Multi-Omic Analysis Identifies Key Genes Driving Testicular Fusion in Spodoptera litura
Abstract
1. Introduction
2. Results
2.1. Transcriptomic Analysis of Spodoptera litura Testicular Sheath
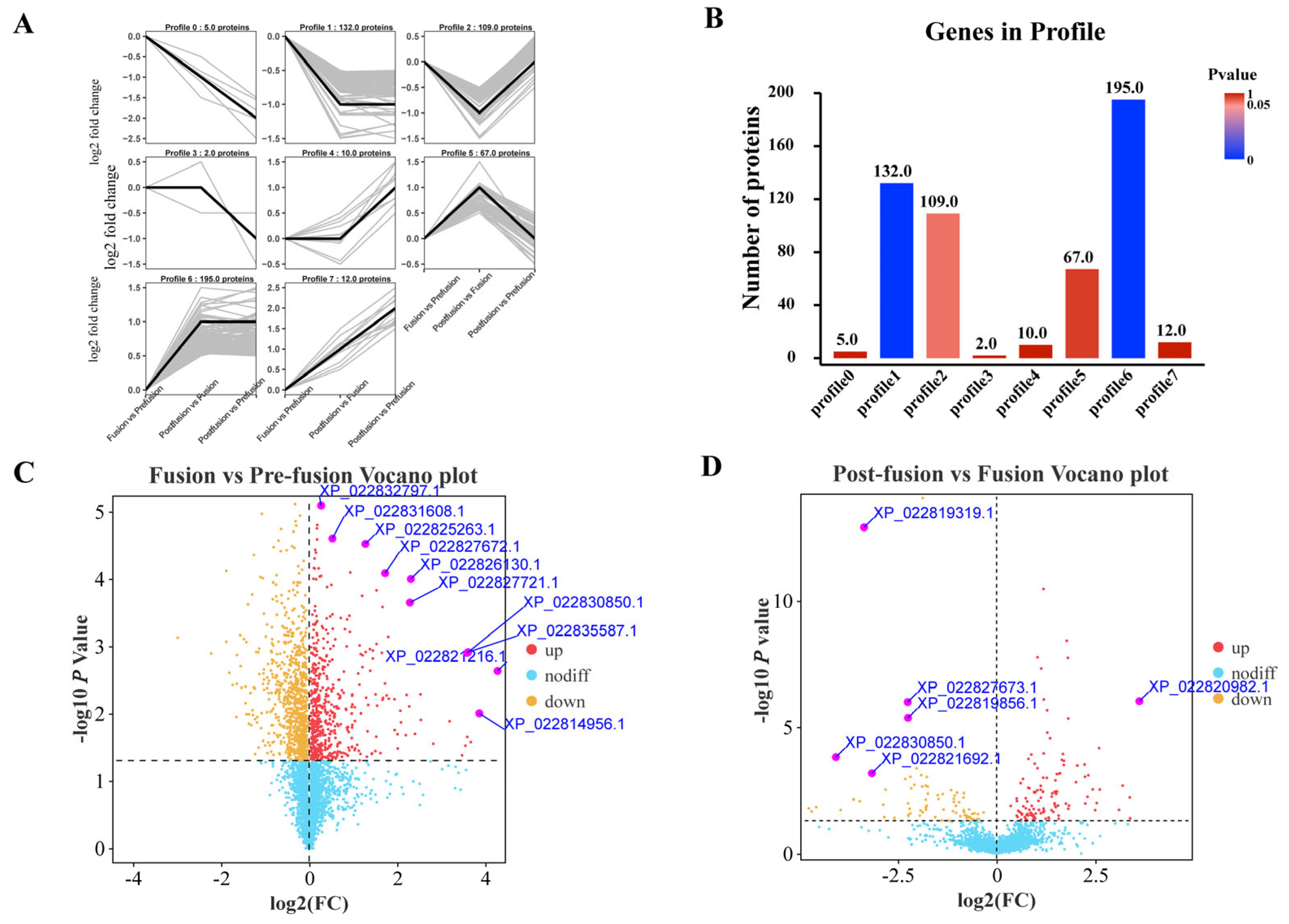
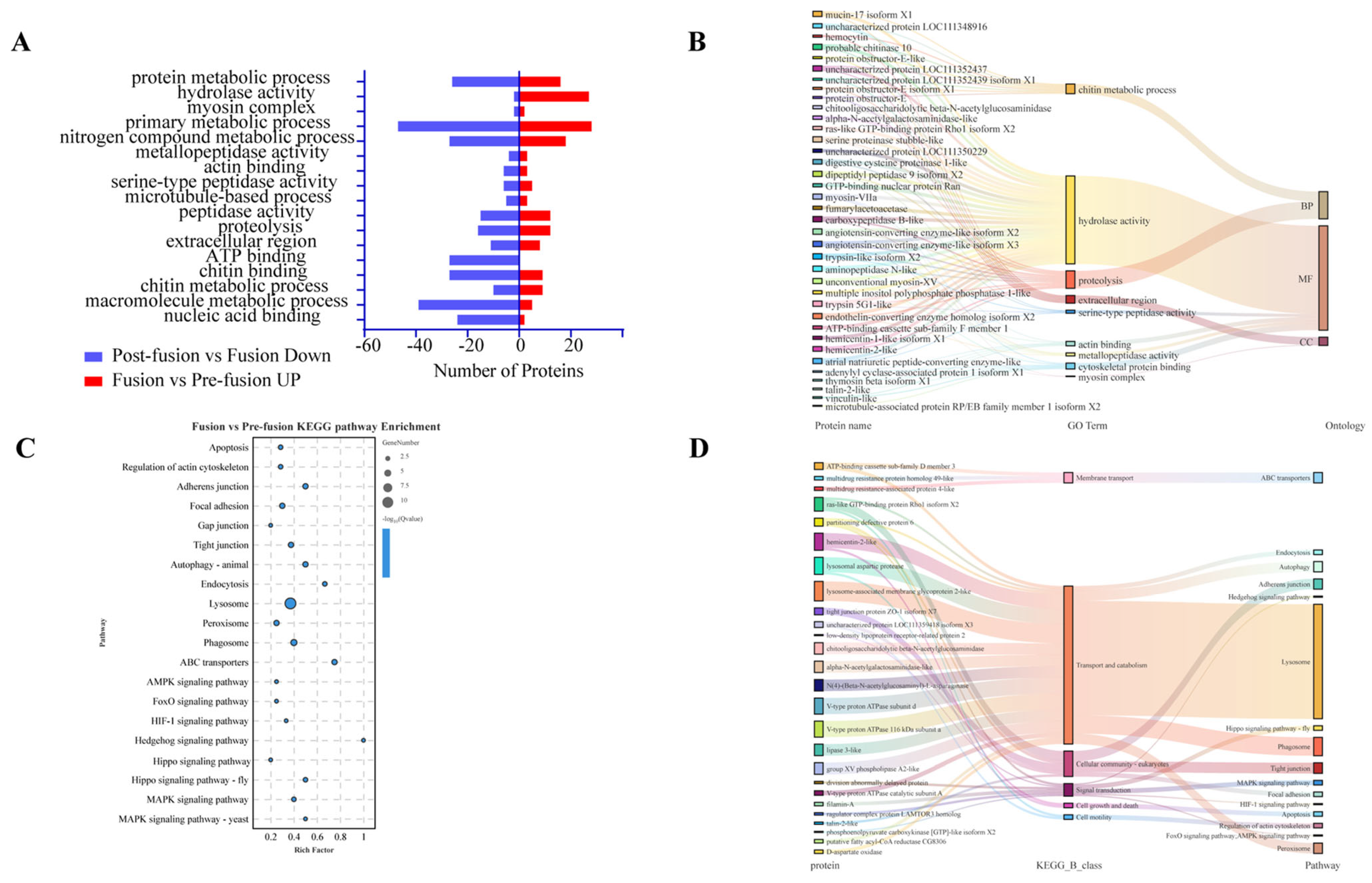
2.2. Proteomic Analysis of Spodoptera litura Testicular Sheath
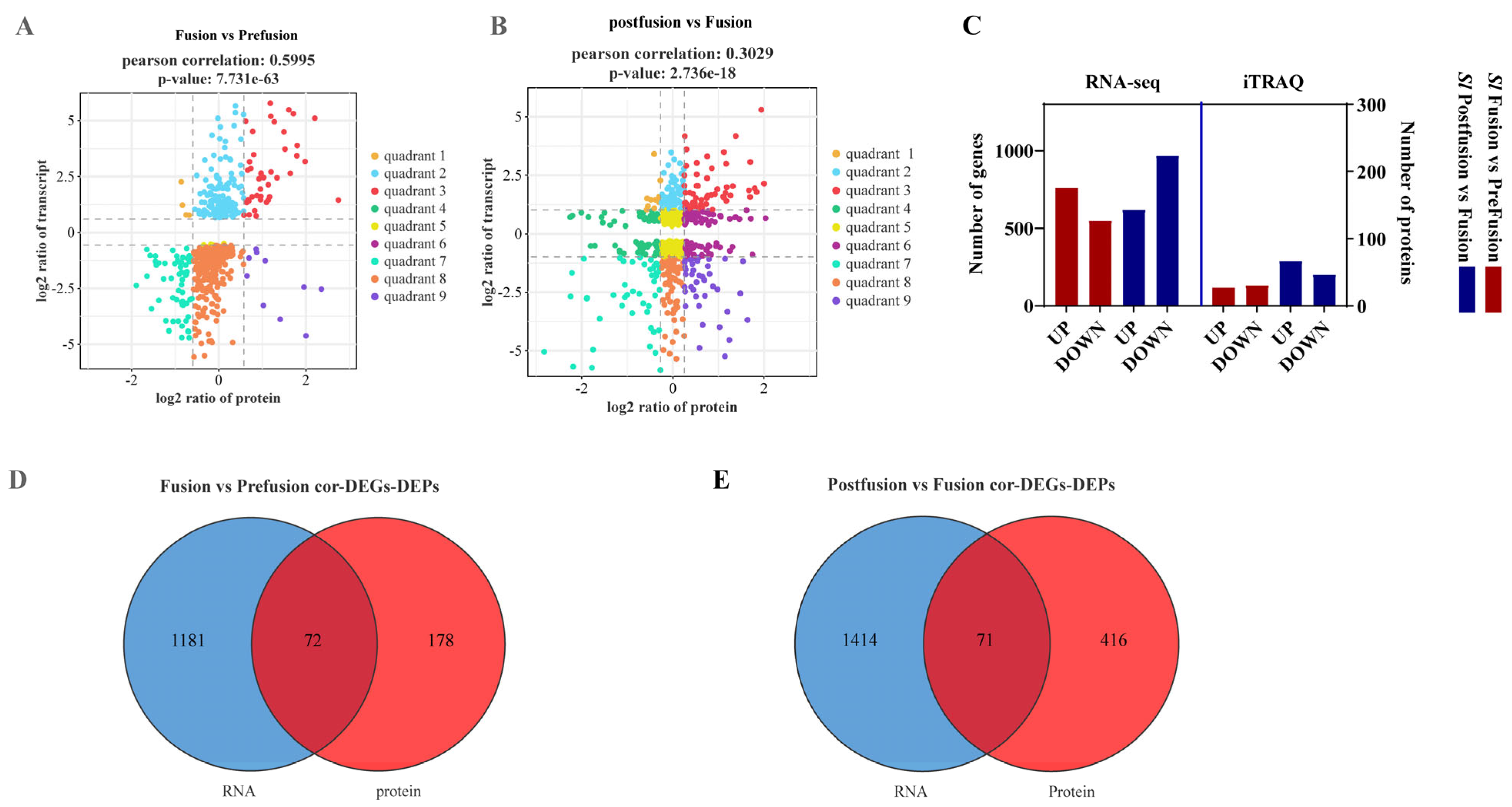
2.3. Combined Transcriptomic and Proteomic Analysis of the Testis Sheath
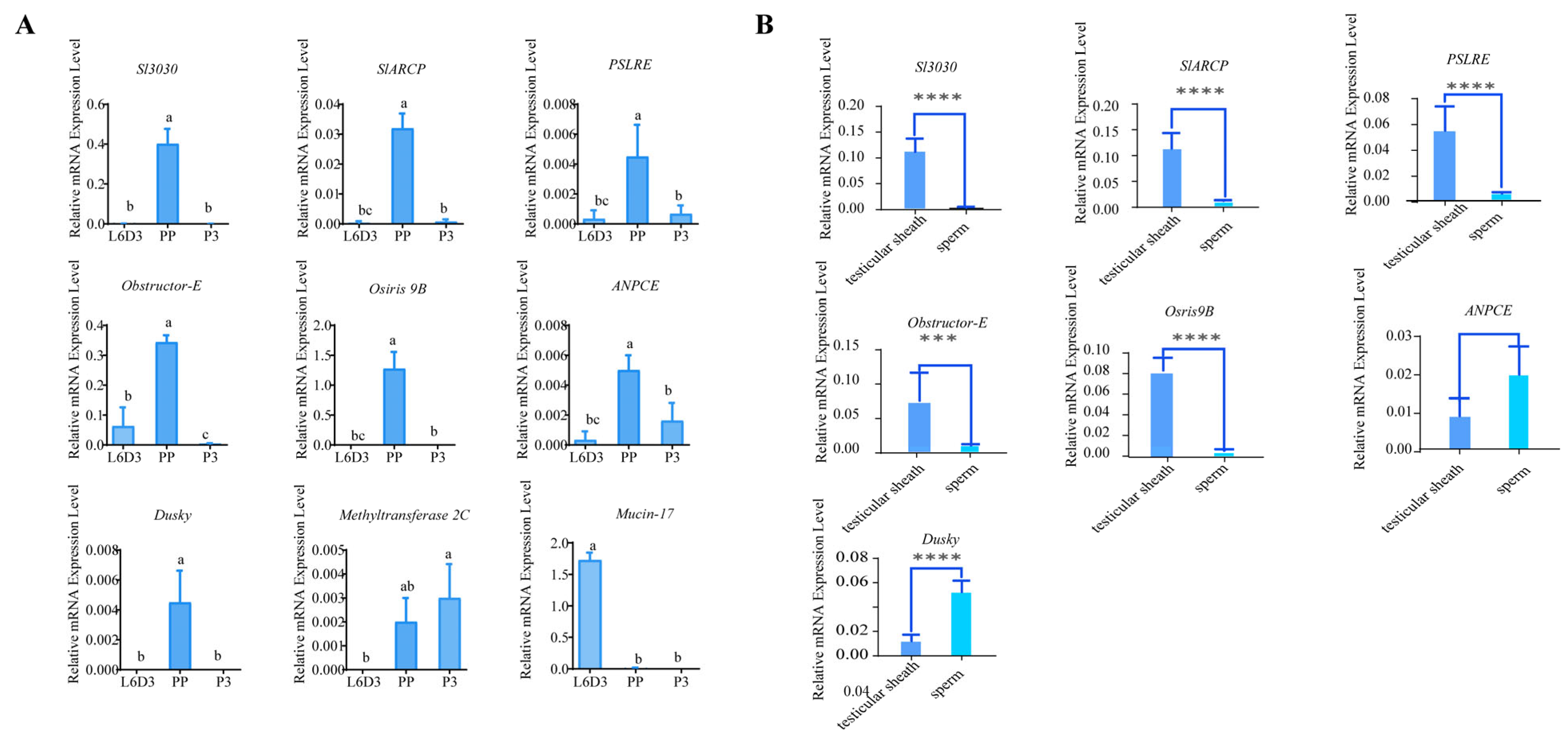
2.4. Phenotypic Analysis of CRISPR/Cas 9 Gene Knockout
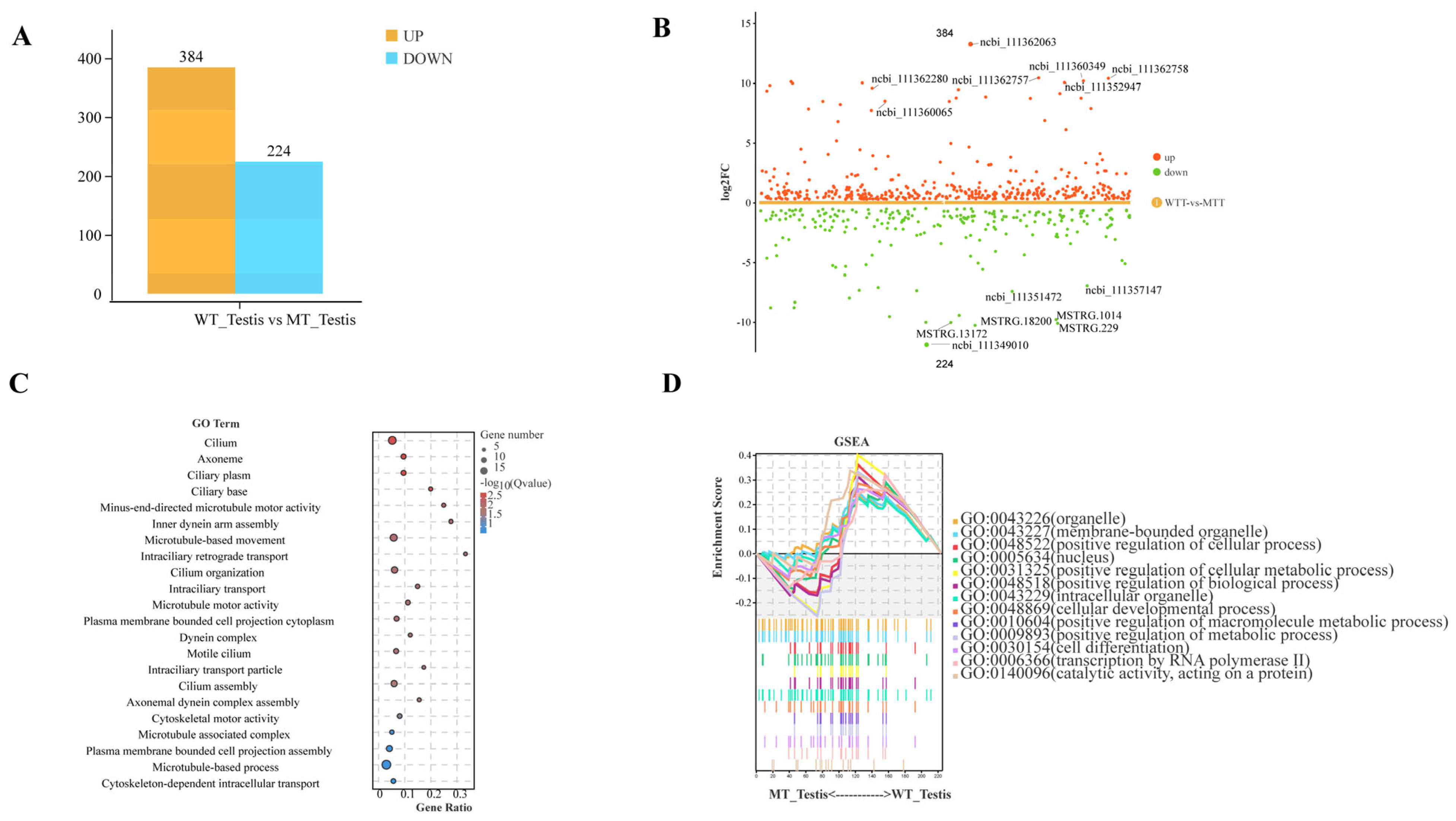
2.5. Transcriptomic Analysis of Spodoptera litura Testis Sheath: Mechanistic Investigation of Sl3030 Mutant Testis Fusion
3. Discussion
4. Materials and Methods
4.1. Spodoptera litura Feeding and Sample Collection for Transcriptome Analysis
4.2. Spodoptera litura Sample Collection for Protomic Analysis
4.3. Protein Digestion and iTRAQ Labeling
4.4. Designing the Target Site and Crafting the Single Guide RNA (sgRNA) for Injection Purposes
4.5. Injection of Cas9/sgRNA Complexes and Subsequent Mutation Analysis
4.6. Data Analysis Methods
4.6.1. Fertility Statistics of CRISPR/Cas9 Gene Knockout Mutants
4.6.2. Measurement of Testicular Size and Thickness of the Peritoneal Sheath
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, S.-W.; Elzaki, M.E.A.; Staehelin, C.; Ma, Z.-H.; Qin, Z.; Wang, R.-L. Exposure to Herbicides Reduces Larval Sensitivity to Insecticides in Spodoptera litura (Lepidoptera: Noctuidae). Insect Sci 2019, 26, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Ranga Rao, G.V.; Wightman, J.A.; Ranga Rao, D.V. World Review of the Natural Enemies and Diseases of Spodoptera litura (F.) (Lepidoptera: Noctuidae). Int. J. Trop. Insect Sci. 1993, 14, 273–284. [Google Scholar] [CrossRef]
- Bragard, C.; Dehnen-Schmutz, K.; Di Serio, F.; Gonthier, P.; Jacques, M.; Jaques Miret, J.A.; Justesen, A.F.; Magnusson, C.S.; Milonas, P.; Navas-Cortes, J.A.; et al. Pest Categorisation of Spodoptera litura. EFSA J. 2019, 17. [Google Scholar] [CrossRef]
- Moussa, M.A.; Zaher, M.A.; Kotby, F.A. Abundance of the Cotton Leaf Worm, Prodenia litura (F.), in Relation to Host Plants. I. Host Plants and Their Effect on Biology (Lepidoptera: Agrotidae-Zenobiinae). Bull. Soc. Entomol. Egypte 1960, 44, 241–251. [Google Scholar]
- Ahmad, M.; Mehmood, R. Monitoring of Resistance to New Chemistry Insecticides in Spodoptera litura (Lepidoptera: Noctuidae) in Pakistan. J. Econ. Entomol. 2015, 108, 1279–1288. [Google Scholar] [CrossRef]
- Simoni, A.; Hammond, A.M.; Beaghton, A.K.; Galizi, R.; Taxiarchi, C.; Kyrou, K.; Meacci, D.; Gribble, M.; Morselli, G.; Burt, A.; et al. A Male-Biased Sex-Distorter Gene Drive for the Human Malaria Vector Anopheles Gambiae. Nat. Biotechnol. 2020, 38, 1054–1060. [Google Scholar] [CrossRef]
- Chen, Y.; Ou, J.; Liu, Y.; Wu, Q.; Wen, L.; Zheng, S.; Li, S.; Feng, Q.; Liu, L. Transcriptomic Analysis of the Testicular Fusion in Spodoptera litura. BMC Genom. 2020, 21, 171. [Google Scholar] [CrossRef]
- Du, Q.; Wen, L.; Zheng, S.; Bi, H.; Huang, Y.; Feng, Q.; Liu, L. Identification and Functional Characterization of Doublesex Gene in the Testis of Spodoptera litura. Insect Sci. 2019, 26, 1000–1010. [Google Scholar] [CrossRef]
- Sutter, A.; Price, T.A.; Wedell, N. The Impact of Female Mating Strategies on the Success of Insect Control Technologies. Curr. Opin. Insect Sci. 2021, 45, 75–83. [Google Scholar] [CrossRef]
- He, X.; Ma, Q.; Jian, B.; Liu, Y.; Wu, Q.; Chen, M.; Feng, Q.; Zhao, P.; Liu, L. Microsurgical Obstruction of Testes Fusion in Spodoptera litura. JoVE 2021, 62524. [Google Scholar] [CrossRef]
- Bilha, J.K.; Brancalhão, R.M.C.; Conte, H. Testicles Fusion of Diatraea Saccharalis F. (Lepidoptera; Crambidae) During Post-Embryonic Development. J. Entomol. Res. Soc. 2012, 14, 1–7. [Google Scholar]
- Loeb, M.J.; De Loof, A.; Gelman, D.B.; Hakim, R.S.; Jaffe, H.; Kochansky, J.P.; Meola, S.M.; Schoofs, L.; Steel, C.; Vafopoulou, X.; et al. Testis Ecdysiotropin, an Insect Gonadotropin That Induces Synthesis of Ecdysteroid. Arch. Insect Biochem. Physiol. 2001, 47, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Nowock, J. Growth and Metamorphosis in the Testes of Ephestia Kühniella in Vitro. J. Insect Physiol. 1973, 19, 941–949. [Google Scholar] [CrossRef]
- Polanska, M.A.; Ciuk, M.A.; Cymborowski, B.; Bebas, P. Germ Cell Death in the Testis and Its Relation to Spermatogenesis in the Wax Moth, Galleria mellonella (Lepidoptera: Pyralidae), Effects of Facultative Diapause. J. Exp. Zool. 2005, 303A, 1013–1029. [Google Scholar] [CrossRef]
- Reinecke, L.H.; Reinecke, J.P.; Adams, T.S. Morphology of the Male Reproductive Tract of Mature Larval, Pupal, and Adult Tobacco Hornworms (Lepidoptera: Sphingidae), Manduca sexta. Ann. Entomol. Soc. Am. 1983, 76, 365–375. [Google Scholar] [CrossRef]
- Salama, H.S. Spermatogenesis and Testicular Development in the Gypsy Moth Porthetria dispar L. Z. Für Angew. Entomol. 1976, 81, 102–110. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Jian, B.; Feng, Q.; Liu, L. Identification and Expression of Integrins during Testicular Fusion in Spodoptera litura. Genes (Basel) 2023, 14, 1452. [Google Scholar] [CrossRef]
- Dong, Y.; Huang, L.; Liu, L. Comparative Analysis of Testicular Fusion in Spodoptera litura (Cutworm) and Bombyx mori (Silkworm): Histological and Transcriptomic Insights. General. Comp. Endocrinol. 2024, 356, 114562. [Google Scholar] [CrossRef]
- Wen, L.; Gong, Q.; Du, Q.; Yu, X.; Feng, Q.; Liu, L. Lacking of Sex-Lethal Gene Lowers the Fertility of Male Reproduction in Spodoptera litura (Lepidoptera). Pestic. Biochem. Physiol. 2022, 184, 105087. [Google Scholar] [CrossRef]
- Bi, H.; Xu, X.; Li, X.; Zhang, Y.; Huang, Y.; Li, K.; Xu, J. CRISPR Disruption of BmOvo Resulted in the Failure of Emergence and Affected the Wing and Gonad Development in the Silkworm Bombyx mori. Insects 2019, 10, 254. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass Spectrometry-Based Proteomics. Nature 2003, 422, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Cheng, C.; Zhang, L. High-Throughput Proteomics: A Methodological Mini-Review. Lab. Investig. 2022, 102, 1170–1181. [Google Scholar] [CrossRef] [PubMed]
- Vidova, V.; Spacil, Z. A Review on Mass Spectrometry-Based Quantitative Proteomics: Targeted and Data Independent Acquisition. Anal. Chim. Acta 2017, 964, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.L.; Huang, Y.N.; Marchese, J.N.; Williamson, B.; Parker, K.; Hattan, S.; Khainovski, N.; Pillai, S.; Dey, S.; Daniels, S.; et al. Multiplexed Protein Quantitation in Saccharomyces Cerevisiae Using Amine-Reactive Isobaric Tagging Reagents. Mol. Cell. Proteom. 2004, 3, 1154–1169. [Google Scholar] [CrossRef]
- Whittington, E.; Zhao, Q.; Borziak, K.; Walters, J.R.; Dorus, S. Characterisation of the Manduca sexta Sperm Proteome: Genetic Novelty Underlying Sperm Composition in Lepidoptera. Insect Biochem. Mol. Biol. 2015, 62, 183–193. [Google Scholar] [CrossRef]
- Barkovits, K.; Chen, W.; Kohl, M.; Bracht, T. Targeted Protein Quantification Using Parallel Reaction Monitoring (PRM). Methods Mol. Biol. 2021, 2228, 145–157. [Google Scholar] [CrossRef]
- Bourmaud, A.; Gallien, S.; Domon, B. Parallel Reaction Monitoring Using Quadrupole-Orbitrap Mass Spectrometer: Principle and Applications. Proteomics 2016, 16, 2146–2159. [Google Scholar] [CrossRef]
- Gallien, S.; Kim, S.Y.; Domon, B. Large-Scale Targeted Proteomics Using Internal Standard Triggered-Parallel Reaction Monitoring (IS-PRM). Mol. Cell Proteom. 2015, 14, 1630–1644. [Google Scholar] [CrossRef]
- Stolze, S.C.; Nakagami, H. Targeted Quantification of Phosphopeptides by Parallel Reaction Monitoring (PRM). Methods Mol. Biol. 2020, 2139, 213–224. [Google Scholar] [CrossRef]
- Jia, Q.; Liu, Y.; Liu, H.; Li, S. Mmp1 and Mmp2 Cooperatively Induce Drosophila Fat Body Cell Dissociation with Distinct Roles. Sci. Rep. 2015, 4, 7535. [Google Scholar] [CrossRef]
- LaFlamme, S.E.; Mathew-Steiner, S.; Singh, N.; Colello-Borges, D.; Nieves, B. Integrin and Microtubule Crosstalk in the Regulation of Cellular Processes. Cell Mol. Life Sci. 2018, 75, 4177–4185. [Google Scholar] [CrossRef] [PubMed]
- Sandbo, N.; Smolyaninova, L.V.; Orlov, S.N.; Dulin, N.O. Control of Myofibroblast Differentiation and Function by Cytoskeletal Signaling. Biochem. (Mosc) 2016, 81, 1698–1708. [Google Scholar] [CrossRef] [PubMed]
- Tuntithavornwat, S.; Shea, D.J.; Wong, B.S.; Guardia, T.; Lee, S.J.; Yankaskas, C.L.; Zheng, L.; Kontrogianni-Konstantopoulos, A.; Konstantopoulos, K. Giant Obscurin Regulates Migration and Metastasis via RhoA-Dependent Cytoskeletal Remodeling in Pancreatic Cancer. Cancer Lett. 2022, 526, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Bullmann, T.; Härtig, W.; Holzer, M.; Arendt, T. Expression of the Embryonal Isoform (0N/3R) of the Microtubule-Associated Protein Tau in the Adult Rat Central Nervous System. J. Comp. Neurol. 2010, 518, 2538–2553. [Google Scholar] [CrossRef]
- Chesta, M.E.; Carbajal, A.; Arce, C.A.; Bisig, C.G. Serum-Induced Neurite Retraction in CAD Cells–Involvement of an ATP-Actin Retractile System and the Lack of Microtubule-Associated Proteins. FEBS J. 2014, 281, 4767–4778. [Google Scholar] [CrossRef]
- Minckley, T.F.; Salvagio, L.A.; Fudge, D.H.; Verhey, K.; Markus, S.M.; Qin, Y. Zn2+ Decoration of Microtubules Arrests Axonal Transport and Displaces Tau, Doublecortin, and MAP2C. J. Cell Biol. 2023, 222, e202208121. [Google Scholar] [CrossRef]
- Reiner, O.; Sapir, T. Mark/Par-1 Marking the Polarity of Migrating Neurons. Adv. Exp. Med. Biol. 2014, 800, 97–111. [Google Scholar] [CrossRef]
- Foster, K.A.; Correia, J.J.; Gilbert, S.P. Equilibrium Binding Studies of Non-Claret Disjunctional Protein (Ncd) Reveal Cooperative Interactions between the Motor Domains. J. Biol. Chem. 1998, 273, 35307–35318. [Google Scholar] [CrossRef]
- Karabasheva, D.; Smyth, J.T. A Novel, Dynein-Independent Mechanism Focuses the Endoplasmic Reticulum around Spindle Poles in Dividing Drosophila Spermatocytes. Sci. Rep. 2019, 9, 12456. [Google Scholar] [CrossRef]
- Komma, D.J.; Horne, A.S.; Endow, S.A. Separation of Meiotic and Mitotic Effects of Claret Non-Disjunctional on Chromosome Segregation in Drosophila. EMBO J. 1991, 10, 419–424. [Google Scholar] [CrossRef]
- Matthies, H.J.; McDonald, H.B.; Goldstein, L.S.; Theurkauf, W.E. Anastral Meiotic Spindle Morphogenesis: Role of the Non-Claret Disjunctional Kinesin-like Protein. J. Cell Biol. 1996, 134, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Ricolo, D.; Araujo, S.J. Coordinated Crosstalk between Microtubules and Actin by a Spectraplakin Regulates Lumen Formation and Branching. eLife 2020, 9, e61111. [Google Scholar] [CrossRef] [PubMed]
- Critchley, D.R. Focal Adhesions—The Cytoskeletal Connection. Curr. Opin. Cell Biol. 2000, 12, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Garcin, C.; Straube, A. Microtubules in Cell Migration. Essays Biochem. 2019, 63, 509–520. [Google Scholar] [CrossRef]
- Hadjisavva, R.; Anastasiou, O.; Ioannou, P.S.; Zheltkova, M.; Skourides, P.A. Adherens Junctions Stimulate and Spatially Guide Integrin Activation and Extracellular Matrix Deposition. Cell Rep. 2022, 40, 111091. [Google Scholar] [CrossRef]
- Citi, S.; Guerrera, D.; Spadaro, D.; Shah, J. Epithelial Junctions and Rho Family GTPases: The Zonular Signalosome. Small GTPases 2014, 5, 1–15. [Google Scholar] [CrossRef]
- Ridley, A.J. Rho GTPases and Actin Dynamics in Membrane Protrusions and Vesicle Trafficking. Trends Cell Biol. 2006, 16, 522–529. [Google Scholar] [CrossRef]
- Steichen, C.; Hervé, C.; Hauet, T.; Bourmeyster, N. Rho GTPases in Kidney Physiology and Diseases. Small GTPases 2022, 13, 141–161. [Google Scholar] [CrossRef]
- Dai, S.; Wu, H.; Chen, H.; Wang, Z.; Yu, X.; Wang, L.; Jia, X.; Qin, C.; Zhu, Y.; Yi, K.; et al. Comparative Transcriptome Analyses under Individual and Combined Nutrient Starvations Provide Insights into N/P/K Interactions in Rice. Plant Physiol. Biochem. 2023, 197, 107642. [Google Scholar] [CrossRef]
- Fukata, M.; Kaibuchi, K. Rho-Family GTPases in Cadherin-Mediated Cell-Cell Adhesion. Nat. Rev. Mol. Cell Biol. 2001, 2, 887–897. [Google Scholar] [CrossRef]
- Kumar, S.; Kapoor, A.; Desai, S.; Inamdar, M.M.; Sen, S. Proteolytic and Non-Proteolytic Regulation of Collective Cell Invasion: Tuning by ECM Density and Organization. Sci. Rep. 2016, 6, 19905. [Google Scholar] [CrossRef]




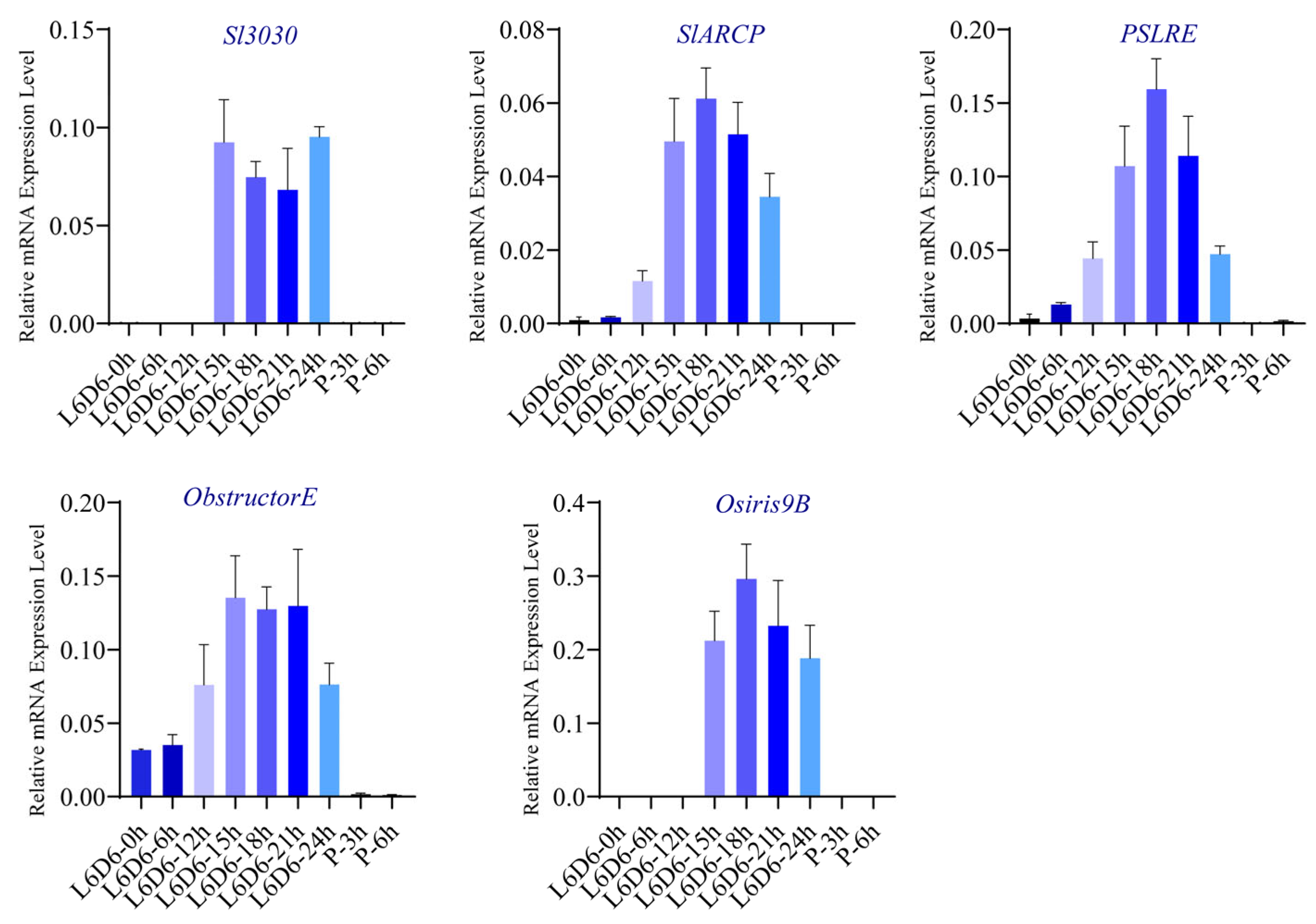

| Accession | Gene_Id | Gene_Description | RNA-Seq_Sl_PP vs. L6D3_log2FC | Padj-1 | RNA-Seq Sl_P3 vs. PP_log2FC | Padj-2 | iTRAQ_log2FC | Padj-3 |
|---|---|---|---|---|---|---|---|---|
| XP_022814918.1 | 111348513 | ATP-dependent RNA helicase DHH1 | 10.83 | 4.29 × 10−25 | −8.95 | 7.81 × 10−31 | 12.73 | 2.64 × 10−2 |
| XP_022837634.1 | 111364820 | serine/arginine repetitive matrix protein | 3.98 | 2.73 × 10−16 | −1.74 | 1.17 × 10−3 | 12.05 | 2.24 × 10−2 |
| XP_022830850.1 | 111359506 | acidic repeat-containing protein-like | 10.80 | 1.50 × 10−29 | −12.61 | 4.35 × 10−32 | 12.05 | 1.26 × 10−3 |
| XP_022825988.1 | 111356021 | uncharacterized LOC111356021 | 6.56 | 9.20 × 10−10 | −7.78 | 3.29 × 10−14 | 5.29 | 2.02 × 10−2 |
| XP_022826940.1 | 111356706 | histone acetyltransferase p300-like | 9.93 | 9.10 × 10−17 | −11.16 | 1.53 × 10−20 | 3.84 | 1.77 × 10−2 |
| XP_022820705.1 | 111352437 | uncharacterized LOC111352437 | 3.87 | 1.37 × 10−19 | −1.70 | 1.80 × 10−4 | 3.51 | 6.01 × 10−3 |
| XP_022836213.1 | 111363605 | atrial natriuretic peptide-converting enzyme-like | 5.29 | 4.40 × 10−8 | −2.20 | 2.11 × 10−2 | 3.30 | 3.60 × 10−3 |
| XP 022822062.1 | 111353322 | apolipoprotein D-like | 5.47 | 9.86 × 10−9 | −3.41 | 7.64 × 10−5 | 3.07 | 3.87 × 10−2 |
| XP_022833045.1 | 111360972 | glycine-rich cell wall structural protein-like | 11.90 | 0.00 | −12.61 | 1.40 × 10−44 | 2.84 | 3.61 × 10−2 |
| XP_022816887.1 | 111349849 | alpha-N-acetylgalactosaminidase-like | 7.28 | 2.84 × 10−9 | −3.92 | 7.47 × 10−5 | 2.60 | 1.10 × 10−3 |
| XP_022817085.1 | 111349954 | serine proteinase stubble-like | 4.93 | 9.38 × 10−6 | −3.65 | 6.49 × 10−4 | 2.44 | 1.22 × 10−3 |
| XP_022819474.1 | 111351669 | uncharacterized LOC111351669 | 1.57 | 4.84 × 10−3 | −1.19 | 3.28 × 10−2 | 2.26 | 9.99 × 10−3 |
| XP_022816962.1 | 111349883 | NAD kinase 2C mitochondrial | 2.12 | 5.43 × 10−7 | −2.41 | 1.42 × 10−10 | 2.21 | 9.72 × 10−4 |
| XP_022814916.1 | 111348511 | polypeptide N-acetylgalactosaminyltransferase 2-like | 3.46 | 5.18 × 10−19 | −1.50 | 4.17 × 10−12 | 1.76 | 5.58 × 10−4 |
| XP_022825632.1 | 111355800 | uncharacterized LOC111355800 | 0.75 | 2.47 × 10−2 | −0.72 | 1.69 × 10−3 | 1.50 | 1.55 × 10−2 |
| XP_022826190.1 | 111356160 | uncharacterized LOC111356160 | 7.70 | 1.39 × 10−9 | −7.65 | 1.23 × 10−9 | 2.85 | 5.18 × 10−3 |
| XP_022826130.1 | 111356111 | uncharacterized LOC111356111 | 7.69 | 2.26 × 10−6 | −10.88 | 1.83 × 10−5 | 4.96 | 8.52 × 10−5 |
| XP_022822850.1 | 111353879 | carboxypeptidase B-like | 8.69 | 1.08 × 10−10 | −7.47 | 9.86 × 10−10 | 9.12 | 1.30 × 10−2 |
| XP_022822903.1 | 111353918 | uncharacterized LOC111353918 | 7.53 | 2.37 × 10−9 | −6.63 | 4.88 × 10−8 | 11.70 | 3.00 × 10−2 |
| XP_022837898.1 | 111365016 | protein split ends-like | 6.01 | 6.48 × 10−14 | −5.07 | 2.63 × 10−11 | 7.06 | 2.80 × 10−2 |
| XP_022814956.1 | 111348538 | zonadhesin | 9.71 | 5.20 × 10−19 | −9.45 | 1.23 × 10−22 | 14.57 | 9.95 × 10−3 |
| XP_022835587.1 | 111363030 | Sl3030 | 10.48 | 5.54 × 10−16 | −10.74 | 3.34 × 10−21 | 12.21 | 1.22 × 10−3 |
| XP_022828128.1 | 111357598 | pollen-specific leucine-rich repeat extensin-like protein 1 | 7.27 | 0.00 | −6.09 | 0.00 | 11.10 | 4.18 × 10−2 |
| XP_022822561.1 | 111353686 | protein obstructor-E | 5.09 | 0.00 | −5.75 | 0.00 | 4.64 | 5.29 × 10−3 |
| XP_022827721.1 | 111357321 | uncharacterized LOC111357321 | 11.34 | 2.14 × 10−18 | −15.27 | 2.47 × 10−17 | 4.89 | 2.25 × 10−4 |
| XP_022820236.1 | 111352109 | protein obstructor-E-like | 2.84 | 9.61 × 10−25 | −2.49 | 7.28 × 10−25 | 1.68 | 4.39 × 10−2 |
| XP_022830682.1 | 111359386 | unconventional myosin-XV | 1.43 | 1.96 × 10−3 | −1.03 | 1.03 × 10−2 | 2.21 | 4.98 × 10−2 |
| XP_022824936.1 | 111355348 | mucin-3A | 8.16 | 0.00E | −1.40 | 8.98 × 10−3 | 1.97 | 5.42 × 10−3 |
| XP_022827651.1 | 111357266 | uncharacterized LOC111357266 | 14.48 | 2.92 × 10−10 | −14.12 | 2.07 × 10−18 | 4.02 | 2.04 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, Y.; Luo, H.; Huang, L.; Liu, L. Multi-Omic Analysis Identifies Key Genes Driving Testicular Fusion in Spodoptera litura. Int. J. Mol. Sci. 2025, 26, 5564. https://doi.org/10.3390/ijms26125564
Dong Y, Luo H, Huang L, Liu L. Multi-Omic Analysis Identifies Key Genes Driving Testicular Fusion in Spodoptera litura. International Journal of Molecular Sciences. 2025; 26(12):5564. https://doi.org/10.3390/ijms26125564
Chicago/Turabian StyleDong, Yaqun, Haoyun Luo, Lihua Huang, and Lin Liu. 2025. "Multi-Omic Analysis Identifies Key Genes Driving Testicular Fusion in Spodoptera litura" International Journal of Molecular Sciences 26, no. 12: 5564. https://doi.org/10.3390/ijms26125564
APA StyleDong, Y., Luo, H., Huang, L., & Liu, L. (2025). Multi-Omic Analysis Identifies Key Genes Driving Testicular Fusion in Spodoptera litura. International Journal of Molecular Sciences, 26(12), 5564. https://doi.org/10.3390/ijms26125564





