Transglutaminase 2 Stimulates Cell Proliferation and Modulates Transforming Growth Factor-Beta Signaling Pathway Independently of Epithelial–Mesenchymal Transition in Hepatocellular Carcinoma Cells
Abstract
1. Introduction
2. Results
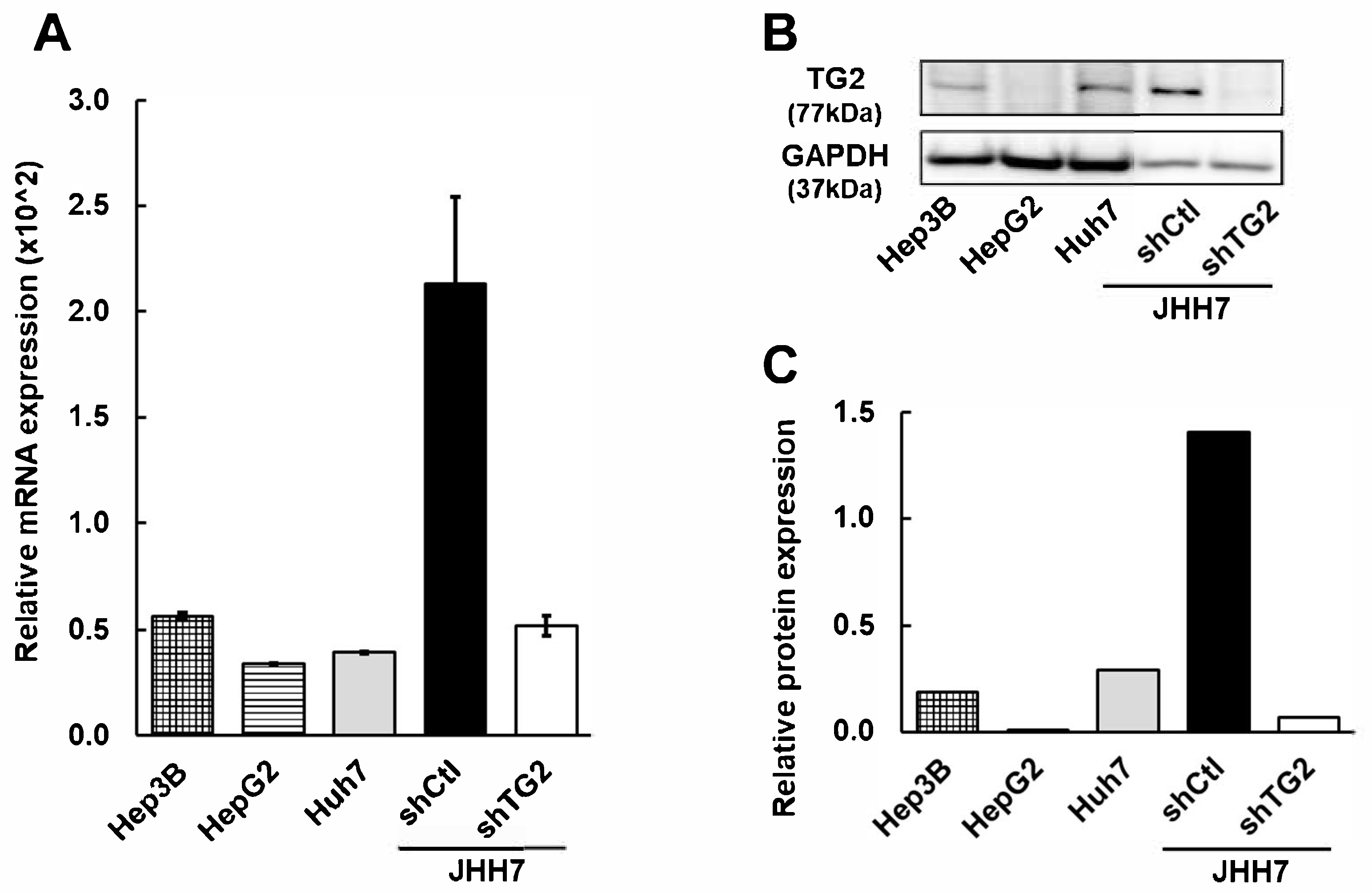
2.1. TG2 Is Differentially Expressed in HCC Cell Lines
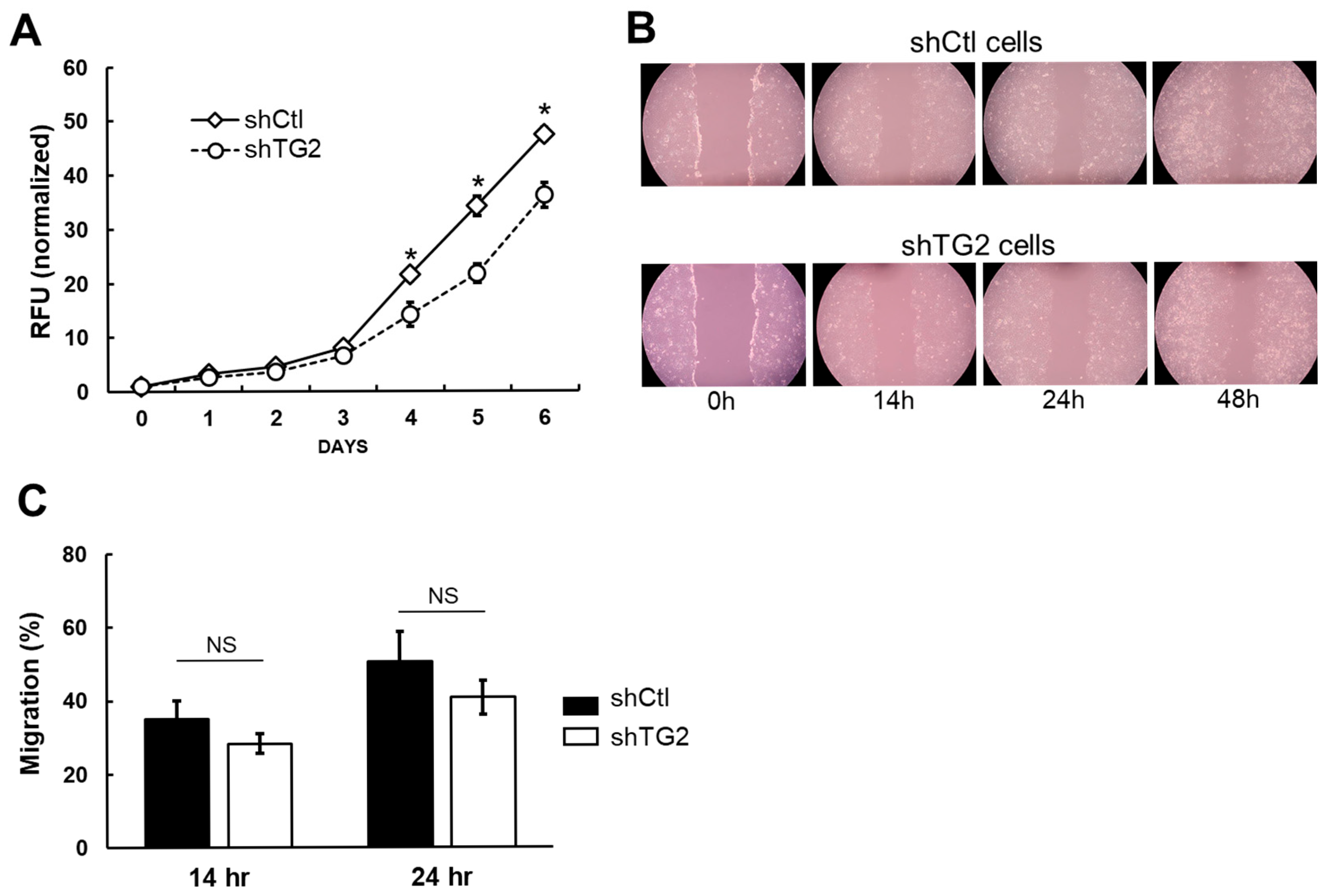
2.2. TG2 Knockdown Reduces Cell Proliferation but Does Not Affect Migration Ability of JHH7 Cells
2.3. Gene Expression Signatures of High-TG2 JHH7 Cells
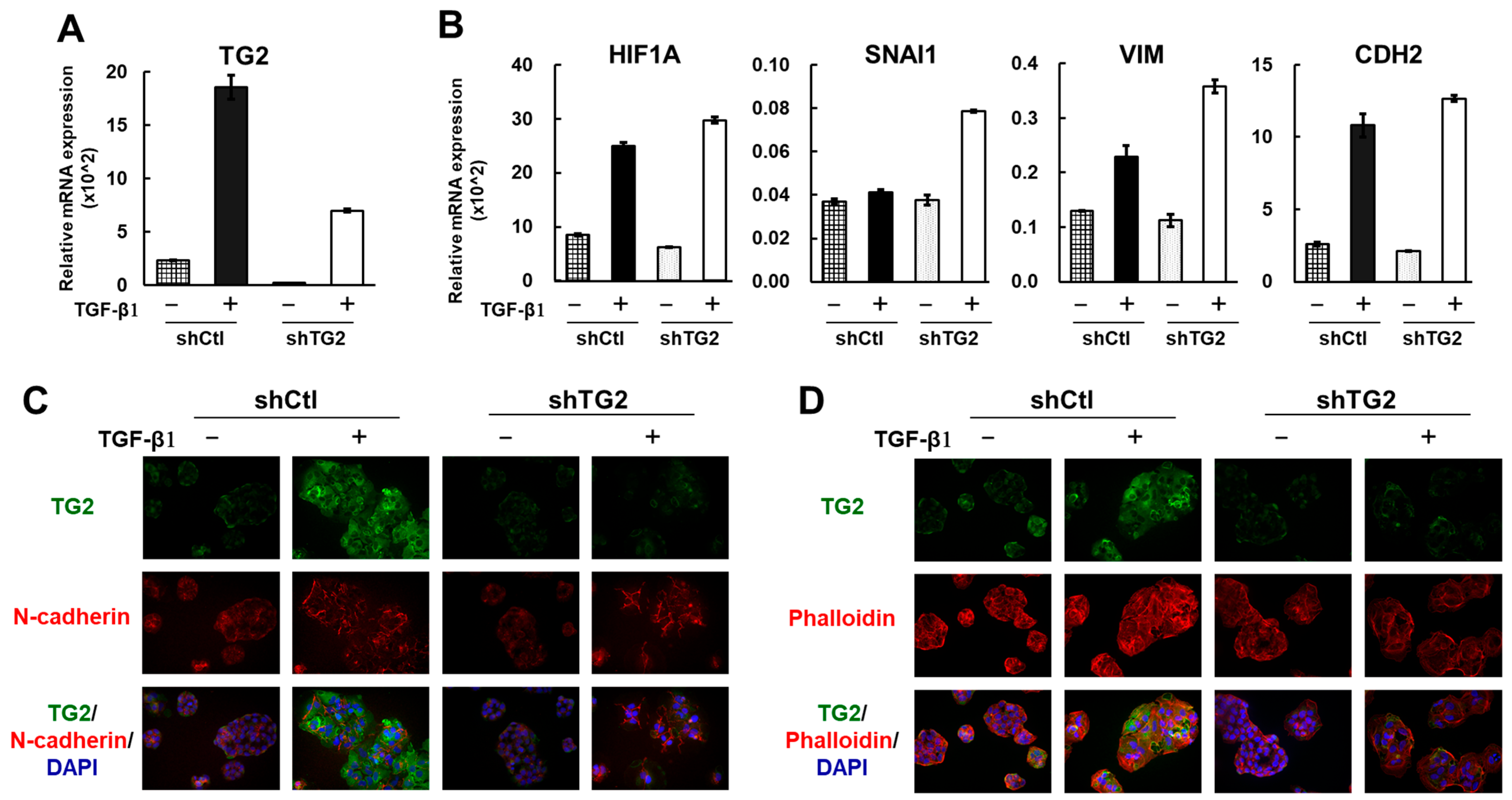
2.4. TG2 Is Induced by TGF-β1 but Is Not a Key Player in the Epithelial–Mesenchymal Transition (EMT)
2.5. TCGA Cohort Study on Characteristics of TG2 and TGF-β1 in Clinical HCC
3. Discussion
4. Materials and Methods
4.1. HCC Cell Lines and Cell Culture
4.2. In Vitro Cell Proliferation Assay
4.3. In Vitro Cell Migration Assay
4.4. GeneChip Transcriptome Analysis
4.5. Quantitative PCR (qPCR) Assay of mRNA
4.6. Western Blot Analysis
4.7. Immunofluorescence Analysis
4.8. Statistical Analysis
4.9. Data Mining from Clinical HCC Database
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| TG2 | Transglutaminase 2 |
| HCC | Hepatocellular carcinoma |
| EMT | Epithelial–mesenchymal transition |
| TGF-β1 | Transforming growth factor-beta 1 |
| DAVID | Database for annotation, visualization, and integrated discovery |
| GSEA | Gene set enrichment analysis |
| FDR | False discovery rate |
| TCGA | The cancer genome atlas |
| OS | Overall survival |
| NES | Normalized enrichment score |
| FWER | Family-wise error rate |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Singal, A.G.; Lampertico, P.; Nahon, P. Epidemiology and Surveillance for Hepatocellular Carcinoma: New Trends. J. Hepatol. 2020, 72, 250–261. [Google Scholar] [CrossRef]
- Teng, W.; Wu, T.-C.; Lin, S.-M. Hepatocellular Carcinoma Systemic Treatment 2024 Update: From Early to Advanced Stage. Biomed. J. 2024, 540, 100815. [Google Scholar]
- Yamaguchi, H.; Kuroda, K.; Sugitani, M.; Takayama, T.; Hasegawa, K.; Esumi, M. Transglutaminase 2 Is Upregulated in Primary Hepatocellular Carcinoma with Early Recurrence as Determined by Proteomic Profiles. Int. J. Oncol. 2017, 50, 1749–1759. [Google Scholar] [CrossRef]
- Nurminskaya, M.V.; Belkin, A.M. Cellular Functions of Tissue Transglutaminase. Int. Rev. Cell Mol. Biol. 2012, 294, 1–97. [Google Scholar] [PubMed]
- Tatsukawa, H.; Hitomi, K. Role of Transglutaminase 2 in Cell Death, Survival, and Fibrosis. Cells 2021, 10, 1842. [Google Scholar] [CrossRef] [PubMed]
- Kuo, T.-F.; Tatsukawa, H.; Kojima, S. New Insights into the Functions and Localization of Nuclear Transglutaminase 2. FEBS J. 2011, 278, 4756–4767. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Xu, A.-M.; Liu, W. Transglutaminase 2 in Cancer. Am. J. Cancer Res. 2015, 5, 2756–2776. [Google Scholar]
- Eckert, R.L.; Fisher, M.L.; Grun, D.; Adhikary, G.; Xu, W.; Kerr, C. Transglutaminase Is a Tumor Cell and Cancer Stem Cell Survival Factor. Mol. Carcinog. 2015, 54, 947–958. [Google Scholar] [CrossRef]
- Sun, Y.; Mi, W.; Cai, J.; Ying, W.; Liu, F.; Lu, H.; Qiao, Y.; Jia, W.; Bi, X.; Lu, N.; et al. Quantitative Proteomic Signature of Liver Cancer Cells: Tissue Transglutaminase 2 Could Be a Novel Protein Candidate of Human Hepatocellular Carcinoma. J. Proteome Res. 2008, 7, 3847–3859. [Google Scholar] [CrossRef]
- Yu, C.; Cao, Q.; Chen, P.; Yang, S.; Gong, X.; Deng, M.; Ruan, B.; Li, L. Tissue Transglutaminase 2 Exerts a Tumor-Promoting Role in Hepatitis B Virus-Related Hepatocellular Carcinoma. Tumor Biol. 2016, 37, 16269–16274. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.-Y.; Lu, J.; Cai, M.; Kojima, S. Arachidonic Acid Suppresses Hepatic Cell Growth through ROS-Mediated Activation of Transglutaminase. FEBS Open Bio 2018, 8, 1703–1710. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Resaul, J.; Owen, S.; Ye, L.; Jiang, W.G. Clinical and Therapeutic Implications of Follistatin in Solid Tumours. Cancer Genom. Proteom. 2016, 13, 425–435. [Google Scholar] [CrossRef]
- Niida, A.; Hiroko, T.; Kasai, M.; Furukawa, Y.; Nakamura, Y.; Suzuki, Y.; Sugano, S.; Akiyama, T. DKK1, a Negative Regulator of Wnt Signaling, Is a Target of the β-Catenin/TCF Pathway. Oncogene 2004, 23, 8520–8526. [Google Scholar] [CrossRef]
- Ren, Q.; Chen, J.; Liu, Y. LRP5 and LRP6 in Wnt Signaling: Similarity and Divergence. Front. Cell Dev. Biol. 2021, 9, 670960. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, Y.; Wang, C.; Zhao, Y.; Cheng, Y.; Yu, S.; Li, X.; Zhang, W.; Zhang, Y.; Quan, H. HCV Core Protein Represses DKK3 Expression via Epigenetic Silencing and Activates the Wnt/β-Catenin Signaling Pathway during the Progression of HCC. Clin. Transl. Oncol. 2022, 24, 1998–2009. [Google Scholar] [CrossRef]
- Xiang, T.; Li, L.; Yin, X.; Zhong, L.; Peng, W.; Qiu, Z.; Ren, G.; Tao, Q. Epigenetic Silencing of the WNT Antagonist Dickkopf 3 Disrupts Normal Wnt/β-Catenin Signalling and Apoptosis Regulation in Breast Cancer Cells. J. Cell Mol. Med. 2013, 17, 1236–1246. [Google Scholar] [CrossRef]
- Wei, T.; Bi, G.; Bian, Y.; Ruan, S.; Yuan, G.; Xie, H.; Zhao, M.; Shen, R.; Zhu, Y.; Wang, Q.; et al. The Significance of Secreted Phosphoprotein 1 in Multiple Human Cancers. Front. Mol. Biosci. 2020, 7, 565383. [Google Scholar] [CrossRef]
- Zheng, S.; Zhou, C.; Wang, Y.; Li, H.; Sun, Y.; Shen, Z. TRIM6 Promotes Colorectal Cancer Cells Proliferation and Response to Thiostrepton by TIS21/FoxM1. J. Exp. Clin. Cancer Res. 2020, 39, 23. [Google Scholar] [CrossRef]
- Zhang, Y.; Yuan, L.; Cui, S.; Wu, S. Tripartite Motif Protein 6 Promotes Hepatocellular Carcinoma Progression via Multiple Pathways. Turk. J. Med. Sci. 2023, 53, 1032–1044. [Google Scholar] [CrossRef]
- Kumar, A.; Xu, J.; Brady, S.; Gao, H.; Yu, D.; Reuben, J.; Mehta, K. Tissue Transglutaminase Promotes Drug Resistance and Invasion by Inducing Mesenchymal Transition in Mammary Epithelial Cells. PLoS ONE 2010, 5, e13390. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.-Y.; Furutani, Y.; Yonezawa, K.; Shimizu, N.; Kato-Murayama, M.; Shirouzu, M.; Xu, Y.; Yamano, Y.; Wada, A.; Gailhouste, L.; et al. Targeting Transglutaminase 2 Mediated Exostosin Glycosyltransferase 1 Signaling in Liver Cancer Stem Cells with Acyclic Retinoid. Cell Death Dis. 2023, 14, 358. [Google Scholar] [CrossRef]
- Jambrovics, K.; Botó, P.; Pap, A.; Sarang, Z.; Kolostyák, Z.; Czimmerer, Z.; Szatmari, I.; Fésüs, L.; Uray, I.P.; Balajthy, Z. Transglutaminase 2 Associated with PI3K and PTEN in a Membrane-Bound Signalosome Platform Blunts Cell Death. Cell Death Dis. 2023, 14, 217. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Guha, S.; Wang, H.; Fok, J.Y.; Koul, D.; Abbruzzese, J.; Mehta, K. Tissue Transglutaminase Regulates Focal Adhesion Kinase/AKT Activation by Modulating PTEN Expression in Pancreatic Cancer Cells. Clin. Cancer Res. 2008, 14, 1997–2005. [Google Scholar] [CrossRef]
- Shimada, S.; Mogushi, K.; Akiyama, Y.; Furuyama, T.; Watanabe, S.; Ogura, T.; Ogawa, K.; Ono, H.; Mitsunori, Y.; Ban, D.; et al. Comprehensive Molecular and Immunological Characterization of Hepatocellular Carcinoma. EBioMedicine 2019, 40, 457–470. [Google Scholar] [CrossRef]
- Dong, R.; Wang, T.; Dong, W.; Zhang, H.; Li, Y.; Tao, R.; Liu, Q.; Liang, H.; Chen, X.; Zhang, B.; et al. TGM2-Mediated Histone Serotonylation Promotes HCC Progression via MYC Signalling Pathway. J. Hepatol. 2025. Epub ahead of print. [Google Scholar] [CrossRef]
- Deasey, S.; Nurminsky, D.; Shanmugasundaram, S.; Lima, F.; Nurminskaya, M. Transglutaminase 2 as a Novel Activator of LRP6/β-Catenin Signaling. Cell. Signal. 2013, 25, 2646–2651. [Google Scholar] [CrossRef]
- Xu, C.; Xu, Z.; Zhang, Y.; Evert, M.; Calvisi, D.F.; Chen, X. β-Catenin Signaling in Hepatocellular Carcinoma. J. Clin. Investig. 2022, 132, e154515. [Google Scholar] [CrossRef]
- Cao, L.; Shao, M.; Schilder, J.; Guise, T.; Mohammad, K.S.; Matei, D. Tissue Transglutaminase Links TGF-β, Epithelial to Mesenchymal Transition and a Stem Cell Phenotype in Ovarian Cancer. Oncogene 2012, 31, 2521–2534. [Google Scholar] [CrossRef]
- Ayinde, O.; Wang, Z.; Griffin, M. Tissue Transglutaminase Induces Epithelial-Mesenchymal-Transition and the Acquisition of Stem Cell like Characteristics in Colorectal Cancer Cells. Oncotarget 2017, 8, 20025–20041. [Google Scholar] [CrossRef]
- Kubo, N.; Araki, K.; Kuwano, H.; Shirabe, K. Cancer-Associated Fibroblasts in Hepatocellular Carcinoma. World J. Gastroenterol. 2016, 22, 6841–6850. [Google Scholar] [CrossRef] [PubMed]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor Microenvironment Complexity and Therapeutic Implications at a Glance. Cell Commun. Signal. 2020, 18, 59. [Google Scholar] [CrossRef] [PubMed]
- Bussard, K.M.; Mutkus, L.; Stumpf, K.; Gomez-Manzano, C.; Marini, F.C. Tumor-Associated Stromal Cells as Key Contributors to the Tumor Microenvironment. Breast Cancer Res. 2016, 18, 84. [Google Scholar] [CrossRef]
- Henke, E.; Nandigama, R.; Ergün, S. Extracellular Matrix in the Tumor Microenvironment and Its Impact on Cancer Therapy. Front. Mol. Biosci. 2020, 6, 160. [Google Scholar] [CrossRef]
- Tempest, R.; Guarnerio, S.; Maani, R.; Cooper, J.; Peake, N. The Biological and Biomechanical Role of Transglutaminase-2 in the Tumour Microenvironment. Cancers 2021, 13, 2788. [Google Scholar] [CrossRef]
- Furini, G.; Verderio, E.A.M. Spotlight on the Transglutaminase 2-Heparan Sulfate Interaction. Med. Sci. 2019, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Lockhart-Cairns, M.P.; Cain, S.A.; Dajani, R.; Steer, R.; Thomson, J.; Alanazi, Y.F.; Kielty, C.M.; Baldock, C. Latent TGFβ Complexes Are Transglutaminase Cross-Linked to Fibrillin to Facilitate TGFβ Activation. Matrix Biol. 2022, 107, 24–39. [Google Scholar] [CrossRef]
- Liao, J.; Chen, R.; Lin, B.; Deng, R.; Liang, Y.; Zeng, J.; Ma, S.; Qiu, X. Cross-Talk between the TGF-β and Cell Adhesion Signaling Pathways in Cancer. Int. J. Med. Sci. 2024, 21, 1307–1320. [Google Scholar] [CrossRef]
- Liang, C.-C.; Park, A.Y.; Guan, J.-L. In Vitro Scratch Assay: A Convenient and Inexpensive Method for Analysis of Cell Migration in Vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics Enrichment Tools: Paths toward the Comprehensive Functional Analysis of Large Gene Lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.-F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1α-Responsive Genes Involved in Oxidative Phosphorylation Are Coordinately Downregulated in Human Diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Kainuma, M.; Takada, I.; Makishima, M.; Sano, K. Farnesoid X Receptor Activation Enhances Transforming Growth Factor β-Induced Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma Cells. Int. J. Mol. Sci. 2018, 19, 1898. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and Interpreting Cancer Genomics Data via the Xena Platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
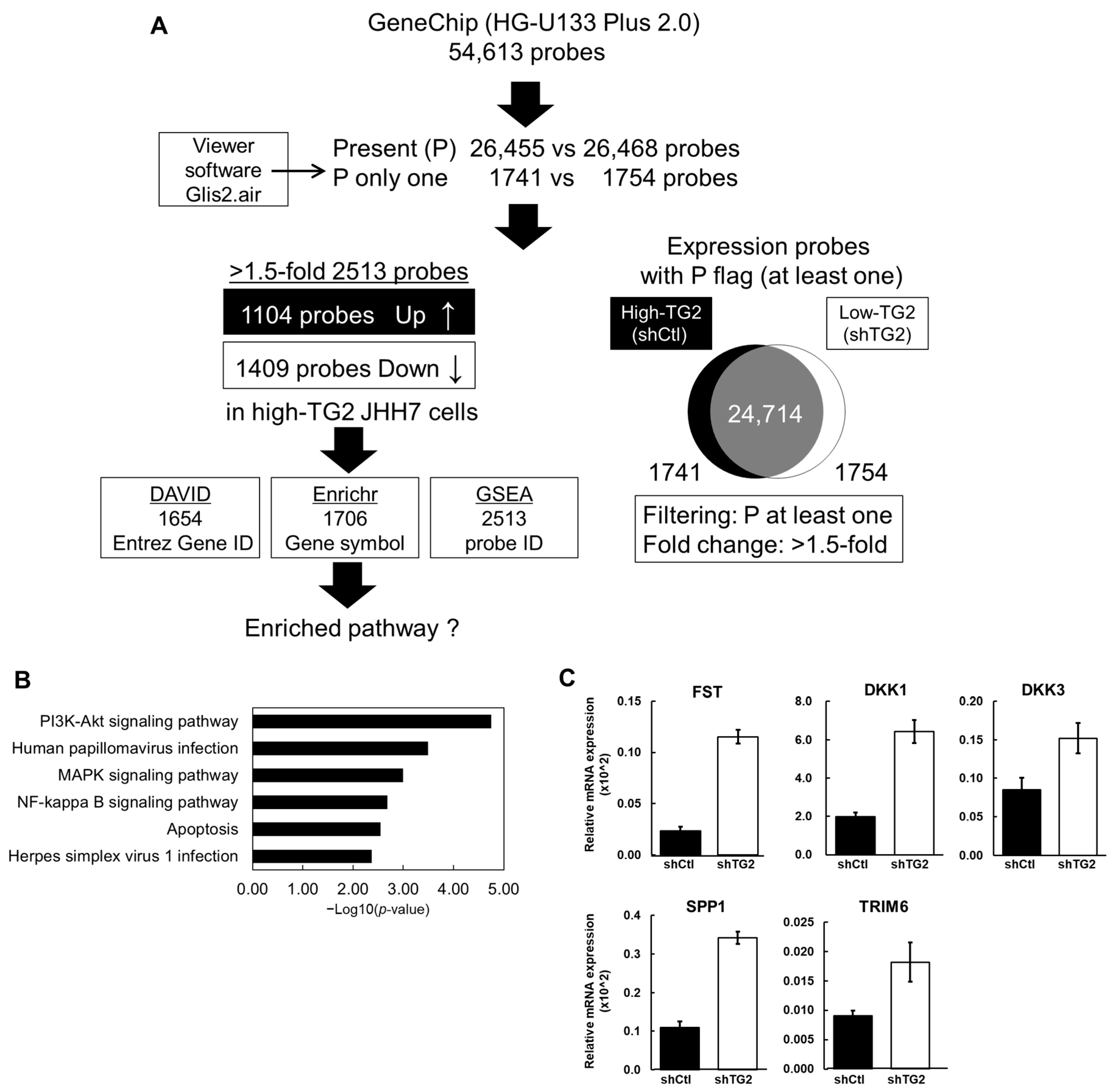

| Gene Name | Gene Symbol | GeneChip | Gene Functions Relevant to Cancer | Ref. | ||
|---|---|---|---|---|---|---|
| Fold Change | shCtl Signal | shTG2 Signal | ||||
| Follistatin | FST | 0.442 | 50.4 | 114.0 | Tumor progression through antagonism of TGF-β family members | [13] |
| Dickkopf WNT signaling pathway inhibitor 1 | DKK1 | 0.395 | 1313.3 | 3325.2 | Negative regulator of Wnt signaling targeting β-catenin/TCF pathway | [14,15] |
| Dickkopf WNT signaling pathway inhibitor 3 | DKK3 | 0.557 | 89.9 | 161.3 | HCC suppressor and Wnt modulator | [16,17] |
| Secreted phosphoprotein 1 | SPP1 | 0.252 | 64.0 | 254.5 | Tumor promoter through interaction with carcinogenic genes and facilitator of immune cell infiltration | [18] |
| Tripartite motif-containing 6 | TRIM6 | 0.258 | 72.7 | 281.4 | Potential prognostic marker for colorectal cancer and hepatocellular carcinoma | [19,20] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamaguchi, H.; González-Duarte, R.J.; Qin, X.-Y.; Abe, Y.; Takada, I.; Charroy, B.; Cázares-Ordoñez, V.; Uno, S.; Makishima, M.; Esumi, M. Transglutaminase 2 Stimulates Cell Proliferation and Modulates Transforming Growth Factor-Beta Signaling Pathway Independently of Epithelial–Mesenchymal Transition in Hepatocellular Carcinoma Cells. Int. J. Mol. Sci. 2025, 26, 5497. https://doi.org/10.3390/ijms26125497
Yamaguchi H, González-Duarte RJ, Qin X-Y, Abe Y, Takada I, Charroy B, Cázares-Ordoñez V, Uno S, Makishima M, Esumi M. Transglutaminase 2 Stimulates Cell Proliferation and Modulates Transforming Growth Factor-Beta Signaling Pathway Independently of Epithelial–Mesenchymal Transition in Hepatocellular Carcinoma Cells. International Journal of Molecular Sciences. 2025; 26(12):5497. https://doi.org/10.3390/ijms26125497
Chicago/Turabian StyleYamaguchi, Hiromi, Ramiro José González-Duarte, Xian-Yang Qin, Yuriko Abe, Ichiro Takada, Benjamin Charroy, Verna Cázares-Ordoñez, Shigeyuki Uno, Makoto Makishima, and Mariko Esumi. 2025. "Transglutaminase 2 Stimulates Cell Proliferation and Modulates Transforming Growth Factor-Beta Signaling Pathway Independently of Epithelial–Mesenchymal Transition in Hepatocellular Carcinoma Cells" International Journal of Molecular Sciences 26, no. 12: 5497. https://doi.org/10.3390/ijms26125497
APA StyleYamaguchi, H., González-Duarte, R. J., Qin, X.-Y., Abe, Y., Takada, I., Charroy, B., Cázares-Ordoñez, V., Uno, S., Makishima, M., & Esumi, M. (2025). Transglutaminase 2 Stimulates Cell Proliferation and Modulates Transforming Growth Factor-Beta Signaling Pathway Independently of Epithelial–Mesenchymal Transition in Hepatocellular Carcinoma Cells. International Journal of Molecular Sciences, 26(12), 5497. https://doi.org/10.3390/ijms26125497









