulates the osteogenic differentiation of mesenchymal stem cells through PRMT6. Cell Death Dis. 2021, 12, 578. [Google Scholar] [CrossRef]
- Feng, L.; Fan, Y.; Zhou, J.; Li, S.; Zhang, X. The RNA demethylase ALKBH5 promotes osteoblast differentiation by modulating Runx2 mRNA stability. FEBS Lett. 2021, 595, 2007–2014. [Google Scholar] [CrossRef]
- Song, Y.; Gao, H.; Pan, Y.; Gu, Y.; Sun, W.; Wang, Y.; Liu, J. ALKBH5 Regulates Osteogenic Differentiation via the lncRNA/mRNA Complex. J. Dent. Res. 2024, 103, 1119–1129. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.; Fan, H.; Liu, H.; Tang, S.; Zheng, Y. Abnormal Decrease of Macrophage ALKBH5 Expression Causes Abnormal Polarization and Inhibits Osteoblast Differentiation. Stem Cells Int. 2023, 2023, 9974098. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.P.; Liu, Y.L.; Luo, L.P.; Xiao, Y.; Jiang, T.J.; Yuan, J.; Wang, M. Alkbh1-mediated DNA N6-methyladenine modification regulates bone marrow mesenchymal stem cell fate during skeletal aging. Cell Prolif. 2022, 55, e13178. [Google Scholar] [CrossRef] [PubMed]
- Frye, M.; Harada, B.T.; Behm, M.; He, C. RNA modifications modulate gene expression during development. Science 2018, 361, 1346–1349. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef]
- Hou, J.; Zhang, H.; Liu, J.; Zhao, Z.; Wang, J.; Lu, Z.; Hu, B.; Zhou, J.; Zhao, Z.; Feng, M.; et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol. Cancer 2019, 18, 163. [Google Scholar] [CrossRef]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef]
- Luxton, H.J.; Simpson, B.S.; Mills, I.G.; Brindle, N.R.; Ahmed, Z.; Stavrinides, V.; Heavey, S.; Stamm, S.; Whitaker, H.C. The Oncogene Metadherin Interacts with the Known Splicing Proteins YTHDC1, Sam68 and T-STAR and Plays a Novel Role in Alternative mRNA Splicing. Cancers 2019, 11, 1233. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5′ UTR m6A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Choe, J.; Du, P.; Triboulet, R.; Gregory, R.I. The m6A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 2016, 62, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Arguello, A.E.; DeLiberto, A.N.; Kleiner, R.E. RNA Chemical Proteomics Reveals the N6-Methyladenosine (m6A)-Regulated Protein-RNA Interactome. J. Am. Chem. Soc. 2017, 139, 17249–17252. [Google Scholar] [CrossRef]
- Ma, B.; Cao, P.; Zhang, L.; Zhu, H.; Ye, X.; Wang, L.; Chen, L. YTHDC2 inhibits rat bone mesenchymal stem cells osteogenic differentiation by accelerating RUNX2 mRNA degradation via m6A methylation. Heliyon 2023, 9, e18876. [Google Scholar] [CrossRef]
- Yuan, T.; Liu, H.; Abudoukadier, M.; Yang, Z.; Zhou, Z.; Cui, Y. YTHDF2-Mediated m6A methylation inhibition by miR27a as a protective mechanism against hormonal osteonecrosis in BMSCs. BMC Musculoskelet. Disord. 2024, 25, 359. [Google Scholar] [CrossRef]
- Feng, Z.W.; Xiao, H.F.; Wang, X.W.; Niu, Y.K.; Zhao, D.C.; Tian, C.; Wang, S.H.; Peng, B.; Yang, F.; Geng, B.; et al. Unraveling Key m6A Modification Regulators Signatures in Postmenopausal Osteoporosis through Bioinformatics and Experimental Verification. Orthop. Surg. 2024, 16, 1418–1433. [Google Scholar] [CrossRef]
- Zhu, H.; Cai, C.; Yu, Y.; Zhou, Y.; Yang, S.; Hu, Y.; Zhu, Y.; Zhou, J.; Zhao, J.; Ma, H.; et al. Quercetin-Loaded Bioglass Injectable Hydrogel Promotes m6A Alteration of Per1 to Alleviate Oxidative Stress for Periodontal Bone Defects. Adv. Sci. 2024, 11, e2403412. [Google Scholar] [CrossRef]
- Deepika, B.A.; Ramamurthy, J.; Kannan, B.; Jayaseelan, V.P.; Arumugam, P. Overexpression of insulin-like growth factor-2 mRNA-binding protein 1 is associated with periodontal disease. J. Oral. Biol. Craniofac. Res. 2024, 14, 494–499. [Google Scholar] [CrossRef]
- Ma, X.X.; Zhou, X.Y.; Feng, M.G.; Ji, Y.T.; Song, F.F.; Tang, Q.C.; He, Q.; Zhang, Y.F. Dual Role of IGF2BP2 in Osteoimmunomodulation during Periodontitis. J. Dent. Res. 2024, 103, 208–217. [Google Scholar] [CrossRef]
- Burra Anand, D.; Ramamurthy, J.; Kannan, B.; Jayaseelan, V.P.; Arumugam, P. N6-methyladenosine-mediated overexpression of TREM-1 is associated with periodontal disease. Odontology 2024, 113, 834–843. [Google Scholar] [CrossRef]
- Gao, L.; Lee, H.; Goodman, J.H.; Ding, L. Hematopoietic stem cell niche generation and maintenance are distinguishable by an epitranscriptomic program. Cell 2024, 187, 2801–2816.e17. [Google Scholar] [CrossRef] [PubMed]
- Fink, H.A.; MacDonald, R.; Forte, M.L.; Rosebush, C.E.; Ensrud, K.E.; Schousboe, J.T.; Nelson, V.A.; Ullman, K.; Butler, M.; Olson, C.M.; et al. Long-Term Drug Therapy and Drug Discontinuations and Holidays for Osteoporosis Fracture Prevention: A Systematic Review. Ann. Intern. Med. 2019, 171, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Adler, R.A.; El-Hajj Fuleihan, G.; Bauer, D.C.; Camacho, P.M.; Clarke, B.L.; Clines, G.A.; Compston, J.E.; Drake, M.T.; Edwards, B.J.; Favus, M.J.; et al. Managing Osteoporosis in Patients on Long-Term Bisphosphonate Treatment: Report of a Task Force of the American Society for Bone and Mineral Research. J. Bone Miner. Res. 2016, 31, 1910. [Google Scholar] [CrossRef] [PubMed]
- Sellmeyer, D.E. Atypical fractures as a potential complication of long-term bisphosphonate therapy. JAMA 2010, 304, 1480–1484. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, Q.; Zhao, X.; Zhu, C.; Ruan, J.; Chen, C. N6-Methyladenosine Modification of the Three Components “Writers”, “Erasers”, and “Readers” in Relation to Osteogenesis. Int. J. Mol. Sci. 2025, 26, 5620. https://doi.org/10.3390/ijms26125620
Dong Q, Zhao X, Zhu C, Ruan J, Chen C. N6-Methyladenosine Modification of the Three Components “Writers”, “Erasers”, and “Readers” in Relation to Osteogenesis. International Journal of Molecular Sciences. 2025; 26(12):5620. https://doi.org/10.3390/ijms26125620
Chicago/Turabian StyleDong, Qiannan, Xubin Zhao, Changze Zhu, Jianping Ruan, and Cheng Chen. 2025. "N6-Methyladenosine Modification of the Three Components “Writers”, “Erasers”, and “Readers” in Relation to Osteogenesis" International Journal of Molecular Sciences 26, no. 12: 5620. https://doi.org/10.3390/ijms26125620
APA StyleDong, Q., Zhao, X., Zhu, C., Ruan, J., & Chen, C. (2025). N6-Methyladenosine Modification of the Three Components “Writers”, “Erasers”, and “Readers” in Relation to Osteogenesis. International Journal of Molecular Sciences, 26(12), 5620. https://doi.org/10.3390/ijms26125620






