Molecular Features Accompanying Richter’s Transformation in Patients with Chronic Lymphocytic Leukemia
Abstract
1. Introduction
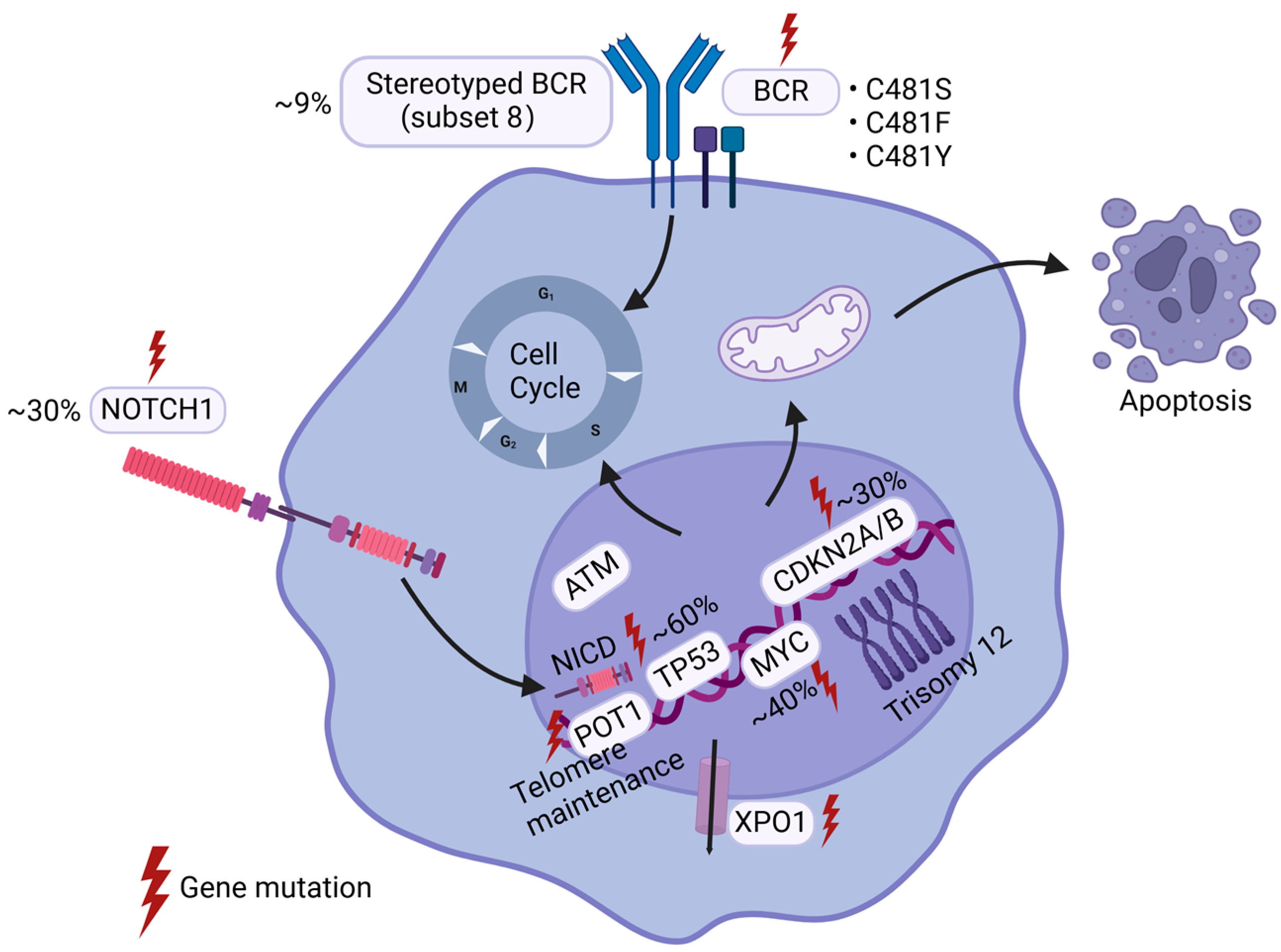
2. Genetic Alterations
2.1. TP53
2.2. CDKN2A/B
2.3. MYC
2.4. NOTCH1
2.5. BCR-Related Gene Alterations
2.6. Other Gene Alterations
3. Gene Instability
4. Signaling Pathways
4.1. PI3K/ATK Pathway
4.2. NF-κB Signaling
4.3. MAPK/RAS/ERK Pathway
4.4. mTOR Pathway
4.5. Other Pathways
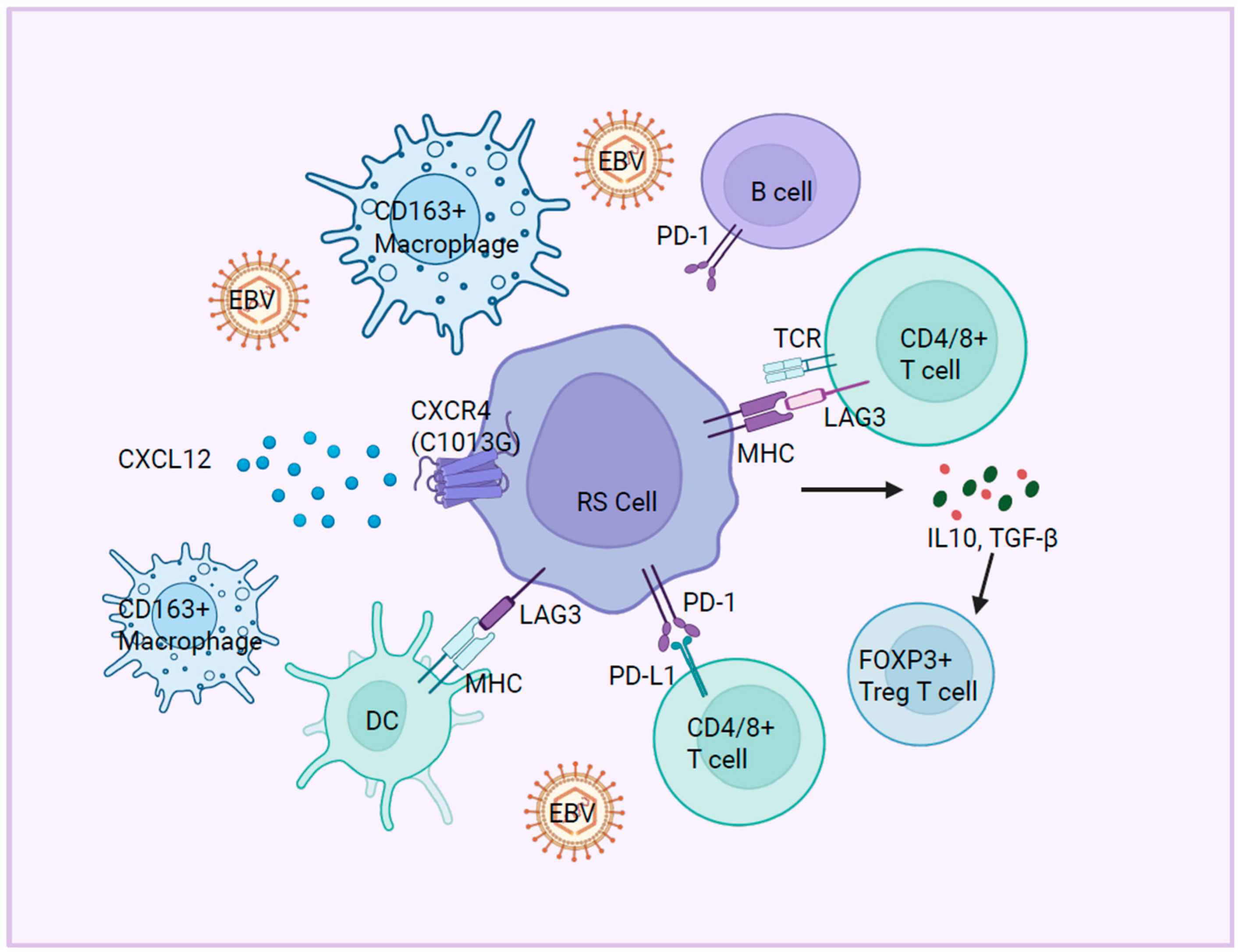
5. Immune Microenvironment
5.1. Immune Checkpoint Molecules
5.2. Immune Cell Dysfunction
5.3. Cytokine and Chemokine Signaling
6. Epigenetic Modifications
6.1. DNA Methylation
6.2. Histone Modifications and Chromatin Remodeling
6.3. Non-Coding RNA Regulation
7. Current Advances in Models for Studying Molecular Pathogenesis of Richter’s Transformation
7.1. Cell Line Models
7.2. Animal Models
7.3. Challenges and Future Directions
8. Conclusion and Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef] [PubMed]
- Shadman, M. Diagnosis and Treatment of Chronic Lymphocytic Leukemia A Review. JAMA 2023, 329, 918–932. [Google Scholar] [CrossRef]
- Hemminki, K.; Hemminki, J.; Försti, A.; Sud, A. Survival in hematological malignancies in the Nordic countries through a half century with correlation to treatment. Leukemia 2023, 37, 854–863. [Google Scholar] [CrossRef]
- Jain, N.; Wierda, W.G.; O’Brien, S. Chronic lymphocytic leukaemia. Lancet 2024, 404, 694–706. [Google Scholar] [CrossRef]
- Smyth, E.; Eyre, T.A.; Cheah, C.Y. Emerging Therapies for the Management of Richter Transformation. J. Clin. Oncol. 2023, 41, 395–409. [Google Scholar] [CrossRef] [PubMed]
- Parikh, S.A.; Kay, N.E.; Shanafelt, T.D. How we treat Richter syndrome. Blood 2014, 123, 1647–1657. [Google Scholar] [CrossRef]
- Fraser, C.R.; Wang, W.; Gomez, M.; Zhang, T.; Mathew, S.; Furman, R.R.; Knowles, D.M.; Orazi, A.; Tam, W. Transformation of chronic lymphocytic leukemia/small lymphocytic lymphoma to interdigitating dendritic cell sarcoma: Evidence for transdifferentiation of the lymphoma clone. Am. J. Clin. Pathol. 2009, 132, 928–939. [Google Scholar] [CrossRef]
- Shao, H.P.; Xi, L.Q.; Raffeld, M.; Feldman, A.L.; Ketterling, R.P.; Knudson, R.; Rodriguez-Canales, J.; Hanson, J.; Pittaluga, S.; Jaffe, E.S. Clonally related histiocytic/dendritic cell sarcoma and chronic lymphocytic leukemia/small lymphocytic lymphoma: A study of seven cases. Mod. Pathol. 2011, 24, 1421–1432. [Google Scholar] [CrossRef] [PubMed]
- Shahidi, H.; Leslie, W.T.; Wool, N.L.; Gregory, S.A. Transformation of chronic lymphocytic leukemia to immunoblastic lymphoma (Richter’s syndrome). Med. Pediatr. Oncol. 1997, 29, 146–151. [Google Scholar] [CrossRef]
- Rossi, D.; Spina, V.; Deambrogi, C.; Rasi, S.; Laurenti, L.; Stamatopoulos, K.; Arcaini, L.; Lucioni, M.; Rocque, G.B.; Xu-Monette, Z.Y.; et al. The genetics of Richter syndrome reveals disease heterogeneity and predicts survival after transformation. Blood 2011, 117, 3391–3401. [Google Scholar] [CrossRef]
- Favini, C.; Talotta, D.; Almasri, M.; Andorno, A.; Rasi, S.; Adhinaveni, R.; Kogila, S.; Awikeh, B.; Schipani, M.; Boggione, P.; et al. Clonally unrelated Richter syndrome are truly de novo diffuse large B-cell lymphomas with a mutational profile reminiscent of clonally related Richter syndrome. Br. J. Haematol. 2022, 198, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, A.M.; Gaidano, G.; Mouhssine, S. Immunological Aspects of Richter Syndrome: From Immune Dysfunction to Immunotherapy. Cancers 2023, 15, 1015. [Google Scholar] [CrossRef]
- Hess, B.; Kalmuk, J.; Znoyko, I.; Schandl, C.A.; Wagner-Johnston, N.; Mazzoni, S.; Hendrickson, L.; Chiad, Z.; Greenwell, I.B.; Wolff, D.J. Clinical utility of chromosomal microarray in establishing clonality and high risk features in patients with Richter transformation. Cancer Genet. 2022, 260, 18–22. [Google Scholar] [CrossRef]
- Eyre, T.A.; Clifford, R.; Bloor, A.; Boyle, L.; Roberts, C.; Cabes, M.; Collins, G.P.; Devereux, S.; Follows, G.; Fox, C.P.; et al. NCRI phase II study of CHOP in combination with ofatumumab in induction and maintenance in newly diagnosed Richter syndrome. Br. J. Haematol. 2016, 175, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Rogers, K.A.; Huang, Y.; Ruppert, A.S.; Salem, G.; Stephens, D.M.; Heerema, N.A.; Andritsos, L.A.; Awan, F.T.; Byrd, J.C.; Flynn, J.M.; et al. A single-institution retrospective cohort study of first-line R-EPOCH chemoimmunotherapy for Richter syndrome demonstrating complex chronic lymphocytic leukaemia karyotype as an adverse prognostic factor. Br. J. Haematol. 2018, 180, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Thompson, P.A.; Siddiqi, T. Treatment of Richter’s syndrome. Hematol. Am. Soc. Hematol. Educ. Program 2022, 2022, 329–336. [Google Scholar] [CrossRef]
- Hampel, P.J.; Rabe, K.G.; Wang, Y.C.; Hwang, S.R.; Kenderian, S.S.; Muchtar, E.; Leis, J.F.; Koehler, A.B.; Tsang, M.; Hilal, T.; et al. Incidence of Richter transformation of chronic lymphocytic leukemia/small lymphocytic lymphoma in the targeted therapy era. Leukemia 2025, 39, 503–507. [Google Scholar] [CrossRef]
- Ramadan, R.; Alzaki, A.; Leitch, H.; Ramadan, K. PB1881: Ibrutinib Reduces the Risk of Richter’s Transformation in Previously Treated Cll Patients: A Population-Based Study in British Columbia. Hemasphere 2022, 6, 1761–1762. [Google Scholar] [CrossRef]
- Hampel, P.J.; Swaminathan, M.; Rogers, K.A.; Parry, E.M.; Burger, J.A.; Davids, M.S.; Ding, W.; Ferrajoli, A.; Hyak, J.M.; Jain, N.; et al. A multicenter study of venetoclax-based treatment for patients with Richter transformation of chronic lymphocytic leukemia. Blood Adv. 2024, 8, 2342–2350. [Google Scholar] [CrossRef]
- Crombie, J.L.; Ryan, C.E.; Ren, Y.; Tyekucheva, S.; Carey, C.; Zou, A.R.; Normilus, S.; Montegaard, J.; Soumerai, J.D.; Bhandari, S.; et al. A Phase 2 Study of Duvelisib and Venetoclax in Patients with Relapsed or Refractory (R/R) Chronic Lymphocytic Leukemia (CLL) or Richter’s Syndrome (RS). Blood 2024, 144, 4628–4629. [Google Scholar] [CrossRef]
- Kittai, A.S.; Huang, Y.; Miller, S.; Allan, J.N.; Bhat, S.A.; Bond, D.A.; Brander, D.M.; Byrd, J.C.; Chavez, J.C.; Chong, E.L.S.; et al. Outcomes of patients with Richter transformation who received no prior chemoimmunotherapy for their CLL. Blood Cancer J. 2025, 15, 23. [Google Scholar] [CrossRef]
- Pallasch, C.P. Cell cycle control in Richter transformation. Blood 2021, 138, 1005–1007. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, G.; Rasi, S.; Rossi, D.; Trifonov, V.; Khiabanian, H.; Ma, J.; Grunn, A.; Fangazio, M.; Capello, D.; Monti, S.; et al. Analysis of the chronic lymphocytic leukemia coding genome: Role of mutational activation. J. Exp. Med. 2011, 208, 1389–1401. [Google Scholar] [CrossRef] [PubMed]
- Audil, H.Y.; Kosydar, S.R.; Larson, D.P.; Parikh, S.A. Richter Transformation of Chronic Lymphocytic Leukemia-Are We Making Progress? Curr. Hematol. Malig. Rep. 2023, 18, 144–157. [Google Scholar] [CrossRef]
- Penter, L.; Gohil, S.H.; Lareau, C.; Ludwig, L.S.; Parry, E.M.; Huang, T.; Li, S.Q.; Zhang, W.D.; Livitz, D.; Leshchiner, I.; et al. Longitudinal Single-Cell Dynamics of Chromatin Accessibility and Mitochondrial Mutations in Chronic Lymphocytic Leukemia Mirror Disease History. Cancer Discov. 2021, 11, 3048–3063. [Google Scholar] [CrossRef]
- Kuruvilla, J.; Savona, M.; Baz, R.; Mau-Sorensen, P.M.; Gabrail, N.; Garzon, R.; Stone, R.; Wang, M.; Savoie, L.; Martin, P.; et al. Selective inhibition of nuclear export with selinexor in patients with non-Hodgkin lymphoma. Blood 2017, 129, 3175–3183. [Google Scholar] [CrossRef] [PubMed]
- Woroniecka, R.; Rymkiewicz, G.; Bystydzienski, Z.; Pienkowska-Grela, B.; Rygier, J.; Malawska, N.; Wojtkowska, K.; Goral, N.; Blachnio, K.; Chmielewski, M.; et al. Cytogenomic features of Richter transformation. Mol. Cytogenet. 2023, 16, 31. [Google Scholar] [CrossRef]
- Jiao, Y.; Feng, Y.P.; Wang, X.L. Regulation of Tumor Suppressor Gene and Encoded p16-INK4a Protein by Covalent Modifications. Biochemistry 2018, 83, 1289–1298. [Google Scholar] [CrossRef]
- Krimpenfort, P.; IJpenberg, A.; Song, J.Y.; van der Valk, M.; Nawijn, M.; Zevenhoven, J.; Berns, A. p15 is a critical tumour suppressor in the absence of p16. Nature 2007, 448, 943–946. [Google Scholar] [CrossRef]
- Suzuki, H.; Zhou, X.L.; Yin, J.; Lei, J.Y.; Jiang, H.Y.; Suzuki, Y.; Chan, T.; Hannon, G.J.; Mergner, W.J.; Abraham, J.M.; et al. Intragenic Mutations of Cdkn2b and Cdkn2a in Primary Human Esophageal Cancers. Hum. Mol. Genet. 1995, 4, 1883–1887. [Google Scholar] [CrossRef]
- Chigrinova, E.; Rinaldi, A.; Kwee, I.; Rossi, D.; Rancoita, P.M.V.; Strefford, J.C.; Oscier, D.; Stamatopoulos, K.; Papadaki, T.; Berger, F.; et al. Two main genetic pathways lead to the transformation of chronic lymphocytic leukemia to Richter syndrome. Blood 2013, 122, 2673–2682. [Google Scholar] [CrossRef]
- Teierle, S.M.; Huang, Y.; Kittai, A.S.; Bhat, S.A.; Grever, M.; Rogers, K.A.; Zhao, W.; Jones, D.; Byrd, J.C.; Avenarius, M.R.; et al. Characteristics and outcomes of patients with CLL and CDKN2A/B deletion by fluorescence in situ hybridization. Blood Adv. 2023, 7, 7239–7242. [Google Scholar] [CrossRef]
- Teierle, S.M.; Huang, Y.; Kittai, A.S.; Bhat, S.A.; Grever, M.R.; Rogers, K.A.; Zhao, W.Q.; Jones, D.; Byrd, J.C.; Avenarius, M.R.; et al. FISH Analysis of CDKN2A/B Deletion in Chronic Lymphocytic Leukemia. Blood 2022, 140, 7016–7017. [Google Scholar] [CrossRef]
- Chakraborty, S.; Martines, C.; Porro, F.; Fortunati, I.; Bonato, A.; Dimishkovska, M.; Piazza, S.; Yadav, B.S.; Innocenti, I.; Fazio, R.; et al. B-cell receptor signaling and genetic lesions in TP53 and CDKN2A/CDKN2B cooperate in Richter transformation. Blood 2021, 138, 1053–1066. [Google Scholar] [CrossRef]
- Chakraborty, S.; Fortunati, I.; Martines, C.; Yadav, B.; Dimishkovska, M.; Innocenti, I.; Fazio, R.; D’Arena, G.; Dimovski, A.; Laurenti, L.J.B. A CRISPR/Cas9-Generated Murine Model Reveals Cooperation between BCR Signaling and CDKN2A/2B and TP53 Disruption in Richter Syndrome. Blood 2019, 134, 4278. [Google Scholar] [CrossRef]
- Lucas, F.; Rogers, K.A.; Harrington, B.K.; Pan, A.; Yu, L.; Breitbach, J.; Bundschuh, R.; Goettl, V.M.; Hing, Z.A.; Kanga, P.; et al. Eμ-TCL1xMyc: A Novel Mouse Model for Concurrent CLL and B-Cell Lymphoma. Clin. Cancer Res. 2019, 25, 6260–6273. [Google Scholar] [CrossRef]
- Fabbri, G.; Khiabanian, H.; Holmes, A.B.; Wang, J.G.; Messina, M.; Mullighan, C.G.; Pasqualucci, L.; Rabadan, R.; Dalla-Favera, R. Genetic lesions associated with chronic lymphocytic leukemia transformation to Richter syndrome. J. Exp. Med. 2013, 210, 2273–2288. [Google Scholar] [CrossRef]
- Parry, E.M.; Leshchiner, I.; Guiéze, R.; Johnson, C.; Tausch, E.; Parikh, S.A.; Lemvigh, C.; Broséus, J.; Hergalant, S.; Messer, C.; et al. Evolutionary history of transformation from chronic lymphocytic leukemia to Richter syndrome. Nat. Med. 2023, 29, 158–169. [Google Scholar] [CrossRef]
- Nadeu, F.; Royo, R.; Massoni-Badosa, R.; Garcia-Torre, B.; Duran-Ferrer, M.; Dawson, K.J.; Kulis, M.; Diaz-Navarro, A.; Villamor, N.; Melero, J.L.; et al. Detection of early seeding of Richter transformation in chronic lymphocytic leukemia. Nat. Med. 2022, 28, 1662–1671. [Google Scholar] [CrossRef]
- Iyer, P.; Zhang, B.; Liu, T.T.; Jin, M.L.; Hart, K.; Zhang, J.B.; Siegert, V.; Remke, M.; Wang, X.S.; Yu, L.; et al. MGA deletion leads to Richter’s transformation by modulating mitochondrial OXPHOS. Sci. Transl. Med. 2024, 16, eadg7915. [Google Scholar] [CrossRef]
- Iyer, P.; Jiang, B.; Venkataraman, G.; Song, J.Y.; Siddiqi, T.; Rosen, S.T.; Danilov, A.; Wang, L.J.B. Glycerolipid Metabolism Via the MGA-MYC-NME1-PGP Axis As a Key Regulator of Oxidative Phosphorylation in Richter’s Transformation. Blood 2023, 142, 79. [Google Scholar] [CrossRef]
- Benedetti, D.; Tissino, E.; Pozzo, F.; Bittolo, T.; Caldana, C.; Perini, C.; Martorelli, D.; Bravin, V.; D’Agaro, T.; Rossi, F.M.; et al. NOTCH1 mutations are associated with high CD49d expression in chronic lymphocytic leukemia: Link between the NOTCH1 and the NF-κB pathways. Leukemia 2018, 32, 654–662. [Google Scholar] [CrossRef]
- Xu, Z.S.; Zhang, J.S.; Zhang, J.Y.; Wu, S.Q.; Xiong, D.L.; Chen, H.J.; Chen, Z.Z.; Zhan, R. Constitutive activation of NF-κB signaling by NOTCH1 mutations in chronic lymphocytic leukemia. Oncol. Rep. 2015, 33, 1609–1614. [Google Scholar] [CrossRef][Green Version]
- Rossi, D.; Rasi, S.; Spina, V.; Fangazio, M.; Monti, S.; Greco, M.; Ciardullo, C.; Famà, R.; Cresta, S.; Bruscaggin, A.; et al. Different impact of NOTCH1 and SF3B1 mutations on the risk of chronic lymphocytic leukemia transformation to Richter syndrome. Br. J. Haematol. 2012, 158, 426–429. [Google Scholar] [CrossRef]
- Arruga, F.; Bracciamà, V.; Vitale, N.; Vaisitti, T.; Gizzi, K.; Yeomans, A.; Coscia, M.; D’Arena, G.; Gaidano, G.; Allan, J.N.; et al. Bidirectional linkage between the B-cell receptor and NOTCH1 in chronic lymphocytic leukemia and in Richter’s syndrome: Therapeutic implications. Leukemia 2020, 34, 462–477. [Google Scholar] [CrossRef]
- Hales, E.C.; Taub, J.W.; Matherly, L.H. New insights into Notch1 regulation of the PI3K-AKT-mTOR1 signaling axis: Targeted therapy of γ-secretase inhibitor resistant T-cell acute lymphoblastic leukemia. Cell. Signal. 2014, 26, 149–161. [Google Scholar] [CrossRef]
- Platonova, N.; Manzo, T.; Mirandola, L.; Colombo, M.; Calzavara, E.; Vigolo, E.; Cermisoni, G.C.; De Simone, D.; Garavelli, S.; Cecchinato, V.; et al. PI3K/AKT signaling inhibits NOTCH1 lysosome-mediated degradation. Genes Chromosomes Cancer 2015, 54, 516–526. [Google Scholar] [CrossRef]
- Villegas, S.N.; Gombos, R.; García-López, L.; Gutiérrez-Pérez, I.; García-Castillo, J.; Vallejo, D.M.; Da Ros, V.G.; Ballesta-Illán, E.; Mihály, J.; Dominguez, M. PI3K/Akt Cooperates with Oncogenic Notch by Inducing Nitric Oxide-Dependent Inflammation. Cell Rep. 2018, 22, 2541–2549. [Google Scholar] [CrossRef]
- Palomero, T.; Lim, W.K.; Odom, D.T.; Sulis, M.L.; Real, P.J.; Margolin, A.; Barnes, K.C.; O’Neil, J.; Neuberg, D.; Weng, A.P.; et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. USA 2006, 103, 18261–18266. [Google Scholar] [CrossRef]
- Kohlhaas, V.; Blakemore, S.J.; Al-Maarri, M.; Nickel, N.; Pal, M.; Roth, A.; Hövelmeyer, N.; Schäfer, S.C.; Knittel, G.; Lohneis, P.; et al. Active Akt signaling triggers CLL toward Richter transformation via overactivation of Notch1. Blood 2021, 137, 646–660. [Google Scholar] [CrossRef]
- Rossi, D.; Spina, V.; Gaidano, G. Biology and treatment of Richter syndrome. Blood 2018, 131, 2761–2772. [Google Scholar] [CrossRef]
- Rossi, D.; Spina, V.; Cerri, M.; Rasi, S.; Deambrogi, C.; De Paoli, L.; Laurenti, L.; Maffei, R.; Forconi, F.; Bertoni, F.; et al. Stereotyped B-Cell Receptor Is an Independent Risk Factor of Chronic Lymphocytic Leukemia Transformation to Richter Syndrome. Clin. Cancer Res. 2009, 15, 4415–4422. [Google Scholar] [CrossRef]
- Yosifov, D.Y.; Stilgenbauer, S. Richter transformation: Epigenetics to blame? Blood 2023, 141, 2915–2917. [Google Scholar] [CrossRef]
- Chu, C.C.; Catera, R.; Hatzi, K.; Yan, X.J.; Zhang, L.; Wang, X.B.; Fales, H.M.; Allen, S.L.; Kolitz, J.E.; Rai, K.R.; et al. Chronic lymphocytic leukemia antibodies with a common stereotypic rearrangement recognize nonmuscle myosin heavy chain IIA. Blood 2008, 112, 5122–5129. [Google Scholar] [CrossRef]
- Klintman, J.; Appleby, N.; Stamatopoulos, B.; Ridout, K.; Eyre, T.A.; Robbe, P.; Pascua, L.L.; Knight, S.J.L.; Dreau, H.; Cabes, M.; et al. Genomic and transcriptomic correlates of Richter transformation in chronic lymphocytic leukemia. Blood 2021, 137, 2800–2816. [Google Scholar] [CrossRef]
- Kanagal-Shamanna, R.; Jain, P.; Patel, K.P.; Routbort, M.; Bueso-Ramos, C.; Alhalouli, T.; Khoury, J.D.; Luthra, R.; Ferrajoli, A.; Keating, M.; et al. Targeted multigene deep sequencing of Bruton tyrosine kinase inhibitor-resistant chronic lymphocytic leukemia with disease progression and Richter transformation. Cancer 2019, 125, 559–574. [Google Scholar] [CrossRef]
- Schweighoffer, E.; Vanes, L.; Mathiot, A.; Nakamura, T.; Tybulewicz, V.L.J. Unexpected requirement for ZAP-70 in pre-B cell development and allelic exclusion. Immunity 2003, 18, 523–533. [Google Scholar] [CrossRef]
- Parikh, S.A.; Rabe, K.G.; Call, T.G.; Zent, C.S.; Habermann, T.M.; Ding, W.; Leis, J.F.; Schwager, S.M.; Hanson, C.A.; Macon, W.R.; et al. Diffuse large B-cell lymphoma (Richter syndrome) in patients with chronic lymphocytic leukaemia (CLL): A cohort study of newly diagnosed patients. Br. J. Haematol. 2013, 162, 774–782. [Google Scholar] [CrossRef]
- Villamor, N.; Conde, L.; Martínez-Trillos, A.; Cazorla, M.; Navarro, A.; Beà, S.; López, C.; Colomer, D.; Pinyol, M.; Aymerich, M.; et al. mutations identify a genetic subgroup of chronic lymphocytic leukemia patients with high risk of transformation and poor outcome. Leukemia 2013, 27, 1100–1106. [Google Scholar] [CrossRef]
- Jelloul, F.Z.; Yang, R.C.K.; Wang, P.; Garces, S.; Kanagal-Shamanna, R.; Ok, C.Y.; Loghavi, S.; Routbort, M.J.; Zuo, Z.; Yin, C.C.; et al. Non-coding mutations in chronic lymphocytic leukemia negatively impact prognosis. Am. J. Hematol. 2022, 97, E100–E102. [Google Scholar] [CrossRef]
- Chen, J.Y.; Moore, A.; Ringshausen, I. ZAP-70 Shapes the Immune Microenvironment in B Cell Malignancies. Front. Oncol. 2020, 10, 595832. [Google Scholar] [CrossRef]
- Müller, D.J.; Wirths, S.; Fuchs, A.R.; Märklin, M.; Heitmann, J.S.; Sturm, M.; Haap, M.; Kirschniak, A.; Sasaki, Y.; Kanz, L.; et al. Loss of NFAT2 expression results in the acceleration of clonal evolution in chronic lymphocytic leukemia. J. Leukoc. Biol. 2019, 105, 531–538. [Google Scholar] [CrossRef]
- Märklin, M.; Fuchs, A.R.; Tandler, C.; Heitmann, J.S.; Salih, H.R.; Kauer, J.; Quintanilla-Martinez, L.; Wirths, S.; Kopp, H.G.; Müller, M.R. Genetic Loss of LCK Kinase Leads to Acceleration of Chronic Lymphocytic Leukemia. Front. Immunol. 2020, 11, 1995. [Google Scholar] [CrossRef]
- Rompietti, C.; Adamo, F.M.; Sorcini, D.; De Falco, F.; Stella, A.; Martino, G.; Bigerna, B.; Dorillo, E.; Barcelos, E.C.S.; Esposito, A.; et al. Bcor loss promotes Richter transformation of chronic lymphocytic leukemia associated with Notch1 activation in mice. Leukemia 2025, 39, 1157–1168. [Google Scholar] [CrossRef]
- Reiniger, L.; Bödör, C.; Bognár, A.; Balogh, Z.; Csomor, J.; Szepesi, A.; Kopper, L.; Matolcsy, A. Richter’s and prolymphocytic transformation of chronic lymphocytic leukemia are associated with high mRNA expression of activation-induced cytidine deaminase and aberrant somatic hypermutation. Leukemia 2006, 20, 1089–1095. [Google Scholar] [CrossRef]
- De Silva, N.S.; Klein, U. Dynamics of B cells in germinal centres. Nat. Rev. Immunol. 2015, 15, 137–148. [Google Scholar] [CrossRef]
- Groelly, F.J.; Fawkes, M.; Dagg, R.A.; Blackford, A.N.; Tarsounas, M. Targeting DNA damage response pathways in cancer. Nat. Rev. Cancer 2023, 23, 78–94. [Google Scholar] [CrossRef]
- Rossi, D.; Bodoni, C.L.; Genuardi, E.; Monitillo, L.; Drandi, D.; Cerri, M.; Deambrogi, C.; Ricca, I.; Rocci, A.; Ferrero, S.; et al. Telomere length is an independent predictor of survival, treatment requirement and Richter’s syndrome transformation in chronic lymphocytic leukemia. Leukemia 2009, 23, 1062–1072. [Google Scholar] [CrossRef]
- Ramsay, A.J.; Quesada, V.; Foronda, M.; Conde, L.; Martínez-Trillos, A.; Villamor, N.; Rodríguez, D.; Kwarciak, A.; Garabaya, C.; Gallardo, M.; et al. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat. Genet. 2013, 45, 526–530. [Google Scholar] [CrossRef]
- Visentin, A.; Bonaldi, L.; Rigolin, G.M.; Mauro, F.R.; Martines, A.; Frezzato, F.; Pravato, S.; Gargarella, L.R.; Bardi, M.A.; Cavallari, M.; et al. The complex karyotype landscape in chronic lymphocytic leukemia allows the refinement of the risk of Richter syndrome transformation. Haematologica 2022, 107, 868–876. [Google Scholar] [CrossRef]
- Cavallari, M.; Cavazzini, F.; Bardi, A.; Volta, E.; Melandri, A.; Tammiso, E.; Saccenti, E.; Lista, E.; Quaglia, F.M.; Urso, A.; et al. Biological significance and prognostic/predictive impact of complex karyotype in chronic lymphocytic leukemia. Oncotarget 2018, 9, 34398–34412. [Google Scholar] [CrossRef]
- Fidai, S.; Sukhanova, M.; Chiu, B.C.H.; Wang, Y.L.L.; Stock, W.; Riedell, P.A.; Smith, S.M.; Hyjek, E.; Venkataraman, G. TP53 Aberrations By FISH in CLL and Complex Karyotype at Transformation Predict for Worse Outcome in Diffuse Large B-Cell Lymphoma—Richter Transformation: A Single Institution Series of 75 DLBCL-RT Cases. Blood 2018, 132 (Suppl. S1), 2984. [Google Scholar] [CrossRef]
- Woroniecka, R.; Rymkiewicz, G.; Grygalewicz, B.; Blachnio, K.; Rygier, J.; Jarmuz-Szymczak, M.; Ratajczak, B.; Pienkowska-Grela, B. Cytogenetic and Flow Cytometry Evaluation of Richter Syndrome Reveals MYC, CDKN2A, IGH Alterations With Loss of CD52, CD62L and Increase of CD71 Antigen Expression as the Most Frequent Recurrent Abnormalities. Am. J. Clin. Pathol. 2015, 143, 25–35. [Google Scholar] [CrossRef]
- Patton, J.T.; Woyach, J.A. Targeting the B cell receptor signaling pathway in chronic lymphocytic leukemia. Semin. Hematol. 2024, 61, 100–108. [Google Scholar] [CrossRef]
- Woyach, J.A.; Johnson, A.J.; Byrd, J.C. The B-cell receptor signaling pathway as a therapeutic target in CLL. Blood 2012, 120, 1175–1184. [Google Scholar] [CrossRef]
- Messana, V.G.; Fasci, A.; Vitale, N.; Micillo, M.; Rovere, M.; Pesce, N.A.; Martines, C.; Efremov, D.G.; Vaisitti, T.; Deaglio, S. A molecular circuit linking the BCR to the NAD biosynthetic enzyme NAMPT is an actionable target in Richter syndrome. Blood Adv. 2024, 8, 1920–1933. [Google Scholar] [CrossRef]
- Ten Hacken, E.; Sewastianik, T.; Yin, S.; Hoffmann, G.B.; Gruber, M.; Clement, K.; Penter, L.; Redd, R.A.; Ruthen, N.; Hergalant, S.; et al. In Vivo Modeling of CLL Transformation to Richter Syndrome Reveals Convergent Evolutionary Paths and Therapeutic Vulnerabilities. Blood Cancer Discov. 2023, 4, 150–169. [Google Scholar] [CrossRef]
- Rohan, P.; Binato, R.; Abdelhay, E. NF-ΚB Activation as a Key Driver in Chronic Lymphocytic Leukemia Evolution to Richter’s Syndrome: Unraveling the Influence of Immune Microenvironment Dynamics. Genes 2024, 15, 1434. [Google Scholar] [CrossRef]
- Vaisitti, T.; Gaudino, F.; Ouk, S.; Moscvin, M.; Vitale, N.; Serra, S.; Arruga, F.; Zakrzewski, J.L.; Liou, H.C.; Allan, J.N.; et al. Targeting metabolism and survival in chronic lymphocytic leukemia and Richter syndrome cells by a novel NF-κB inhibitor. Haematologica 2017, 102, 1878–1889. [Google Scholar] [CrossRef]
- Maier, J.; Lechel, A.; Marienfeld, R.; Barth, T.F.E.; Moeller, P.; Mellert, K. CARD9 Forms an Alternative CBM Complex in Richter Syndrome. Cancers 2022, 14, 531. [Google Scholar] [CrossRef]
- Parigger, T.; Drothler, S.; Scherhäufl, C.; Gassner, F.J.; Schubert, M.; Steiner, M.; Höpner, J.P.; Hödlmoser, A.; Schultheis, L.; Abu Bakar, A.; et al. Oncogenic MTOR Signaling Axis Compensates BTK Inhibition in a Chronic Lymphocytic Leukemia Patient with Richter Transformation: A Case Report and Review of the Literature. Acta Haematol. 2024, 147, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Rigo, A.; Vaisitti, T.; Laudanna, C.; Terrabuio, E.; Micillo, M.; Frusteri, C.; D’Ulivo, B.; Merigo, F.; Sbarbati, A.; Mellert, K.; et al. Decreased apoptotic priming and loss of BCL-2 dependence are functional hallmarks of Richter’s syndrome. Cell Death Dis. 2024, 15, 323. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.F.; Glytsou, C.; Zhou, H.; Narang, S.; Reyna, D.E.; Lopez, A.; Sakellaropoulos, T.; Gong, Y.X.; Kloetgen, A.; Yap, Y.S.; et al. Targeting Mitochondrial Structure Sensitizes Acute Myeloid Leukemia to Venetoclax Treatment. Cancer Discov. 2019, 9, 890–909. [Google Scholar] [CrossRef]
- Ten Hacken, E.; Yin, S.Y.; Sewastianik, T.; Penter, L.; Ruthen, N.; Gruber, M.; Parry, E.M.; Guieze, R.; Redd, R.A.; Uduman, M.; et al. Dissecting Richter’s Syndrome in a Multiplexed CRISPR-Based Mouse Model Reveals Key Changes in MYC, Interferon and BCR Signaling Underlying Transformation. Blood 2021, 138, 251. [Google Scholar] [CrossRef]
- Lewis, R.; Maurer, H.C.; Singh, N.; Gonzalez-Menendez, I.; Wirth, M.; Schick, M.; Zhang, L.; Isaakidis, K.; Scherger, A.K.; Schulze, V.; et al. CXCR4 hyperactivation cooperates with TCL1 in CLL development and aggressiveness. Leukemia 2021, 35, 2895–2905. [Google Scholar] [CrossRef]
- Lutzny, G.; Kocher, T.; Schmidt-Supprian, M.; Rudelius, M.; Klein-Hitpass, L.; Finch, A.J.; Dürig, J.; Wagner, M.; Haferlach, C.; Kohlmann, A.; et al. Protein Kinase C-β-Dependent Activation of NF-κB in Stromal Cells Is Indispensable for the Survival of Chronic Lymphocytic Leukemia B Cells In Vivo. Cancer Cell 2013, 23, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Condoluci, A.; Rossi, D. Biology and Treatment of Richter Transformation. Front. Oncol. 2022, 12, 829983. [Google Scholar] [CrossRef]
- Wang, Y.C.; Sinha, S.; Wellik, L.E.; Secreto, C.R.; Rech, K.L.; Call, T.G.; Parikh, S.A.; Kenderian, S.S.; Muchtar, E.; Hayman, S.R.; et al. Distinct immune signatures in chronic lymphocytic leukemia and Richter syndrome. Blood Cancer J. 2021, 11, 86. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; International Agency for Research on Cancer: Lyon, France, 2017; Volume 2. [Google Scholar]
- He, R.; Ding, W.; Viswanatha, D.S.; Chen, D.; Shi, M.; Van Dyke, D.; Tian, S.L.; Dao, L.N.; Parikh, S.A.; Shanafelt, T.D.; et al. PD-1 Expression in Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma (CLL/SLL) and Large B-cell Richter Transformation (DLBCL-RT): A Characteristic Feature of DLBCL-RT and Potential Surrogate Marker for Clonal Relatedness. Am. J. Surg. Pathol. 2018, 42, 843–854. [Google Scholar] [CrossRef]
- Sinha, S.; Price-Troska, T.; Tian, S.; Secreto, C.R.; Wu, X.S.; Call, T.G.; He, R.; Parikh, S.A.; Kenderian, S.S.; Leis, J.F.; et al. PD-1 Overexpression in Richter’s Transformation (RT) and Aggressive Chronic Lymphocytic Leukemia (CLL) after Progression on Ibrutinib Increases Bcl-2 Expression Via Akt/mTOR Pathway. Blood 2018, 132, 586. [Google Scholar] [CrossRef]
- Parry, E.M.; Lemvigh, C.K.; Deng, S.; Dangle, N.; Ruthen, N.; Knisbacher, B.A.; Broséus, J.; Hergalant, S.; Guièze, R.; Li, S.Q.; et al. ZNF683 marks a CD8+T cell population associated with anti-tumor immunity following anti-PD-1 therapy for Richter syndrome. Cancer Cell 2023, 41, 1803–1816.e8. [Google Scholar] [CrossRef]
- Parry, E.; Lemvigh, C.K.; Anandappa, A.J.; Li, S.Q.; Werner, L.; Lako, A.; Purroy, N.; Gohil, S.H.; Oliveira, G.; Bachireddy, P.; et al. T Cell Determinants of Response and Resistance to PD-1 Blockade in Richter’s Transformation. Blood 2019, 134, 680. [Google Scholar] [CrossRef]
- Iyer, P.; Yang, L.; Yang, Z.Z.; Secreto, C.R.; Sinha, S.; Kay, N.E.; Ansell, S.M.; Wang, L.L.; Ding, W.; Kim, H. Pre-Existing T Cell Subsets Determine Anti-PD1 Blockade Response in Richter’s Transformation. Blood 2020, 136, 42–43. [Google Scholar] [CrossRef]
- García-Barchino, M.J.; Sarasquete, M.E.; Panizo, C.; Morscio, J.; Martinez, A.; Alcoceba, M.; Fresquet, V.; Gonzalez-Farre, B.; Paiva, B.; Young, K.H.; et al. Richter transformation driven by Epstein-Barr virus reactivation during therapy-related immunosuppression in chronic lymphocytic leukaemia. J. Pathol. 2018, 245, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Younesian, S.; Mohammadi, M.H.; Younesian, O.; Momeny, M.; Ghaffari, S.H.; Bashash, D. DNA methylation in human diseases. Heliyon 2024, 10, e32366. [Google Scholar] [CrossRef]
- Zhu, D.; Zeng, S.; Su, C.; Li, J.; Xuan, Y.; Lin, Y.; Xu, E.; Fan, Q. The interaction between DNA methylation and tumor immune microenvironment: From the laboratory to clinical applications. Clin. Epigenetics 2024, 16, 24. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Chen, L. CRISPR/Cas-mediated macromolecular DNA methylation editing: Precision targeting of DNA methyltransferases in cancer therapy. Int. J. Biol. Macromol. 2025, 308 Pt 2, 142401. [Google Scholar] [CrossRef]
- Rinaldi, A.; Mensah, A.A.; Kwee, I.; Forconi, F.; Orlandi, E.M.; Lucioni, M.; Gattei, V.; Marasca, R.; Berger, F.; Cogliatti, S.; et al. Promoter methylation patterns in Richter syndrome affect stem-cell maintenance and cell cycle regulation and differ from de novo diffuse large B-cell lymphoma. Br. J. Haematol. 2013, 163, 194–204. [Google Scholar] [CrossRef]
- Parry, E.M.; Ten Hacken, E.; Wu, C.J. Richter syndrome: Novel insights into the biology of transformation. Blood 2023, 142, 11–22. [Google Scholar] [CrossRef]
- Nadeu, F.; Royo, R.; Clot, G.; Duran-Ferrer, M.; Navarro, A.; Martín, S.; Lu, J.Y.; Zenz, T.; Baumann, T.; Jares, P.; et al. IGLV3-21 identifies an aggressive biological subtype of chronic lymphocytic leukemia with intermediate epigenetics. Blood 2021, 137, 2935–2946. [Google Scholar] [CrossRef]
- Broséus, J.; Hergalant, S.; Vogt, J.; Tausch, E.; Kreuz, M.; Mottok, A.; Schneider, C.; Dartigeas, C.; Roos-Weil, D.; Quinquenel, A.; et al. Molecular characterization of Richter syndrome identifies de novo diffuse large B-cell lymphomas with poor prognosis. Nat. Commun. 2023, 14, 309. [Google Scholar] [CrossRef] [PubMed]
- Fülöp, Z.; Csernus, B.; Tímár, B.; Szepesi, Á.; Matolcsy, A. Microsatellite instability and hMLH1 promoter hypermethylation in Richter’s transformation of chronic lymphocytic leukemia. Leukemia 2003, 17, 411–415. [Google Scholar] [CrossRef]
- Policarpi, C.; Munafò, M.; Tsagkris, S.; Carlini, V.; Hackett, J.A. Systematic epigenome editing captures the context-dependent instructive function of chromatin modifications. Nat. Genet. 2024, 56, 1168–1180. [Google Scholar] [CrossRef]
- Kawaf, R.R.; Ramadan, W.S.; El-Awady, R. Deciphering the interplay of histone post-translational modifications in cancer: Co-targeting histone modulators for precision therapy. Life Sci. 2024, 346, 122639. [Google Scholar] [CrossRef]
- Gourisankar, S.; Krokhotin, A.; Wenderski, W.; Crabtree, G.R. Context-specific functions of chromatin remodellers in development and disease. Nat. Rev. Genet. 2024, 25, 340–361. [Google Scholar] [CrossRef] [PubMed]
- Narlikar, G.J.; Sundaramoorthy, R.; Owen-Hughes, T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 2013, 154, 490–503. [Google Scholar] [CrossRef]
- Hing, Z.A.; Walker, J.S.; Whipp, E.C.; Brinton, L.; Cannon, M.; Zhang, P.; Sher, S.; Cempre, C.B.; Brown, F.; Smith, P.L.; et al. Dysregulation of PRMT5 in chronic lymphocytic leukemia promotes progression with high risk of Richter’s transformation. Nat. Commun. 2023, 14, 97. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. Micrornas: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Giovannetti, E.; Erozenci, A.; Smit, J.; Danesi, R.; Peters, G.J. Molecular mechanisms underlying the role of microRNAs (miRNAs) in anticancer drug resistance and implications for clinical practice. Crit. Rev. Oncol. Hematol. 2012, 81, 103–122. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes and at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.V.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435, 839–843. [Google Scholar] [CrossRef]
- Van Roosbroeck, K.; Bayraktar, R.; Calin, S.; Bloehdorn, J.; Dragomir, M.P.; Okubo, K.; Bertilaccio, M.T.S.; Zupo, S.; You, M.J.; Gaidano, G.; et al. The involvement of microRNA in the pathogenesis of Richter syndrome. Haematologica 2019, 104, 1004–1015. [Google Scholar] [CrossRef] [PubMed]
- Fulci, V.; Chiaretti, S.; Goldoni, M.; Azzalin, G.; Carucci, N.; Tavolaro, S.; Castellano, L.; Magrelli, A.; Citarella, F.; Messina, M.; et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood 2007, 109, 4944–4951. [Google Scholar] [CrossRef]
- Moussay, E.; Wang, K.; Cho, J.H.; van Moer, K.; Pierson, S.; Paggetti, J.; Nazarov, P.V.; Palissot, V.; Hood, L.E.; Berchem, G.; et al. MicroRNA as biomarkers and regulators in B-cell chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2011, 108, 6573–6578. [Google Scholar] [CrossRef]
- Jurj, A.; Pop, L.; Petrushev, B.; Pasca, S.; Dima, D.; Frinc, I.; Deak, D.; Desmirean, M.; Trifa, A.; Fetica, B.; et al. Exosome-carried microRNA-based signature as a cellular trigger for the evolution of chronic lymphocytic leukemia into Richter syndrome. Crit. Rev. Clin. Lab. Sci. 2018, 55, 501–515. [Google Scholar] [CrossRef]
- Balatti, V.; Tomasello, L.; Rassenti, L.Z.; Veneziano, D.; Nigita, G.; Wang, H.Y.; Thorson, J.A.; Kipps, T.J.; Pekarsky, Y.; Croce, C.M. miR-125a and miR-34a expression predicts Richter syndrome in chronic lymphocytic leukemia patients. Blood 2018, 132, 2179–2182. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.C.; Srinivasan, L.; Calado, D.P.; Patterson, H.C.; Zhang, B.C.; Wang, J.; Henderson, J.M.; Kutok, J.L.; Rajewsky, K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 2008, 9, 405–414. [Google Scholar] [CrossRef]
- Schmid, T.; Maier, J.; Martin, M.; Tasdogan, A.; Tausch, E.; Barth, T.F.E.; Stilgenbauer, S.; Bloehdorn, J.; Möller, P.; Mellert, K. U-RT1-A new model for Richter transformation. Neoplasia 2021, 23, 140–148. [Google Scholar] [CrossRef]
- Bichi, R.; Shinton, S.A.; Martin, E.S.; Koval, A.; Calin, G.A.; Cesari, R.; Russo, G.; Hardy, R.R.; Croce, C.M. Human chronic lymphocytic leukemia modeled in mouse by targeted expression. Proc. Natl. Acad. Sci. USA 2002, 99, 6955–6960. [Google Scholar] [CrossRef]
- Knittel, G.; Rehkämper, T.; Korovkina, D.; Liedgens, P.; Fritz, C.; Torgovnick, A.; Al-Baldawi, Y.; Al-Maarri, M.; Cun, Y.P.; Fedorchenko, O.; et al. Two mouse models reveal an actionable PARP1 dependence in aggressive chronic lymphocytic leukemia. Nat. Commun. 2017, 8, 153. [Google Scholar] [CrossRef] [PubMed]
- Pekarsky, Y.; Hallas, C.; Croce, C.M. The role of in human T-cell leukemia. Oncogene 2001, 20, 5638–5643. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.Y.; Fang, W.J. Current understandings on T-cell prolymphocytic leukemia and its association with TCL1 proto-oncogene. Biomed. Pharmacother. 2020, 126, 110107. [Google Scholar] [CrossRef] [PubMed]
- Bresin, A.; D’Abundo, L.; Narducci, M.G.; Fiorenza, M.T.; Croce, C.M.; Negrini, M.; Russo, G. TCL1 transgenic mouse model as a tool for the study of therapeutic targets and microenvironment in human B-cell chronic lymphocytic leukemia. Cell Death Dis. 2016, 7, e2071. [Google Scholar] [CrossRef] [PubMed]
- Vaisitti, T.; Braggio, E.; Allan, J.N.; Arruga, F.; Serra, S.; Zamò, A.; Tam, W.; Chadburn, A.; Furman, R.R.; Deaglio, S. Novel Richter Syndrome Xenograft Models to Study Genetic Architecture, Biology, and Therapy Responses. Cancer Res. 2018, 78, 3413–3420. [Google Scholar] [CrossRef]
- Fiskus, W.; Mill, C.P.; Perera, D.; Birdwell, C.; Deng, Q.; Yang, H.P.; Lara, B.H.; Jain, N.; Burger, J.; Ferrajoli, A.; et al. BET proteolysis targeted chimera-based therapy of novel models of Richter Transformation-diffuse large B-cell lymphoma. Leukemia 2021, 35, 2621–2634. [Google Scholar] [CrossRef]

| Signaling Pathways | Regulators | Functions |
|---|---|---|
| PI3K/AKT | BTK↑ * [76] | Cell survival, proliferation, and metabolic reprogramming |
| PI3K↑ [50,77] | Cell survival, proliferation, and metabolic reprogramming | |
| AKT↑ [50] | Cell survival, proliferation, and metabolic reprogramming | |
| NOTCH1↑ [46,47,48] | Tumor microenvironment and RT progression | |
| MYC↑ [77] | Hallmark of RT and RT progression | |
| PTEN↓ [77] | Key negative regulator of AKT activity | |
| NAMPT↑ [76] | Cell survival and metabolism | |
| NF-κB | BTK↑ | Cell survival, proliferation, and metabolic reprogramming |
| Major components of the NF-κB pathway↑ [78,79] | Cell survival, apoptosis, and tumor microenvironment | |
| CARD9↑ [80] | Cell activity | |
| MAPK/RAS/ERK | PTPN11↑ [55] | Positive regulator of the MAPK-RAS-ERK cascade |
| KRAS↑ and B-Raf↓ [55] | A selective pressure for RAS-driven signaling in RT, which may promote cell survival and proliferation | |
| mTOR | Major components of the mTOR pathway↑ [81] | Cell survival, drug resistance, and cell cycle |
| TP53↑ [34] | Cell cycle | |
| Others | Bcl-2↑ [82,83] | Cell apoptosis |
| MGA↓ [40] | A functional MYC suppressor, which regulates oxidative phosphorylation, metabolic reprogramming, and cell aggressive proliferation | |
| JAK/STAT↑ [77,84] | Tumor microenvironment | |
| CXCL12/CXCR4↑ [85] | Tumor microenvironment | |
| PLK1/FOXM1↑ [85] | Cell cycle and tumor microenvironment |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Chen, J. Molecular Features Accompanying Richter’s Transformation in Patients with Chronic Lymphocytic Leukemia. Int. J. Mol. Sci. 2025, 26, 5563. https://doi.org/10.3390/ijms26125563
Wang X, Chen J. Molecular Features Accompanying Richter’s Transformation in Patients with Chronic Lymphocytic Leukemia. International Journal of Molecular Sciences. 2025; 26(12):5563. https://doi.org/10.3390/ijms26125563
Chicago/Turabian StyleWang, Xiaole, and Jingyu Chen. 2025. "Molecular Features Accompanying Richter’s Transformation in Patients with Chronic Lymphocytic Leukemia" International Journal of Molecular Sciences 26, no. 12: 5563. https://doi.org/10.3390/ijms26125563
APA StyleWang, X., & Chen, J. (2025). Molecular Features Accompanying Richter’s Transformation in Patients with Chronic Lymphocytic Leukemia. International Journal of Molecular Sciences, 26(12), 5563. https://doi.org/10.3390/ijms26125563







