PKA-Mediated Phosphorylation of SFRP4 Promotes Wnt/β-Catenin Activation and Cancer Stemness in Gastric Cancer
Abstract
1. Introduction
2. Results
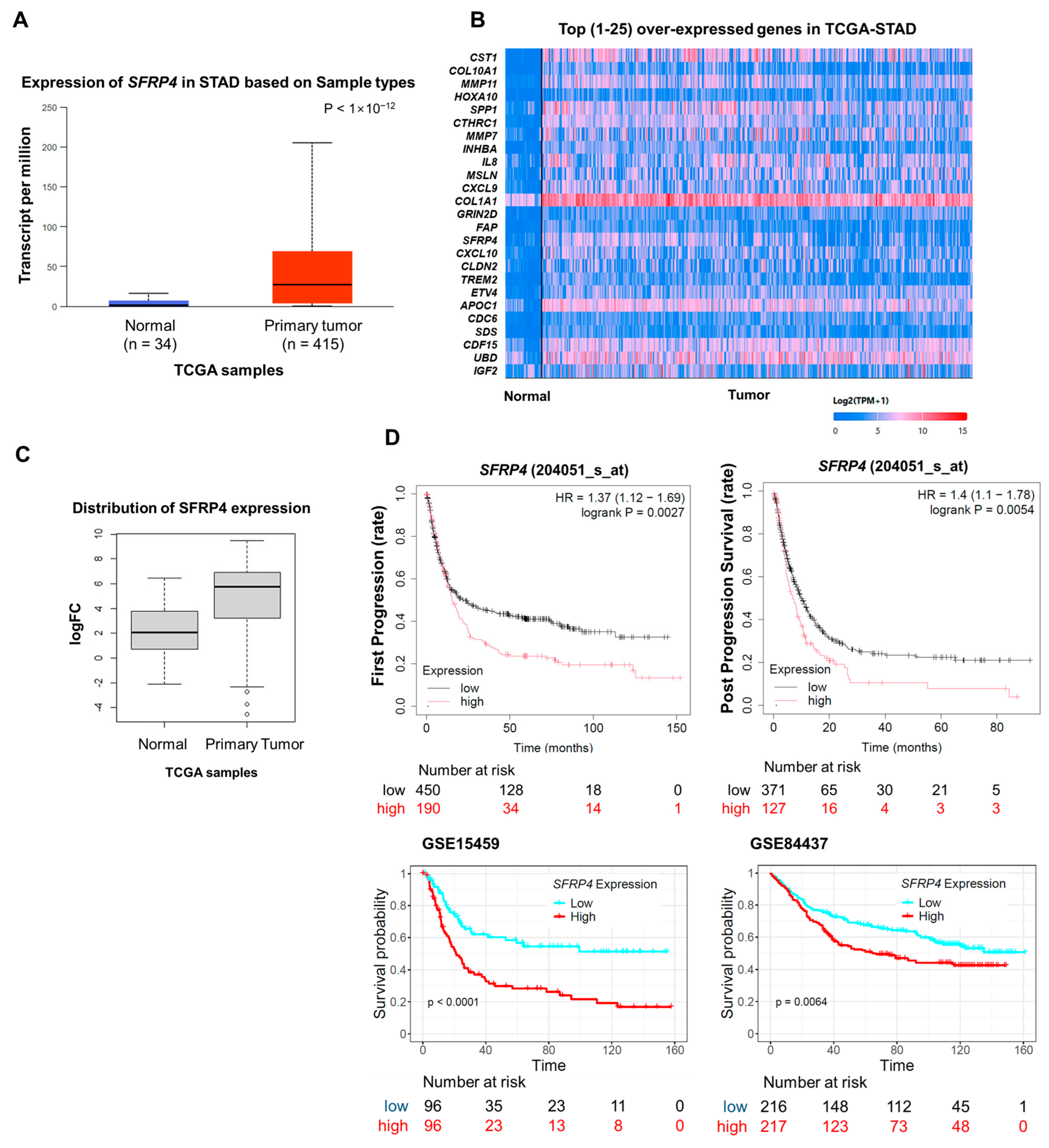
2.1. Overexpression of SFRP4 Correlates with Poor Prognosis in Gastric Cancer
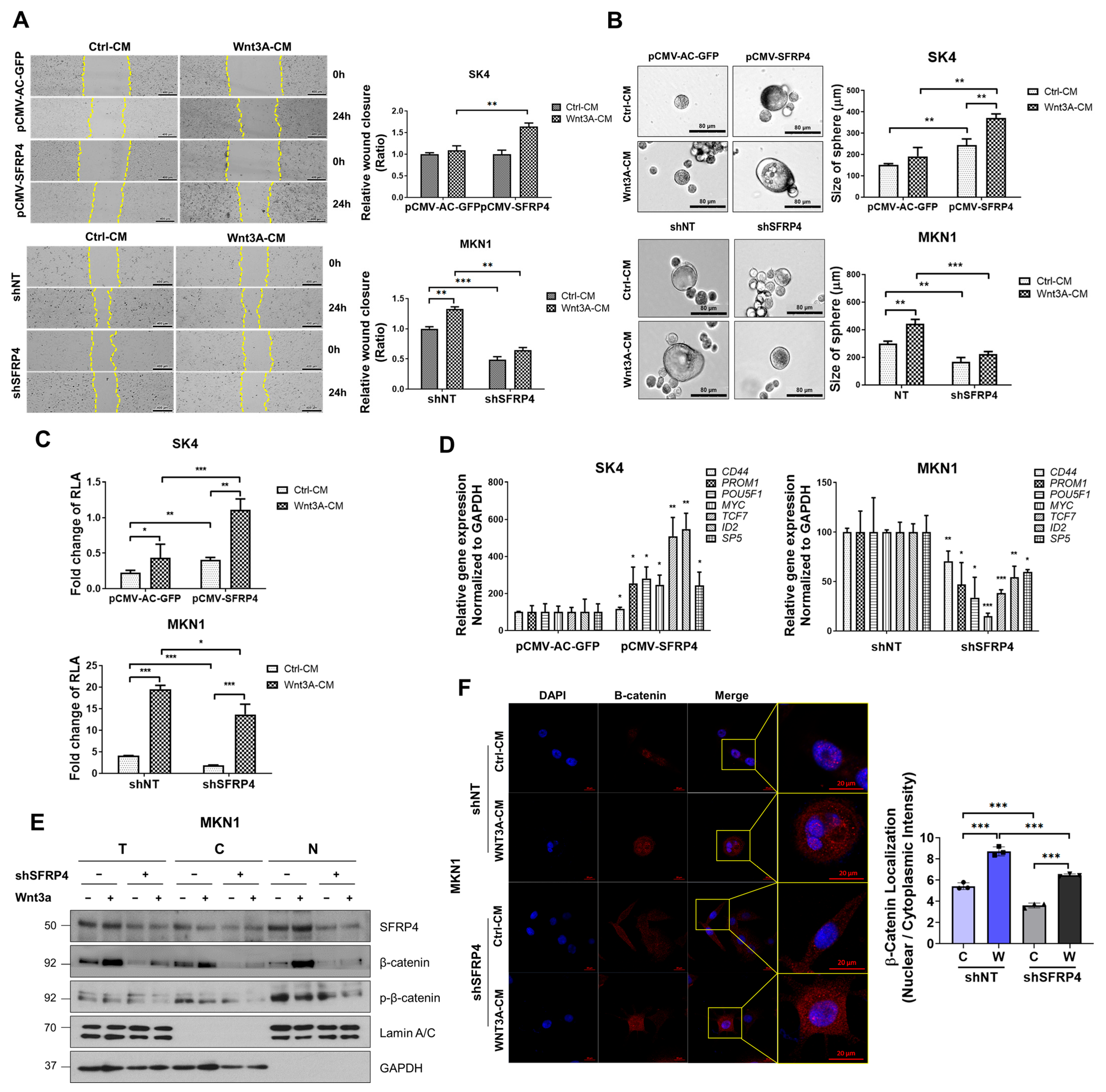
2.2. SFRP4 Induces Stemness-Related Properties in Gastric Cancer
2.3. SFRP4 Induces Stemness-Related Properties Dependent on Wnt Signaling
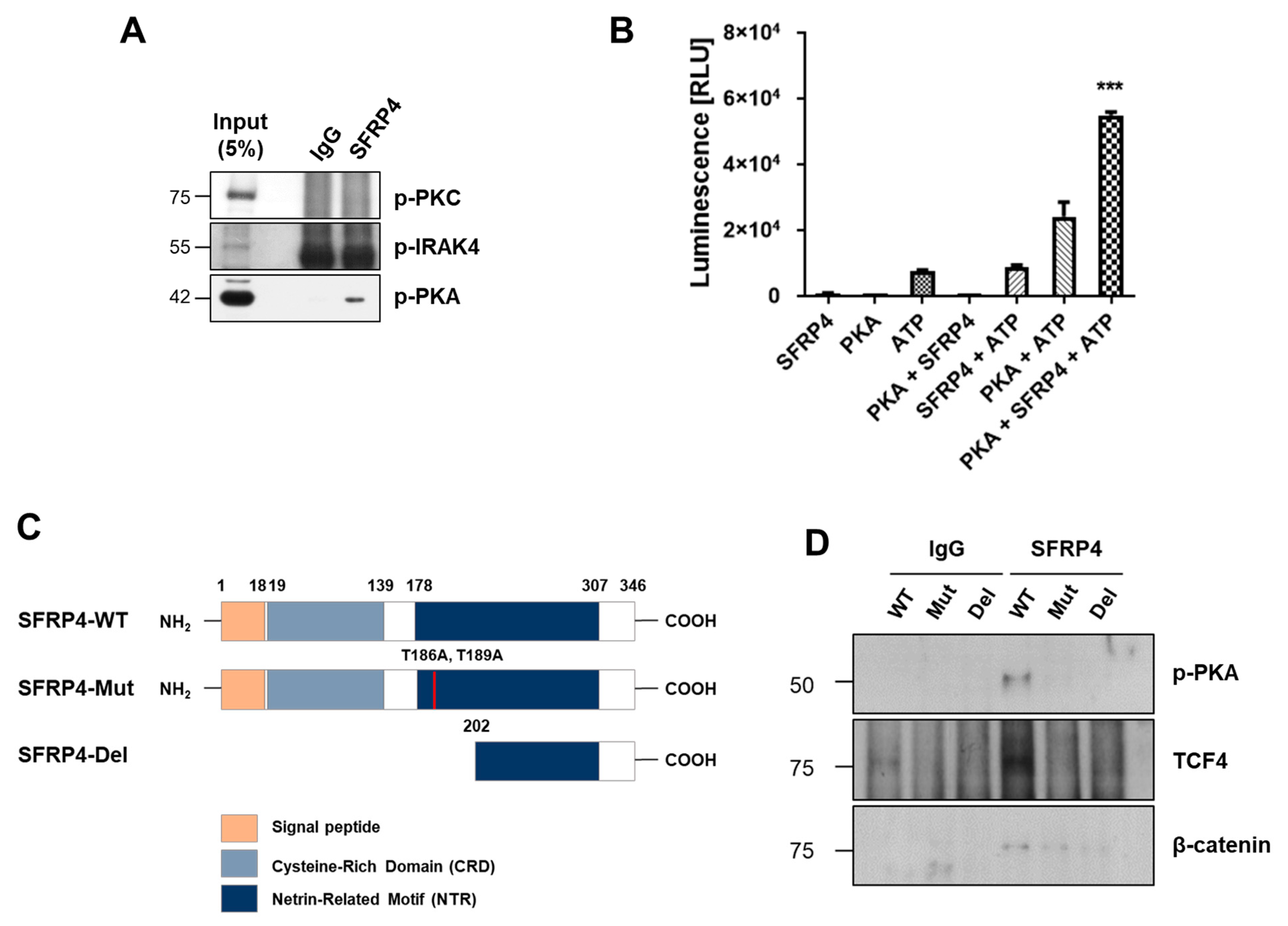
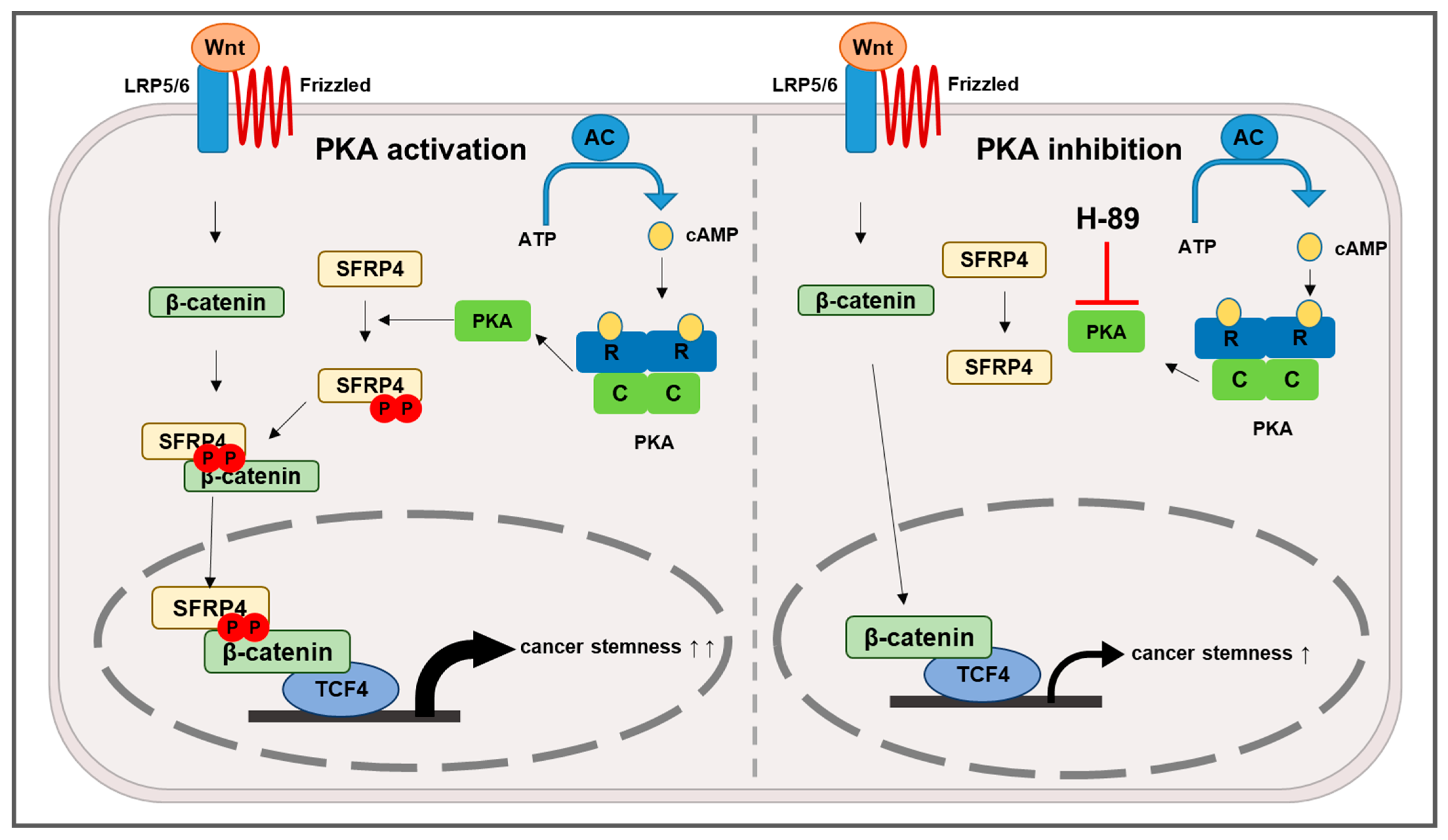
2.4. PKA Is a Putative Kinase for Phosphorylation of SFRP4
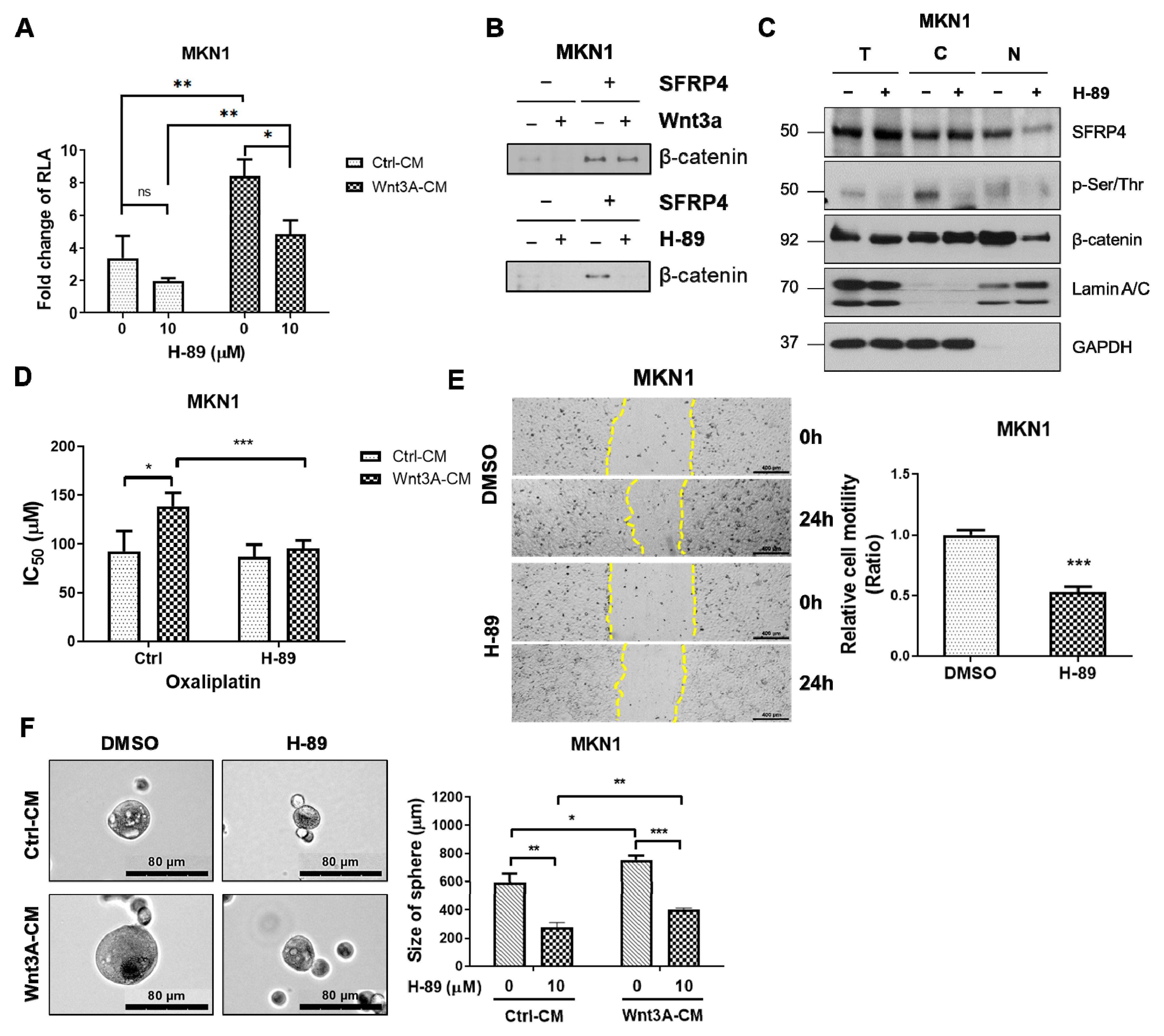
2.5. Inhibition of PKA Decreases Wnt Signaling-Dependent Stemness in Gastric Cancer
3. Discussion
4. Materials and Methods
4.1. Cell Cultures, Reagents, and Antibodies
4.2. Western Blotting
4.3. Wnt Luciferase Activity Assay
4.4. Co-Immunoprecipitation
4.5. In Vitro Kinase Assay
4.6. Nuclear/Cytoplasmic Fractionation
4.7. Sphere-Forming Assay
4.8. Wound-Healing Assay
4.9. Chemoresistance Test
4.10. Phosphoproteomics Database Analysis
4.11. Confocal Microscopy
4.12. UALCAN and Kaplan–Meier Plot Analysis
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 5-FU | 5-fluorouracil |
| ACRG | Asian Cancer Research Group |
| CM | Conditioned media |
| DKK1 | Dickkopf-1 |
| GC | Gastric cancer |
| HCC | Hepatocellular carcinoma |
| IC50 | Half maximal inhibitory concentration |
| PKA | Protein kinase A |
| p-PKA | Phosphorylated PKA |
| p-Ser/Thr | Phosphorylated serine/threonine |
| p-β-catenin | Phosphorylated β-catenin |
| SEM | Stem-like/EMT/Mesenchymal |
| SFRP4 | Secreted Frizzled-related protein 4 |
| STAD | Stomach adenocarcinoma |
| TCGA | The Cancer Genome Atlas |
| YCC | Yonsei Cancer Center |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Marusyk, A.; Almendro, V.; Polyak, K. Intra-tumour heterogeneity: A looking glass for cancer? Nat. Rev. Cancer 2012, 12, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Tan, I.B.; Das, K.; Deng, N.; Zouridis, H.; Pattison, S.; Chua, C.; Feng, Z.; Guan, Y.K.; Ooi, C.H.; et al. Identification of molecular subtypes of gastric cancer with different responses to PI3-kinase inhibitors and 5-fluorouracil. Gastroenterology 2013, 145, 554–565. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Cheong, J.H.; Yang, H.K.; Kim, H.; Kim, W.H.; Kim, Y.W.; Kook, M.C.; Park, Y.K.; Kim, H.H.; Lee, H.S.; Lee, K.H.; et al. Predictive test for chemotherapy response in resectable gastric cancer: A multi-cohort, retrospective analysis. Lancet Oncol. 2018, 19, 629–638. [Google Scholar] [CrossRef]
- Jeon, J.; Cheong, J.H. Clinical Implementation of Precision Medicine in Gastric Cancer. J. Gastric Cancer 2019, 19, 235–253. [Google Scholar] [CrossRef] [PubMed]
- Yoon, B.K.; Kim, H.; Oh, T.G.; Oh, S.K.; Jo, S.; Kim, M.; Chun, K.H.; Hwang, N.; Lee, S.; Jin, S.; et al. PHGDH preserves one-carbon cycle to confer metabolic plasticity in chemoresistant gastric cancer during nutrient stress. Proc. Natl. Acad. Sci. USA 2023, 120, e2217826120. [Google Scholar] [CrossRef]
- Jang, E.; Shin, M.K.; Kim, H.; Lim, J.Y.; Lee, J.E.; Park, J.; Kim, J.; Kim, H.; Shin, Y.; Son, H.Y.; et al. Author Correction: Clinical molecular subtyping reveals intrinsic mesenchymal reprogramming in gastric cancer cells. Exp. Mol. Med. 2023, 55, 1277. [Google Scholar] [CrossRef]
- Lee, E.; Yang, J.; Ku, M.; Kim, N.H.; Park, Y.; Park, C.B.; Suh, J.S.; Park, E.S.; Yook, J.I.; Mills, G.B.; et al. Metabolic stress induces a Wnt-dependent cancer stem cell-like state transition. Cell Death Dis. 2015, 6, e1805. [Google Scholar] [CrossRef]
- MacDonald, B.T.; He, X. Frizzled and LRP5/6 receptors for Wnt/beta-catenin signaling. Cold Spring Harb. Perspect. Biol. 2012, 4, a007880. [Google Scholar] [CrossRef]
- Van Raay, T.J.; Moore, K.B.; Iordanova, I.; Steele, M.; Jamrich, M.; Harris, W.A.; Vetter, M.L. Frizzled 5 signaling governs the neural potential of progenitors in the developing Xenopus retina. Neuron 2005, 46, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Cole, M.F.; Johnstone, S.E.; Newman, J.J.; Kagey, M.H.; Young, R.A. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes. Dev. 2008, 22, 746–755. [Google Scholar] [CrossRef]
- Pereira, L.; Yi, F.; Merrill, B.J. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol. Cell Biol. 2006, 26, 7479–7491. [Google Scholar] [CrossRef]
- Ruiz de Galarreta, M.; Bresnahan, E.; Molina-Sanchez, P.; Lindblad, K.E.; Maier, B.; Sia, D.; Puigvehi, M.; Miguela, V.; Casanova-Acebes, M.; Dhainaut, M.; et al. beta-Catenin Activation Promotes Immune Escape and Resistance to Anti-PD-1 Therapy in Hepatocellular Carcinoma. Cancer Discov. 2019, 9, 1124–1141. [Google Scholar] [CrossRef] [PubMed]
- Tomar, V.S.; Patil, V.; Somasundaram, K. Temozolomide induces activation of Wnt/beta-catenin signaling in glioma cells via PI3K/Akt pathway: Implications in glioma therapy. Cell Biol. Toxicol. 2020, 36, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Webster, M.R.; Xu, M.; Kinzler, K.A.; Kaur, A.; Appleton, J.; O’Connell, M.P.; Marchbank, K.; Valiga, A.; Dang, V.M.; Perego, M.; et al. Wnt5A promotes an adaptive, senescent-like stress response, while continuing to drive invasion in melanoma cells. Pigment. Cell Melanoma Res. 2015, 28, 184–195. [Google Scholar] [CrossRef]
- Martin-Orozco, E.; Sanchez-Fernandez, A.; Ortiz-Parra, I.; Ayala-San Nicolas, M. WNT Signaling in Tumors: The Way to Evade Drugs and Immunity. Front. Immunol. 2019, 10, 2854. [Google Scholar] [CrossRef]
- Zeng, L.; Fagotto, F.; Zhang, T.; Hsu, W.; Vasicek, T.J.; Perry, W.L., 3rd; Lee, J.J.; Tilghman, S.M.; Gumbiner, B.M.; Costantini, F. The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell 1997, 90, 181–192. [Google Scholar] [CrossRef]
- Niehrs, C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene 2006, 25, 7469–7481. [Google Scholar] [CrossRef]
- Uren, A.; Reichsman, F.; Anest, V.; Taylor, W.G.; Muraiso, K.; Bottaro, D.P.; Cumberledge, S.; Rubin, J.S. Secreted frizzled-related protein-1 binds directly to Wingless and is a biphasic modulator of Wnt signaling. J. Biol. Chem. 2000, 275, 4374–4382. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.J.; Wang, Z.W.; Chang, Y.W.; Lee, K.C.; Lin, W.H.; Lee, J.L. SFRPs Are Biphasic Modulators of Wnt-Signaling-Elicited Cancer Stem Cell Properties beyond Extracellular Control. Cell Rep. 2019, 28, 1511–1525 e1515. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Watkins, D.N.; Jair, K.W.; Schuebel, K.E.; Markowitz, S.D.; Chen, W.D.; Pretlow, T.P.; Yang, B.; Akiyama, Y.; Van Engeland, M.; et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat. Genet. 2004, 36, 417–422. [Google Scholar] [CrossRef]
- Jacob, F.; Ukegjini, K.; Nixdorf, S.; Ford, C.E.; Olivier, J.; Caduff, R.; Scurry, J.P.; Guertler, R.; Hornung, D.; Mueller, R.; et al. Loss of secreted frizzled-related protein 4 correlates with an aggressive phenotype and predicts poor outcome in ovarian cancer patients. PLoS ONE 2012, 7, e31885. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Xu, C.; Zeng, X.; Zhang, Z.; Yang, B.; Rao, Z. Tumor suppressor role of sFRP-4 in hepatocellular carcinoma via the Wnt/beta-catenin signaling pathway. Mol. Med. Rep. 2021, 23, 336. [Google Scholar] [CrossRef]
- Busuttil, R.A.; George, J.; House, C.M.; Lade, S.; Mitchell, C.; Di Costanzo, N.S.; Pattison, S.; Huang, Y.K.; Tan, P.; Cheong, J.H.; et al. SFRP4 drives invasion in gastric cancer and is an early predictor of recurrence. Gastric Cancer 2021, 24, 589–601. [Google Scholar] [CrossRef]
- Vincent, K.M.; Postovit, L.M. A pan-cancer analysis of secreted Frizzled-related proteins: Re-examining their proposed tumour suppressive function. Sci. Rep. 2017, 7, 42719. [Google Scholar] [CrossRef]
- Hino, S.; Tanji, C.; Nakayama, K.I.; Kikuchi, A. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase stabilizes beta-catenin through inhibition of its ubiquitination. Mol. Cell. Biol. 2005, 25, 9063–9072. [Google Scholar] [CrossRef]
- Cho-Chung, Y.S.; Nesterova, M.; Becker, K.G.; Srivastava, R.; Park, Y.G.; Lee, Y.N.; Cho, Y.S.; Kim, M.K.; Neary, C.; Cheadle, C. Dissecting the circuitry of protein kinase A and cAMP signaling in cancer genesis: Antisense, microarray, gene overexpression, and transcription factor decoy. Ann. N. Y. Acad. Sci. 2002, 968, 22–36. [Google Scholar] [CrossRef]
- Li, F.; Li, C.; Marquez-Lago, T.T.; Leier, A.; Akutsu, T.; Purcell, A.W.; Ian Smith, A.; Lithgow, T.; Daly, R.J.; Song, J.; et al. Quokka: A comprehensive tool for rapid and accurate prediction of kinase family-specific phosphorylation sites in the human proteome. Bioinformatics 2018, 34, 4223–4231. [Google Scholar] [CrossRef]
- Wang, C.; Xu, H.; Lin, S.; Deng, W.; Zhou, J.; Zhang, Y.; Shi, Y.; Peng, D.; Xue, Y. GPS 5.0: An Update on the Prediction of Kinase-specific Phosphorylation Sites in Proteins. Genom. Proteom. Bioinform. 2020, 18, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Goel, R.; Harsha, H.C.; Pandey, A.; Prasad, T.S. Human Protein Reference Database and Human Proteinpedia as resources for phosphoproteome analysis. Mol. Biosyst. 2012, 8, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Chijiwa, T.; Mishima, A.; Hagiwara, M.; Sano, M.; Hayashi, K.; Inoue, T.; Naito, K.; Toshioka, T.; Hidaka, H. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J. Biol. Chem. 1990, 265, 5267–5272. [Google Scholar]
- Yu, F.; Yu, C.; Li, F.; Zuo, Y.; Wang, Y.; Yao, L.; Wu, C.; Wang, C.; Ye, L. Wnt/beta-catenin signaling in cancers and targeted therapies. Signal Transduct. Target. Ther. 2021, 6, 307. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Yeom, J.; Cho, H.J.; Kim, J.H.; Yoon, S.J.; Kim, H.; Sa, J.K.; Ju, S.; Lee, H.; Oh, M.J.; et al. Integrated pharmaco-proteogenomics defines two subgroups in isocitrate dehydrogenase wild-type glioblastoma with prognostic and therapeutic opportunities. Nat. Commun. 2020, 11, 3288. [Google Scholar] [CrossRef]
- Caretta, A.; Mucignat-Caretta, C. Protein kinase a in cancer. Cancers 2011, 3, 913–926. [Google Scholar] [CrossRef]
- Riitano, G.; Capozzi, A.; Recalchi, S.; Caissutti, D.; Longo, A.; Mattei, V.; Conti, F.; Misasi, R.; Garofalo, T.; Sorice, M.; et al. Anti-beta2-GPI Antibodies Induce Endothelial Cell Expression of Tissue Factor by LRP6 Signal Transduction Pathway Involving Lipid Rafts. Cells 2022, 11, 1288. [Google Scholar] [CrossRef]
- Goyal, L.; Sirard, C.; Schrag, M.; Kagey, M.H.; Eads, J.R.; Stein, S.; El-Khoueiry, A.B.; Manji, G.A.; Abrams, T.A.; Khorana, A.A.; et al. Phase I and Biomarker Study of the Wnt Pathway Modulator DKN-01 in Combination with Gemcitabine/Cisplatin in Advanced Biliary Tract Cancer. Clin. Cancer Res. 2020, 26, 6158–6167. [Google Scholar] [CrossRef]
- Ahmed, R.A.; Alawin, O.A.; Sylvester, P.W. gamma-Tocotrienol reversal of epithelial-to-mesenchymal transition in human breast cancer cells is associated with inhibition of canonical Wnt signalling. Cell Prolif. 2016, 49, 460–470. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jhe, Y.-L.; Lee, S.; Jung, Y.; Cheong, J.-H. PKA-Mediated Phosphorylation of SFRP4 Promotes Wnt/β-Catenin Activation and Cancer Stemness in Gastric Cancer. Int. J. Mol. Sci. 2025, 26, 5572. https://doi.org/10.3390/ijms26125572
Jhe Y-L, Lee S, Jung Y, Cheong J-H. PKA-Mediated Phosphorylation of SFRP4 Promotes Wnt/β-Catenin Activation and Cancer Stemness in Gastric Cancer. International Journal of Molecular Sciences. 2025; 26(12):5572. https://doi.org/10.3390/ijms26125572
Chicago/Turabian StyleJhe, Yoo-Lim, Suji Lee, Youjin Jung, and Jae-Ho Cheong. 2025. "PKA-Mediated Phosphorylation of SFRP4 Promotes Wnt/β-Catenin Activation and Cancer Stemness in Gastric Cancer" International Journal of Molecular Sciences 26, no. 12: 5572. https://doi.org/10.3390/ijms26125572
APA StyleJhe, Y.-L., Lee, S., Jung, Y., & Cheong, J.-H. (2025). PKA-Mediated Phosphorylation of SFRP4 Promotes Wnt/β-Catenin Activation and Cancer Stemness in Gastric Cancer. International Journal of Molecular Sciences, 26(12), 5572. https://doi.org/10.3390/ijms26125572





