Diversity of Effects of Mechanical Influences on Living Systems and Aqueous Solutions
Abstract
1. Introduction
2. Diversity of Effects of Mechanical Influences on Living Systems and Aqueous Solutions
2.1. Impact Types
2.2. Effects of Mechanical Influences
- The whole organism in vivo;
- Cell cultures;
- Aqueous solutions or deionized water.
2.2.1. Mechanical Impact on Living Systems
2.2.2. Mechanical Impact on Water Solution
2.3. Ultrasound Impact
2.3.1. US Impact on Living Systems
2.3.2. US Impact on Water Solutions
3. Mechanisms of the Mechanical Impact Effects
3.1. Receptor Response of Living Systems. Mechanotransduction
- “elements of cytoskeleton → creation of internal force on the mechanoreceptor from the intracellular matrix → activation of mechanoreceptor → detection of changes in the extracellular matrix under external mechanical stimulation” (inside out).
- “absence of internal force on the mechanoreceptor from the intracellular matrix → inhibition of mechanoreceptor → inhibition of detection of changes in the extracellular matrix under external mechanical stimulation”
3.2. ROS Generation in Water Solutions
- Activation of oxygen molecules dissolved in water;
- Dissociation of water molecules and hydroxyl anions.
4. Dependence of the Magnitude of the Effects of Mechanical Impacts on Their Characteristics
4.1. Mechanical Influence
4.1.1. Principal Conditions of Realisation of Effects
4.1.2. Dependences of Mechanical Effects on the Combination of Factors
4.1.3. Correlations of Effects from Individual Parameters
4.2. Dependence of the Magnitude of Ultrasound Effects on Its Characteristics
Correlations of Ultrasound Effects on Individual Parameters
5. Limitations and Prospects
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACAN | Aggrecan |
| AJ | Adherens junctions |
| Akt | RAC-alpha serine/threonine-protein kinase |
| ALP | Alkaline phosphatase |
| ASICs | Acid-sensing ion channels |
| BMDCs | Bone Marrow-Derived Dendritic Cell |
| BrdU | 5-bromo-2′-deoxyuridine |
| BSA | bovine serum albumin |
| CD | Cluster of differentiation |
| Cenpf | Centromere protein F |
| COL2A1 | collagen II type gene |
| CPK | creatine phosphokinase |
| CYP1B1 | cytochrome P450 1B1 gene |
| DKK1 | Dickkopf-related protein 1 |
| DLS | Dynamic light scattering |
| DMEM | Dulbecco’s Modified Eagle Medium |
| DN | double-network |
| E.G7-OVA | T lymphoblast cancer cell line |
| EGF | Epidermal Growth Factor |
| EMR | Electromagnetic radiation |
| ER | Endoplasmic reticulum |
| ERK | Extracellular signal-regulated kinase |
| fMLF | N-Formylmethionine-leucyl-phenylalanine |
| GPCRs | G-protein coupled receptors |
| hAD-MSC | Human amnion-derived mesenchymal stem cells |
| hESC | Human embryonic stem cells |
| HEWL | Hen egg white lysozyme |
| Hk2 | Hexokinase II gene |
| hPDLSC | Human periodontal ligament stem cells |
| IDO | Indoleamine 2,3-dioxygenase |
| IFN | Interferon |
| IGF-I | insulin-like growth factor 1 |
| IgG | Immunoglobulin G |
| IL | Interleukin |
| LINC | Linker of Nucleoskeleton and Cytoskeleton |
| MAPK | Mitogen-activated protein kinase |
| MMP2 | Matrix metalloproteinase-2 |
| MSCs | Mesenchymal stem cells |
| MT-1a | Metallothionein 1A |
| NFAT | Nuclear factor of activated T-cells |
| OCN | Osteocalcin |
| OPG | Osteoprotegerin |
| OSX | Osterix protein |
| PAAm | polyacrylamide |
| Pcdh17 | Protocadherin 17 |
| PEG | poly(ethylene glycol) |
| PGE-2 | Prostaglandin E 2 |
| PHEMA | poly(2-hydroxyethyl methacrylate) |
| PI3K | Phosphoinositide 3-kinases |
| PLA2 | phospholipase A2 |
| PNaAMPS | poly(2-acrylamido-2-methylpropanesulfonic acid sodium salt) |
| PVS | poly(vinyl alcohol) |
| RANKL | Receptor activator of nuclear factor kappa-B ligand |
| ROS | Reactive oxygen species |
| RPMI | Roswell Park Memorial Institute |
| Runx2 | Runt-related transcription factor 2 |
| SACs | stress-activated channels |
| Slc2a1 | major glucose transporter GLUT1 gene |
| SOX | Superoxide dismutase |
| TGF | Transforming growth factor |
| TNF | Tumor necrosis factor |
| TREK | Two-Pore Domain K+ channels |
| TRP | Transient receptor potential channels |
| US | Ultrasound |
| VEGF | Vascular Endothelial Growth Factor |
Appendix A
| # | Solution | Object | Impact Type | Concetration (If Applied) | Frequency. Hz | Amplitude (Value) | Amplitude (Units) | Mass. g | Time. s | Mesured Characterisctic | Effect. % | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Culture medium DMEM | human fibroblasts | Ultrasound | - | 45,000 | 15 | mW/cm2 | 5 | 300 | Cells proliferation | +30% | [102] |
| 2 | Culture medium DMEM | human fibroblasts | Ultrasound | - | 45,000 | 50 | mW/cm2 | 5 | 300 | Cells proliferation | +45% | [102] |
| 3 | Culture medium DMEM | human fibroblasts | Ultrasound | - | 1,000,000 | 0.7 | mW/cm2 | 5 | 300 | Cells proliferation | +45% | [102] |
| 4 | Culture medium DMEM | human fibroblasts | Ultrasound | - | 1,000,000 | 1 | mW/cm2 | 5 | 300 | Cells proliferation | +55% | [102] |
| 5 | Culture medium DMEM | human fibroblasts | Ultrasound | - | 45,000 | 15 | mW/cm2 | 5 | 300 | Collagen expression | +45% | [102] |
| 6 | Culture medium DMEM | human fibroblasts | Ultrasound | - | 45,000 | 50 | mW/cm2 | 5 | 300 | Collagen expression | +38% | [102] |
| 7 | Culture medium DMEM | human osteoblasts | Ultrasound | - | 45,000 | 5 | mW/cm2 | 5 | 300 | Cells proliferation | +32% | [102] |
| 8 | Culture medium DMEM | human fibroblasts | Ultrasound | - | 1,000,000 | 0.1 | mW/cm2 | 5 | 300 | Collagen expression | +49% | [102] |
| 9 | Culture medium DMEM | human fibroblasts | Ultrasound | - | 1,000,000 | 0.4 | mW/cm2 | 5 | 300 | Collagen expression | +58% | [102] |
| 10 | Culture medium DMEM | human fibroblasts | Ultrasound | - | 1,000,000 | 0.7 | mW/cm2 | 5 | 300 | Collagen expression | +53% | [102] |
| 11 | Culture medium DMEM | human osteoblasts | Ultrasound | - | 45,000 | 30 | mW/cm2 | 5 | 300 | Cells proliferation | +36% | [102] |
| 12 | Culture medium DMEM | human osteoblasts | Ultrasound | - | 1,000,000 | 0.7 | mW/cm2 | 5 | 300 | Cells proliferation | +46% | [102] |
| 13 | Culture medium DMEM | human osteoblasts | Ultrasound | - | 1,000,000 | 1 | mW/cm2 | 5 | 300 | Cells proliferation | +38% | [102] |
| 14 | Culture medium DMEM | human osteoblasts | Ultrasound | 45,000 | 15 | mW/cm2 | 5 | 300 | Collagen expression | +45% | [102] | |
| 15 | Culture medium DMEM | human osteoblasts | Ultrasound | - | 45,000 | 50 | mW/cm2 | 5 | 300 | Collagen expression | +37% | [102] |
| 16 | Culture medium DMEM | human osteoblasts | Ultrasound | - | 1,000,000 | 0.1 | mW/cm2 | 5 | 300 | Collagen expression | +53% | [102] |
| 17 | Culture medium DMEM | human osteoblasts | Ultrasound | - | 1,000,000 | 0.4 | mW/cm2 | 5 | 300 | Collagen expression | +38% | [102] |
| 18 | Culture medium DMEM | human osteoblasts | Ultrasound | - | 1,000,000 | 0.4 | mW/cm2 | 5 | 75 | Secretion of IL-1β | +16 | [102] |
| 19 | Culture medium DMEM | human osteoblasts | Ultrasound | - | 1,000,000 | 0.7 | mW/cm2 | 5 | 75 | Secretion of IL-1β | +20% | [102] |
| 20 | Culture medium DMEM | human osteoblasts | Ultrasound | - | 1,000,000 | 1 | mW/cm2 | 5 | 75 | Secretion of IL-1β | +17% | [102] |
| 21 | Culture medium DMEM | human osteoblasts | Ultrasound | - | 450,000 | 30 | mW/cm2 | 5 | 300 | Secretion of IL-8 | +55% | [102] |
| 22 | Culture medium DMEM | human osteoblasts | Ultrasound | - | 1,000,000 | 0.4 | mW/cm2 | 5 | 300 | Secretion of IL-8 | +60% | [102] |
| 23 | Culture medium DMEM | human osteoblasts | Ultrasound | - | 1,000,000 | 0.7 | mW/cm2 | 5 | 300 | Secretion of IL-8 | +70% | [102] |
| 24 | Culture medium DMEM | human osteoblasts | Ultrasound | - | 45,0000 | 5 | mW/cm2 | 5 | 300 | Secretion of EGFb | +3000% | [102] |
| 25 | Culture medium DMEM | human osteoblasts | Ultrasound | - | 45,0000 | 15 | mW/cm2 | 5 | 300 | Secretion of EGFb | +1500% | [102] |
| 26 | Culture medium DMEM | human osteoblasts | Ultrasound | - | 45,0000 | 50 | mW/cm2 | 5 | 300 | Secretion of EGFb | +750% | [102] |
| 27 | Culture medium DMEM | human osteoblasts | Ultrasound | - | 1,000,000 | 0.1 | mW/cm2 | 5 | 300 | Secretion of EGFb | +300% | [102] |
| 28 | Culture medium DMEM | human osteoblasts | Ultrasound | 450,000 | 15 | mW/cm2 | 5 | 300 | Secretion of VEGF | +25% | [102] | |
| 29 | Culture medium DMEM | human osteoblasts | Ultrasound | 1,000,000 | 0.1 | mW/cm2 | 5 | 300 | Secretion of VEGF | +40% | [102] | |
| 30 | Culture medium RPMI 1640 | human monocytes | Ultrasound | - | 45,000 | 15 | mW/cm2 | 5 | 300 | Secretion of IL-1β | +25% | [102] |
| 31 | Culture medium RPMI 1640 | human monocytes | Ultrasound | - | 45,000 | 30 | mW/cm2 | 5 | 300 | Secretion of IL-1β | +103% | [102] |
| 32 | Culture medium RPMI 1640 | human monocytes | Ultrasound | - | 45,000 | 50 | mW/cm2 | 5 | 300 | Secretion of IL-1β | +80% | [102] |
| 33 | Culture medium RPMI 1640 | human monocytes | Ultrasound | - | 45,000 | 15 | mW/cm2 | 5 | 300 | Secretion of VEGF | +25% | [102] |
| 34 | Culture medium RPMI 1640 | human monocytes | Ultrasound | - | 45,000 | 30 | mW/cm2 | 5 | 300 | Secretion of VEGF | +30% | [102] |
| 35 | Culture medium RPMI 1640 | human monocytes | Ultrasound | - | 45,000 | 50 | mW/cm2 | 5 | 300 | Secretion of VEGF | +35% | [102] |
| 36 | Culture medium RPMI 1640 | human monocytes | Ultrasound | - | 1,000,000 | 0.1 | mW/cm2 | 5 | 300 | Secretion of VEGF | +35% | [102] |
| 37 | Culture medium RPMI 1640 | human monocytes | Ultrasound | - | 1,000,000 | 0.4 | mW/cm2 | 5 | 300 | Secretion of VEGF | +15% | [102] |
| 38 | Culture medium RPMI 1640 | human monocytes | Ultrasound | - | 1,000,000 | 1 | mW/cm2 | 5 | 300 | Secretion of VEGF | +10% | [102] |
| 39 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 100 | mW/cm2 | 2 | 600 | Cells differentiation | +10% | [103] |
| 40 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 200 | mW/cm2 | 2 | 600 | Cells differentiation | +15% | [103] |
| 41 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 300 | mW/cm2 | 2 | 600 | Cells differentiation | +15% | [103] |
| 42 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 60 | mW/cm2 | 2 | 1200 | Cells differentiation | +15% | [103] |
| 43 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 100 | mW/cm2 | 2 | 1200 | Cells differentiation | +15% | [103] |
| 44 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 200 | mW/cm2 | 2 | 1200 | Cells differentiation | +10% | [103] |
| 45 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 300 | mW/cm2 | 2 | 1200 | Cells differentiation | +11% | [103] |
| 46 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 30 | mW/cm2 | 2 | 1800 | Cells differentiation | +15% | [103] |
| 47 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 100 | mW/cm2 | 2 | 600 | Concentration of Cyclin D1/β | +60% | [103] |
| 48 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 200 | mW/cm2 | 2 | 600 | Concentration of Cyclin D1/β | +40% | [103] |
| 49 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 300 | mW/cm2 | 2 | 600 | Concentration of Cyclin D1/β | +45% | [103] |
| 50 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 60 | mW/cm2 | 2 | 1200 | Concentration of Cyclin D1/β | +100% | [103] |
| 51 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 100 | mW/cm2 | 2 | 1200 | Concentration of Cyclin D1/β | +90% | [103] |
| 52 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 200 | mW/cm2 | 2 | 1200 | Concentration of Cyclin D1/β | +80% | [103] |
| 53 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 300 | mW/cm2 | 2 | 1200 | Concentration of Cyclin D1/β | +70% | [103] |
| 54 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 30 | mW/cm2 | 2 | 1800 | Concentration of Cyclin D1/β | +110% | [103] |
| 55 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 100 | mW/cm2 | 2 | 600 | Concentration of Cyclin В1/β | +200% | [103] |
| 56 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 200 | mW/cm2 | 2 | 600 | Concentration of Cyclin В1/β | +380% | [103] |
| 57 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 300 | mW/cm2 | 2 | 600 | Concentration of Cyclin В1/β | +500% | [103] |
| 58 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 60 | mW/cm2 | 2 | 1200 | Concentration of Cyclin В1/β | +500% | [103] |
| 59 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 100 | mW/cm2 | 2 | 1200 | Concentration of Cyclin В1/β | +400% | [103] |
| 60 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 200 | mW/cm2 | 2 | 1200 | Concentration of Cyclin В1/β | +530% | [103] |
| 61 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 300 | mW/cm2 | 2 | 1200 | Concentration of Cyclin В1/β | +490% | [103] |
| 62 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 30 | mW/cm2 | 2 | 1800 | Concentration of Cyclin В1/β | +490% | [103] |
| 63 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 100 | mW/cm2 | 2 | 600 | Concentration of Cyclin Е1/β | +80% | [103] |
| 64 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 200 | mW/cm2 | 2 | 600 | Concentration of Cyclin Е1/β | +110% | [103] |
| 65 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 300 | mW/cm2 | 2 | 600 | Concentration of Cyclin Е1/β | +150% | [103] |
| 66 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 60 | mW/cm2 | 2 | 1200 | Concentration of Cyclin Е1/β | +200% | [103] |
| 67 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 100 | mW/cm2 | 2 | 1200 | Concentration of Cyclin Е1/β | +250% | [103] |
| 68 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 200 | mW/cm2 | 2 | 1200 | Concentration of Cyclin Е1/β | +200% | [103] |
| 69 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 300 | mW/cm2 | 2 | 1200 | Concentration of Cyclin Е1/β | +180% | [103] |
| 70 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 30 | mW/cm2 | 2 | 1800 | Concentration of Cyclin Е1/β | +190% | [103] |
| 71 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 100 | mW/cm2 | 2 | 600 | Concentration of Cyclin A1/β | +250% | [103] |
| 72 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 200 | mW/cm2 | 2 | 600 | Concentration of Cyclin A1/β | +100% | [103] |
| 73 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 300 | mW/cm2 | 2 | 600 | Concentration of Cyclin A1/β | +90% | [103] |
| 74 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 60 | mW/cm2 | 2 | 1200 | Concentration of Cyclin A1/β | +250% | [103] |
| 75 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 100 | mW/cm2 | 2 | 1200 | Concentration of Cyclin A1/β | +245% | [103] |
| 76 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 200 | mW/cm2 | 2 | 1200 | Concentration of Cyclin A1/β | +240% | [103] |
| 77 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 300 | mW/cm2 | 2 | 1200 | Concentration of Cyclin A1/β | +70% | [103] |
| 78 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 30 | mW/cm2 | 2 | 1800 | Concentration of Cyclin A1/β | +270% | [103] |
| 79 | Culture medium DMEM | Human amnion-derived mesenchymal stem cells (hAD-MSC) | Ultrasound | - | 250,000 | 30 | mW/cm2 | 2 | 1800 | Cells proliferation | +180% | [103] |
| 80 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 500,000 | 500 | mW/cm2 | 1 | 86,400 | Cells proliferation | +90% | [104] |
| 81 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 1,000,000 | 500 | mW/cm2 | 1 | 86,400 | Cells proliferation | +95% | [104] |
| 82 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 3,000,000 | 500 | mW/cm2 | 1 | 86,400 | Cells proliferation | +150% | [104] |
| 83 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 3,000,000 | 1000 | mW/cm2 | 1 | 86,400 | Cells proliferation | +180% | [104] |
| 84 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 3,000,000 | 250 | mW/cm2 | 1 | 86,400 | Cells proliferation | +145% | [104] |
| 85 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 1,000,000 | 500 | mW/cm2 | 1 | 2400 | myotube width | +20% | [104] |
| 86 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 1,000,000 | 500 | mW/cm2 | 1 | 2400 | myotube length | +50% | [104] |
| 87 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 5,000,000 | 500 | mW/cm2 | 1 | 2400 | myotube length | +10% | [104] |
| 88 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 1,000,000 | 500 | mW/cm2 | 1 | 2400 | fusion index | +100% | [104] |
| 89 | Rat (trauma model) | in vivo | Ultrasound | - | 870,000 | 1000 | mW/cm2 | 250 | 1500 | Total tissue protein content | −10% | [97] |
| 90 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 1,500,000 | 30 | mW/cm2 | 10 | 7200 | Cells proliferation | +18% | [113] |
| 91 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 1,500,000 | 30 | mW/cm2 | 10 | 9600 | Cells proliferation | +12% | [113] |
| 92 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 1,500,000 | 30 | mW/cm2 | 10 | 7200 | Expression of myogenin | +47% | [113] |
| 93 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 1,500,000 | 30 | mW/cm2 | 10 | 9600 | Expression of myogenin | +34% | [113] |
| 94 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 1,500,000 | 30 | mW/cm2 | 10 | 16,800 | Numbers of regenerating myofibers | +90% | [113] |
| 95 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 1,500,000 | 30 | mW/cm2 | 10 | 25,200 | Numbers of regenerating myofibers | +250% | [113] |
| 96 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 1,500,000 | 30 | mW/cm2 | 10 | 33,600 | Numbers of regenerating myofibers | +320% | [113] |
| 97 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 1,500,000 | 30 | mW/cm2 | 10 | 25,200 | Restoration of fast twitch strength | +150% | [113] |
| 98 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 1,500,000 | 30 | mW/cm2 | 10 | 33,600 | Restoration of fast twitch strength | +300% | [113] |
| 99 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 1,500,000 | 30 | mW/cm2 | 10 | 25200 | Restoration of tetanus strength strength | +90% | [113] |
| 100 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 1,500,000 | 30 | mW/cm2 | 10 | 33600 | Restoration of fast twitch strength | +100% | [113] |
| 101 | in vivo | Wistar rats | Ultrasound | - | 1,000,000 | 1 | mW/cm2 | 220 | 900 | Muscle fibre diameter | +10% | [98] |
| 102 | in vivo | Wistar rats | Ultrasound | - | 1,000,000 | 1 | mW/cm2 | 220 | 900 | BrdU positive cells count | +30% | [98] |
| 103 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 3,000,000 | 30 | mW/cm2 | 5 | 900 | Pax7 positive cells count | +60% | [115] |
| 104 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 3,000,000 | 30 | mW/cm2 | 5 | 900 | Expression of gene COX2 | +80% | [115] |
| 105 | Culture medium DMEM | Mouse myoblasts (C2C12) | Ultrasound | - | 3,000,000 | 30 | mW/cm2 | 5 | 900 | Proiflammatory cells count | −40% | [115] |
| 106 | Culture medium DMEM | BMDCs | Constant pressure from below | - | 0 | 50,00050,000 | Pa | 5 | 950,400 | Efficiency of glycolysis | −30% | [72] |
| 107 | Culture medium DMEM | BMDCs | Constant pressure from below | - | 0 | 2000 | Pa | 5 | 950,400 | Efficiency of glycolysis | −50% | [72] |
| 108 | Culture medium DMEM | BMDCs | Constant pressure from below | - | 0 | 50,000 | Pa | 5 | 950,400 | Lactase activity | −5% | [72] |
| 109 | Culture medium DMEM | BMDCs | Constant pressure from below | - | 0 | 2000 | Pa | 5 | 950,400 | Lactase activity | −60% | [72] |
| 110 | Culture medium DMEM | BMDCs | Constant pressure from below | - | 0 | 50,000 | Pa | 5 | 950,400 | overall rate curves of oxygen consumption rate | −70% | [72] |
| 111 | Culture medium DMEM | BMDCs | Constant pressure from below | - | 0 | 2000 | Pa | 5 | 950,400 | overall rate curves of oxygen consumption rate | −80% | [72] |
| 112 | Culture medium DMEM | BMDCs | Constant pressure from below | - | 0 | 50,000 | Pa | 5 | 950,400 | Expression of gene Hk2 | −55% | [72] |
| 113 | Culture medium DMEM | BMDCs | Constant pressure from below | - | 0 | 2000 | Pa | 5 | 950,400 | Expression of gene Slc2a1 | −96% | [72] |
| 114 | Culture medium DMEM | BMDCs | Constant pressure from below | - | 0 | 2000 | Pa | 5 | 950,400 | Cells proliferation | −95% | [72] |
| 115 | Culture medium DMEM | BMDCs | Constant pressure from below | - | 0 | 50,000 | Pa | 5 | 950,400 | Tumor size (11 days post E.G7-OVA injection) in WT mice injected with E.G7-OVA cells and WT BMDCs | −50% | [72] |
| 116 | Culture medium DMEM | BMDCs | Constant pressure from below | - | 0 | 50,000 | Pa | 5 | 950,400 | tumor weight (14 days post E.G7-OVA injection) in WT mice injected with E.G7-OVA cells and WT BMDCs | −85% | [72] |
| 117 | Culture medium DMEM | CD8+ T cells | Constant pressure from below | - | 0 | 50,000 | Pa | 5 | 950,400 | Immunology memory | +120% | [72] |
| 118 | Culture medium DMEM | CD4+ T cells | Constant pressure from below | - | 0 | 50,000 | Pa | 5 | 950,400 | Immunology memory | +110% | [72] |
| 119 | Water | Water | Shaking | - | 30 | 12 | mm | 10 | 60 | Chemiluminescence intensity | −20% | [21] |
| 120 | Water | Water | Shaking | - | 30 | 2.3 | mm | 10 | 60 | Chemiluminescence intensity | +25% | [21] |
| 121 | Water | Water | Shaking | - | 10 | 12 | mm | 10 | 15 | Decline rate of chemiluminescence intensity | −20% | [21] |
| 122 | Water | Water | Shaking | - | 10 | 12 | mm | 10 | 60 | Decline rate of chemiluminescence intensity | −25% | [21] |
| 123 | Water | Water | Shaking | - | 30 | 2.3 | mm | 10 | 60 | Decline rate of chemiluminescence intensity | +30% | [21] |
| 124 | Water | Water | Shaking | - | 30 | 2.3 | mm | 10 | 60 | Standard deviation of chemiluminescence intensity | +10% | [21] |
| 125 | Water | BSA | Shaking | 1 mg/mL | 30 | 2.3 | mm | 10 | 15 | Chemiluminescence intensity | −20% | [21] |
| 126 | Water | BSA | Shaking | 1 mg/mL | 10 | 12 | mm | 10 | 15 | Chemiluminescence intensity | −15% | [21] |
| 127 | Water | BSA | Shaking | 1 mg/mL | 10 | 12 | mm | 10 | 60 | Chemiluminescence intensity | −10% | [21] |
| 128 | Water | BSA | Shaking | 1 mg/mL | 30 | 2.3 | mm | 10 | 15 | Chemiluminescence intensity | −8% | [21] |
| 129 | Water | BSA | Shaking | 1 mg/mL | 30 | 2.3 | mm | 10 | 60 | Chemiluminescence intensity | −20% | [21] |
| 130 | Water | BSA | Shaking | 1 mg/mL | 30 | 2.3 | mm | 10 | 15 | Chemiluminescence intensity | −12% | [21] |
| 131 | Water | BSA | Shaking | 1 mg/mL | 30 | 2.3 | mm | 10 | 60 | Chemiluminescence intensity | −20% | [21] |
| 132 | Water | BSA | Shaking | 1 mg/mL | 10 | 2.3 | mm | 10 | 15 | Decline rate of chemiluminescence intensity | −18% | [21] |
| 133 | Water | BSA | Shaking | 1 mg/mL | 10 | 12 | mm | 10 | 15 | Decline rate of chemiluminescence intensity | −17% | [21] |
| 134 | Water | BSA | Shaking | 1 mg/mL | 10 | 12 | mm | 10 | 60 | Decline rate of chemiluminescence intensity | −16% | [21] |
| 135 | Water | BSA | Shaking | 1 mg/mL | 30 | 2.3 | mm | 10 | 15 | Decline rate of chemiluminescence intensity | −25% | [21] |
| 136 | Water | BSA | Shaking | 1 mg/mL | 30 | 2.3 | mm | 10 | 60 | Decline rate of chemiluminescence intensity | −16% | [21] |
| 137 | Water | BSA | Shaking | 1 mg/mL | 30 | 2.3 | mm | 10 | 60 | Standard deviation of chemiluminescence intensity | −10% | [21] |
| 138 | Water | Human IgG | Alternating magnetic field | 1 mg/mL | 50 | 50 | мкТл | 10 | 300 | Chemiluminescence intensity | +10% | [21] |
| 139 | Water | Human IgG | Alternating magnetic field | 1 mg/mL | 8 | 50 | мкТл | 10 | 300 | Standard deviation of chemiluminescence intensity | +5% | [21] |
| 140 | Hanks’ balanced salts solution | Murine neutrophils (Male Balb/c mice weighing 22–25 g) | Alternating magnetic field | 2.5 × 105 cells/mL | 12.6 | 0.01 | мкТл | 5400 | fMLF-induced ROS generation | +45% | [32] | |
| 141 | Hanks’ balanced salts solution | Murine neutrophils (Male Balb/c mice weighing 22–25 g) | Alternating magnetic field | 2.5 × 105 cells/mL | 12.6 | 0.05 | мкТл | 5400 | fMLF-induced ROS generation | +45% | [32] | |
| 142 | Water | Viscum album Quercus L. 3. plant extract | Turbulent mixing | 0.1% (10−3) | 10 | 2 | sm | 10 | 150 | local connected fractal dimension | −27% | [22] |
| 143 | Water | Viscum album Quercus L. 3. plant extract | Circular mixing | 0.1% (10−3) | 10 | 2 | sm | 10 | 150 | local connected fractal dimension | +3% | [22] |
| 144 | Water | Viscum album Quercus L. 3. plant extract | Turbulent mixing | 0.1% (10−3) | 10 | 2 | sm | 10 | 150 | mass fractal dimension | −9% | [22] |
| 145 | Water | Viscum album Quercus L. 3. plant extract | Circular mixing | 0.1% (10−3) | 10 | 2 | sm | 10 | 150 | mass fractal dimension | +2% | [22] |
| 146 | Water | Viscum album Quercus L. 3. plant extract | Turbulent mixing | 0.1% (10−3) | 10 | 2 | sm | 10 | 150 | lacunarity | +37% | [22] |
| 147 | Water | Viscum album Quercus L. 3. plant extract | Circular mixing | 0.1% (10−3) | 10 | 2 | sm | 10 | 150 | lacunarity | +4% | [22] |
| 148 | Water | Viscum album Quercus L. 3. plant extract | Turbulent mixing | 0.1% (10−3) | 10 | 2 | sm | 10 | 150 | ascending second moment | +30% | [22] |
| 149 | Water | Viscum album Quercus L. 3. plant extract | Circular mixing | 0.1% (10−3) | 10 | 2 | sm | 10 | 150 | ascending second moment | −15% | [22] |
| 150 | Water | Viscum album Quercus L. 3. plant extract | Turbulent mixing | 0.1% (10−3) | 10 | 2 | sm | 10 | 150 | contrast | −30% | [22] |
| 151 | Water | Viscum album Quercus L. 3. plant extract | Circular mixing | 0.1% (10−3) | 10 | 2 | sm | 10 | 150 | contrast | −12% | [22] |
| 152 | Water | Viscum album Quercus L. 3. plant extract | Turbulent mixing | 0.1% (10−3) | 10 | 2 | sm | 10 | 150 | Correlation | +30% | [22] |
| 153 | Water | Viscum album Quercus L. 3. plant extract | Circular mixing | 0.1% (10−3) | 10 | 2 | sm | 10 | 150 | Correlation | +90% | [22] |
| 154 | Water | Viscum album Quercus L. 3. plant extract | Turbulent mixing | 0.1% (10−3) | 10 | 2 | sm | 10 | 150 | Entropy | −3% | [22] |
| 155 | Water | Viscum album Quercus L. 3. plant extract | Circular mixing | 0.1% (10−3) | 10 | 2 | sm | 10 | 150 | Entropy | +2% | [22] |
| 156 | Hanks’ balanced salts solution | Murine neutrophils (Male Balb/c mice weighing 22–25 g) | Alternating magnetic field | 2.5 × 105 cells/mL | 0 | 0 | 0 | 3600 | fMLF-induced ROS production primered by PMA | −21% | [33] | |
| 157 | Hanks’ balanced salts solution | Murine neutrophils (Male Balb/c mice weighing 22–25 g) | Alternating magnetic field | 2.5 × 105 cells/mL | 12.6 | 0.1 | мкТл | 3600 | fMLF-induced ROS production primered by PMA | +67% | [33] | |
| 158 | Hanks’ balanced salts solution | Murine neutrophils (Male Balb/c mice weighing 22–25 g) | Alternating magnetic field | 2.5 × 105 cells/mL | 48.5 | 0.1 | мкТл | 3600 | fMLF-induced ROS production primered by PMA | +635% | [33] | |
| 159 | Hanks’ balanced salts solution | Murine neutrophils (Male Balb/c mice weighing 22–25 g) | Alternating magnetic field | 2.5 × 105 cells/mL | 12.6 | 0.1 | мкТл | 2400 | fMLF-induced ROS generation | +36% | [34] | |
| 160 | Hanks’ balanced salts solution | Murine neutrophils (Male Balb/c mice weighing 22–25 g) | Alternating magnetic field | 2.5 × 105 cells/mL | 48.5 | 0.1 | мкТл | 2400 | fMLF-induced ROS generation | −19% | [34] | |
| 161 | Water | IFNγ | Circular mixing | 0.1 М | 50 | 1 | sm | 10 | 50 | Conductivity of the solution | +20% | [16] |
| 162 | Water | polyclonal antibodies to human IFNγ | Circular mixing | 26 μg/mL | 50 | 1 | sm | 10 | 50 | Solurion EMR | +150% | [17] |
| 163 | Water | Lactose | Strokes | 820 nM | 21 | 300 (20) | g (CM) | 24 | 1050 | Luminescence at 550 nm | +100% | [24] |
| 164 | Water | Lactose | Strokes | 820 нМ | 21 | 300 (20) | g (CM) | 24 | 525 | Luminescence at 550 nm | +200% | [24] |
| 165 | 20 mM histidine buffer. | antistreptavidin IgG1 | Downfall | 1 mg/mL | 0 | 25.6 | sm | 1 | 0.025 | Particle counts (protein aggregation?) | +150% | [91] |
| 166 | 20 mM histidine buffer. | antistreptavidin IgG1 | Downfall | 1 mg/mL | 0 | 51.2 | sm | 1 | 0.025 | Particle counts | +300% | [91] |
| 167 | 20 mM histidine buffer. | antistreptavidin IgG1 | Downfall | 1 mg/mL | 0 | 76.8 | sm | 1 | 0.025 | Particle counts | +1000% | [91] |
| 168 | 20 mM histidine buffer. | antistreptavidin IgG1 | Downfall | 1 mg/mL | 0 | 102.4 | sm | 1 | 0.025 | Particle counts | +350% | [91] |
| 169 | 20 mM histidine buffer. | antistreptavidin IgG1 | Downfall | 35 mg/mL | 0 | 25.6 | sm | 1 | 0.025 | Particle counts | +900% | [91] |
| 170 | 20 mM histidine buffer. | antistreptavidin IgG1 | Downfall | 35 mg/mL | 0 | 51.2 | sm | 1 | 0.025 | Particle counts | +1100% | [91] |
| 171 | 20 mM histidine buffer. | antistreptavidin IgG1 | Downfall | 35 mg/mL | 0 | 76.8 | sm | 1 | 0.025 | Particle counts | +1000% | [91] |
| 172 | 20 mM histidine buffer. | human growth hormone | Downfall | 35 mg/mL | 0 | 102.4 | sm | 1 | 0.025 | Particle counts | +800% | [91] |
| 173 | 20 mM histidine buffer | human growth hormone | Downfall | 1.75 mg/mL | 0 | 102.4 | sm | 1 | 0.025 | Particle counts | +60% | [91] |
| 174 | 20 mM histidine buffer. | antistreptavidin IgG1 | Downfall | 1 mg/mL | 0 | 25.6 | sm | 1 | 0.025 | recovered from the walls of dropped vials following treatment with urea | +20% | [91] |
| 175 | 20 mM histidine buffer | antistreptavidin IgG1 | Downfall | 1 mg/mL | 0 | 76.8 | sm | 1 | 0.025 | recovered from the walls of dropped vials following treatment with urea | +110% | [91] |
| 176 | 20 mM histidine buffer. | antistreptavidin IgG1 | Downfall | 1 mg/mL | 0 | 102.4 | sm | 1 | 0.025 | recovered from the walls of dropped vials following treatment with urea | +180% | [91] |
| 177 | 20 mM histidine buffer | antistreptavidin IgG1 | Downfall | 35 mg/mL | 0 | 51.2 | sm | 1 | 0.025 | recovered from the walls of dropped vials following treatment with urea | +60% | [91] |
| 178 | 20 mM histidine buffer. | antistreptavidin IgG1 | Downfall | 35 mg/mL | 0 | 76.8 | sm | 1 | 0.025 | recovered from the walls of dropped vials following treatment with urea | +120% | [91] |
| 179 | 20 mM histidine buffer | antistreptavidin IgG1 | Downfall | 35 mg/mL | 0 | 102.4 | sm | 1 | 0.025 | recovered from the walls of dropped vials following treatment with urea | +90% | [91] |
| 180 | 20 mM histidine buffer | antistreptavidin IgG1 | Downfall | 1 mg/mL | 0 | 25.6 | sm | 1 | 0.025 | recovered from the walls of dropped vials following treatment with guanidine hydrochloride | +35% | [91] |
| 181 | 20 mM histidine buffer. | antistreptavidin IgG1 | Downfall | 1 mg/mL | 0 | 76.8 | sm | 1 | 0.025 | recovered from the walls of dropped vials following treatment with guanidine hydrochloride | +30% | [91] |
| 182 | 20 mM histidine buffer | antistreptavidin IgG1 | Downfall | 35 mg/mL | 0 | 51.2 | sm | 1 | 0.025 | recovered from the walls of dropped vials following treatment with guanidine hydrochloride | +200% | [91] |
| 183 | Water | Echinacea | succussion strokes | 0.8 × 10−2 g/mL | 1 | 20 | sm | 0.05 | 100 | Characteristics of crystal shape after evaporation | +10% | [23] |
| 184 | Water | Echinacea | succussion strokes | 0.8 × 10−2 g/mL | 1 | 20 | sm | 0.05 | 10 | Characteristics of crystal shape after evaporation | +8% | [23] |
| 185 | Water | Baptisia | succussion strokes | 0.8 × 10−3 g/mL | 1 | 20 | sm | 0.05 | 100 | Characteristics of crystal shape after evaporation | −10% | [23] |
| 186 | Water | Baptisia | succussion strokes | 0.8 × 10−3 g/mL | 1 | 20 | sm | 0.05 | 10 | Characteristics of crystal shape after evaporation | −10% | [23] |
| 187 | Water | Baptisia | succussion strokes | 0.8 × 10−4 g/mL | 1 | 20 | sm | 0.05 | 100 | Characteristics of crystal shape after evaporation | +60% | [23] |
| 188 | Water | Baptisia | succussion strokes | 0.8 × 10−4 g/mL | 1 | 20 | sm | 0.05 | 10 | Characteristics of crystal shape after evaporation | +50% | [23] |
| 189 | Water | Luffa | succussion strokes | 0.8 × 10−4 g/mL | 1 | 20 | sm | 0.05 | 100 | Characteristics of crystal shape after evaporation | −20% | [23] |
| 190 | Water | Luffa | succussion strokes | 0.8 × 10−4 g/mL | 1 | 20 | sm | 0.05 | 10 | Characteristics of crystal shape after evaporation | −20% | [23] |
| 191 | Water | Spongia | succussion strokes | 0.8 × 10−6 g/mL | 1 | 20 | sm | 0.05 | 100 | Characteristics of crystal shape after evaporation | +5% | [23] |
| 192 | Water | Spongia | succussion strokes | 0.8 × 10−6 g/mL | 1 | 20 | sm | 0.05 | 10 | Characteristics of crystal shape after evaporation | +10% | [23] |
| 193 | Water | IgG | Vertical shaking | IgG molecules 3 × 1012 cm−3 | 5 | 10 | mm | 20 | 30 | DLS peak intensity ~200 nm (bubble count) | +300% | [6] |
| 194 | Water +36.7% ethanol | IgG | Vertical shaking | IgG molecules 3 × 1012 cm−3 | 5 | 10 | mm | 20 | 30 | DLS peak intensity ~200 nm (bubble count) | +20% | [6] |
| 195 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 300 | Concentration of molecular oxigen | −4% | [80] |
| 196 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 600 | Concentration of molecular oxigen | −5% | [80] |
| 197 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 900 | Concentration of molecular oxigen | −7% | [80] |
| 198 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 1800 | Concentration of molecular oxigen | −8% | [80] |
| 199 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 2700 | Concentration of molecular oxigen | −9% | [80] |
| 200 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 600 | Water temperature | +2% | [80] |
| 201 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 900 | Water temperature | +15% | [80] |
| 202 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 1800 | Water temperature | +20% | [80] |
| 203 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 2700 | Water temperature | +24% | [80] |
| 204 | Water | - | Shaking | - | 15 | 5 | mm | 10 | 300 | Concentration of molecular oxigen | −10% | [80] |
| 205 | Water | - | Shaking | - | 45 | 5 | mm | 10 | 300 | Concentration of molecular oxigen | −13% | [80] |
| 206 | Water | - | Shaking | - | 60 | 5 | mm | 10 | 300 | Concentration of molecular oxigen | −15% | [80] |
| 207 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 300 | light scattering intensity | +50% | [80] |
| 208 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 900 | pH | +12% | [80] |
| 209 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 1800 | pH | +16% | [80] |
| 210 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 2700 | pH | +17% | [80] |
| 211 | Water | - | Shaking | - | 45 | 5 | mm | 10 | 300 | pH | +6% | [80] |
| 212 | Water | - | Shaking | - | 60 | 5 | mm | 10 | 300 | pH | +8% | [80] |
| 213 | Water | - | Shaking | - | 15 | 5 | mm | 10 | 300 | Concentration of H2O2 | +2000% | [80] |
| 214 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 300 | Concentration of H2O2 | +5000% | [80] |
| 215 | Water | - | Shaking | - | 45 | 5 | mm | 10 | 300 | Concentration of H2O2 | +14000% | [80] |
| 216 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 60 | Concentration of H2O2 | +1000% | [80] |
| 217 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 150 | Concentration of H2O2 | +2500% | [80] |
| 218 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 210 | Concentration of H2O2 | +3500% | [80] |
| 219 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 300 | Concentration of H2O2 | +4500% | [80] |
| 220 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 180 | Concentration of OH-radicals | +1500% | [80] |
| 221 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 300 | Concentration of OH-radicals | +2500% | [80] |
| 222 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 420 | Concentration of OH-radicals | +4000% | [80] |
| 223 | Water | - | Shaking | - | 30 | 5 | mm | 10 | 600 | Concentration of OH-radicals | +4300% | [80] |
| 224 | Water | - | Shaking | - | 15 | 5 | mm | 10 | 300 | Concentration of OH-radicals | +900% | [80] |
| 225 | Water | - | Shaking | - | 45 | 5 | mm | 10 | 300 | Concentration of OH-radicals | +5000% | [80] |
| 226 | Water | - | Shaking | - | 60 | 5 | mm | 10 | 300 | Concentration of OH-radicals | +9000% | [80] |
| 227 | Water | O2 gas | Creating an aerosol with N2 | - | Constant | 5 | μL/min | 0.011 | 60 | Concentration of H2O2 | +50 | [90] |
| 228 | Water | O2 gas | Creating an aerosol with N2 | - | Constant | 5 | μL/min | 0.011 | 120 | Concentration of H2O2 | +50% | [90] |
| 229 | Water | O2 gas | Creating an aerosol with N2 | - | Constant | 5 | μL/min | 0.011 | 180 | Concentration of H2O2 | +100% | [90] |
| 230 | Water | O2 gas | Creating an aerosol with N2 | - | Constant | 5 | μL/min | 0.011 | 240 | Concentration of H2O2 | +500% | [90] |
| 231 | Water | O2 gas | Creating an aerosol with N2 | - | Constant | 5 | μL/min | 0.011 | 300 | Concentration of H2O2 | +600% | [90] |
| 232 | Water | O2 gas | Creating an aerosol with N2 | - | Constant | 5 | μL/min | 0.011 | 420 | Concentration of H2O2 | +1000% | [90] |
| 233 | Water | O2 gas | Creating an aerosol with N2 | - | Constant | 5 | μL/min | 0.011 | 1800 | Concentration of H2O2 | −15% | [90] |
| 234 | Water | O2 gas | Creating an aerosol with N2 | - | Constant | 5 | μL/min | 0.011 | 3600 | Concentration of H2O2 | −35% | [90] |
| 235 | Water | O2 gas | Creating an aerosol with N2 | - | Constant | 5 | μL/min | 0.011 | 14,400 | Concentration of H2O2 | −45% | [90] |
| 236 | Water | O2 gas | Creating an aerosol with N2 | - | Constant | 5 | μL/min | 0.011 | 21,600 | Concentration of H2O2 | −50% | [90] |
| 237 | Water | - | Mixing | - | 12 | 1 | sm | 10 | 500 | Absorption at 180–220 sm−1 | +30% | [82] |
| 238 | Water | - | Mixing | - | 12 | 1 | sm | 10 | 50 | Wavelength of maximum absorption | +80% (280–340 нм) | [83] |
| 239 | Water | - | Mixing | - | 12 | 1 | sm | 10 | 50 | Wavelength of maximum absorption | +18% (280–330 нм) | [83] |
| 240 | Water | - | Mixing | - | 12 | 1 | sm | 10 | 50 | Fluorescence intensity | +400% | [83] |
| 240 | Water | - | Vibration | 1,000,000 | 1 | sm | 10 | 500 | Redox potentials ΔE | +1 mV (+100%) | [89] | |
| 240 | Water coated by oil layes 10 mm | Vibration | 1,000,000 | 1 | sm | 10 | 500 | Redox potentials ΔE | +2.5 mV (+250%) | [89] | ||
| 241 | Water | AcEu solutions | Microfluidic mixing | - | Constant | 400 35.5–250.6 | Reynolds number | 10 | 30 | Position of the IR band maximum of the OH groups’ stretching vibrations on Raman spectrum | +0.1% | [94] |
| 242 | Water | AcEu solutions | Microfluidic mixing | - | Constant | 400 35.5–250.6 | Reynolds number | 10 | 250 | Position of the IR band maximum of the OH groups’ stretching vibrations on Raman spectrum | 0.3 | [94] |
| 243 | 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid sodium buffer (pH 7.0) | HEWL | Shaking | 12.5 mg/mL | 10 | 99 | мкм | 0.15 | 172,800 | Protein crystallisation success | −25% | [92] |
| 244 | 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid sodium buffer (pH 7.0) | HEWL | Shaking | 12.5 mg/mL | 15 | 94.2 | мкм | 0.15 | 172,800 | Protein crystallisation success | −25% | [92] |
| 245 | 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid sodium buffer (pH 7.0) | HEWL | Shaking | 12.5 mg/mL | 30 | 60 | мкм | 0.15 | 172,800 | Protein crystallisation success | −25% | [92] |
| 246 | 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid sodium buffer (pH 7.0) | HEWL | Shaking | 12.5 mg/mL | 50 | 34.8 | мкм | 0.15 | 172,800 | Protein crystallisation success | −25% | [92] |
| 247 | 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid sodium buffer (pH 7.0) | HEWL | Shaking | 12.5 mg/mL | 60 | 4 | мкм | 0.15 | 172,800 | Protein crystallisation success | −25% | [92] |
| 248 | 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid sodium buffer (pH 7.0) | HEWL | Shaking | 12.5 mg/mL | 70 | 2.4 | мкм | 0.15 | 172,800 | Protein crystallisation success | −25% | [92] |
| 249 | 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid sodium buffer (pH 7.0) | HEWL | Shaking | 12.5 mg/mL | 80 | 0.85 | мкм | 0.15 | 172,800 | Protein crystallisation success | −25% | [92] |
| 250 | 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid sodium buffer (pH 7.0) | HEWL | Shaking | 12.5 mg/mL | 90 | 0.6 | мкм | 0.15 | 172,800 | Protein crystallisation success | −25% | [92] |
| 251 | 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid sodium buffer (pH 7.0) | HEWL | Shaking | 12.5 mg/mL | 100 | 0.4 | мкм | 0.15 | 172,800 | Protein crystallisation success | −25% | [92] |
| 252 | 0.1 M acetate buffer pH 4.5 | HEWL | Ultrasound | 25 mg/mL | 100,000 | 100,000 | mW/cm2 | 9.4 | 40 | Number of protein crystals | +150% | [93] |
| 253 | 0.1 M acetate buffer pH 4.5 | HEWL | Ultrasound | 25 mg/mL | 100,000 | 100,000 | mW/cm2 | 9.4 | 70 | Number of protein crystals | +600% | [93] |
| 254 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 50 mg/mL | 1.6 | 20 | mm | 0.5 | 3600 | Concentration of protein after precipitation in 100 mg/mL NaCl | −5% | [209] |
| 255 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 50 mg/mL | 2.5 | 20 | mm | 0.5 | 3600 | Concentration of protein after precipitation in 100 mg/mL NaCl | −10% | [209] |
| 256 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 50 mg/mL | 3.3 | 20 | mm | 0.5 | 3600 | Concentration of protein after precipitation in 100 mg/mL NaCl | −15% | [209] |
| 257 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 50 mg/mL | 1.6 | 20 | mm | 0.5 | 4800 | Concentration of protein after precipitation in 100 mg/mL NaCl | −10% | [209] |
| 258 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 50 mg/mL | 2.5 | 20 | mm | 0.5 | 4800 | Concentration of protein after precipitation in 100 mg/mL NaCl | −20% | [209] |
| 259 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 50 mg/mL | 3.3 | 20 | mm | 0.5 | 4800 | Concentration of protein after precipitation in 100 mg/mL NaCl | −40% | [209] |
| 260 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 50 mg/mL | 1.6 | 20 | mm | 0.5 | 10,800 | Concentration of protein after precipitation in 100 mg/mL NaCl | −35% | [209] |
| 261 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 50 mg/mL | 2.5 | 20 | mm | 0.5 | 10,800 | Concentration of protein after precipitation in 100 mg/mL NaCl | −40% | [209] |
| 262 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 50 mg/mL | 3.3 | 20 | mm | 0.5 | 10,800 | Concentration of protein after precipitation in 100 mg/mL NaCl | −50% | [209] |
| 263 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 90 mg/mL | 1.6 | 20 | mm | 0.5 | 1800 | Concentration of protein after precipitation in 100 mg/mL NaCl | −5% | [209] |
| 264 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 90 mg/mL | 2.5 | 20 | mm | 0.5 | 1800 | Concentration of protein after precipitation in 100 mg/mL NaCl | −5% | [209] |
| 265 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 90 mg/mL | 3.3 | 20 | mm | 0.5 | 1800 | Concentration of protein after precipitation in 100 mg/mL NaCl | −60% | [209] |
| 266 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 90 mg/mL | 1.6 | 20 | mm | 0.5 | 3600 | Concentration of protein after precipitation in 100 mg/mL NaCl | −50% | [209] |
| 267 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 90 mg/mL | 2.5 | 20 | mm | 0.5 | 3600 | Concentration of protein after precipitation in 100 mg/mL NaCl | −70% | [209] |
| 268 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 90 mg/mL | 3.3 | 20 | mm | 0.5 | 3600 | Concentration of protein after precipitation in 100 mg/mL NaCl | −80% | [209] |
| 269 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 90 mg/mL | 1.6 | 20 | mm | 0.5 | 5400 | Concentration of protein after precipitation in 100 mg/mL NaCl | −75% | [209] |
| 270 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 90 mg/mL | 2.5 | 20 | mm | 0.5 | 5400 | Concentration of protein after precipitation in 100 mg/mL NaCl | −78% | [209] |
| 271 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 90 mg/mL | 3.3 | 20 | mm | 0.5 | 5400 | Concentration of protein after precipitation in 100 mg/mL NaCl | −80% | [209] |
| 272 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 50 mg/mL | 0.1 | 20 | mm | 0.5 | 5400 | Concentration of protein after precipitation in 100 mg/mL NaCl | −5% | [209] |
| 273 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 50 mg/mL | 0.2 | 20 | mm | 0.5 | 5400 | Concentration of protein after precipitation in 100 mg/mL NaCl | −10% | [209] |
| 274 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 50 mg/mL | 0.5 | 20 | mm | 0.5 | 5400 | Concentration of protein after precipitation in 100 mg/mL NaCl | −45% | [209] |
| 275 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 50 mg/mL | 1 | 20 | mm | 0.5 | 5400 | Concentration of protein after precipitation in 100 mg/mL NaCl | −75% | [209] |
| 276 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 50 mg/mL | 0.5 | 5 | mm | 0.5 | 5400 | Concentration of protein after precipitation in 100 mg/mL NaCl | −10% | [209] |
| 277 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 50 mg/mL | 0.5 | 10 | mm | 0.5 | 5400 | Concentration of protein after precipitation in 100 mg/mL NaCl | −20% | [209] |
| 278 | 0.1 M acetate buffer pH 4.8 | HEWL 40,000 U/mg | Flow rate oscillations | 50 mg/mL | 0.5 | 30 | mm | 0.5 | 5400 | Concentration of protein after precipitation in 100 mg/mL NaCl | −80% | [209] |
| 279 | Water | 1 mmol (110 μL) benzaldehyde was with 1 mmol (145 μL) aminoacetaldehyde diethyl | Atomization (formations of nanobubbles in the flow N2) | 1 mmol | 0 | 30 | mL/min | 0.00000002 | 0.01 | Pomeranz–Fritsch reaction rate | +588% | [29] |
| 280 | Methanol | 1 mmol (110 μL) benzaldehyde was with 1 mmol (145 μL) aminoacetaldehyde diethyl | Atomization (formations of nanobubbles in the flow N2) | 1 mmol | 0 | 30 | mL/min | 0.00000002 | 0.01 | Pomeranz–Fritsch reaction rate | +644% | [29] |
| 281 | ACN/DMF (1:1; v/v) | 1 mmol (110 μL) benzaldehyde was with 1 mmol (145 μL) aminoacetaldehyde diethyl | Atomization (formations of nanobubbles in the flow N2) | 1 mmol | 0 | 30 | mL/min | 0.00000002 | 0.01 | Pomeranz–Fritsch reaction rate | +233% | [29] |
| 282 | 1% (v/v) m-NBA in water | 1 mmol (110 μL) benzaldehyde was with 1 mmol (145 μL) aminoacetaldehyde diethyl | Atomization (formations of nanobubbles in the flow N2) | 1 mmol | 0 | 30 | mL/min | 0.00000002 | 0.01 | Pomeranz–Fritsch reaction rate | +736% | [29] |
| 283 | Water | Bi3TiNbO9 | Ultrasound | not applicable. solid catalyst | 40,000 | 50,000 | mW/cmm2 | 10 | 18,000 | H2O2 production | +40,705% | [126] |
| 284 | Water | - | Shaking | - | 30 | 1 | sm | 10 | 60 | H2O2 production | +100% | [84] |
| 285 | Water | - | Shaking | - | 30 | 1 | sm | 10 | 150 | H2O2 production | +200% | [84] |
| 286 | Water | - | Shaking | - | 30 | 1 | sm | 10 | 210 | H2O2 production | +280% | [84] |
| 287 | Water | - | Shaking | - | 30 | 1 | sm | 10 | 300 | H2O2 production | +300% | [84] |
| 288 | Water | - | Shaking | - | 30 | 0.5 | sm | 10 | 60 | H2O2 production | +20% | [86] |
| 289 | Water | - | Shaking | - | 30 | 0.5 | sm | 10 | 150 | H2O2 production | +50% | [86] |
| 290 | Water | - | Shaking | - | 30 | 0.5 | sm | 10 | 210 | H2O2 production | +60% | [86] |
| 291 | Water | - | Shaking | - | 30 | 0.5 | sm | 10 | 300 | H2O2 production | +80% | [86] |
| 292 | Water | - | Shaking (turbulent) | - | 15 | 1 | sm | 10 | 300 | H2O2 production | +150% | [85] |
| 293 | Water | - | Shaking (turbulent) | - | 30 | 1 | sm | 10 | 300 | H2O2 production | +250% | [85] |
| 294 | Water | - | Shaking (turbulent) | - | 60 | 1 | sm | 10 | 300 | H2O2 production | +1500% | [85] |
| 295 | Water | - | Shaking (turbulent) | - | 30 | 1 | sm | 10 | 900 | [O2] concentration | +100% | [85] |
| 296 | Water | - | Shaking (turbulent) | - | 30 | 1 | sm | 10 | 1500 | [O2] concentration | +100% | [85] |
| 297 | Water | - | Shaking (turbulent) | - | 30 | 1 | sm | 10 | 1800 | [O2] concentration | +150% | [85] |
| 298 | Water | - | Shaking (turbulent) | - | 30 | 1 | sm | 10 | 2400 | [O2] concentration | +160% | [85] |
| 299 | Water | - | Shaking (turbulent) | - | 30 | 1 | sm | 10 | 2700 | [O2] concentration | +200% | [85] |
| 300 | Water | - | Shaking (turbulent) | - | 30 | 1 | sm | 10 | 4200 | [O2] concentration | +200% | [85] |
| 301 | Water | - | Shaking (laminar) | - | 30 | 1 | sm | 10 | 900 | [O2] concentration | +10% | [85] |
| 302 | Water | - | Shaking (laminar) | - | 30 | 1 | sm | 10 | 1500 | [O2] concentration | +10% | [85] |
| 303 | Water | - | Shaking (laminar) | - | 30 | 1 | sm | 10 | 1800 | [O2] concentration | +20% | [85] |
| 304 | Water | - | Shaking (laminar) | - | 30 | 1 | sm | 10 | 2400 | [O2] concentration | +20% | [85] |
| 305 | Water | - | Shaking (laminar) | - | 30 | 1 | sm | 10 | 2700 | [O2] concentration | +25% | [85] |
| 306 | Water | - | Shaking (laminar) | - | 30 | 1 | sm | 10 | 4200 | [O2] concentration | +25% | [85] |
| 307 | Hedysarum laeve seedlings | - | Shaking 45° | - | 2 | 6.4 | m/sm2 | 1 | 60 | Primary stem length | −15% | [55] |
| 308 | Hedysarum laeve seedlings | - | Shaking 45° | - | 2 | 6.4 | m/sm2 | 1 | 60 | Leaf numbe | −15% | [55] |
| 309 | Hedysarum laeve seedlings | - | Shaking 45° | - | 2 | 6.4 | m/sm2 | 1 | 60 | Basal diameter | −12% | [55] |
| 310 | Hedysarum laeve seedlings | - | Shaking 45° | - | 2 | 6.4 | m/sm2 | 1 | 60 | Total biomass | −30% | [55] |
| 311 | Water | - | Downfall of drops | 0 | 0 | 1 | m | 0.05 | 0.1 | [H2O2] concentration | +300% | [27] |
| 312 | Water | - | Downfall of drops | 0 | 0 | 2 | m | 0.05 | 0.1 | [H2O2] concentration | +550% | [27] |
| 313 | Water | - | Downfall of drops | 0 | 0 | 3 | m | 0.05 | 0.1 | [H2O2] concentration | +750% | [27] |
| 314 | Water | - | Downfall of drops | 0 | 0 | 4 | m | 0.05 | 0.1 | [H2O2] concentration | +900% | [27] |
| 315 | Water | - | Downfall of drops | 0 | 0 | 1 | m | 0.05 | 0.1 | [OH] concentration | +200% | [27] |
| 316 | Water | - | Downfall of drops | 0 | 0 | 2 | m | 0.05 | 0.1 | [OH] concentration | +350% | [27] |
| 317 | Water | - | Downfall of drops | 0 | 0 | 3 | m | 0.05 | 0.1 | [OH] concentration | +500% | [27] |
| 318 | Water | - | Downfall of drops | 0 | 0 | 4 | m | 0.05 | 0.1 | [OH] concentration | +550% | [27] |
| 319 | Water | PNaAMPS/PAAm double-network (DN) hydrogels | Stretching | 1.4 М | 0 | 1.07 | Value of the first Lame parameter λ | 20 | 1800 | Normalised width w(t)/w(0) | +2% | [95] |
| 321 | Water | PNaAMPS/PAAm double-network (DN) hydrogels | Stretching | 1.4 М | 0 | 1.14 | Value of the first Lame parameter λ | 20 | 1800 | Normalised width w(t)/w(0) | +2% | [95] |
| 322 | Water | PNaAMPS/PAAm double-network (DN) hydrogels | Stretching | 1.4 М | 0 | 1.21 | Value of the first Lame parameter λ | 20 | 1800 | Normalised width w(t)/w(0) | +1% | [95] |
| 323 | Water | PNaAMPS/PAAm double-network (DN) hydrogels | Stretching | 1.4 М | 0 | 1.23 | Value of the first Lame parameter λ | 20 | 1800 | Normalised width w(t)/w(0) | +0% | [95] |
| 324 | Water | PNaAMPS/PAAm double-network (DN) hydrogels | Stretching | 1.4 М | 0 | 1.27 | Value of the first Lame parameter λ | 20 | 1800 | Normalised width w(t)/w(0) | −1.5% | [95] |
| 325 | Water | PNaAMPS/PAAm double-network (DN) hydrogels | Stretching | 1.4 М | 0 | 1.32 | Value of the first Lame parameter λ | 20 | 1800 | Normalised width w(t)/w(0) | −2% | [95] |
| 326 | Water | PNaAMPS/PAAm double-network (DN) hydrogels | Stretching | 1.4 М | 0 | 1.38 | Value of the first Lame parameter λ | 20 | 1800 | Normalised width w(t)/w(0) | −3% | [95] |
| 327 | Water | PNaAMPS/PAAm double-network (DN) hydrogels | Stretching | 1.4 М | 0 | 1.07 | Value of the first Lame parameter λ | 20 | 1800 | normalized engineering stress σ(t)/σ(0) | −6% | [95] |
| 328 | Water | PNaAMPS/PAAm double-network (DN) hydrogels | Stretching | 1.4 М | 0 | 1.14 | Value of the first Lame parameter λ | 20 | 1800 | normalized engineering stress σ(t)/σ(0) | −0% | [95] |
| 329 | Water | PNaAMPS/PAAm double-network (DN) hydrogels | Stretching | 1.4 М | 0 | 1.21 | Value of the first Lame parameter λ | 20 | 1800 | normalized engineering stress σ(t)/σ(0) | −0% | [95] |
| 330 | Water | PNaAMPS/PAAm double-network (DN) hydrogels | Stretching | 1.4 М | 0 | 1.23 | Value of the first Lame parameter λ | 20 | 1800 | normalized engineering stress σ(t)/σ(0) | −1% | [95] |
| 331 | Water | PNaAMPS/PAAm double-network (DN) hydrogels | Stretching | 1.4 М | 0 | 1.27 | Value of the first Lame parameter λ | 20 | 1800 | normalized engineering stress σ(t)/σ(0) | −3% | [95] |
| 332 | Water | PNaAMPS/PAAm double-network (DN) hydrogels | Stretching | 1.4 М | 0 | 1.32 | Value of the first Lame parameter λ | 20 | 1800 | normalized engineering stress σ(t)/σ(0) | −14% | [95] |
| 333 | Water | PNaAMPS/PAAm double-network (DN) hydrogels | Stretching | 1.4 М | 0 | 1.38 | Value of the first Lame parameter λ | 20 | 1800 | normalized engineering stress σ(t)/σ(0) | −17% | [95] |
| 334 | waste water after blowing by 99.99% N2 | T. denitrificans. immobilised on electrod | Mixing | 99.99% N2 | 10 | 5 | mm | 26.2 | 259,200 | Concntration of Na+ | −20% | [79] |
| 335 | waste water after blowing by 99.99% N2 | T. denitrificans. immobilised on electrod | Mixing | 99.99% N2 | 10 | 5 | mm | 26.2 | 259,200 | Electrode voltage | +80% | [79] |
| 336 | waste water after blowing by 99.99% N2 | T. denitrificans. immobilised on electrod | Mixing | 99.99% N2 | 10 | 5 | mm | 26.2 | 259,200 | Consumption of NO3- | +35% | [79] |
| 337 | Water | - | Mixing | - | 10 | 37 | mm | 30 | 180 | Concentration of bulk nanobubbles | +150% | [96] |
| 338 | Water | - | Mixing | - | 5 | 37 | mm | 30 | 180 | Concentration of bulk nanobubbles | +80% | [96] |
| 339 | Water | - | Mixing | - | 15 | 37 | mm | 30 | 180 | Concentration of bulk nanobubbles | +200% | [96] |
| 340 | Water | - | Mixing | - | 15 | 37 | mm | 30 | 180 | Concentration of bulk nanobubbles | +30% | [96] |
| 341 | Water | - | Mixing | - | 15 | 37 | mm | 30 | 60 | Concentration of bulk nanobubbles | +200% | [96] |
| 342 | Water | - | Mixing | - | 15 | 37 | mm | 30 | 360 | Concentration of bulk nanobubbles | +1500% | [96] |
| 343 | Water | - | Mixing | - | 15 | 37 | mm | 30 | 600 | Concentration of bulk nanobubbles | +1100% | [96] |
| 344 | Water | - | Ultrasound | - | 22,000 | 2000 | mW/cm2 | 5000 | 300 | pH | +6% | [121] |
| 345 | Water | - | Ultrasound | - | 100,0001,000,000 | 2000 | mW/cm2 | 5000 | 300 | pH | −17% | [121] |
| 346 | Water (artesian) | total hardness. carbonate hardness; Ca+2 content; Mg+2 content; pH 7. sulfates. chlorides | Ultrasound | 16.6 mg-eq/L. 4.8 mg-eq/. 17.7 mg/L. 8.9 mg/L. 7.8 mg/L. 576 mg/L. 12.0 mg/L | 22,000 | 2000 | mW/cm2 | 5000 | 300 | pH | +12% | [121] |
| 347 | Water | Clay particles | Ultrasound | 20 mg/mL | 20,000 | 2000 | mW/cm2 | 1000 | 10 | Precipitation rate | −30% | [124] |
| 348 | Water | Clay particles | Ultrasound | 20 mg/mL | 20,000 | 2000 | mW/cm2 | 1000 | 30 | Precipitation rate | +1% | [124] |
| 349 | Water | Clay particles | Ultrasound | 20 mg/mL | 20,000 | 2000 | mW/cm2 | 1000 | 60 | Precipitation rate | +50% | [124] |
| 350 | Water | Clay particles | Ultrasound | 20 mg/mL | 20,000 | 2000 | mW/cm2 | 1000 | 120 | Precipitation rate | +60% | [124] |
| 351 | Water | Clay particles | Ultrasound | 20 mg/mL | 20,000 | 2000 | mW/cm2 | 1000 | 300 | Precipitation rate | +101% | [124] |
| 352 | Mouse | in vivo | Sinusoidal vibrations | - | 45 | 0.3 | g | 21 | 37,800 | Number of lectin-positive vessels | −29% | [20] |
| 353 | Mouse | in vivo | Sinusoidal vibrations | - | 45 | 0.3 | g | 21 | 37,800 | α-actin-positive vessels | −36% | [20] |
| 355 | Rabbit | in vivo | Shaking | - | 60 | 5 | g | 4900 | 2,160,000 | Myelin fibre diameter | +80% | [53] |
| 356 | Human | in vivo | Vibration | - | 25 | 3 | m/s2 | 92,900 | 60 | vibration intensity transmission | +90% | [51] |
| 357 | Human | in vivo | Vibration | - | 200 | 3 | m/s2 | 92,900 | 60 | vibration intensity transmission | +900% | [51] |
| 358 | Human | in vivo | Vibration | - | 25 | 3 | m/s2 | 92,900 | 60 | Apparent mass | −50% | [51] |
| 359 | Human | in vivo | Vibration | - | 200 | 3 | m/s2 | 92,900 | 60 | Apparent mass | −80% | [51] |
| 360 | Human | in vivo | Vibration | - | 25 | 3 | m/s2 | 92,900 | 60 | Mechanical impedance | +25% | [51] |
| 361 | Human | in vivo | Vibration | - | 200 | 3 | m/s2 | 92,900 | 60 | Mechanical impedance | +150% | [51] |
| 362 | Rat | in vivo | Vibration | - | 80 | 32 | m/s2 | 205 | 90,000 | Length of nerve regeneration by 6 days | +57% | [54] |
| 363 | Rat | in vivo | Vibration | - | 80 | 32 | m/s2 | 205 | 90,000 | Length of nerve regeneration by 6 days | +23% | [54] |
| 364 | Rat | in vivo | Vibration | - | 81 | 0.5 | mm | 240 | 28,800 | Expression of IGF-I in nerve fibers | +100% | [210] |
| 365 | Rat. tail | in vivo | Vertical shaking | - | 62.5 | 49 | m/s2 | 6 | 144,00 | Expression of MT-1a | +700% | [40] |
| 366 | Rat. tail | in vivo | Vertical shaking | - | 125 | 49 | m/s2 | 6 | 144,00 | Expression of MT-1a | +660% | [40] |
| 367 | Rat. tail | in vivo | Vertical shaking | - | 250 | 49 | m/s2 | 6 | 144,00 | Expression of MT-1a | +630% | [40] |
| 368 | Rat. tail | in vivo | Vertical shaking | - | 62.5 | 49 | m/s2 | 6 | 144,00 | Expression of IL-6 | +900% | [40] |
| 369 | Rat. tail | in vivo | Vertical shaking | - | 125 | 49 | m/s2 | 6 | 144,00 | Expression of IL-6 | +500% | [40] |
| 370 | Rat. tail | in vivo | Vertical shaking | - | 250 | 49 | m/s2 | 6 | 14,400 | Expression of IL-6 | +300% | [40] |
| 371 | Rat. tail | in vivo | Vertical shaking | - | 62.5 | 49 | m/s2 | 6 | 144,00 | Expression of IL-6 | +400% | [40] |
| 372 | Rat. tail | in vivo | Vertical shaking | - | 125 | 49 | m/s2 | 6 | 144,00 | Expression of IL-6 | +220% | [40] |
| 373 | Rat. tail | in vivo | Vertical shaking | - | 250 | 49 | m/s2 | 6 | 14,400 | Internal diameter of the caudal vein | −70% | [40] |
| 374 | Rat. tail | in vivo | Vertical shaking | - | 250 | 49 | m/s2 | 6 | 144,00 | Vascular smooth muscle thickness | +35% | [40] |
| 375 | Rat. tail | in vivo | Vertical shaking | - | 125 | 49 | m/s2 | 6 | 14,400 | Synthesis of nitrotyrosine | +125% | [40] |
| 376 | Rat. tail | in vivo | Vertical shaking | - | 250 | 49 | m/s2 | 6 | 144,00 | Synthesis of nitrotyrosine | +150% | [40] |
| 377 | Rat. tail | in vivo | Vertical shaking | - | 125 | 49 | m/s2 | 6 | 144,00 | Synthesis of IL-6 | +50% | [40] |
| 378 | Rat. tail | in vivo | Vertical shaking | - | 250 | 49 | m/s2 | 6 | 14,400 | Synthesis of IL-6 | +70% | [40] |
| 379 | Rat. tail | in vivo | Vertical shaking | - | 60 | 49 | m/s2 | 5 | 14,000 | Number of vacuoles in endothelial cells | +2900% | [41] |
| 380 | Rat. tail | in vivo | Vertical shaking | - | 120 | 49 | m/s2 | 5 | 14,000 | Number of vacuoles in endothelial cells | +1300% | [41] |
| 381 | Rat. tail | in vivo | Vertical shaking | - | 30 | 49 | m/s2 | 5 | 144,00 | Number of ruptures of the inner vessel wall | +1600: | [41] |
| 382 | Rat. tail | in vivo | Vertical shaking | - | 69 | 49 | m/s2 | 5 | 144,00 | Number of ruptures of the inner vessel wall | +2100% | [41] |
| 383 | Rat. tail | in vivo | Vertical shaking | - | 120 | 49 | m/s2 | 5 | 14,400 | Number of ruptures of the inner vessel wall | +1500% | [41] |
| 384 | Rat. tail | in vivo | Vertical shaking | - | 800 | 49 | m/s2 | 5 | 14,400 | Number of ruptures of the inner vessel wall | +1600% | [41] |
| 385 | Rat. tail | in vivo | Vertical shaking | - | 30 | 49 | m/s2 | 5 | 14,400 | Expression of NFATc3 | +100% | [41] |
| 386 | Rat. tail | in vivo | Vertical shaking | - | 60 | 49 | m/s2 | 5 | 14,400 | Expression of NFATc3 | +200% | [41] |
| 387 | Rat. tail | in vivo | Vertical shaking | - | 120 | 49 | m/s2 | 5 | 14,400 | Expression of NFATc3 | +200% | [41] |
| 388 | Rat. lower limbs | in vivo | Vertical shaking | - | 30 | 49 | m/s2 | 15 | 57,600 | Total plasma creatine phosphokinase (t-CPK) activity | +1830% | [42] |
| 389 | Rat. lower limbs | in vivo | Vertical shaking | - | 60 | 49 | m/s2 | 15 | 57,600 | Total plasma creatine phosphokinase (t-CPK) activity | +760% | [42] |
| 390 | Rat. lower limbs | in vivo | Vertical shaking | - | 120 | 49 | m/s2 | 15 | 57,600 | Total plasma creatine phosphokinase (t-CPK) activity | +700% | [42] |
| 391 | Rat. lower limbs | in vivo | Vertical shaking | - | 240 | 49 | m/s2 | 15 | 57,600 | Total plasma creatine phosphokinase (t-CPK) activity | +400% | [42] |
| 392 | Rat. lower limbs | in vivo | Vertical shaking | - | 480 | 49 | m/s2 | 15 | 57,600 | Total plasma creatine phosphokinase (t-CPK) activity | +200% | [42] |
| 393 | Culture medium DMEM | Mouse Fibroblast Cells | Sinusoidal shaking | - | 1250 | 16.5 | m/s2 | 5 | 900 | Cells viability | −59% | [57] |
| 394 | Culture medium DMEM | Mouse Fibroblast Cells | Shock shaking | - | 1250 | 12.9 | m/s2 | 5 | 900 | Cells viability | −99% | [57] |
| 395 | Culture medium DMEM | Mouse red blood cells | Shock shaking | - | 1250 | 30,000 | m/s2 | 5 | 900 | Erythrocyte lysis | +100% | [57] |
| 396 | Culture medium DMEM | Mouse red blood cells | Shock shaking | - | 1250 | 15,000 | m/s2 | 5 | 900 | Erythrocyte lysis | +0.4% | [57] |
| 397 | Culture medium DMEM | Mouse red blood cells | Shock shaking | - | 1250 | 2000 | m/s2 | 5 | 900 | Erythrocyte lysis | +0.07% | [57] |
| 398 | Culture medium DMEM | Mouse red blood cells | Shock shaking | - | 1250 | 1000 | m/s2 | 5 | 900 | Erythrocyte lysis | +0.1% | [57] |
| 399 | Culture medium DMEM | Mouse red blood cells | Shock shaking | - | 1250 | 500 | m/s2 | 5 | 900 | Erythrocyte lysis | +0.1% | [57] |
| 400 | Human | in vivo | Vibration | - | 5.5 | 74.4 | N | 60,000 | 600 | Peak inspiratory flow rate | +136% | [52] |
| 401 | Human | in vivo | Vibration | - | 5.5 | 74.4 | N | 60,000 | 600 | Inspired volume | +200% | [52] |
| 402 | Human | in vivo | Vibration | - | 5.5 | 74.4 | N | 60,000 | 600 | expired volume | +290% | [52] |
| 403 | Human. healthy pre-pubertal boys | in vivo | Vibration (platform) | - | 40 | 2.1 | g | 34,000 | 600 | Legs’ skin temperature | +8% | [46] |
| 404 | Human. healthy pre-pubertal boys | in vivo | Vibration (platform) | - | 40 | 2.1 | g | 34,000 | 3000 | P1NP plasma level after 8 days | +18% | [46] |
| 405 | Human. healthy pre-pubertal boys | in vivo | Vibration (platform) | - | 40 | 2.1 | g | 34,000 | 3000 | CTx plasma level after 8 days | +10% | [46] |
| 406 | Rat with titanium hip implants | in vivo | Vibration (whole body) | - | 80 | 0.3 | g | 353 | 300 | Bone-to-implant contact | +63% | [48] |
| 407 | Rat with titanium hip implants | in vivo | Vibration (whole body) | - | 140 | 0.3 | g | 353 | 300 | Bone-to-implant contact | +65% | [48] |
| 408 | Rat with titanium hip implants | in vivo | Vibration (whole body) | - | 26 | 0.3 | g | 353 | 300 | Peri-implant bone formation | +20% | [48] |
| 409 | Rat with titanium hip implants | in vivo | Vibration (whole body) | - | 80 | 0.3 | g | 353 | 300 | Peri-implant bone formation | +25% | [48] |
| 410 | Rat with titanium hip implants | in vivo | Vibration (whole body) | - | 140 | 0.043 | g | 353 | 300 | Peri-implant bone formation | +19% | [48] |
| 411 | Rat with titanium hip implants | in vivo | Vibration (whole body) | - | 140 | 0.3 | g | 353 | 300 | Peri-implant bone formation | +25% | [48] |
| 412 | Rat with removed ovaries (osteoporosis model) | in vivo | Vibration (whole body) | - | 30 | 0.3 | g | 285 | 134,400 | Obesity | −50% | [28] |
| 413 | Rat with removed ovaries (osteoporosis model) | in vivo | Vibration (whole body) | 30 | 0.3 | g | 285 | 67,200 | Bone mineral densities | +15% | [28] | |
| 414 | Rat with removed ovaries (osteoporosis model) | in vivo | Vibration (whole body) | - | 30 | 0.3 | g | 285 | 134,400 | Vertebra compression test (maximum load) | −18% | [28] |
| 415 | Rat (healthy) | in vivo | Vibration (whole body) | - | 30 | 0.3 | g | 285 | 134,400 | vertebra compression test (rigidity) | −45% | [28] |
| 416 | Rat with removed ovaries (osteoporosis model) | in vivo | Vibration (whole body) | - | 30 | 0.3 | g | 285 | 134,400 | Osteoblastic Cell Viability | −10% | [28] |
| 417 | Rat (healthy) | in vivo | Vibration (whole body) | - | 30 | 0.3 | g | 285 | 134,400 | Osteoblastic Cell Viability | −12% | [28] |
| 418 | Rat (healthy) | in vivo | Vibration (whole body) | - | 30 | 0.3 | g | 285 | 134,400 | Alkaline phosphatase (ALP) activity 4 day | +15% | [28] |
| 419 | Rat (healthy) | in vivo | Vibration (whole body) | - | 30 | 0.3 | g | 285 | 134,400 | alkaline phosphatase (ALP) activity 7 day | −10% | [28] |
| 420 | Rat (healthy) | in vivo | Vibration (whole body) | - | 30 | 0.3 | g | 285 | 134,400 | Expression of ALP | −66% | [28] |
| 421 | Rat with removed ovaries (osteoporosis model) | in vivo | Vibration (whole body) | - | 30 | 0.3 | g | 285 | 134,400 | Expression of ALP | −50% | [28] |
| 422 | Rat (healthy) | in vivo | Vibration (whole body) | - | 30 | 0.3 | g | 285 | 134,400 | Expression of MMP2 | −40% | [28] |
| 423 | Rat with removed ovaries (osteoporosis model) | in vivo | Vibration (whole body) | - | 30 | 0.3 | g | 285 | 134,400 | Expression of MMP2 | −40% | [28] |
| 424 | Rat (healthy) | in vivo | Vibration (whole body) | - | 30 | 0.3 | g | 285 | 134,400 | Expression of OSX | −10% | [28] |
| 425 | Rat with removed ovaries (osteoporosis model) | in vivo | Vibration (whole body) | - | 30 | 0.3 | g | 285 | 134,400 | Expression of OSX | +20% | [28] |
| 426 | C57BL/6 mouse | in vivo | Vibration (whole body) | - | 90 | 0.75 | m/s2 | 27.5 | 300 | Arterial blood pressure | +25% | [39] |
| 427 | C57BL/6 mouse | in vivo | Vibration (whole body) | - | 80 | 0.75 | m/s2 | 27.5 | 300 | Heart beat rate | +19% | [39] |
| 428 | C57BL/6 mouse | in vivo | Vibration (whole body) | - | 90 | 0.75 | m/s2 | 27.5 | 300 | Heart beat rate | +17% | [39] |
| 429 | Human | in vivo | Vibration (whole body) | - | 30 | 0.3 | m/s2 | 61500 | 14,400 | Bone mineralisation density | +40% | [44] |
| 430 | Human | in vivo | Vibration (whole body) | - | 30 | 0.3 | m/s2 | 61500 | 14,400 | TRAP synthesis | −20% | [44] |
| 431 | Culture medium DMEM | Human embryonic stem cells (hESC) | Ultrasound | - | 19,690,000 | 24,000 | mW | 5 | 7200 | Expression of markers of neuronal differentiation and Alk | +250% | [116] |
| 432 | Culture medium DMEM | Human embryonic stem cells (hESC) | Ultrasound | - | 19,690,000 | 310,000 | mW | 5 | 7200 | Expression of markers of neuronal differentiation and Alk | +200% | [116] |
| 433 | Culture medium DMEM | Human embryonic stem cells (hESC) | Ultrasound | - | 19,690,000 | 24,000 | mW | 5 | 7200 | Expression of Cenpf | +40% | [116] |
| 434 | Culture medium DMEM | Human embryonic stem cells (hESC) | Ultrasound | - | 19,690,000 | 31,000 | mW | 5 | 7200 | Expression of Cenpf | +20% | [116] |
| 435 | Culture medium DMEM | Human embryonic stem cells (hESC) | Ultrasound | - | 19,690,000 | 39,000 | mW | 5 | 7200 | Expression of Cenpf | −30% | [116] |
| 436 | Culture medium DMEM | Human embryonic stem cells (hESC) | Ultrasound | - | 19,690,000 | 24,000 | mW | 5 | 7200 | Expression of actin | −30% | [116] |
| 437 | Culture medium DMEM | Human embryonic stem cells (hESC) | Ultrasound | - | 19,690,000 | 31,000 | mW | 5 | 7200 | Expression of actin | −40% | [116] |
| 438 | Culture medium DMEM | Human embryonic stem cells (hESC) | Ultrasound | - | 19,690,000 | 39,000 | mW | 5 | 7200 | Expression of actin | −80% | [116] |
| 439 | Culture medium DMEM | Human embryonic stem cells (hESC) | Ultrasound | - | 19,690,000 | 24,000 | mW | 5 | 7200 | Expression of Pcdh17 | +60% | [116] |
| 440 | Culture medium DMEM | Human embryonic stem cells (hESC) | Ultrasound | - | 19,690,000 | 31,000 | mW | 5 | 7200 | Expression of Pcdh17 | +70% | [116] |
| 441 | Culture medium DMEM | Human embryonic stem cells (hESC) | Ultrasound | - | 19,690,000 | 39,000 | mW | 5 | 7200 | Expression of Pcdh17 | +20% | [116] |
| 442 | Culture medium DMEM | SHED: stem cells from human exfoliated deciduous teeth | Centrifugation | - | - | 100 | g | 5 | 30 min. every 24 h.. 7 дней | Cells proliferation | −16.32% | [25] |
| 443 | Culture medium DMEM | SHED: stem cells from human exfoliated deciduous teeth | Centrifugation | - | - | 200 | g | 5 | 30 min. every 24 h.. 7 дней | Cells proliferation | −18.36% | [25] |
| 444 | Culture medium DMEM | SHED: stem cells from human exfoliated deciduous teeth | Centrifugation | - | - | 300 | g | 5 | 30 min. every 24 h.. 7 дней | Cells proliferation | −16.32% | [25] |
| 445 | Culture medium α-MEM | MC3T3-E1 | Vibration | - | 12.5 | 0.5 | G | 5 | 1,382,400 | Confluency | +19.4% | [26] |
| 446 | Culture medium α-MEM | MC3T3-E1 | Vibration | - | 12.5 | 0.5 | G | 4 | 1,123,200 | Confluency | +15.6% | [26] |
| 447 | Culture medium α-MEM | MC3T3-E1 | Constant flow | - | 0 | 0.28 ± 0.02 | mL/min | 4 | 1,382,400 | Confluency | −22.2% | [26] |
| 448 | Culture medium α-MEM | MC3T3-E1 | Constant flow | - | 0 | 0.28 ± 0.02 | mL/min | 4 | 1,123,200 | Confluency | +12.5% | [26] |
| 449 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 50 | 0.3 | G | 10 | 30 min. every 24 h. | Alkaline phosphatase activity | +123.8% | [211] |
| 450 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 60 | 0.3 | G | 10 | 30 min. every 24 h. | Alkaline phosphatase activity | +96.23% | [211] |
| 451 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 10 | 0.3 | G | 10 | 30 min. every 24 h. | Expression of gene Runx2 | −43.75% | [211] |
| 452 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 40 | 0.3 | G | 10 | 30 min. every 24 h. | Expression of gene Runx2 | +175% | [211] |
| 453 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 50 | 0.3 | G | 10 | 30 min. every 24 h. | Expression of gene Runx2 | +275% | [211] |
| 454 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 60 | 0.3 | G | 10 | 30 min. every 24 h. | Expression of gene Runx2 | +68.75% | [211] |
| 455 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 90 | 0.3 | G | 10 | 30 min. every 24 h. | Expression of gene Runx2 | +34.37% | [211] |
| 456 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 180 | 0.3 | G | 10 | 30 min. every 24 h. | Expression of gene Runx2 | −25% | [211] |
| 457 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 20 | 0.3 | G | 10 | 30 min. every 24 h. | Expression of gene Osx | −50% | [211] |
| 458 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 40 | 0.3 | G | 10 | 30 min. every 24 h. | Expression of gene Osx | +62.5% | [211] |
| 459 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 50 | 0.3 | G | 10 | 30 min. every 24 h. | Expression of gene Osx | +109.3% | [211] |
| 460 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 60 | 0.3 | G | 10 | 30 min. every 24 h. | Expression of gene Osx | −43.75% | [211] |
| 461 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 120 | 0.3 | G | 10 | 30 min. every 24 h. | Expression of gene Osx | −43.75% | [211] |
| 462 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 150 | 0.3 | G | 10 | 30 min. every 24 h. | Expression of gene Osx | −56.25% | [211] |
| 463 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 180 | 0.3 | G | 10 | 30 min. every 24 h. | Expression of gene Osx | +37.5% | [211] |
| 464 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 40 | 0.3 | G | 10 | 30 min. every 24 h. | Osteocalcin levels (OCN) | +340.9% | [211] |
| 465 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 50 | 0.3 | G | 10 | 30 min. every 24 h. | Osteocalcin levels (OCN) | +390.9% | [211] |
| 466 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 60 | 0.3 | G | 10 | 30 min. every 24 h. | Osteocalcin levels (OCN) | +318.18% | [211] |
| 467 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 90 | 0.3 | G | 10 | 30 min. every 24 h. | Osteocalcin levels (OCN) | +122.72% | [211] |
| 468 | Culture medium α-MEM | human PDL stem cells | Vibration | - | 120 | 0.3 | G | 10 | 30 min. every 24 h. | Osteocalcin levels (OCN) | +154.54% | [211] |
| 469 | Culture medium DMEM low glucose | mesenchymal stem cells (MSCs) | Stretching | - | 0.03 | 5 | % | 2 | 240 min/day | Collagen content | +26.22% | [67] |
| 470 | StemFit AK02N | iPSC | Vibration | - | 100 | 0.02 | mm | 2 | 3000 | Cells differentiation | −28.5% | [68] |
| 471 | StemFit AK02N | iPSC | Vibration | - | 150 | 0.04 | mm | 2 | 3000 | Cells differentiation | −22.85% | [68] |
| 472 | (DMEM)/Ham’s F12 | human adipose-derived stem cells (hASCs) | Cyclic stretching | - | 0.5 | 5% | % | 2 | 864,000 | Expression of CYP1B1 | +68.18% | [69] |
| 473 | (DMEM)/Ham’s F12 | human adipose-derived stem cells (hASCs) | Cyclic stretching | - | 0.5 | 5% | % | 2 | 864,000 | Metabolic activity | −30.6% | [69] |
| 474 | Ham’s F–12 K | Chinese Hamster Ovary (CHO)-adherent cells | Vibration | - | 10–500 | <1 | g | 10 | 345,600 | Cells proliferation | +79% | [56] |
| 475 | Freestyle CHO Expression | Chinese Hamster Ovary (CHO)- suspension cells | Vibration | - | 10–500 | <1 | g | 10 | 345,600 | Cells proliferation | −13.0% | [56] |
| 476 | Freestyle CHO Expression | Chinese Hamster Ovary (CHO)- suspension cells | Vibration | - | 30 | 0.7 | g | 10 | 345,600 | Cells proliferation | +210% | [56] |
| 477 | RPMI | Е-cells | Vibration | - | 30 | 0.7 | g | 10 | 345,600 | Cells proliferation | +20.3% | [56] |
| 478 | DMEM | Primary fibrochondrocytes | Cyclic stretching | - | 1 | 8 | % | 2 | 28,800 | Expression of COL2A1 | +110% | [59] |
| 479 | DMEM | Primary fibrochondrocytes | Cyclic stretching | - | 1 | 8 | % | 2 | 28,800 | Expression of SOX9 | +200% | [59] |
| 480 | DMEM | Primary fibrochondrocytes | Cyclic stretching | - | 1 | 8 | % | 2 | 28,800 | Expression of ACAN | +215.78% | [59] |
| 481 | DMEM | Primary fibrochondrocytes | Cyclic stretching | - | 1 | 8 | % | 2 | 28,800 | Proportion of BrdU-positive cells | +46.6% | [59] |
| 482 | DMEM | Human osteoblast-like cells primary explant cultures | Vibration | - | 60 | 30 | µm | 5 | 96 h | Confluency | +23.07% | [60] |
| 483 | DMEM | Human osteoblast-like cells primary explant cultures | Vibration | - | 60 | 30 | µm | 5 | 96 h | Alkaline phosphatase activity | −29.82% | [60] |
| 484 | Culture medium α-MEM | GD25 cells | Vibration | - | 12.5 | 0.5 | G | 5 | 12 days | Cells culture density | +50% | [61] |
| 485 | Culture medium α-MEM | GD25 cells | Vibration | - | 12.5 | 0.5 | G | 5 | 15 days | Cells culture density | +60% | [61] |
| 486 | Culture medium α-MEM | GD25 cells | Vibration | - | 12.5 | 0.5 | G | 5 | 18 days | Cells culture density | +83.67% | [61] |
| 487 | Culture medium α-MEM | GD25 cells | Vibration | - | 12.5 | 0.5 | G | 5 | 21 day | Cells culture density | +73% | [61] |
| 488 | Culture medium α-MEM | GD25 cells | Vibration | - | 12.5 | 0.5 | G | 5 | 24 days | Cells culture density | +45.8% | [61] |
| 489 | Culture medium α-MEM | GD25 cells | Vibration | - | 12.5 | 0.5 | G | 5 | 15 days | Cell layer thickness | +46.15% | [61] |
| 490 | Culture medium DMEM | Human periodontal ligament stem cell (hPDLSC) | Shear stress | - | - | 0.5 | dyn/cm2 | 0.5 | 10,800 | Expression of gene IDO | +80% | [62] |
| 491 | Culture medium DMEM | Human periodontal ligament stem cells (hPDLSC) | Shear stress | - | - | 5 | dyn/cm2 | 0.5 | 10,800 | Expression of gene IDO | +140% | [62] |
| 492 | Culture medium DMEM | Human periodontal ligament stem cells (hPDLSC) | Shear stress | - | - | 0.5 | dyn/cm2 | 0.5 | 10,800 | Expression of gene COX2 | +83.3% | [62] |
| 493 | Culture medium DMEM | Human periodontal ligament stem cells (hPDLSC) | Shear stress | - | - | 5 | dyn/cm2 | 0.5 | 10,800 | Expression of gene COX2 | +266.6% | [62] |
| 494 | Culture medium DMEM | Human periodontal ligament stem cells (hPDLSC) | Shear stress | - | - | 0.5 | dyn/cm2 | 0.5 | 10,800 | Activity of IDO | −18.18% | [62] |
| 495 | Culture medium DMEM | Human periodontal ligament stem cells (hPDLSC) | Shear stress | - | - | 5 | dyn/cm2 | 0.5 | 10,800 | Activity of IDO | +106% | [62] |
| 496 | Culture medium DMEM | Human periodontal ligament stem cells (hPDLSC) | Shear stress | - | - | 10 | dyn/cm2 | 0.5 | 10,800 | Activity of IDO | +78.78% | [62] |
| 497 | Culture medium DMEM | Human periodontal ligament stem cells (hPDLSC) | Shear stress | - | - | 0.5 | dyn/cm2 | 0.5 | 10,800 | Kynurenine syntethis | +58.85% | [62] |
| 498 | Culture medium DMEM | Human periodontal ligament stem cells (hPDLSC) | Shear stress | - | - | 5 | dyn/cm2 | 0.5 | 10,800 | Kynurenine syntethis | +170.5% | [62] |
| 499 | Culture medium DMEM | Human periodontal ligament stem cells (hPDLSC) | Shear stress | - | - | 0.5 | dyn/cm2 | 0.5 | 10,800 | General concentration of TGF-β1 in medium | +17.3% | [62] |
| 500 | Culture medium DMEM | Human periodontal ligament stem cells (hPDLSC) | Shear stress | - | - | 5 | dyn/cm2 | 0.5 | 10,800 | General concentration of TGF-β1 in medium | −9.6% | [62] |
| 501 | Culture medium DMEM | Human periodontal ligament stem cells (hPDLSC) | Shear stress | - | - | 0.5 | dyn/cm2 | 0.5 | 10,800 | Concentration of active TGF-β1 in medium | +2.87% | [62] |
| 502 | Culture medium DMEM | Human periodontal ligament stem cells (hPDLSC) | Shear stress | - | - | 5 | dyn/cm2 | 0.5 | 10,800 | Concentration of active TGF-β1 in medium | +125% | [62] |
| 503 | Culture medium DMEM | Human periodontal ligament stem cells (hPDLSC) | Shear stress | - | - | 10 | dyn/cm2 | 0.5 | 10,800 | Concentration of active TGF-β1 in medium | +125% | [62] |
| 504 | Culture medium DMEM | Human periodontal ligament stem cells (hPDLSC) | Shear stress | - | - | 5 | dyn/cm2 | 0.5 | 10,800 | Concentration of mature TGF-β1/β-ACTIN | −6.66% | [62] |
| 505 | Culture medium DMEM | Human periodontal ligament stem cells (hPDLSC) | Shear stress | - | - | 10 | dyn/cm2 | 0.5 | 10,800 | Concentration of mature TGF-β1/β-ACTIN | −43.3% | [62] |
| 506 | Culture medium DMEM | Human periodontal ligament stem cells (hPDLSC) | Shear stress | - | - | 0.5 | dyn/cm2 | 0.5 | 10,800 | Concentration of IFN-γ in lysate of cells | −51.21% | [62] |
| 507 | Culture medium DMEM | Human periodontal ligament stem cells (hPDLSC) | Shear stress | - | - | 5 | dyn/cm2 | 0.5 | 10,800 | Concentration of IFN-γ in lysate of cells | −51% | [62] |
| 508 | Culture medium DMEM | Human periodontal ligament stem cells (hPDLSC) | Shear stress | - | - | 10 | dyn/cm2 | 0.5 | 10,800 | Concentration of IFN-γ in lysate of cells | −65.85% | [62] |
| 509 | Culture medium DMEM | Chondrocytes from pig | Vibration | - | 25 | 0.5 | G | 0.5 | 86,400 | Type II collagen expression | +308.3% | [64] |
| 510 | Culture medium DMEM | Chondrocytes from pig | Vibration | - | 25 | 0.5 | G | 0.5 | 86,400 | Aggrecan expression | +225% | [64] |
| 511 | Culture medium DMEM | Chondrocytes from pig | Vibration | - | 25 | 0.5 | G | 0.5 | 86,400 | Type I collagen expression | +372.72% | [64] |
| 512 | Culture medium DMEM | Chondrocytes from pig | Vibration | - | 25 | 0.5 | G | 0.5 | 86,400 | Fibronectin expression | +206.25% | [64] |
| 513 | Culture medium α-MEM | Osteocytic MLO-Y4 cells | Vibration | - | 45 | 0.5 | g | 0.5 | 259,200 | Cells viability | +47.36% | [66] |
| 514 | Culture medium α-MEM | Osteocytic MLO-Y4 cells | Vibration | - | 45 | 0.5 | g | 0.5 | 259,200 | Proportion of apoptosis events | −47.36% | [66] |
| 515 | Culture medium α-MEM | Osteocytic MLO-Y4 cells | Vibration | - | 45 | 0.5 | g | 0.5 | 259,200 | Relative cell area | +52.17% | [66] |
| 516 | Culture medium α-MEM | Osteocytic MLO-Y4 cells | Vibration | - | 45 | 0.5 | g | 0.5 | 259,200 | Relative fluorescence intensity | +37% | [66] |
| 517 | Culture medium α-MEM | Osteocytic MLO-Y4 cells | Vibration | - | 45 | 0.5 | g | 0.5 | 259,200 | Cell anisotropy | +111% | [66] |
| 518 | Culture medium α-MEM | Osteocytic MLO-Y4 cells | Vibration | - | 45 | 0.5 | g | 0.5 | 259,200 | Expression of Wnt3a | +37.9% | [66] |
| 519 | Culture medium α-MEM | Osteocytic MLO-Y4 cells | Vibration | - | 45 | 0.5 | g | 0.5 | 259,200 | Expression of β-catenin | +52% | [66] |
| 520 | Culture medium α-MEM | Osteocytic MLO-Y4 cells | Vibration | - | 45 | 0.5 | g | 0.5 | 259,200 | Expression of Sost | −36.53% | [66] |
| 521 | Culture medium α-MEM | Osteocytic MLO-Y4 cells | Vibration | - | 45 | 0.5 | g | 0.5 | 259,200 | Expression of DKK1 | −29.5% | [66] |
| 522 | Culture medium α-MEM | Osteocytic MLO-Y4 cells | Vibration | - | 45 | 0.5 | g | 0.5 | 259,200 | Expression of RANKL | −33.33% | [66] |
| 523 | Culture medium α-MEM | Osteocytic MLO-Y4 cells | Vibration | - | 45 | 0.5 | g | 0.5 | 259,200 | Expression of OPG | +62.2% | [66] |
| 524 | Culture medium α-MEM | Osteocytic MLO-Y4 cells | Vibration | - | 45 | 0.5 | g | 0.5 | 259,200 | Expression of PGE-2 | +40% | [66] |
| 525 | Culture medium α-MEM | Osteocytic MLO-Y4 cells | Vibration | - | 45 | 0.5 | g | 0.5 | 259,200 | Expression of TNF- α | −31% | [66] |
| 526 | Culture medium α-MEM | Osteocytic MLO-Y4 cells | Vibration | - | 45 | 0.5 | g | 0.5 | 259,200 | Expression of RANKL | −20.5% | [66] |
| 527 | Culture medium α-MEM | Osteocytic MLO-Y4 cells | Vibration | - | 45 | 0.5 | g | 0.5 | 259,200 | Expression of OPG | +16.6% | [66] |
| 528 | Holtfreter’s saline | Newt Triturus alpestri notochord (tails) | US | - | 1,000,000 | 8000 | mW/cm2 | - | 300 | Summ of cells with abnorman endoplasmatic reticulum | +7000% | [100] |
| 529 | TC culture mediun | Whole blood cells | US | - | 2,250,000 | 30 | mW | 5 | 3600 | Cell with chromosomal aberrations | +560 | [111] |
| 530 | TC culture mediun | Whole blood cells | US | - | 2,250,000 | 30 | mW | 5 | 7200 | Cell with chromosomal aberrations | +620 | [111] |
| 531 | TC culture mediun | Whole blood cells | US | - | 2,250,000 | 30 | mW | 5 | 10,800 | Cell with chromosomal aberrations | +360 | [111] |
| 532 | TC culture mediun | Whole blood cells | US | - | 2,250,000 | 30 | mW | 5 | 3600 | Chromotid aberrations per 100 cells | +375 | [111] |
| 533 | TC culture mediun | Whole blood cells | US | - | 2,250,000 | 30 | mW | 5 | 7200 | Chromotid aberrations per 100 cells | +800 | [111] |
| 534 | TC culture mediun | Whole blood cells | US | - | 2,250,000 | 30 | mW | 5 | 10,800 | Chromotid aberrations per 100 cells | +375 | [111] |
| 535 | TC culture mediun | Whole blood cells | US | - | 2,250,000 | 30 | mW | 5 | 3600 | Chromosome aberrations per 100 cells | +1100 | [111] |
| 536 | TC culture mediun | Whole blood cells | US | - | 2,250,000 | 30 | mW | 5 | 7200 | Chromosome aberrations per 100 cells | +1800 | [111] |
| 537 | TC culture mediun | Whole blood cells | US | - | 2,250,000 | 30 | mW | 5 | 10,800 | Chromosome aberrations per 100 cells | +700 | [111] |
| 538 | TC culture mediun | Whole blood cells | US | - | 2,250,000 | 30 | mW | 5 | 3600 | Total aberrations | +520 | [111] |
| 539 | TC culture mediun | Whole blood cells | US | - | 2,250,000 | 30 | mW | 5 | 7200 | Total aberrations | +1000 | [111] |
| 540 | TC culture mediun | Whole blood cells | US | - | 2,250,000 | 30 | mW | 5 | 10,800 | Total aberrations | +440 | [111] |
| 541 | Water | Elodea leaves cells with gas bodies sizes 10 × 14.7 μm | US | - | 500,000 | 80 | mW/cm2 | 0.5 | 100 | Cell death | +100 | [212] |
| 542 | Water | Elodea leaves cells with gas bodies sizes 10 × 14.7 μm | US | - | 1,000,000 | 200 | mW/cm2 | 0.5 | 100 | Cell death | +100 | [212] |
| 543 | Water | Elodea leaves cells with gas bodies sizes 10 × 14.7 μm= | US | - | 400,000 | 140 | mW/cm2 | 0.5 | 100 | Cell death | +100 | [212] |
| 544 | Water | Elodea leaves cells with gas bodies sizes 10 × 14.7 μm | US | - | 1,500,000 | 400 | mW/cm2 | 0.5 | 100 | Cell death | +100 | [212] |
| 545 | Water | Elodea leaves cells with gas bodies sizes 10 × 14.7 μm | US | - | 200,000 | 500 | mW/cm2 | 0.5 | 100 | Cell death | +100 | [212] |
| 546 | Water | Elodea leaves cells with gas bodies sizes 10 × 14.7 μm | US | - | 250,000 | 450 | mW/cm2 | 0.5 | 100 | Cell death | +100 | [212] |
| 547 | Water | Elodea leaves cells with gas bodies sizes 10 × 14.7 μm | US | - | 500,0000 | 400 | mW/cm2 | 0.5 | 100 | Cell death | +100 | [212] |
| 548 | Water | Elodea leaves cells with gas bodies sizes 10 × 14.7 μm | US | - | 7,500,000 | 750 | mW/cm2 | 0.5 | 100 | Cell death | +100 | [212] |
| 549 | Water | Elodea leaves cells with gas bodies sizes 10 × 14.7 μm | US | - | 1,000,0000 | 1500 | mW/cm2 | 0.5 | 100 | Cell death | +100 | [212] |
| 550 | Water | Elodea leaves cells with gas bodies sizes less than 5.3 × 7.4 μm | US | - | 500,000 | 1100 | mW/cm2 | 0.5 | 100 | Cell death | +100 | [212] |
| 551 | Water | Elodea leaves cells with gas bodies sizes less than 5.3 × 7.4 μm | US | - | 1,000,000 | 1300 | mW/cm2 | 0.5 | 100 | Cell death | +100 | [112] |
| 552 | Water | Elodea leaves cells with gas bodies sizes less than 5.3 × 7.4 μm | US | - | 1,500,000 | 400 | mW/cm2 | 0.5 | 100 | Cell death | +100 | [212] |
| 553 | Water | Elodea leaves cells with gas bodies sizes less than 5.3 × 7.4 μm | US | - | 2,000,000 | 300 | mW/cm2 | 0.5 | 100 | Cell death | +100 | [212] |
| 554 | Water | Elodea leaves cells with gas bodies sizes less than 5.3 × 7.4 μm | US | - | 500,0000 | 200 | mW/cm2 | 0.5 | 100 | Cell death | +100 | [212] |
| 555 | Water | Elodea leaves cells with gas bodies sizes less than 5.3 × 7.4 μm | US | - | 1,000,000 | 900 | mW/cm2 | 0.5 | 100 | Cell death | +100 | [212] |
| 556 | Isotonic saline | Healthy human latelets after incubation at 22 °C for 30 min | US | - | 1,000,000 | 200 | mW/cm2 | 1.5 | 300 | Floculation during incubation | +10 | [213] |
| 557 | Isotonic saline | Healthy human latelets after incubation at 22 °C for 30 min | US | - | 1,000,000 | 600 | mW/cm2 | 1.5 | 300 | Floculation during incubation | +100 | [213] |
| 558 | Isotonic saline | Red blood cells of healthy human | US | - | 1,000,000 | 1000 | mW/cm2 | 0.45 | 60 | Hemolysis (percent) | +1900 | [110] |
| 559 | Isotonic saline | Red blood cells of healthy human | US | - | 1,000,000 | 2000 | mW/cm2 | 0.45 | 60 | Hemolysis (percent) | +2300 | [110] |
| 560 | Isotonic saline | Red blood cells of healthy human | US | - | 1,000,000 | 3000 | mW/cm2 | 0.45 | 60 | Hemolysis (percent) | +3200 | [110] |
| 561 | Isotonic saline | Red blood cells of healthy human | US | - | 1,000,000 | 4000 | mW/cm2 | 0.45 | 60 | Hemolysis (percent) | +1600 | [110] |
| 562 | Isotonic saline | Red blood cells of healthy human | US | - | 1,000,000 | 5000 | mW/cm2 | 0.45 | 60 | Hemolysis (percent) | +1400 | [110] |
| 563 | Albunex solution | Red blood cells of healthy human | US | 33 μL/mL | 1,000,000 | 1000 | mW/cm2 | 0.45 | 60 | Hemolysis (percent) | +2600 | [110] |
| 564 | Albunex solution | Red blood cells of healthy human | US | 33 μL/mL | 1,000,000 | 2000 | mW/cm2 | 0.45 | 60 | Hemolysis (percent) | +4500 | [110] |
| 565 | Albunex solution | Red blood cells of healthy human | US | 33 μL/mL | 1,000,000 | 3000 | mW/cm2 | 0.45 | 60 | Hemolysis (percent) | +5600 | [110] |
| 566 | Albunex solution | Red blood cells of healthy human | US | 33 μL/mL | 1,000,000 | 4000 | mW/cm2 | 0.45 | 60 | Hemolysis (percent) | +5100 | [110] |
| 567 | Albunex solution | Red blood cells of healthy human | US | 33 μL/mL | 1,000,000 | 5000 | mW/cm2 | 0.45 | 60 | Hemolysis (percent) | +4800 | [110] |
| 568 | photosensitive drug merocyanine 540 (MC 540) | HL-60 cell line | US | 15 μg/mL | 225,000 | 400 | mW/cm2 | 5 | 30 | Cell death | +49.5 | [214] |
| 569 | Culture medium | Bacillus subtilis spores | US | - | 67,000 | 50,000 | mW | 0.125 | 120 | DNA releazing from spores | +800 | [117] |
| 570 | Culture medium | Bacillus subtilis spores | US | - | 67,000 | 50,000 | mW | 0.125 | 120 | CFU counts growth from spores | −99 | [117] |
| 571 | Culture medium | Bacillus subtilis spores | US | - | 67,000 | 50,000 | mW | 0.125 | 120 | DNA releazing from spores | +800 | [117] |
| 572 | Culture medium | Bacillus subtilis spores | US | - | 67,000 | 50,000 | mW | 0.125 | 120 | DNA releazing from spores | +12800 | [117] |
| 573 | Culture medium | Bacillus subtilis spores | US | - | 67,000 | 50,000 | mW | 0.125 | 120 | DNA releazing from spores | +25600 | [117] |
| 574 | Culture medium | Bacillus subtilis spores | US | - | 67,000 | 50,000 | mW | 0.125 | 120 | DNA releazing from spores | +51200 | [117] |
| 575 | Culture medium | Bacillus subtilis spores | US | - | 67,000 | 50,000 | mW | 0.125 | 120 | DNA releazing from spores | +204800 | [117] |
| 576 | Culture medium | Bacillus subtilis spores | US | - | 67,000 | 50,000 | mW | 0.125 | 120 | DNA releazing from spores | +204800 | [117] |
| 577 | RPMI1640 | leukemic cells HL-60 | US | - | 750,000 | 103,700 | mW/cm−2 | 15 | 30 | DNA repair synthesis | +60 | [109] |
| 578 | RPMI1640 | leukemic cells HL-60 | US | - | 750,000 | 103,700 | mW/cm−2 | 15 | 30 | Cell viability | −45 | [109] |
| 579 | RPMI1640 | leukemic cells HL-60 | US | - | 750,000 | 103,700 | mW/cm−2 | 15 | 30 | Apoptose percantage | +200 | [109] |
| 580 | RPMI1640 | leukemic cells HL-60 | US | - | 750,000 | 103,700 | mW/cm−2 | 15 | 30 | Proliferation rate | −60 | [109] |
| 581 | RPMI1640 | leukemic cells K562 | US | - | 750,000 | 54,600 | mW/cm−2 | 15 | 30 | Apoptose percantage | +300 | [109] |
| 582 | RPMI1640 | leukemic cells U937 | US | - | 750,000 | 54,600 | mW/cm−2 | 15 | 30 | Apoptose percantage | +400 | [109] |
| 583 | RPMI1640 | leukemic cells M1/2 | US | - | 750,000 | 54,600 | mW/cm−2 | 15 | 30 | Apoptose percantage | +110 | [109] |
| 584 | RPMI1640 | leukemic cells HL-60 | US | - | 750,000 | 103,700 | mW/cm−2 | 15 | 30 | Late apoptose percantage | +300 | [109] |
| 585 | RPMI1640 | leukemic cells HL-60 | US | - | 750,000 | 22,400 | mW/cm−2 | 15 | 30 | Early apoptose percantage | +300 | [109] |
| 586 | RPMI1640 | leukemic cells HL-60 | US | - | 750,000 | 103,700 | mW/cm−2 | 15 | 30 | Early apoptose percantage | +400 | [109] |
| 587 | RPMI1640 | Healthy human platelet | US | - | 22,000 | 1600 | mW | 5 | 10 | Agregation rate | +15 | [108] |
| 588 | RPMI1640 | Healthy human platelet | US | - | 22,000 | 1600 | mW | 5 | 30 | Agregation rate | +35 | [108] |
| 589 | RPMI1640 | Healthy human platelet | US | - | 22,000 | 1600 | mW | 5 | 60 | Agregation rate | +55 | [108] |
| 590 | RPMI1640 | Healthy human platelet | US | - | 22,000 | 1400 | mW | 5 | 15 | Ca2+ concentration is cytoplasm | +100 | [108] |
| 591 | RPMI1640 | Healthy human platelet | US | - | 22,000 | 1400 | mW | 5 | 30 | Ca2+ concentration is cytoplasm | +180 | [108] |
| 592 | RPMI1640 | Healthy human platelet | US | - | 22000 | 16,000 | mW | 5 | 60 | Ca2+ concentration is cytoplasm | +900 | [108] |
| 593 | RPMI-1640 | K562 cells | US | - | 1,800,000 | 220 | mW/cm−2 | 2.5 | 1800 | Early apoptic cells count | +410 | [215] |
| 594 | RPMI-1640 | K562 cells | US | - | 1,800,000 | 220 | mW/cm−2 | 2.5 | 7200 | Early apoptic cells count | +740 | [215] |
| 595 | RPMI-1640 | K562 cells | US | - | 1,800,000 | 220 | mW/cm−2 | 2.5 | 18,000 | Early apoptic cells count | +1450 | [215] |
| 596 | RPMI-1640 | K562 cells | US | - | 1,800,000 | 220 | mW/cm−2 | 2.5 | 7200 | bcl-2 protein expression | +10000 | [215] |
| 597 | RPMI-1640 | K562 cells | US | - | 1,800,000 | 220 | mW/cm−2 | 2.5 | 7200 | bax protein expression | +1000 | [215] |
| 598 | YEPD medium | E. coli strain DH5-alpha | US | - | 1,000,000 | 5200 | mW/cm−2 | 0.002 | 30 | Cell viability | −99 | [118] |
| 599 | YEPD medium | Saccharomyces cerevisiae | US | - | 1,000,000 | 5200 | mW/cm−2 | 0.002 | 30 | Cell viability | −99 | [118] |
| 600 | Tryptic soy broth | S. epidermidis | US | - | 70,000 | 3000 | mW/cm−2 | 2 | 3600 | Proliferation rate | +20 | [216] |
| 601 | Tryptic soy broth | S. epidermidis | US | - | 70,000 | 3000 | mW/cm−2 | 2 | 7200 | Proliferation rate | +20 | [216] |
| 602 | Tryptic soy broth | S. epidermidis | US | - | 70,000 | 3000 | mW/cm2 | 2 | 10,800 | Proliferation rate | +15 | [216] |
| 603 | Healthy rabbits’ eyes | In vivo | US | - | 880,000 | 190 | mW/cm2 | 1 | 300 | Sodium fluorescein concentration in the aqueous humor | +240 | [99] |
| 604 | Healthy rabbits’ eyes | In vivo | US | - | 880,000 | 340 | mW/cm2 | 1 | 300 | Sodium fluorescein concentration in the aqueous humor | +380 | [99] |
| 605 | Healthy rabbits’ eyes | In vivo | US | - | 880,000 | 560 | mW/cm2 | 1 | 300 | Sodium fluorescein concentration in the aqueous humor | +1060 | [99] |
| 606 | Healthy rabbits’ eyes | In vivo | US | - | 880,000 | 190 | mW/cm2 | 1 | 300 | Damaged cells | +179 | [99] |
| 607 | Healthy rabbits’ eyes | In vivo | US | - | 880,000 | 340 | mW/cm2 | 1 | 300 | Damaged cells | +174 | [99] |
| 608 | Healthy rabbits’ eyes | In vivo | US | - | 880,000 | 560 | mW/cm2 | 1 | 300 | Damaged cells | +171 | [99] |
| 609 | MCDB-131 medium | Bovine aortic endothelial cells | US (continuous wave) | - | 1,000,000 | 1200 | mW/cm2 | 5 | 900 | Cell proliferation after 48 h | +162 | [114] |
| 610 | MCDB-131 medium | Bovine aortic endothelial cells | US (continuous wave) | - | 3500,000 | 1200 | mW/cm2 | 5 | 900 | Cell proliferation after 48 h | +154 | [114] |
| 611 | MCDB-131 medium | Bovine aortic endothelial cells | US (pulse wave) | - | 1,000,000 | 1200 | mW/cm2 | 5 | 900 | Cell proliferation after 48 h | +135 | [114] |
| 612 | MCDB-131 medium | Bovine aortic endothelial cells | US (pulse wave) | - | 3500,000 | 1200 | mW/cm2 | 5 | 900 | Cell proliferation after 48 h | +123 | [114] |
| 613 | 50 mM CaCl2 | Pseudomonas putida UWC1 | US | - | 40,000 | 240 | mW/cm−2 | 0.5 | 10 | Ultrasound DNA transfer | +100 | [119] |
| 614 | 50 mM CaCl2 | Escherichia coli DH5α | US | - | 40,000 | 240 | mW/cm−2 | 0.5 | 10 | Ultrasound DNA transfer | +100 | [119] |
| 615 | 50 mM CaCl2 | Pseudomonas fluorescens SBW25 | US | - | 40,000 | 240 | mW/cm−2 | 0.5 | 10 | Ultrasound DNA transfer | +100 | [119] |
| 616 | BG11 medium | Microcystis aeruginosa cyanobacteria | US | - | 25,000 | 320 | mW/cm−2 | 250 | 300 | Proliferation | −90 | [120] |
| 617 | BG11 medium | Microcystis aeruginosa cyanobacteria | US | - | 25,000 | 320 | mW/cm−2 | 250 | 300 | Chlorophyll a concentration | −21 | [120] |
| 618 | BG11 medium | Microcystis aeruginosa cyanobacteria | US | - | 25,000 | 320 | mW/cm−2 | 250 | 300 | PC absorbance | −45 | [120] |
| 619 | BG11 medium | Microcystis aeruginosa cyanobacteria | US | - | 25,000 | 320 | mW/cm−2 | 250 | 300 | Extracellular microcystins | −17 | [120] |
| 620 | BG11 medium | Microcystis aeruginosa cyanobacteria | US | - | 25,000 | 320 | mW/cm−2 | 250 | 300 | Oxygen evolution rate | −40 | [120] |
| 621 | F12 medium | Human Airway Smooth Muscle (HASM) cells | US | - | 1,000,000 | 1000 | mW/cm−2 | 0.8 | 300 | Cell contractile moment | −40 | [217] |
| 622 | F12 medium | Human Airway Smooth Muscle (HASM) cells | US | - | 1,000,000 | 2000 | mW/cm−2 | 0.8 | 300 | Cell contractile moment | −65 | [217] |
References
- Nasser, A.; Mingelgrin, U. Mechanochemistry: A review of surface reactions and environmental applications. Appl. Clay Sci. 2012, 67–68, 141–150. [Google Scholar] [CrossRef]
- Stunzhas, P.A. Mechanochemical Instability of Water and Its Applications. Phys. Wave Phenom. 2020, 28, 111–115. [Google Scholar] [CrossRef]
- Yin, Z.; Zhang, Q.; Li, S.; Cagnetta, G.; Huang, J.; Deng, S.; Yu, G. Mechanochemical synthesis of catalysts and reagents for water decontamination: Recent advances and perspective. Sci. Total Environ. 2022, 825, 153992. [Google Scholar] [CrossRef]
- Gudkov, S.V.; Sarimov, R.M.; Astashev, M.E.; Pishchalnikov, R.Y.; Yanykin, D.V.; Simakin, A.V.; Shkirin, A.V.; Serov, D.A.; Konchekov, E.M.; Gusein-zade, N.G.O.; et al. Modern physical methods and technologies in agriculture. Physics-Uspekhi 2023, 67, 194–210. [Google Scholar] [CrossRef]
- Males, R.M.; Clark, R.M.; Wehrman, P.J.; Gates, W.E. Algorithm for Mixing Problems in Water Systems. J. Hydraul. Eng. 1985, 111, 206–219. [Google Scholar] [CrossRef]
- Bunkin, N.F.; Shkirin, A.V.; Ninham, B.W.; Chirikov, S.N.; Chaikov, L.L.; Penkov, N.V.; Kozlov, V.A.; Gudkov, S.V. Shaking-Induced Aggregation and Flotation in Immunoglobulin Dispersions: Differences between Water and Water–Ethanol Mixtures. ACS Omega 2020, 5, 14689–14701. [Google Scholar] [CrossRef]
- Metreveli, G.; Philippe, A.; Schaumann, G.E. Disaggregation of silver nanoparticle homoaggregates in a river water matrix. Sci. Total Environ. 2015, 535, 35–44. [Google Scholar] [CrossRef]
- Bull, H.B. Protein Denaturation. Cold Spring Harb. Symp. Quant. Biol. 1938, 6, 140–149. [Google Scholar] [CrossRef]
- Kiese, S.; Papppenberger, A.; Friess, W.; Mahler, H.-C. Shaken, Not Stirred: Mechanical Stress Testing of an IgG1 Antibody. J. Pharm. Sci. 2008, 97, 4347–4366. [Google Scholar] [CrossRef]
- Boldyrev, V.V. Mechanochemistry and mechanical activation of solids. Russ. Chem. Rev. 2006, 75, 177–189. [Google Scholar] [CrossRef]
- Kákosy, T. 2 Vibration disease. Baillière’s Clin. Rheumatol. 1989, 3, 25–50. [Google Scholar] [CrossRef] [PubMed]
- de Souza, R.J.; Nilsson, T.; Wahlström, J.; Burström, L. Hand-arm vibration and the risk of vascular and neurological diseases—A systematic review and meta-analysis. PLoS ONE 2017, 12, e0180795. [Google Scholar] [CrossRef]
- Heaver, C.; Goonetilleke, K.S.; Ferguson, H.; Shiralkar, S. Hand–arm vibration syndrome: A common occupational hazard in industrialized countries. J. Hand Surg. Eur. Vol. 2011, 36, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.P.; Saez, N.J.; Cristofori-Armstrong, B.; Anangi, R.; King, G.F.; Smith, M.T.; Rash, L.D. Inhibition of acid-sensing ion channels by diminazene and APETx2 evoke partial and highly variable antihyperalgesia in a rat model of inflammatory pain. Br. J. Pharmacol. 2018, 175, 2204–2218. [Google Scholar] [CrossRef]
- Giudice, E.D.; Spinetti, P.R.; Tedeschi, A. Water Dynamics at the Root of Metamorphosis in Living Organisms. Water 2010, 2, 566–586. [Google Scholar] [CrossRef]
- Petrova, A.; Tarasov, S.; Gorbunov, E.; Stepanov, G.; Fartushnaya, O.; Zubkov, E.; Molodtsova, I.; Boriskin, V.; Zatykina, A.; Smirnov, A.; et al. Phenomenon of Post-Vibration Interactions. Symmetry 2024, 16, 958. [Google Scholar] [CrossRef]
- Stepanov, G.O.; Boriskin, V.S.; Rodionova, N.N.; Fedorova, M.M.; Petrova, A.O.; Novikov, V.V.; Yablokova, E.V.; Chernikov, D.Y.; Minakov, A.V.; Tarasov, S.A. The Effect of Aqueous Solutions Processed Using Gradual Technology on the Emission of a Carbohydrate (Lactose) in the RF Range. Phys. Wave Phenom. 2024, 32, 43–47. [Google Scholar] [CrossRef]
- Subbotin, K.A.; Didenko, Y.S.; Titov, A.I.; Lis, D.A.; Pavlov, S.K.; Volkov, P.A.; Runina, K.I.; Voronov, V.V.; Chernova, E.V.; Lis, O.N.; et al. Influence of Ultrahigh Dilution Treatment of the Charge on the Growth and Spectroscopic Properties of Nd:MgMoO4 Potential Laser Crystal. Crystals 2024, 14, 100. [Google Scholar] [CrossRef]
- Don, E.S.; Stepanov, G.O.; Tarasov, S.A. The effects of physical treatment on physicochemical and biological properties of water and aqueous solutions. Fine Chem. Technol. 2023, 18, 426–445. [Google Scholar] [CrossRef]
- Murfee, W.L.; Hammett, L.A.; Evans, C.; Xie, L.; Squire, M.; Rubin, C.; Judex, S.; Skalak, T.C. High-frequency, low-magnitude vibrations suppress the number of blood vessels per muscle fiber in mouse soleus muscle. J. Appl. Physiol. 2005, 98, 2376–2380. [Google Scholar] [CrossRef]
- Astashev, M.E.; Serov, D.A.; Sarimov, R.M.; Gudkov, S.V. Influence of the Vibration Impact Mode on the Spontaneous Chemiluminescence of Aqueous Protein Solutions. Phys. Wave Phenom. 2023, 31, 189–199. [Google Scholar] [CrossRef]
- Kokornaczyk, M.O.; Acuña, C.; Mier y Terán, A.; Castelán, M.; Baumgartner, S. Vortex-like vs. turbulent mixing of a Viscum album preparation affects crystalline structures formed in dried droplets. Sci. Rep. 2024, 14, 12965. [Google Scholar] [CrossRef] [PubMed]
- Kokornaczyk, M.O.; Würtenberger, S.; Baumgartner, S. Impact of succussion on pharmaceutical preparations analyzed by means of patterns from evaporated droplets. Sci. Rep. 2020, 10, 570. [Google Scholar] [CrossRef]
- Tytik, D.L.; Kuzmin, V.I.; Souvorova, O.V.; Revina, A.A. Dynamics of optical properties of sequentially diluted lucigenin aqueous solutions according to luminescence data. Front. Chem. 2024, 12, 1439250. [Google Scholar] [CrossRef]
- Liu, Q.; Qian, H.; Yu, H.; Ren, F.; Fang, J.; Liu, F.; Liu, H.; Liang, J. Effects of mechanical force on proliferation and apoptosis of stem cells from human exfoliated deciduous teeth. Clin. Oral Investig. 2022, 26, 5205–5213. [Google Scholar] [CrossRef]
- Yamamoto, T.; Morishita, S. A study on the effect of medium sloshing caused by mechanical vibration on cell proliferation. In Proceedings of the Conference of Kanto Branch, Tokyo, Japan, 17–18 March 2018; Volume 2018.24. [Google Scholar] [CrossRef]
- Baimler, I.V.; Gudkov, S.V.; Matveeva, T.A.; Simakin, A.V.; Shcherbakov, I.A. Generation of Reactive Oxygen Species during Water Drops Fall on a Solid Surface. Phys. Wave Phenom. 2024, 32, 187–189. [Google Scholar] [CrossRef]
- Qing, F.; Xie, P.; Liem, Y.S.; Chen, Y.; Chen, X.; Zhu, X.; Fan, Y.; Yang, X.; Zhang, X. Administration duration influences the effects of low-magnitude, high-frequency vibration on ovariectomized rat bone. J. Orthop. Res. 2015, 34, 1147–1157. [Google Scholar] [CrossRef]
- Lee, J.K.; Banerjee, S.; Nam, H.G.; Zare, R.N. Acceleration of reaction in charged microdroplets. Q. Rev. Biophys. 2015, 48, 437–444. [Google Scholar] [CrossRef]
- Robertson, V.J.; Baker, K.G. A Review of Therapeutic Ultrasound: Effectiveness Studies. Phys. Ther. 2001, 81, 1339–1350. [Google Scholar] [CrossRef]
- Ambattu, L.A.; Yeo, L.Y. Sonomechanobiology: Vibrational stimulation of cells and its therapeutic implications. Biophys. Rev. 2023, 4, 021301. [Google Scholar] [CrossRef]
- Fesenko, E.E.; Yablokova, E.V.; Novikov, V.V. Weak Magnetic Fields Regulate the Ability of High Dilutions of Water to Enhance ROS Production by Neutrophils. Appl. Sci. 2024, 14, 3290. [Google Scholar] [CrossRef]
- Novikov, V.V.; Yablokova, E.V. Interaction between Highly Diluted Samples, Protein Solutions and Water in a Controlled Magnetic Field. Appl. Sci. 2022, 12, 5185. [Google Scholar] [CrossRef]
- Novikov, V.V.; Yablokova, E.V.; Fesenko, E.E. The Role of Water in the Effect of Weak Combined Magnetic Fields on Production of Reactive Oxygen Species (ROS) by Neutrophils. Appl. Sci. 2020, 10, 3326. [Google Scholar] [CrossRef]
- Petrov, S.I.; Epstein, O.I. Effect of Potentiated Solutions on Mercury(II) Signal in Inversion Voltammetry. Bull. Exp. Biol. Med. 2003, 135, 99–101. [Google Scholar] [CrossRef]
- Novikov, V.V. Effect of vibrational iterations of magnetized water on the physico-chemical properties of intact water. Russ. Phys. J. 2025, 67, 1718–1727. [Google Scholar] [CrossRef]
- Tarasov, S.A.; Gorbunov, E.A.; Don, E.S.; Emelyanova, A.G.; Kovalchuk, A.L.; Yanamala, N.; Schleker, A.S.S.; Klein-Seetharaman, J.; Groenestein, R.; Tafani, J.-P.; et al. Insights into the Mechanism of Action of Highly Diluted Biologics. J. Immunol. 2020, 205, 1345–1354. [Google Scholar] [CrossRef]
- Epstein, O. The Supramolecular Matrix Concept. Symmetry 2023, 15, 1914. [Google Scholar] [CrossRef]
- Li, Y.; Rabey, K.N.; Schmitt, D.; Norton, J.N.; Reynolds, R.P. Characteristics of Vibration that Alter Cardiovascular Parameters in Mice. J. Am. Assoc. Lab. Anim. Sci. 2015, 54, 372–377. [Google Scholar]
- Krajnak, K.; Miller, G.R.; Waugh, S.; Johnson, C.; Li, S.; Kashon, M.L. Characterization of Frequency-Dependent Responses of the Vascular System to Repetitive Vibration. J. Occup. Environ. Med. 2010, 52, 584–594. [Google Scholar] [CrossRef]
- Curry, B.D.; Govindaraju, S.R.; Bain, J.L.W.; Zhang, L.L.; Yan, J.-G.; Matloub, H.S.; Riley, D.A. Evidence for frequency-dependent arterial damage in vibrated rat tails. Anat. Rec. Part A Discov. Mol. Cell. Evol. Biol. 2005, 284A, 511–521. [Google Scholar] [CrossRef]
- Okada, A.; Okuda, H.; Inaba, R.; Ariizumi, M. Influence of local vibration on plasma creatine phosphokinase (CPK) activity. Occup. Environ. Med. 1985, 42, 678–681. [Google Scholar] [CrossRef] [PubMed]
- Morandi, L.; Angelini, C.; Prelle, A.; Pini, A.; Grassi, B.; Bernardi, G.; Politano, L.; Bruno, C.; De Grandis, D.; Cudia, P.; et al. High plasma creatine kinase: Review of the literature and proposal for a diagnostic algorithm. Neurol. Sci. 2006, 27, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wan, Y.; Tam, K.F.; Ling, S.; Bai, Y.; Deng, Y.; Liu, Y.; Zhang, H.; Cheung, W.H.; Qin, L.; et al. Resistive vibration exercise retards bone loss in weight-bearing skeletons during 60 days bed rest. Osteoporos. Int. 2011, 23, 2169–2178. [Google Scholar] [CrossRef] [PubMed]
- Ballanti, P.; Minisola, S.; Pacitti, M.T.; Scarnecchia, L.; Rosso, R.; Mazzuoli, G.F.; Bonucci, E. Tartrate-resistant acid phosphate activity as osteoclastic marker: Sensitivity of cytochemical assessment and serum assay in comparison with standardized osteoclast histomorphometry. Osteoporos. Int. 1997, 7, 39–43. [Google Scholar] [CrossRef]
- Harrison, R.; Ward, K.; Lee, E.; Razaghi, H.; Horne, C.; Bishop, N.J. Acute bone response to whole body vibration in healthy pre-pubertal boys. J. Musculoskelet. Neuronal Interact. 2015, 15, 112–122. [Google Scholar]
- Szulc, P.; Naylor, K.; Hoyle, N.R.; Eastell, R.; Leary, E.T. Use of CTX-I and PINP as bone turnover markers: National Bone Health Alliance recommendations to standardize sample handling and patient preparation to reduce pre-analytical variability. Osteoporos. Int. 2017, 28, 2541–2556. [Google Scholar] [CrossRef]
- Ogawa, T.; Vandamme, K.; Zhang, X.; Naert, I.; Possemiers, T.; Chaudhari, A.; Sasaki, K.; Duyck, J. Stimulation of Titanium Implant Osseointegration Through High-Frequency Vibration Loading is Enhanced when Applied at High Acceleration. Calcif. Tissue Int. 2014, 95, 467–475. [Google Scholar] [CrossRef]
- Jiang, L.; Sheng, K.; Wang, C.; Xue, D.; Pan, Z. The Effect of MMP-2 Inhibitor 1 on Osteogenesis and Angiogenesis During Bone Regeneration. Front. Cell Dev. Biol. 2021, 8, 596783. [Google Scholar] [CrossRef]
- Liu, Q.; Li, M.; Wang, S.; Xiao, Z.; Xiong, Y.; Wang, G. Recent Advances of Osterix Transcription Factor in Osteoblast Differentiation and Bone Formation. Front. Cell Dev. Biol. 2020, 8, 601224. [Google Scholar] [CrossRef]
- Dong, R.G.; Welcome, D.E.; McDowell, T.W.; Wu, J.Z. Biodynamic Response of Human Fingers in a Power Grip Subjected to a Random Vibration. J. Biomech. Eng. 2004, 126, 447–457. [Google Scholar] [CrossRef]
- McCarren, B.; Alison, J.A.; Herbert, R.D. Vibration and its effect on the respiratory system. Aust. J. Physiother. 2006, 52, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.T.; Yu, H.S. Ultrastructural changes of the peripheral nerve induced by vibration: An experimental study. Occup. Environ. Med. 1989, 46, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Dahlin, L.B.; Necking, L.E.; Lundström, R.; Lundborg, G. Vibration exposure and conditioning lesion effect in nerves: An experimental study in rats. J. Hand Surg. 1992, 17, 858–861. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; He, W.M.; Dong, M.; Yu, F.H.; Zhang, L.L.; Cui, Q.G.; Chu, Y. Effects of Shaking on the Growth and Mechanical Properties of Hedysarum leave May Be Independent of Water Regimes. Int. J. Plant Sci. 2008, 169, 503–508. [Google Scholar] [CrossRef]
- Chan, M.E.; Ashdown, C.; Strait, L.; Pasumarthy, S.; Hassan, A.; Crimarco, S.; Singh, C.; Patel, V.S.; Pagnotti, G.; Khan, O.; et al. Low intensity mechanical signals promote proliferation in a cell-specific manner: Tailoring a non-drug strategy to enhance biomanufacturing yields. Mechanobiol. Med. 2024, 2, 100080. [Google Scholar] [CrossRef]
- Lindell, H.; Peciulyte, A.; Grétarsson, S.L. Physics of Shock and Physiological Effects on Biological Systems. In Proceedings of the 15th International Conference on Hand-Arm Vibration, Nancy, France, 6–9 June 2023. [Google Scholar]
- Chen, S.; Gluhak-Heinrich, J.; Wang, Y.H.; Wu, Y.M.; Chuang, H.H.; Chen, L.; Yuan, G.H.; Dong, J.; Gay, I.; MacDougall, M. Runx2, Osx, and Dspp in Tooth Development. J. Dent. Res. 2009, 88, 904–909. [Google Scholar] [CrossRef]
- Bian, X.; Liu, X.; Zhou, M.; Tang, H.; Wang, R.; Ma, L.; He, G.; Xu, S.; Wang, Y.; Tan, J.; et al. Mechanical stimulation promotes fibrochondrocyte proliferation by activating the TRPV4 signaling pathway during tendon–bone insertion healing: CCN2 plays an important regulatory role. Burn. Trauma 2024, 12, tkae028. [Google Scholar] [CrossRef]
- Rosenberg, N.; Rosenberg, O.; Halevi Politch, J.; Abramovich, H. Optimal parameters for the enhancement of human osteoblast-like cell proliferation in vitro via shear stress induced by high-frequency mechanical vibration. Iberoam. J. Med. 2021, 3, 204–211. [Google Scholar] [CrossRef]
- Nishijima, A.; Shiraishi, T. A Study of Mechanism of Cell Proliferation Under Mechanical Vibration Considering β1 Integrin. In Proceedings of the Dynamics & Design Conference, Yamaguchi, Japan, 23–26 August 2016; Volume 2016. [Google Scholar] [CrossRef]
- Suwittayarak, R.; Klincumhom, N.; Ngaokrajang, U.; Namangkalakul, W.; Ferreira, J.N.; Pavasant, P.; Osathanon, T. Shear Stress Enhances the Paracrine-Mediated Immunoregulatory Function of Human Periodontal Ligament Stem Cells via the ERK Signalling Pathway. Int. J. Mol. Sci. 2022, 23, 7119. [Google Scholar] [CrossRef]
- Park, I.-K.; Letterio, J.J.; Gorham, J.D. TGF-β1 inhibition of IFN-γ-induced signaling and Th1 gene expression in CD4+ T cells is Smad3 independent but MAP kinase dependent. Mol. Immunol. 2007, 44, 3283–3290. [Google Scholar] [CrossRef]
- Watanabe, Y.; Shiraishi, T. Effects of mechanical vibrations on chondrocytes in monolayer culture for regenerative cartilage. Mech. Eng. J. 2024, 11, 24-00102. [Google Scholar] [CrossRef]
- Nham, G.T.H.; Zhang, X.; Asou, Y.; Shinomura, T. Expression of type II collagen and aggrecan genes is regulated through distinct epigenetic modifications of their multiple enhancer elements. Gene 2019, 704, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Yan, Z.; Cai, J.; Shao, X.; Wang, D.; Ding, Y.; Feng, Y.; Yang, J.; Luo, E.; Feng, X.; et al. Effects of mechanical vibration on cell morphology, proliferation, apoptosis, and cytokine expression/secretion in osteocyte-like MLO-Y4 cells exposed to high glucose. Cell Biol. Int. 2019, 44, 216–228. [Google Scholar] [CrossRef]
- Choi, K.-M.; Seo, Y.-K.; Yoon, H.-H.; Song, K.-Y.; Kwon, S.-Y.; Lee, H.-S.; Park, J.-K. Effects of mechanical stimulation on the proliferation of bone marrow-derived human mesenchymal stem cells. Biotechnol. Bioprocess Eng. 2007, 12, 601–609. [Google Scholar] [CrossRef]
- Kanie, K.; Sakai, T.; Imai, Y.; Yoshida, K.; Sugimoto, A.; Makino, H.; Kubo, H.; Kato, R. Effect of mechanical vibration stress in cell culture on human induced pluripotent stem cells. Regen. Ther. 2019, 12, 27–35. [Google Scholar] [CrossRef]
- Paul, N.E.; Denecke, B.; Kim, B.-S.; Dreser, A.; Bernhagen, J.; Pallua, N. The effect of mechanical stress on the proliferation, adipogenic differentiation and gene expression of human adipose-derived stem cells. J. Tissue Eng. Regen. Med. 2018, 12, 276–284. [Google Scholar] [CrossRef]
- Astashev, M.; Serov, D.; Gudkov, S. Application of Spectral Methods of Analysis for Description of Ultradian Biorhythms at the Levels of Physiological Systems, Cells and Molecules (Review). Mathematics 2023, 11, 3307. [Google Scholar] [CrossRef]
- Li, Y.-J.; Yu, M.; Xue, C.-D.; Zhang, H.-J.; Wang, G.-Z.; Chen, X.-M.; Qin, K.-R. Modeling of Endothelial Calcium Responses within a Microfluidic Generator of Spatio-Temporal ATP and Shear Stress Signals. Micromachines 2021, 12, 161. [Google Scholar] [CrossRef]
- Chakraborty, M.; Chu, K.; Shrestha, A.; Revelo, X.S.; Zhang, X.; Gold, M.J.; Khan, S.; Lee, M.; Huang, C.; Akbari, M.; et al. Mechanical Stiffness Controls Dendritic Cell Metabolism and Function. Cell Rep. 2021, 34, 108609. [Google Scholar] [CrossRef]
- Li, K.; Ma, H.; Li, S.; Wang, C. Numerical simulation of three physical fields in counter-current ultrasound. J. Food Process Eng. 2024, 47, e14676. [Google Scholar] [CrossRef]
- Annabi, N.; Tamayol, A.; Uquillas, J.A.; Akbari, M.; Bertassoni, L.E.; Cha, C.; Camci-Unal, G.; Dokmeci, M.R.; Peppas, N.A.; Khademhosseini, A. 25th Anniversary Article: Rational Design and Applications of Hydrogels in Regenerative Medicine. Adv. Mater. 2013, 26, 85–124. [Google Scholar] [CrossRef] [PubMed]
- Vining, K.H.; Stafford, A.; Mooney, D.J. Sequential modes of crosslinking tune viscoelasticity of cell-instructive hydrogels. Biomaterials 2019, 188, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Charrier, E.E.; Pogoda, K.; Wells, R.G.; Janmey, P.A. Control of cell morphology and differentiation by substrates with independently tunable elasticity and viscous dissipation. Nat. Commun. 2018, 9, 449. [Google Scholar] [CrossRef]
- Makisha, N.; Gulshin, I. Sustainable Modernization of Wastewater Treatment Plants. Sustainability 2024, 16, 8757. [Google Scholar] [CrossRef]
- Abdiyev, K.; Azat, S.; Kuldeyev, E.; Ybyraiymkul, D.; Kabdrakhmanova, S.; Berndtsson, R.; Khalkhabai, B.; Kabdrakhmanova, A.; Sultakhan, S. Review of Slow Sand Filtration for Raw Water Treatment with Potential Application in Less-Developed Countries. Water 2023, 15, 2007. [Google Scholar] [CrossRef]
- Ye, J.; Ren, G.; Liu, L.; Zhang, D.; Zeng, R.J.; van Loosdrecht, M.C.M.; Zhou, S. Wastewater denitrification driven by mechanical energy through cellular piezo-sensitization. Nat. Water 2024, 2, 531–540. [Google Scholar] [CrossRef]
- Gudkov, S.V.; Penkov, N.V.; Baimler, I.V.; Lyakhov, G.A.; Pustovoy, V.I.; Simakin, A.V.; Sarimov, R.M.; Scherbakov, I.A. Effect of Mechanical Shaking on the Physicochemical Properties of Aqueous Solutions. Int. J. Mol. Sci. 2020, 21, 8033. [Google Scholar] [CrossRef]
- Lu, G.; Miao, X.; Liu, D. Crosstalk of physiological pH and chemical pKa under the umbrella of physiologically based pharmacokinetic modeling of drug absorption, distribution, metabolism, excretion, and toxicity. Expert Opin. Drug Metab. Toxicol. 2021, 17, 1103–1124. [Google Scholar] [CrossRef]
- Woods, K.N. Modeling of protein hydration dynamics is supported by THz spectroscopy of highly diluted solutions. Front. Chem. 2023, 11, 1131935. [Google Scholar] [CrossRef]
- Belovolova, L.V.; Glushkov, M.V.; Vinogradov, E.A.; Babintsev, V.A.; Golovanov, V.I. Ultraviolet fluorescence of water and highly diluted aqueous media. Phys. Wave Phenom. 2009, 17, 21–31. [Google Scholar] [CrossRef]
- Gudkov, S.V.; Lyakhov, G.A.; Pustovoy, V.I.; Shcherbakov, I.A. Influence of Mechanical Effects on the Hydrogen Peroxide Concentration in Aqueous Solutions. Phys. Wave Phenom. 2019, 27, 141–144. [Google Scholar] [CrossRef]
- Shcherbakov, I.A. Influence of External Impacts on the Properties of Aqueous Solutions. Phys. Wave Phenom. 2021, 29, 89–93. [Google Scholar] [CrossRef]
- Gudkov, S.V.; Lyakhov, G.A.; Pustovoy, V.I.; Shcherbakov, I.A. Vibration–Vortex Mechanism of Radical-Reaction Activation in an Aqueous Solution: Physical Analogies. Phys. Wave Phenom. 2021, 29, 108–113. [Google Scholar] [CrossRef]
- Sharapov, M.G.; Goncharov, R.G.; Parfenyuk, S.B.; Glushkova, O.V.; Novoselov, V.I. The Role of Phospholipase Activity of Peroxiredoxin 6 in Its Transmembrane Transport and Protective Properties. Int. J. Mol. Sci. 2022, 23, 15265. [Google Scholar] [CrossRef]
- Averill-Bates, D. Reactive oxygen species and cell signaling. Review. Biochim. Biophys. Acta BBA—Mol. Cell Res. 2024, 1871, 119573. [Google Scholar] [CrossRef]
- Styrkas, A.D.; Nikishina, N.G. Mechanochemical processes in water. High Energy Chem. 2007, 41, 396–402. [Google Scholar] [CrossRef]
- Lee, J.K.; Walker, K.L.; Han, H.S.; Kang, J.; Prinz, F.B.; Waymouth, R.M.; Nam, H.G.; Zare, R.N. Spontaneous generation of hydrogen peroxide from aqueous microdroplets. Proc. Natl. Acad. Sci. USA 2019, 116, 19294–19298. [Google Scholar] [CrossRef]
- Randolph, T.W.; Schiltz, E.; Sederstrom, D.; Steinmann, D.; Mozziconacci, O.; Schöneich, C.; Freund, E.; Ricci, M.S.; Carpenter, J.F.; Lengsfeld, C.S. Do Not Drop: Mechanical Shock in Vials Causes Cavitation, Protein Aggregation, and Particle Formation. J. Pharm. Sci. 2015, 104, 602–611. [Google Scholar] [CrossRef]
- Lu, Q.-Q.; Yin, D.-C.; Liu, Y.-M.; Wang, X.-K.; Yang, P.-F.; Liu, Z.-T.; Shang, P. Effect of mechanical vibration on protein crystallization. J. Appl. Crystallogr. 2010, 43, 473–482. [Google Scholar] [CrossRef]
- Kakinouchi, K.; Adachi, H.; Matsumura, H.; Inoue, T.; Murakami, S.; Mori, Y.; Koga, Y.; Takano, K.; Kanaya, S. Effect of ultrasonic irradiation on protein crystallization. J. Cryst. Growth 2006, 292, 437–440. [Google Scholar] [CrossRef]
- Slatinskaya, O.V.; Pyrkov, Y.N.; Filatova, S.A.; Guryev, D.A.; Penkov, N.V. Study of the Effect of Europium Acetate on the Intermolecular Properties of Water. Front. Phys. 2021, 9, 641110. [Google Scholar] [CrossRef]
- Imaoka, C.; Nakajima, T.; Indei, T.; Iwata, M.; Hong, W.; Marcellan, A.; Gong, J.P. Inverse mechanical-swelling coupling of a highly deformed double-network gel. Sci. Adv. 2023, 9, eabp8351. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Wang, X.; Zhou, L.; Zhang, L.; Hu, J. Formation and Stability of Bulk Nanobubbles by Vibration. Langmuir 2020, 36, 2264–2270. [Google Scholar] [CrossRef] [PubMed]
- Fisher, B.D.; Hiller, C.M.; Rennie, S.G.A. A Comparison of Continuous Ultrasound and Pulsed Ultrasound on Soft Tissue Injury Markers in the Rat. J. Phys. Ther. Sci. 2003, 15, 65–70. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Nakano, J.; Oga, S.; Kataoka, H.; Honda, Y.; Sakamoto, J.; Okita, M. The Non-Thermal Effects of Pulsed Ultrasound Irradiation on the Development of Disuse Muscle Atrophy in Rat Gastrocnemius Muscle. Ultrasound Med. Biol. 2014, 40, 1578–1586. [Google Scholar] [CrossRef]
- Zderic, V.; Clark, J.I.; Vaezy, S. Drug Delivery Into the Eye With the Use of Ultrasound. J. Ultrasound Med. 2004, 23, 1349–1359. [Google Scholar] [CrossRef]
- Selman, G.G.; Jurand, A. An Electron Microscope Study of the Endoplasmic Reticulum in Newt Notochord Cells after Disturbance with Ultrasonic Treatment and Subsequent Regeneration. J. Cell Biol. 1964, 20, 175–183. [Google Scholar] [CrossRef]
- Miller, M.W. Comparison of micronuclei induction for X-ray and ultrasound exposures of Vicia faba root meristem cells. Ultrasound Med. Biol. 1978, 4, 263–267. [Google Scholar] [CrossRef]
- Doan, N.; Reher, P.; Meghji, S.; Harris, M. In vitro effects of therapeutic ultrasound on cell proliferation, protein synthesis, and cytokine production by human fibroblasts, osteoblasts, and monocytes. J. Oral Maxillofac. Surg. 1999, 57, 409–419. [Google Scholar] [CrossRef]
- Ling, L.; Wei, T.; He, L.; Wang, Y.; Wang, Y.; Feng, X.; Zhang, W.; Xiong, Z. Low-intensity pulsed ultrasound activates ERK1/2 and PI3K-Akt signalling pathways and promotes the proliferation of human amnion-derived mesenchymal stem cells. Cell Prolif. 2017, 50, e12383. [Google Scholar] [CrossRef]
- Salgarella, A.R.; Cafarelli, A.; Ricotti, L.; Capineri, L.; Dario, P.; Menciassi, A. Optimal Ultrasound Exposure Conditions for Maximizing C2C12 Muscle Cell Proliferation and Differentiation. Ultrasound Med. Biol. 2017, 43, 1452–1465. [Google Scholar] [CrossRef] [PubMed]
- Wee, P.; Wang, Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.K.; Ishii, G.; Chiba, H.; Ochiai, A. The VEGF angiogenic switch of fibroblasts is regulated by MMP-7 from cancer cells. Oncogene 2007, 26, 7194–7203. [Google Scholar] [CrossRef] [PubMed]
- Uzbekov, R.; Prigent, C. A Journey through Time on the Discovery of Cell Cycle Regulation. Cells 2022, 11, 704. [Google Scholar] [CrossRef]
- Samal, A.B.; Adzerikho, I.D.; Mrochek, A.G.; Loiko, E.N. Platelet aggregation and change in intracellular Ca2+ induced by low frequency ultrasound in vitro. Eur. J. Ultrasound 2000, 11, 53–59. [Google Scholar] [CrossRef]
- Ashush, H.; Rozenszajn, L.A.; Blass, M.; Barda-Saad, M.; Azimov, D.; Radnay, J.; Zipori, D.; Rosenschein, U. Apoptosis induction of human myeloid leukemic cells by ultrasound exposure. Cancer Res. 2000, 60, 1014–1020. [Google Scholar]
- Miller, M.W.; Azadniv, M.; Doida, Y.; Brayman, A.A. Effect of a Stabilized Microbubble Contrast Agent on CW Ultrasound Induced Red Blood Cell Lysis In Vitro. Echocardiography 2007, 12, 1–11. [Google Scholar] [CrossRef]
- Macintosh, I.J.C.; Davey, D.A. Chromosome Aberrations Induced by an Ultrasonic Fetal Pulse Detector. BMJ 1970, 4, 92–93. [Google Scholar] [CrossRef]
- Trenchard, P.M. Ultrasound-induced orientation of discoid platelets and simultaneous changes in light transmission: Preliminary characterisation of the phenomenon. Ultrasound Med. Biol. 1987, 13, 183–195. [Google Scholar] [CrossRef]
- Chan, Y.-S.; Hsu, K.-Y.; Kuo, C.-H.; Lee, S.-D.; Chen, S.-C.; Chen, W.-J.; Ueng, S.W.-N. Using Low-Intensity Pulsed Ultrasound to Improve Muscle Healing After Laceration Injury: An in vitro and in vivo Study. Ultrasound Med. Biol. 2010, 36, 743–751. [Google Scholar] [CrossRef]
- Raz, D.; Zaretsky, U.; Einav, S.; Elad, D. Cellular Alterations in Cultured Endothelial Cells Exposed to Therapeutic Ultrasound Irradiation. Endothelium 2009, 12, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Nagata, K.; Nakamura, T.; Fujihara, S.; Tanaka, E. Ultrasound Modulates the Inflammatory Response and Promotes Muscle Regeneration in Injured Muscles. Ann. Biomed. Eng. 2013, 41, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Dong, Y.; Wei, J.; Cai, M.; Liang, D.; Fu, Y.; Zhou, Y.; Sui, Y.; Wu, F.; Mikhaylov, R.; et al. Acoustically accelerated neural differentiation of human embryonic stem cells. Acta Biomater. 2022, 151, 333–345. [Google Scholar] [CrossRef]
- Belgrader, P.; Hansford, D.; Kovacs, G.T.A.; Venkateswaran, K.; Mariella, R.; Milanovich, F.; Nasarabadi, S.; Okuzumi, M.; Pourahmadi, F.; Northrup, M.A. A Minisonicator to Rapidly Disrupt Bacterial Spores for DNA Analysis. Anal. Chem. 1999, 71, 4232–4236. [Google Scholar] [CrossRef]
- Li, G.; Xiao, H.; Glo, M.; Cheg, J. Miniaturized cell lysis device using spherically focused ultrasound. Tsinghua Sci. Technol. 2003, 8, 487–492. [Google Scholar]
- Song, Y.; Hahn, T.; Thompson, I.P.; Mason, T.J.; Preston, G.M.; Li, G.; Paniwnyk, L.; Huang, W.E. Ultrasound-mediated DNA transfer for bacteria. Nucleic Acids Res. 2007, 35, e129. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, P.; Liu, H.; Wang, B. Ultrasonic damages on cyanobacterial photosynthesis. Ultrason. Sonochem. 2006, 13, 501–505. [Google Scholar] [CrossRef]
- Vikulin, P.; Vikulina, V. Influence of ultrasound on the concentration of hydrogen ions in water. IOP Conf. Ser. Mater. Sci. Eng. 2020, 869, 042007. [Google Scholar] [CrossRef]
- Villalón-López, D.N.; Martínez-Padilla, L.P. Ultrasound-assisted process to improve proteins recovery from industrial canola and soybean byproducts. J. Food Sci. Technol. 2024, 61, 1–12. [Google Scholar] [CrossRef]
- Yao, S.; Li, W.; Wu, Y.; Martin, G.; Ashokkumar, M. The Impact of High-Intensity Ultrasound-Assisted Extraction on the Structural and Functional Properties of Hempseed Protein Isolate (HPI). Foods 2023, 12, 348. [Google Scholar] [CrossRef]
- Vikulina, V.; Vikulin, P. Effect of ultrasound on sedimentation of suspended solids in water. IOP Conf. Ser. Mater. Sci. Eng. 2018, 365, 032001. [Google Scholar] [CrossRef]
- Serov, D.A.; Gritsaeva, A.V.; Yanbaev, F.M.; Simakin, A.V.; Gudkov, S.V. Review of Antimicrobial Properties of Titanium Dioxide Nanoparticles. Int. J. Mol. Sci. 2024, 25, 10519. [Google Scholar] [CrossRef]
- Cui, Y.; Wang, F.; Yuan, P.; Liu, W.; Fang, B.; Wang, Z.; Pu, Y. Harvesting Vibration Energy to Produce Hydrogen Peroxide with Bi3TiNbO9 Nanosheets through a Water Oxidation Dominated Dual-Channel Pathway. ACS Sustain. Chem. Eng. 2024, 12, 3595–3607. [Google Scholar] [CrossRef]
- Wen, Y.; Zhang, T.; Wang, J.; Pan, Z.; Wang, T.; Yamashita, H.; Qian, X.; Zhao, Y. Electrochemical Reactors for Continuous Decentralized H2O2 Production. Angew. Chem. Int. Ed. 2022, 61, e202205972. [Google Scholar] [CrossRef]
- Martino, F.; Perestrelo, A.R.; Vinarský, V.; Pagliari, S.; Forte, G. Cellular Mechanotransduction: From Tension to Function. Front. Physiol. 2018, 9, 824. [Google Scholar] [CrossRef]
- Burridge, K.; Monaghan-Benson, E.; Graham, D.M. Mechanotransduction: From the cell surface to the nucleus via RhoA. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180229. [Google Scholar] [CrossRef]
- Gillespie, P.G.; Walker, R.G. Molecular basis of mechanosensory transduction. Nature 2001, 413, 194–202. [Google Scholar] [CrossRef]
- Janota, C.S.; Calero-Cuenca, F.J.; Gomes, E.R. The role of the cell nucleus in mechanotransduction. Curr. Opin. Cell Biol. 2020, 63, 204–211. [Google Scholar] [CrossRef]
- Maurer, M.; Lammerding, J. The Driving Force: Nuclear Mechanotransduction in Cellular Function, Fate, and Disease. Annu. Rev. Biomed. Eng. 2019, 21, 443–468. [Google Scholar] [CrossRef]
- Na, S.; Collin, O.; Chowdhury, F.; Tay, B.; Ouyang, M.; Wang, Y.; Wang, N. Rapid signal transduction in living cells is a unique feature of mechanotransduction. Proc. Natl. Acad. Sci. USA 2008, 105, 6626–6631. [Google Scholar] [CrossRef]
- Lin, S.-Y.; Corey, D.P. TRP channels in mechanosensation. Curr. Opin. Neurobiol. 2005, 15, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Ma, Y.; Ye, X.; Zhang, N.; Pan, L.; Wang, B. TRP (transient receptor potential) ion channel family: Structures, biological functions and therapeutic interventions for diseases. Signal Transduct. Target. Ther. 2023, 8, 261. [Google Scholar] [CrossRef] [PubMed]
- King, M.C. Dynamic regulation of LINC complex composition and function across tissues and contexts. FEBS Lett. 2023, 597, 2823–2832. [Google Scholar] [CrossRef] [PubMed]
- Swift, J.; Discher, D.E. The nuclear lamina is mechano-responsive to ECM elasticity in mature tissue. J. Cell Sci. 2014, 127, 3005–3015. [Google Scholar] [CrossRef]
- Uhler, C.; Shivashankar, G.V. Regulation of genome organization and gene expression by nuclear mechanotransduction. Nat. Rev. Mol. Cell Biol. 2017, 18, 717–727. [Google Scholar] [CrossRef]
- Khilan, A.A.; Al-Maslamani, N.A.; Horn, H.F. Cell stretchers and the LINC complex in mechanotransduction. Arch. Biochem. Biophys. 2021, 702, 108829. [Google Scholar] [CrossRef]
- Plant, L.D. A Role for K2P Channels in the Operation of Somatosensory Nociceptors. Front. Mol. Neurosci. 2012, 5, 21. [Google Scholar] [CrossRef]
- Salzer, I.; Ray, S.; Schicker, K.; Boehm, S. Nociceptor Signalling through ion Channel Regulation via GPCRs. Int. J. Mol. Sci. 2019, 20, 2488. [Google Scholar] [CrossRef]
- Lesage, F.; Terrenoire, C.; Romey, G.; Lazdunski, M. Human TREK2, a 2P Domain Mechano-sensitive K+ Channel with Multiple Regulations by Polyunsaturated Fatty Acids, Lysophospholipids, and Gs, Gi, and Gq Protein-coupled Receptors. J. Biol. Chem. 2000, 275, 28398–28405. [Google Scholar] [CrossRef]
- Lauritzen, I.; Chemin, J.; Honoré, E.; Jodar, M.; Guy, N.; Lazdunski, M.; Jane Patel, A. Cross-talk between the mechano-gated K2P channel TREK-1 and the actin cytoskeleton. EMBO Rep. 2005, 6, 642–648. [Google Scholar] [CrossRef]
- Ruan, N.; Tribble, J.; Peterson, A.M.; Jiang, Q.; Wang, J.Q.; Chu, X.-P. Acid-Sensing Ion Channels and Mechanosensation. Int. J. Mol. Sci. 2021, 22, 4810. [Google Scholar] [CrossRef] [PubMed]
- Bornstein, B.; Watkins, B.; Passini, F.S.; Blecher, R.; Assaraf, E.; Sui, X.M.; Brumfeld, V.; Tsoory, M.; Kröger, S.; Zelzer, E. The mechanosensitive ion channel ASIC2 mediates both proprioceptive sensing and spinal alignment. Exp. Physiol. 2023, 109, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Stephan, G.; Huang, L.; Tang, Y.; Vilotti, S.; Fabbretti, E.; Yu, Y.; Nörenberg, W.; Franke, H.; Gölöncsér, F.; Sperlágh, B.; et al. The ASIC3/P2X3 cognate receptor is a pain-relevant and ligand-gated cationic channel. Nat. Commun. 2018, 9, 1354. [Google Scholar] [CrossRef] [PubMed]
- Jaalouk, D.E.; Lammerding, J. Mechanotransduction gone awry. Nat. Rev. Mol. Cell Biol. 2009, 10, 63–73. [Google Scholar] [CrossRef]
- Springer, T.A.; Dustin, M.L. Integrin inside-out signaling and the immunological synapse. Curr. Opin. Cell Biol. 2012, 24, 107–115. [Google Scholar] [CrossRef]
- Anishkin, A.; Loukin, S.H.; Teng, J.; Kung, C. Feeling the hidden mechanical forces in lipid bilayer is an original sense. Proc. Natl. Acad. Sci. USA 2014, 111, 7898–7905. [Google Scholar] [CrossRef]
- Chen, W.; Lou, J.; Evans, E.A.; Zhu, C. Observing force-regulated conformational changes and ligand dissociation from a single integrin on cells. J. Cell Biol. 2012, 199, 497–512. [Google Scholar] [CrossRef]
- Chuang, Y.-C.; Chen, C.-C. Force From Filaments: The Role of the Cytoskeleton and Extracellular Matrix in the Gating of Mechanosensitive Channels. Front. Cell Dev. Biol. 2022, 10, 886048. [Google Scholar] [CrossRef]
- Cox, C.D.; Bavi, N.; Martinac, B. Biophysical Principles of Ion-Channel-Mediated Mechanosensory Transduction. Cell Rep. 2019, 29, 1–12. [Google Scholar] [CrossRef]
- Grevesse, T.; Dabiri, B.E.; Parker, K.K.; Gabriele, S. Opposite rheological properties of neuronal microcompartments predict axonal vulnerability in brain injury. Sci. Rep. 2015, 5, 9475. [Google Scholar] [CrossRef]
- Martinac, B.; Adler, J.; Kung, C. Mechanosensitive ion channels of E. coli activated by amphipaths. Nature 1990, 348, 261–263. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, D.; Versaevel, M.; Bruyère, C.; Alaimo, L.; Luciano, M.; Vercruysse, E.; Procès, A.; Gabriele, S. Innovative Tools for Mechanobiology: Unraveling Outside-In and Inside-Out Mechanotransduction. Front. Bioeng. Biotechnol. 2019, 7, 162. [Google Scholar] [CrossRef] [PubMed]
- Sachs, F. Stretch-Activated Ion Channels: What Are They? Physiology 2010, 25, 50–56. [Google Scholar] [CrossRef]
- Shah, N.; Morsi, Y.; Manasseh, R. From mechanical stimulation to biological pathways in the regulation of stem cell fate. Cell Biochem. Funct. 2014, 32, 309–325. [Google Scholar] [CrossRef]
- Wilde, C.; Mitgau, J.; Suchý, T.; Schöneberg, T.; Liebscher, I. Translating the force—Mechano-sensing GPCRs. Am. J. Physiol.-Cell Physiol. 2022, 322, C1047–C1060. [Google Scholar] [CrossRef]
- Murthy, S.E.; Dubin, A.E.; Patapoutian, A. Piezos thrive under pressure: Mechanically activated ion channels in health and disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 771–783. [Google Scholar] [CrossRef]
- Enyedi, P.; Czirják, G. Molecular Background of Leak K+ Currents: Two-Pore Domain Potassium Channels. Physiol. Rev. 2010, 90, 559–605. [Google Scholar] [CrossRef]
- Venkatachalam, K.; Montell, C. TRP Channels. Annu. Rev. Biochem. 2007, 76, 387–417. [Google Scholar] [CrossRef]
- Millward-Sadler, S.J.; Salter, D.M. Integrin-Dependent Signal Cascades in Chondrocyte Mechanotransduction. Ann. Biomed. Eng. 2004, 32, 435–446. [Google Scholar] [CrossRef]
- Chen, Q.; Yang, D.; Zong, H.; Zhu, L.; Wang, L.; Wang, X.; Zhu, X.; Song, X.; Wang, J. Growth-induced stress enhances epithelial-mesenchymal transition induced by IL-6 in clear cell renal cell carcinoma via the Akt/GSK-3β/β-catenin signaling pathway. Oncogenesis 2017, 6, e375. [Google Scholar] [CrossRef]
- Quiroga, X.; Walani, N.; Disanza, A.; Chavero, A.; Mittens, A.; Tebar, F.; Trepat, X.; Parton, R.G.; Geli, M.I.; Scita, G.; et al. A mechanosensing mechanism controls plasma membrane shape homeostasis at the nanoscale. eLife 2023, 12, 72316. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.R.; Knight, M.M.; Lee, D.A.; Bader, D.L. Mechanical compression influences intracellular Ca2+ signaling in chondrocytes seeded in agarose constructs. J. Appl. Physiol. 2001, 90, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.G.; Miyazu, M.; Matsushita, E.; Sokabe, M.; Naruse, K. Uniaxial Cyclic Stretch Induces Focal Adhesion Kinase (FAK) Tyrosine Phosphorylation Followed by Mitogen-Activated Protein Kinase (MAPK) Activation. Biochem. Biophys. Res. Commun. 2001, 288, 356–361. [Google Scholar] [CrossRef]
- Xu, B.; Song, G.; Ju, Y.; Li, X.; Song, Y.; Watanabe, S. RhoA/ROCK, cytoskeletal dynamics, and focal adhesion kinase are required for mechanical stretch-induced tenogenic differentiation of human mesenchymal stem cells. J. Cell. Physiol. 2012, 227, 2722–2729. [Google Scholar] [CrossRef]
- Abuammah, A.; Maimari, N.; Towhidi, L.; Frueh, J.; Chooi, K.Y.; Warboys, C.; Krams, R. New developments in mechanotransduction: Cross talk of the Wnt, TGF-β and Notch signalling pathways in reaction to shear stress. Curr. Opin. Biomed. Eng. 2018, 5, 96–104. [Google Scholar] [CrossRef]
- Chachisvilis, M.; Zhang, Y.-L.; Frangos, J.A. G protein-coupled receptors sense fluid shear stress in endothelial cells. Proc. Natl. Acad. Sci. USA 2006, 103, 15463–15468. [Google Scholar] [CrossRef]
- Choi, H.Y.; Yang, G.-M.; Dayem, A.A.; Saha, S.K.; Kim, K.; Yoo, Y.; Hong, K.; Kim, J.-H.; Yee, C.; Lee, K.-M.; et al. Hydrodynamic shear stress promotes epithelial-mesenchymal transition by downregulating ERK and GSK3β activities. Breast Cancer Res. 2019, 21, 6. [Google Scholar] [CrossRef]
- Das, J.; Maji, S.; Agarwal, T.; Chakraborty, S.; Maiti, T.K. Hemodynamic shear stress induces protective autophagy in HeLa cells through lipid raft-mediated mechanotransduction. Clin. Exp. Metastasis 2018, 35, 135–148. [Google Scholar] [CrossRef]
- Kuipers, A.J.; Middelbeek, J.; van Leeuwen, F.N. Mechanoregulation of cytoskeletal dynamics by TRP channels. Eur. J. Cell Biol. 2012, 91, 834–846. [Google Scholar] [CrossRef]
- Li, H.; Zhou, W.-Y.; Xia, Y.-Y.; Zhang, J.-X. Endothelial Mechanosensors for Atheroprone and Atheroprotective Shear Stress Signals. J. Inflamm. Res. 2022, 15, 1771–1783. [Google Scholar] [CrossRef]
- Park, M.G.; Jang, H.; Lee, S.-H.; Lee, C.J. Flow Shear Stress Enhances the Proliferative Potential of Cultured Radial Glial Cells Possibly Via an Activation of Mechanosensitive Calcium Channel. Exp. Neurobiol. 2017, 26, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Simfia, I.; Schiavi, J.; McNamara, L.M. Alterations in osteocyte mediated osteoclastogenesis during estrogen deficiency and under ROCK-II inhibition: An in vitro study using a novel postmenopausal multicellular niche model. Exp. Cell Res. 2020, 392, 112005. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Sakamoto, N.; Song, G.; Sato, M. Low-Level Shear Stress Induces Human Mesenchymal Stem Cell Migration Through the SDF-1/CXCR4 Axis Via MAPK Signaling Pathways. Stem Cells Dev. 2013, 22, 2384–2393. [Google Scholar] [CrossRef]
- Belosludtseva, N.V.; Uryupina, T.A.; Pavlik, L.L.; Mikheeva, I.B.; Talanov, E.Y.; Venediktova, N.I.; Serov, D.A.; Stepanov, M.R.; Ananyan, M.A.; Mironova, G.D. Pathological Alterations in Heart Mitochondria in a Rat Model of Isoprenaline-Induced Myocardial Injury and Their Correction with Water-Soluble Taxifolin. Int. J. Mol. Sci. 2024, 25, 11596. [Google Scholar] [CrossRef] [PubMed]
- Rubin, D.M.; Anderton, N.; Smalberger, C.; Polliack, J.; Nathan, M.; Postema, M. On the Behaviour of Living Cells under the Influence of Ultrasound. Fluids 2018, 3, 82. [Google Scholar] [CrossRef]
- Gudkov, S.V.; Ivanov, V.E.; Matveeva, T.A.; Sarimov, R.M.; Lomonosov, A.M.; Astashev, M.E.; Baimler, I.V.; Simakin, A.V.; Bunkin, N.F.; Pustovoy, V.I.; et al. Physical and chemical changes that occur in water when drops fall on a hard surface (rain, waterfall, shower). J. Mol. Liq. 2024, 401, 124686. [Google Scholar] [CrossRef]
- Penkov, N.V. Temporal Dynamics of the Scattering Properties of Deionized Water. Phys. Wave Phenom. 2020, 28, 135–139. [Google Scholar] [CrossRef]
- Chernikov, A.V.; Gudkov, S.V.; Shtarkman, I.N.; Bruskov, V.I. Oxygen effect in heat-mediated damage to DNA. Biofizika 2007, 52, 244–251. [Google Scholar]
- Sharapov, M.G.; Gudkov, S.V.; Lankin, V.Z.; Novoselov, V.I. Role of Glutathione Peroxidases and Peroxiredoxins in Free Radical-Induced Pathologies. Biochemistry 2021, 86, 1418–1433. [Google Scholar] [CrossRef]
- Sharapov, M.G.; Gudkov, S.V.; Lankin, V.Z. Hydroperoxide-Reducing Enzymes in the Regulation of Free-Radical Processes. Biochemistry 2021, 86, 1256–1274. [Google Scholar] [CrossRef]
- Lyakhov, G.A.; Man’ko, V.I.; Suyazov, N.V.; Shcherbakov, I.A.; Shermeneva, M.A. Physical Mechanisms of Activation of Radical Reactions in Aqueous Solutions under Mechanical and Magnetic Effect: Problem of Singlet Oxygen. Phys. Wave Phenom. 2022, 30, 174–181. [Google Scholar] [CrossRef]
- Minaev, B.F. Electronic mechanisms of activation of molecular oxygen. Russ. Chem. Rev. 2007, 76, 1059–1083. [Google Scholar] [CrossRef]
- Lyakhov, G.A.; Man’ko, V.I.; Shcherbakov, I.A.; Suyazov, N.V. Impact of Classical Vibrations and Magnetic Fields on Quantum Objects. Phys. Wave Phenom. 2024, 32, 67–72. [Google Scholar] [CrossRef]
- Lyakhov, G.A.; Man’ko, V.I.; Shcherbakov, I.A. Action of Classical Fields on Quantum Systems within the Schrödinger–Robertson Uncertainty Relation. Phys. Wave Phenom. 2022, 30, 169–173. [Google Scholar] [CrossRef]
- Shcherbakov, I.A. Current Trends in the Studies of Aqueous Solutions. Phys. Wave Phenom. 2022, 30, 129–134. [Google Scholar] [CrossRef]
- Gudkov, S.V.; Karp, O.E.; Garmash, S.A.; Ivanov, V.E.; Chernikov, A.V.; Manokhin, A.A.; Astashev, M.E.; Yaguzhinsky, L.S.; Bruskov, V.I. Generation of reactive oxygen species in water under exposure to visible or infrared irradiation at absorption bands of molecular oxygen. Biophysics 2012, 57, 1–8. [Google Scholar] [CrossRef]
- Gudkov, S.V.; Bruskov, V.I.; Astashev, M.E.; Chernikov, A.V.; Yaguzhinsky, L.S.; Zakharov, S.D. Oxygen-Dependent Auto-Oscillations of Water Luminescence Triggered by the 1264 nm Radiation. J. Phys. Chem. B 2011, 115, 7693–7698. [Google Scholar] [CrossRef]
- Bruskov, V.I. Heat-induced formation of reactive oxygen species and 8-oxoguanine, a biomarker of damage to DNA. Nucleic Acids Res. 2002, 30, 1354–1363. [Google Scholar] [CrossRef]
- Bruskov, V.I.; Chernikov, A.V.; Ivanov, V.E.; Karmanova, E.E.; Gudkov, S.V. Formation of the Reactive Species of Oxygen, Nitrogen, and Carbon Dioxide in Aqueous Solutions under Physical Impacts. Phys. Wave Phenom. 2020, 28, 103–106. [Google Scholar] [CrossRef]
- Kloss, A. Electron-radical dissociation and water activation mechanism. Dokl. Akad. Nauk SSSR 1988, 303, 1403–1407. [Google Scholar]
- Lavrik, N.L.; Bazhin, N.M. On the impossibility of OH radical formation from the hydroxyl ion in water. Biophysics 2011, 56, 535–536. [Google Scholar] [CrossRef]
- Ward, J.F. DNA Damage Produced by Ionizing Radiation in Mammalian Cells: Identities, Mechanisms of Formation, and Reparability. Prog. Nucleic Acid Res. Mol. Biol. 1988, 35, 95–125. [Google Scholar] [PubMed]
- Draganić, I.G. Radiolysis of water: A look at its origin and occurrence in the nature. Radiat. Phys. Chem. 2005, 72, 181–186. [Google Scholar] [CrossRef]
- Sergeichev, K.F.; Lukina, N.A.; Sarimov, R.M.; Smirnov, I.G.; Simakin, A.V.; Dorokhov, A.S.; Gudkov, S.V. Physicochemical Properties of Pure Water Treated by Pure Argon Plasma Jet Generated by Microwave Discharge in Opened Atmosphere. Front. Phys. 2021, 8, 614684. [Google Scholar] [CrossRef]
- Barmina, E.V.; Gudkov, S.V.; Simakin, A.V.; Shafeev, G.A. Stable Products of Laser-Induced Breakdown of Aqueous Colloidal Solutions of Nanoparticles. J. Laser Micro/Nanoeng. 2017, 12, 254–257. [Google Scholar] [CrossRef]
- Novakovskaya, Y.V. Hydrogen-bond Network of Water and Irradiation Effects. Phys. Wave Phenom. 2020, 28, 161–167. [Google Scholar] [CrossRef]
- Colussi, A.J. Mechanism of Hydrogen Peroxide Formation on Sprayed Water Microdroplets. J. Am. Chem. Soc. 2023, 145, 16315–16317. [Google Scholar] [CrossRef]
- Sarimov, R.M.; Nagaev, E.I.; Matveyeva, T.A.; Binhi, V.N.; Burmistrov, D.E.; Serov, D.A.; Astashev, M.E.; Simakin, A.V.; Uvarov, O.V.; Khabatova, V.V.; et al. Investigation of Aggregation and Disaggregation of Self-Assembling Nano-Sized Clusters Consisting of Individual Iron Oxide Nanoparticles upon Interaction with HEWL Protein Molecules. Nanomaterials 2022, 12, 3960. [Google Scholar] [CrossRef]
- Moore, K.D.; Wu, J.Z.; Krajnak, K.; Warren, C.; Dong, R.G. Quantification of mechanical behavior of rat tail under compression. Bio-Med. Mater. Eng. 2024, 35, 337–349. [Google Scholar] [CrossRef]
- Yarasheski, K.E.; Lemon, P.W.; Gilloteaux, J. Effect of heavy-resistance exercise training on muscle fiber composition in young rats. J. Appl. Physiol. 1990, 69, 434–437. [Google Scholar] [CrossRef]
- Odintsov, S.D.; Oikonomou, V.K. Towards Modelling Mechanical Shaking Using Potential Energy Surfaces: A Toy Model Analysis. Symmetry 2024, 16, 572. [Google Scholar] [CrossRef]
- Orgel, L.E. The origin of life—A review of facts and speculations. Trends Biochem. Sci. 1998, 23, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, R.P.; Norton, J.N. Evaluation of Vibration as an Extrinsic Variable in In Vivo Research. J. Am. Assoc. Lab. Anim. Sci. 2024, 63, 107–115. [Google Scholar] [CrossRef]
- Na, S.; Wang, N. Application of Fluorescence Resonance Energy Transfer and Magnetic Twisting Cytometry to Quantify Mechanochemical Signaling Activities in a Living Cell. Sci. Signal. 2008, 1, pl1. [Google Scholar] [CrossRef]
- Cross, R.A.; McAinsh, A.D.; Straube, A. Mechanochemical cell biology. Semin. Cell Dev. Biol. 2011, 22, 913–915. [Google Scholar] [CrossRef]
- Yang, H.; Chen, W.; Peczulis, P.; Heng, J.Y.Y. Development and Workflow of a Continuous Protein Crystallization Process: A Case of Lysozyme. Cryst. Growth Des. 2019, 19, 983–991. [Google Scholar] [CrossRef]
- Hansson, H.A.; Dahlin, L.B.; LÖWenadler, B.; Lundborg, G.; Paleus, S.; Skottneru, A. Transient increase in insulin-like growth factor I immunoreactivity in rat peripheral nerves exposed to vibrations. Acta Physiol. Scand. 2008, 132, 35–41. [Google Scholar] [CrossRef]
- Zhang, C.; Li, J.; Zhang, L.; Zhou, Y.; Hou, W.; Quan, H.; Li, X.; Chen, Y.; Yu, H. Effects of mechanical vibration on proliferation and osteogenic differentiation of human periodontal ligament stem cells. Arch. Oral Biol. 2012, 57, 1395–1407. [Google Scholar] [CrossRef]
- Miller, D.L. Cell death thresholds in Elodea for 0.45–10 MHz ultrasound compared to gas-body resonance theory. Ultrasound Med. Biol. 1979, 5, 351–357. [Google Scholar] [CrossRef]
- Williams, A.R.; Sykes, S.M.; O’Brien, W.D. Ultrasonic exposure modifies platelet morphology and function in vitro. Ultrasound Med. Biol. 1977, 2, 311–317. [Google Scholar] [CrossRef]
- Tachibana, K.; Uchida, T.; Ogawa, K.; Yamashita, N.; Tamura, K. Induction of cell-membrane porosity by ultrasound. Lancet 1999, 353, 1409. [Google Scholar] [CrossRef] [PubMed]
- Lagneaux, L.; de Meulenaer, E.C.; Delforge, A.; Dejeneffe, M.; Massy, M.; Moerman, C.; Hannecart, B.; Canivet, Y.; Lepeltier, M.-F.; Bron, D. Ultrasonic low-energy treatment. Exp. Hematol. 2002, 30, 1293–1301. [Google Scholar] [CrossRef] [PubMed]
- Pitt, W.G.; Ross, S.A. Ultrasound Increases the Rate of Bacterial Cell Growth. Biotechnol. Prog. 2003, 19, 1038–1044. [Google Scholar] [CrossRef] [PubMed]
- Mizrahi, N.; Zhou, E.H.; Lenormand, G.; Krishnan, R.; Weihs, D.; Butler, J.P.; Weitz, D.A.; Fredberg, J.J.; Kimmel, E. Low intensity ultrasound perturbs cytoskeleton dynamics. Soft Matter 2012, 8, 2438–2443. [Google Scholar] [CrossRef]
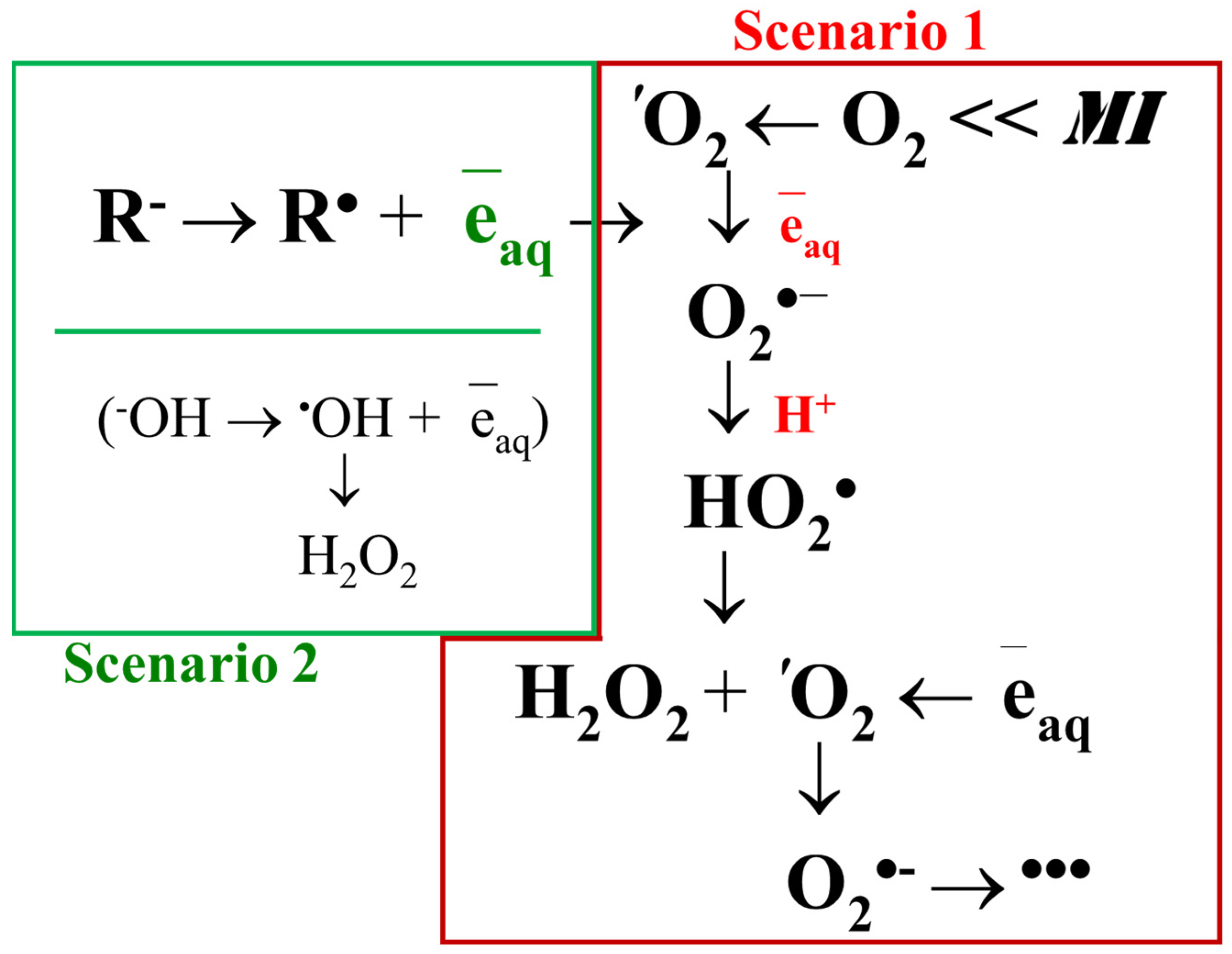

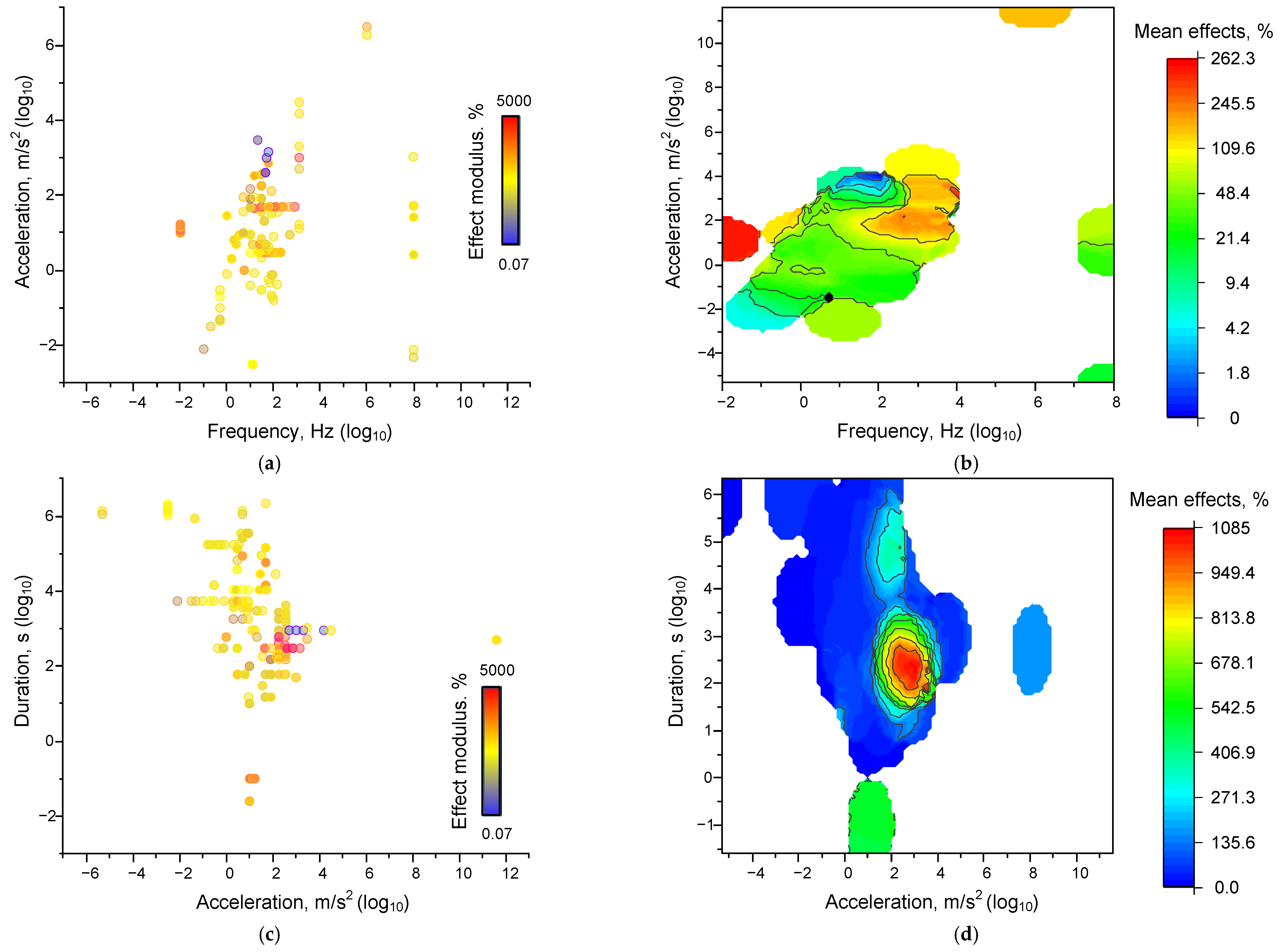

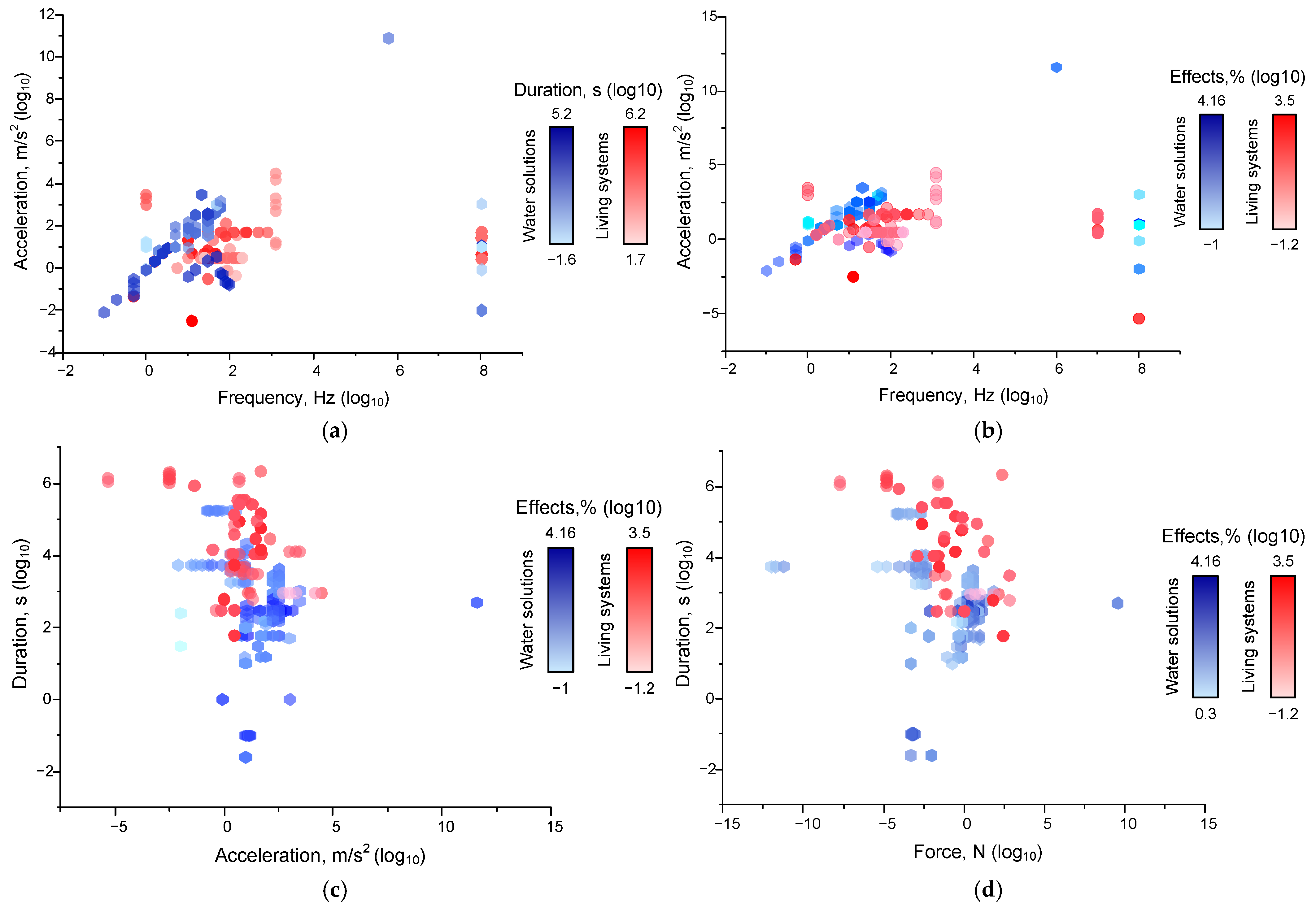
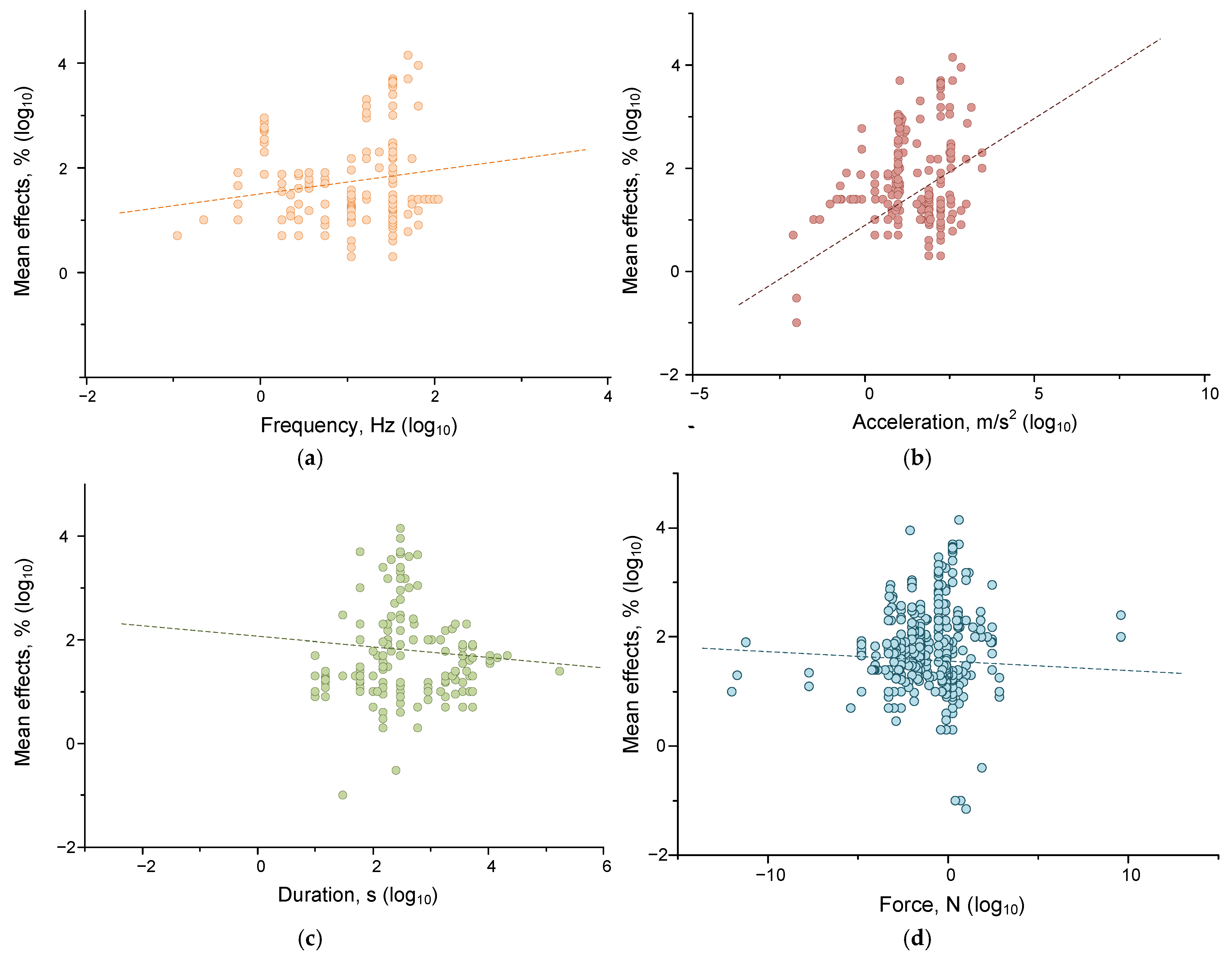
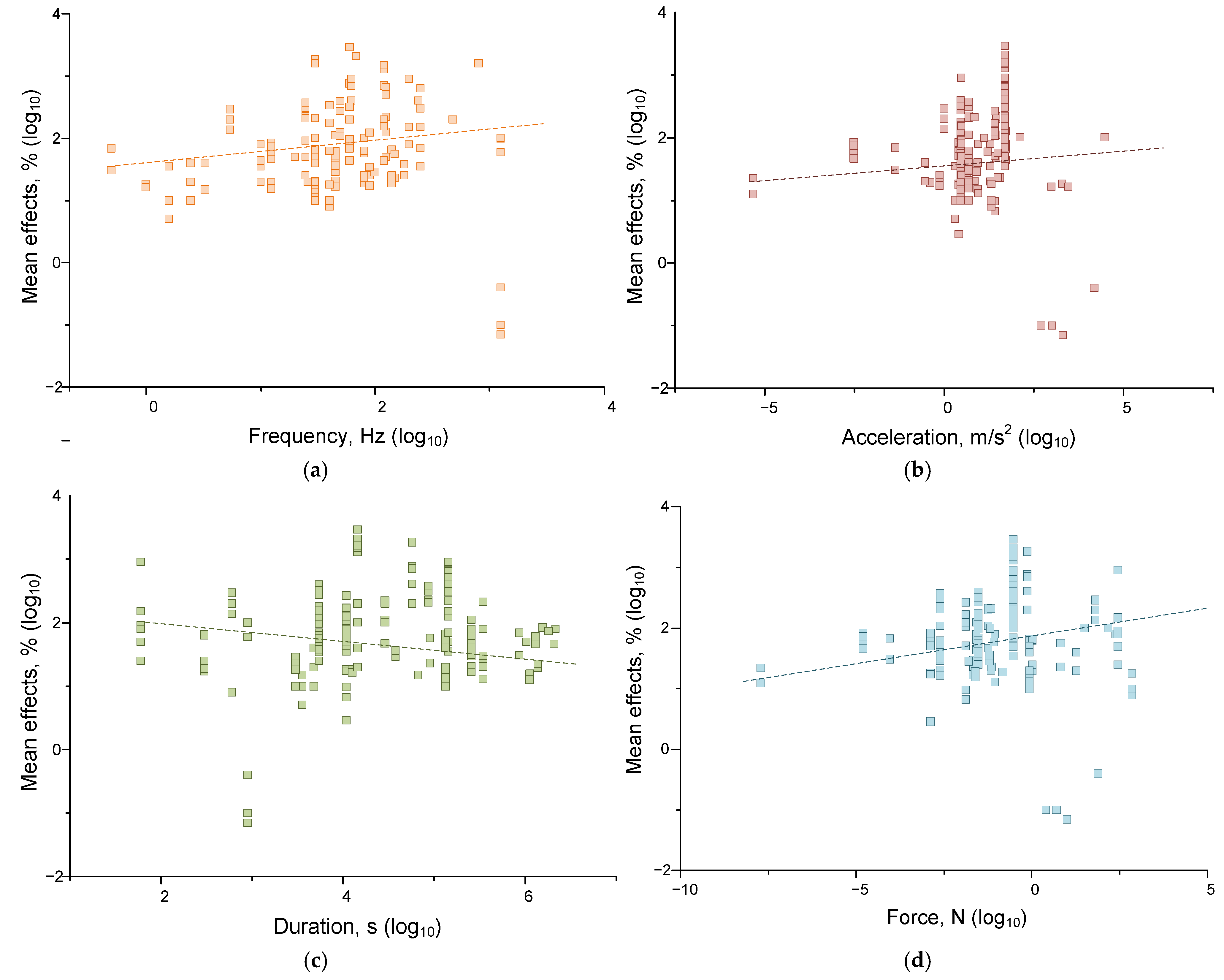

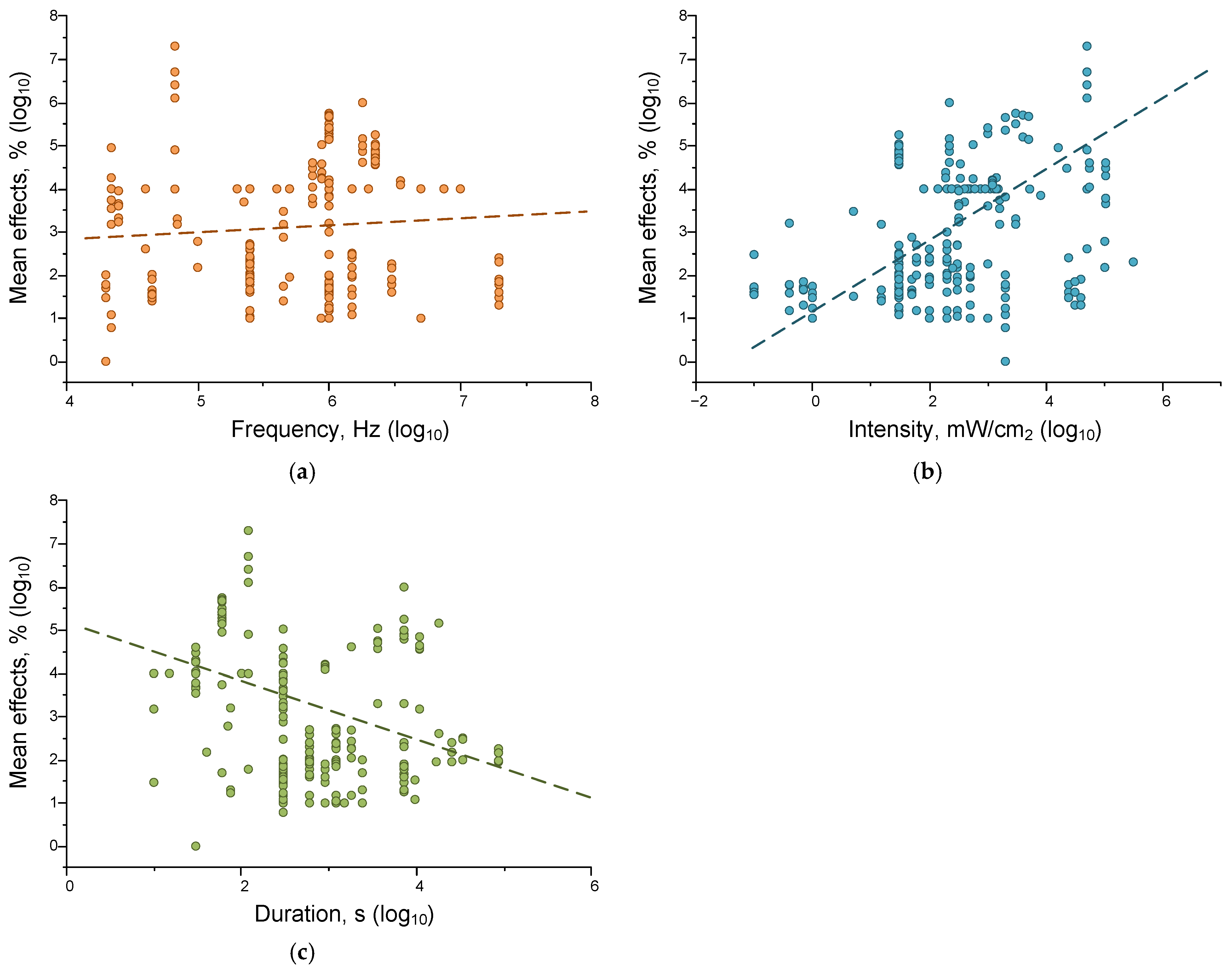
| System | Level | Reaction | Benefit | References |
|---|---|---|---|---|
| Cardiovascular | Molecular | Change Ca2+ responses to AChR ligands | ↓ or ↑ depend on conditions | [70,71] |
| Cells | Micro-damage of endothelial cells damage | ↓ | [41] | |
| Calcium homeostasis of endotheliocytes changes | ↑ | [70,71] | ||
| Tissues | Number of capillaries, venules, and arterioles reduction | ↓ | [20] | |
| Smooth muscle work of microvessels | ↑ | [40] | ||
| Organisms | HR and blood pressure, Raynaud’s syndrome, etc. | ↓ | [11,39] | |
| Immune and blood cells | Molecular | NFAT activity, IL-6 synthesis, ROS production, and t-CPK activity increasing | ↓ | [40,41,42,43] |
| Decreasing the synthesis of pro-inflammatory IFN-β, RANKL, and TNF-α, increased oxygen consumption, and gene expression of glycolysis enzymes and transporters (Hk2, GLUT1) | ↑ | [62,66,72] | ||
| Cells | Erythrocytes hemolysis rate increasing | ↓ | [57] | |
| T-cell proliferation altered, ‘neutrophil respiratory burst’ (ROS productions), reduction of differentiated dendritic cell proliferation | ↓ or ↑ depend on frequency and conditions | [32,33,56,72] | ||
| Tissues | Immune memory and antitumor activity of dendric cells | ↑ | [72] | |
| Organisms | Anti-inflammatory effect | ↑ | [63] | |
| Proinflammatory effect | ↓ or ↑ depend on conditions | [32,33] | ||
| Musculoskeletal | Molecular | TRAP-dependent pathway activation, increasing of ACAN and SOX9 expression, increasing of collagen content | ↑ | [44,45,59,67] |
| P1NP and CTx concentration increase, osteogenesis regulators MMP 2 and OSX expression decreasing. Decreasing of osteocalcin levels, Runx2 and Osx gene expression, synthesis of osteoarthritis markers collagen II and aggrecan | ↓ | [26,28,46,47,49,50,58,64,65] | ||
| Cells | Osteoclast proliferation, differentiations and activity increasing, increasing of differentiation of primary fibro chondrocytes | ↑ | [26,44,45,59,60,61] | |
| The viability of osteoblasts decreases; decrease of osteoblast differentiation | ↓ | [26,58] | ||
| Tissues | Improving bone mechanical characteristics and bone structure | ↑ | [28,44,45] | |
| Bone tissue damage | ↓ | [42,43] | ||
| Organisms | Osteoporosis markers P1NP and CTx levels | ↓ | [46,47] | |
| Improvement bone-implant contact, obesity decreasing, and muscle strength increasing | ↑ | [28,48,51] | ||
| Respiratory | Organisms | Inspiratory flow rate, inspired volume, and expired volume | ↑ | [52] |
| Nervous | Molecular | Increasing the expression of the IGF-I factor | ↑ | [54] |
| Cells | Improve of myelinization | ↑ | [53] | |
| Other | Molecular | Increasing in A cytochrome P450 monooxygenase CYP1B1 expression and metabolic activity changes | ↑ | [69] |
| Cells | Stem cell proliferation inhibition | ↓ | [25] |
| System | Level | Reaction | Benefit | References |
|---|---|---|---|---|
| Cardiovascular | Molecular | Enhancing VEGF secretion | ↑ | [102] |
| Tissues | Enhance angiogenesis | [102,106] | ||
| Immune and blood cells | Molecular | Secretion of proinflammatory cytokines IL-1β, IL-8 | ↑ | [102] |
| Increasing the number of chromosomal aberrations | ↓ | [111] | ||
| Cells | Induction of Ca2+ influx and platelet aggregation; induction of apoptosis of leucocyte cells | ↓ | [108,109,110,111,112] | |
| Organisms | inhibition of pro-inflammatory reactions | ↑ or ↓ depend on situation | [113,115] | |
| Musculoskeletal | Molecular | Increased expression of collagen, expression of myogenin, Pax7, COX2 | ↑ | [102,113,115] |
| Cells | Increasing of myocyte proliferation, myoblast differentiation | ↑ | [97,98,104] | |
| Tissues | Increasing of length of myotubes and the number of cells fused into one myotube, and regeneration of myofibrils | ↑ | [97,98,104,113,114] | |
| Nervous | Molecular | Enhanced expression of markers Alk, Cenpf, Pcdh17 and actin | ↑ | [116] |
| Cells | Promotions of proliferation and differentiation of neurons, formation of outgrowths | ↑ | [116] | |
| Other | Molecular | Acceleration of drug delivery, EGFb regulatory factor expression increasing, change Cyclin D1/β, Cyclin B1/β, Cyclin E1/β, and Cyclin A1/β levels | ↑ | [99,102,103,105] |
| Accelerate DNA plasmid transfer in bacterial cells | ↑ | [119] | ||
| Plants photosynthesis inhibition | ↓ | [120] | ||
| Cells | ER microstructure damage | ↓ | [100] | |
| Stem cell differentiation and proliferation increasing | ↑ | [102,103,104,107] | ||
| Destruction of bacterial spores and cells | ↓ | [117,118] |
| Coefficients | Mechanical Impact Parameters | Effect log10 | |
|---|---|---|---|
| In Vivo | In Vitro | ||
| Spearman’s correlation | frequency, Hz (log10) | 0.173 | 0.196 |
| acceleration, m/s2 (log10) | 0.253 | 0.263 | |
| duration, s (log10) | 0.052 | 0.11407 | |
| force, N (log10) | 0.195 | −0.06187 | |
| p-value | frequency, Hz (log10) | 0.02344 | 0.01622 |
| acceleration, m/s2 (log10) | 7.76 × 10−4 | 0.00113 | |
| duration, s (log10) | 0.49627 | 0.166 | |
| force, N (log10) | 0.01341 | 0.45196 | |
| Coefficients | US Impact Parameters | Effect log10 |
|---|---|---|
| Spearman’s correlation | frequency, Hz (log10) | 0.1086 |
| intensity, mW/cm2 (log10)_ | 0.38302 | |
| duration, s (log10) | −0.27719 | |
| p-value | frequency, Hz (log10) | 0.10657 |
| intensity, mW/cm2 (log10)_ | 3.60925 × 10−9 | |
| duration, s (log10) | 2.8004 × 10−5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gudkov, S.V.; Pustovoy, V.I.; Sarimov, R.M.; Serov, D.A.; Simakin, A.V.; Shcherbakov, I.A. Diversity of Effects of Mechanical Influences on Living Systems and Aqueous Solutions. Int. J. Mol. Sci. 2025, 26, 5556. https://doi.org/10.3390/ijms26125556
Gudkov SV, Pustovoy VI, Sarimov RM, Serov DA, Simakin AV, Shcherbakov IA. Diversity of Effects of Mechanical Influences on Living Systems and Aqueous Solutions. International Journal of Molecular Sciences. 2025; 26(12):5556. https://doi.org/10.3390/ijms26125556
Chicago/Turabian StyleGudkov, Sergey V., Vladimir I. Pustovoy, Ruslan M. Sarimov, Dmitriy A. Serov, Alexander V. Simakin, and Ivan A. Shcherbakov. 2025. "Diversity of Effects of Mechanical Influences on Living Systems and Aqueous Solutions" International Journal of Molecular Sciences 26, no. 12: 5556. https://doi.org/10.3390/ijms26125556
APA StyleGudkov, S. V., Pustovoy, V. I., Sarimov, R. M., Serov, D. A., Simakin, A. V., & Shcherbakov, I. A. (2025). Diversity of Effects of Mechanical Influences on Living Systems and Aqueous Solutions. International Journal of Molecular Sciences, 26(12), 5556. https://doi.org/10.3390/ijms26125556








