Transcriptomic Analysis of Cold-Induced Temporary Cysts in Marine Dinoflagellate Prorocentrum cordatum
Abstract
1. Introduction
2. Results
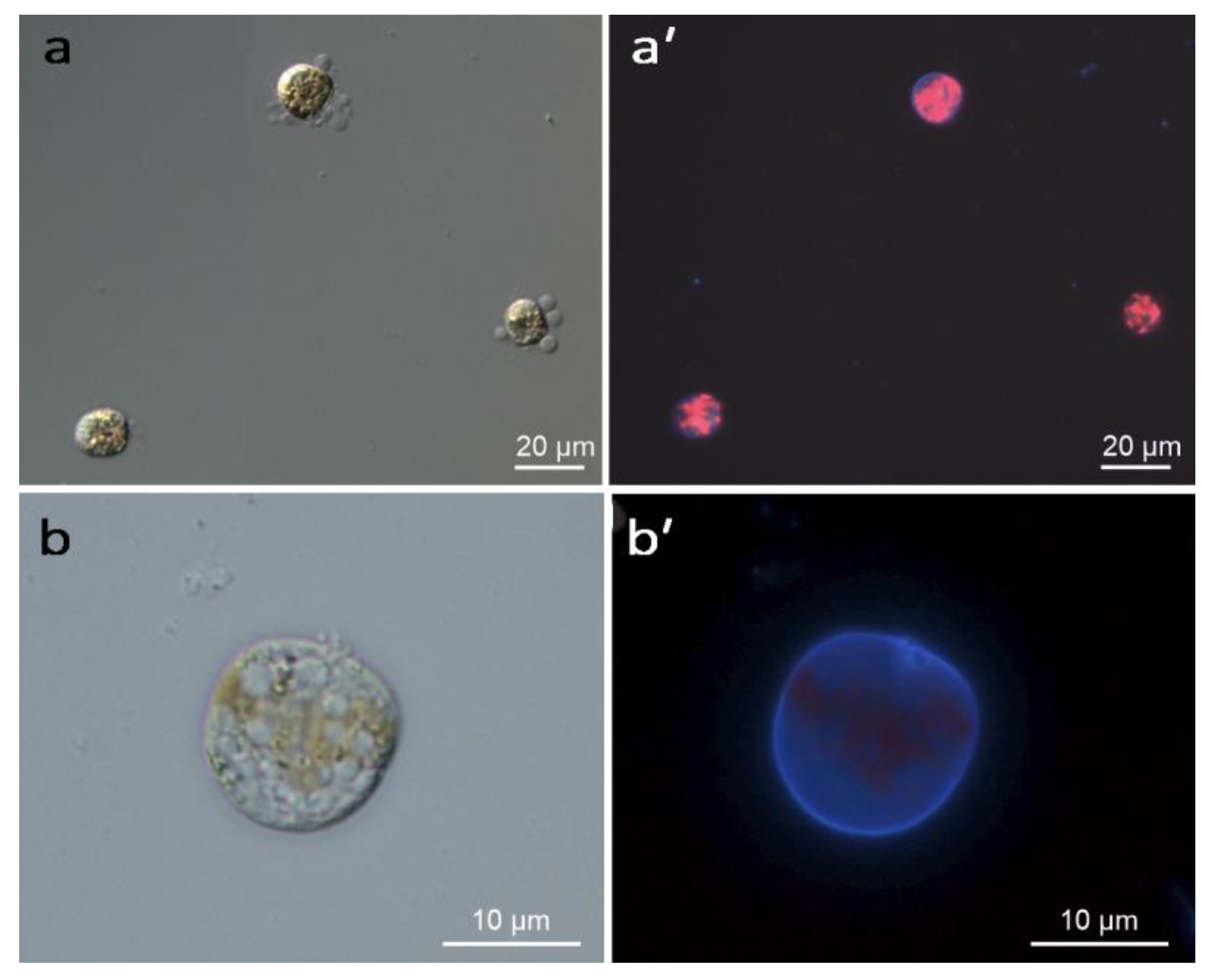
2.1. Induction of Encystment
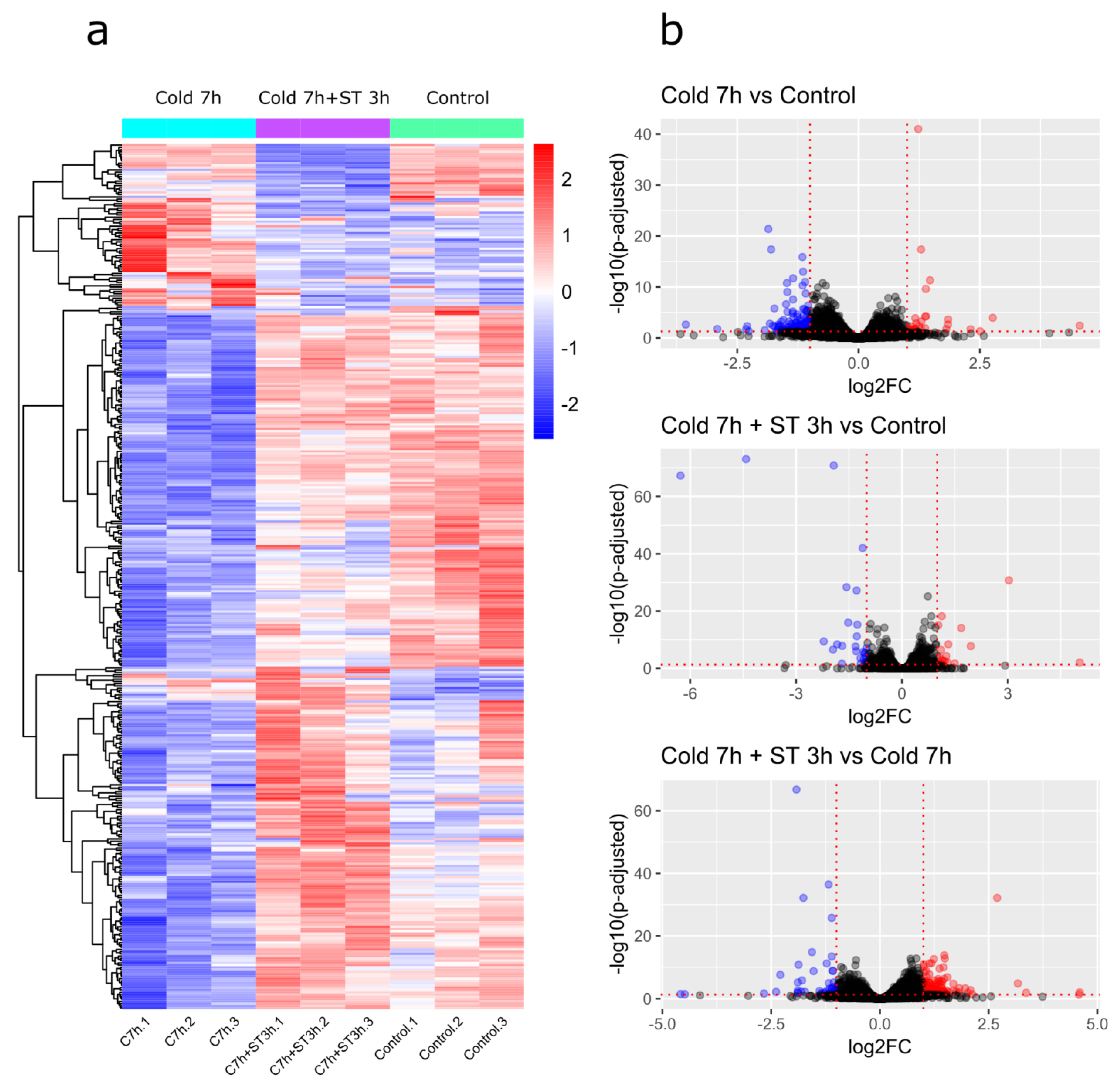
2.2. RNA Sequencing Results
2.3. Differential Gene Expression Analysis
2.3.1. Cold 7 h vs. Control
2.3.2. Cold 7 h + ST 3 h vs. Control
2.3.3. Cold 7 h + ST 3 h vs. Cold 7 h
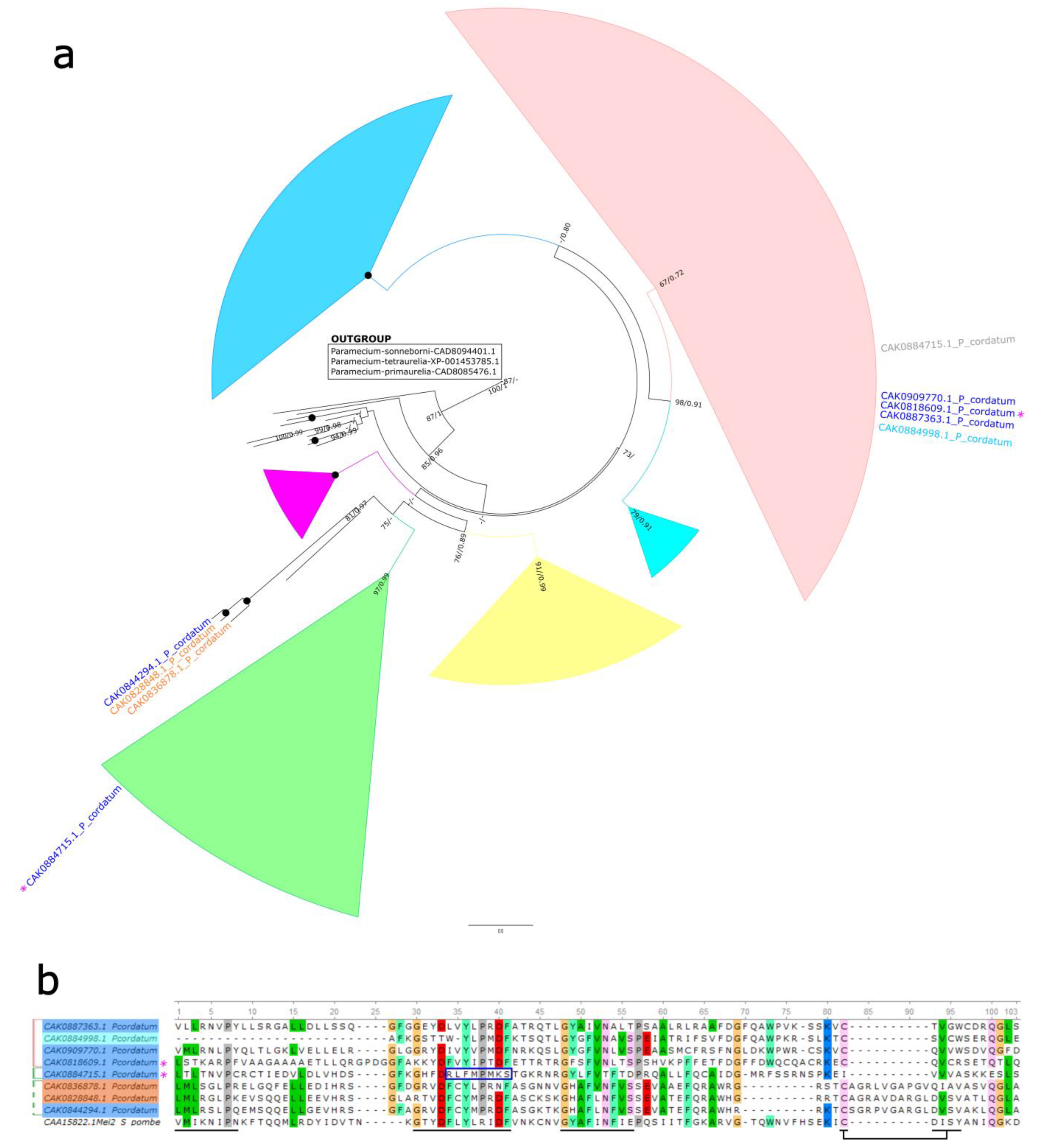
2.4. MEI-like Sequences Analysis
3. Discussion
4. Material and Methods
4.1. Cell Culture and Experimental Design
4.2. Light Microscopy
4.3. RNA Extraction and Sequencing
4.4. Sequence Assembly
4.5. Analysis of Gene Expression
4.6. Bioinformatic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anderson, D.M.; Cembella, A.D.; Hallegraeff, G.M. Progress in Understanding Harmful Algal Blooms: Paradigm Shifts and New Technologies for Research, Monitoring, and Management. Annu. Rev. Mar. Sci. 2012, 4, 143–176. [Google Scholar] [CrossRef]
- Figueroa, R.I.; Estrada, M.; Garcés, E. Life Histories of Microalgal Species Causing Harmful Blooms: Haploids, Diploids and the Relevance of Benthic Stages. Harmful Algae 2018, 73, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Yu, L.; Wu, X.; Li, M.; Zhang, Y.; Luo, H.; Li, H.; Li, T.; Li, L. Active Meiosis during Dinoflagellate Blooms: A ‘Sex for Proliferation’ Hypothesis. Harmful Algae 2022, 118, 102307. [Google Scholar] [CrossRef]
- Bravo, I.; Figueroa, R.I.; Garcés, E.; Fraga, S.; Massanet, A. The Intricacies of Dinoflagellate Pellicle Cysts: The Example of Alexandrium minutum Cysts from a Bloom-Recurrent Area (Bay of Baiona, NW Spain). Deep-Sea Res. II 2010, 57, 166–174. [Google Scholar] [CrossRef]
- Kremp, A.; Heiskanen, A.-S. Sexuality and Cyst Formation of the Spring-Bloom Dinoflagellate Scrippsiella hangoei in the Coastal Northern Baltic Sea. Mar. Biol. 1999, 134, 771–777. [Google Scholar] [CrossRef]
- Gong, W.; Browne, J.; Hall, N.; Schruth, D.; Paerl, H.; Marchetti, A. Molecular Insights into a Dinoflagellate Bloom. ISME J. 2017, 11, 439–452. [Google Scholar] [CrossRef]
- Perini, F.; Bastianini, M.; Capellacci, S.; Pugliese, L.; DiPoi, E.; Cabrini, M.; Buratti, S.; Marini, M.; Penna, A. Molecular Methods for Cost-Efficient Monitoring of HAB (Harmful Algal Bloom) Dinoflagellate Resting Cysts. Mar. Pollut. Bull. 2019, 147, 209–218. [Google Scholar] [CrossRef]
- Wells, M.L.; Karlson, B.; Wulff, A.; Kudela, R.; Trick, C.; Asnaghi, V.; Berdalet, E.; Cochlan, W.; Davidson, K.; De Rijcke, M.; et al. Future HAB Science: Directions and Challenges in a Changing Climate. Harmful Algae 2020, 91, 101632. [Google Scholar] [CrossRef]
- Bravo, I.; Figueroa, R. Towards an Ecological Understanding of Dinoflagellate Cyst Functions. Microorganisms 2014, 2, 11–32. [Google Scholar] [CrossRef]
- Bogus, K.; Mertens, K.N.; Lauwaert, J.; Harding, I.C.; Vrielinck, H.; Zonneveld, K.A.F.; Versteegh, G.J.M. Differences in the Chemical Composition of Organic-Walled Dinoflagellate Resting Cysts from Phototrophic and Heterotrophic Dinoflagellates. J. Phycol. 2014, 50, 254–266. [Google Scholar] [CrossRef]
- Sekida, S.; Horiguchi, T.; Okuda, K. Development of Thecal Plates and Pellicle in the Dinoflagellate Scrippsiella hexapraecingula (Peridiniales, Dinophyceae) Elucidated by Changes in Stainability of the Associated Membranes. Eur. J. Phycol. 2004, 39, 105–114. [Google Scholar] [CrossRef]
- Matantseva, O.; Berdieva, M.; Kalinina, V.; Pozdnyakov, I.; Pechkovskaya, S.; Skarlato, S. Stressor-Induced Ecdysis and Thecate Cyst Formation in the Armoured Dinoflagellates Prorocentrum cordatum. Sci. Rep. 2020, 10, 18322. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, Z.; Liu, L.; Li, Y. Transcriptome and Metabolome Analyses of Cold and Darkness-Induced Pellicle Cysts of Scrippsiella trochoidea. BMC Genom. 2021, 22, 526. [Google Scholar] [CrossRef]
- Roy, S.; Letourneau, L.; Morse, D. Cold-Induced Cysts of the Photosynthetic Dinoflagellate Lingulodinium polyedrum Have an Arrested Circadian Bioluminescence Rhythm and Lower Levels of Protein Phosphorylation. Plant Physiol. 2014, 164, 966–977. [Google Scholar] [CrossRef] [PubMed]
- Hardeland, R. Induction of Cyst Formation by Low Temperature in the Dinoflagellate Gonyaulax polyedra Stein: Dependence on Circadian Phase and Requirement of Light. Experientia 1994, 50, 60–62. [Google Scholar] [CrossRef]
- Grzebyk, D.; Berland, B. Influences of Temperature, Salinity and Irradiance on Growth of Prorocentrum minimum (Dinophyceae) from the Mediterranean Sea. J. Plankton Res. 1996, 18, 1837–1849. [Google Scholar] [CrossRef]
- Lundgren, V.; Granéli, E. Influence of Altered Light Conditions and Grazers on Scrippsiella trochoidea (Dinophyceae) Cyst formation. Aquat. Microb. Ecol. 2011, 63, 231–243. [Google Scholar] [CrossRef]
- Jauzein, C.; Erdner, D.L. Stress-Related Responses in Alexandrium tamarense Cells Exposed to Environmental Changes. J. Eukaryot. Microbiol. 2013, 60, 526–538. [Google Scholar] [CrossRef]
- Warns, A.; Hense, I.; Kremp, A. Modelling the Life Cycle of Dinoflagellates: A Case Study with Biecheleria baltica. J. Plankton Res. 2013, 35, 379–392. [Google Scholar] [CrossRef]
- Heil, C.A.; Glibert, P.M.; Fan, C. Prorocentrum minimum (Pavillard) Schiller: A Review of a Harmful Algal Bloom Species of Growing Worldwide Importance. Harmful Algae 2005, 4, 449–470. [Google Scholar] [CrossRef]
- Watanabe, Y.; Yamamoto, M.S. pombe Mei2+ Encodes an RNA-Binding Protein Essential for Premeiotic DNA Synthesis and Meiosis I, Which Cooperates with a Novel RNA Species meiRNA. Cell 1994, 78, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Sebastian, J.; Siddiqi, I. The Arabidopsis-Mei2-Like Genes Play a Role in Meiosis and Vegetative Growth in Arabidopsis. Plant Cell 2006, 18, 545–559. [Google Scholar] [CrossRef] [PubMed]
- Palii, O.; Safonov, P.; Berdieva, M. MEI2-like Proteins in Dinoflagellates: Diversity and Possible Functions in Life Cycle Transitions. Protistology 2024, 18, 249–262. [Google Scholar] [CrossRef]
- Deng, Y.; Hu, Z.; Shang, L.; Peng, Q.; Tang, Y.Z. Transcriptomic Analyses of Scrippsiella trochoidea Reveals Processes Regulating Encystment and Dormancy in the Life Cycle of a Dinoflagellate, with a Particular Attention to the Role of Abscisic Acid. Front. Microbiol. 2017, 8, 2450. [Google Scholar] [CrossRef] [PubMed]
- Dougan, K.E.; Deng, Z.-L.; Wöhlbrand, L.; Reuse, C.; Bunk, B.; Chen, Y.; Hartlich, J.; Hiller, K.; John, U.; Kalvelage, J.; et al. Multi-Omics Analysis Reveals the Molecular Response to Heat Stress in a “Red Tide” Dinoflagellate. Genome Biol. 2023, 24, 265. [Google Scholar] [CrossRef]
- Kang, H.C.; Jeong, H.J.; Park, S.A.; Ok, J.H.; You, J.H.; Eom, S.H.; Park, E.C.; Jang, S.H.; Lee, S.Y. Comparative Transcriptome Analysis of the Phototrophic Dinoflagellate Biecheleriopsis adriatica Grown Under Optimal Temperature and Cold and Heat Stress. Front. Mar. Sci. 2021, 8, 761095. [Google Scholar] [CrossRef]
- Zhang, H.; Gu, B.; Zhou, Y.; Ma, X.; Liu, T.; Xu, H.; Xie, Z.; Liu, K.; Wang, D.; Xia, X. Multi-Omics Profiling Reveals Resource Allocation and Acclimation Strategies to Temperature Changes in a Marine Dinoflagellate. Appl. Environ. Microbiol. 2022, 88, e01213-22. [Google Scholar] [CrossRef]
- Xu, J.; Wu, X.H.; Yang, Y.; Xu, S.; Kang, Y.; Fu, X.; Yue, H.; Shi, J.; Wu, Z. Changes in Growth, Photosynthesis and Chlorophyll Fluorescence in the Freshwater Dinoflagellate Peridinium umbonatum (Peridiniales, Pyrrophyta) in Response to Different Temperatures. Phycologia 2016, 55, 469–477. [Google Scholar] [CrossRef]
- Kim, D.-I.; Matsuyama, Y.; Nagasoe, S.; Yamaguchi, M.; Yoon, Y.-H.; Oshima, Y.; Imada, N.; Honjo, T. Effects of Temperature, Salinity and Irradiance on the Growth of the Harmful Red Tide Dinoflagellate Cochlodinium polykrikoides Margalef (Dinophyceae). J. Plankton Res. 2004, 26, 61–66. [Google Scholar] [CrossRef]
- Tuteja, N.; Sopory, S.K. Plant Signaling in Stress. Plant Signal. Behav. 2008, 3, 79–86. [Google Scholar] [CrossRef]
- Barkan, A. Expression of Plastid Genes: Organelle-Specific Elaborations on a Prokaryotic Scaffold. Plant Physiol. 2011, 155, 1520–1532. [Google Scholar] [CrossRef]
- Berry, J.O.; Yerramsetty, P.; Zielinski, A.M.; Mure, C.M. Photosynthetic Gene Expression in Higher Plants. Photosynth. Res. 2013, 117, 91–120. [Google Scholar] [CrossRef] [PubMed]
- Johnson, X.; Wostrikoff, K.; Finazzi, G.; Kuras, R.; Schwarz, C.; Bujaldon, S.; Nickelsen, J.; Stern, D.B.; Wollman, F.-A.; Vallon, O. MRL1, a Conserved Pentatricopeptide Repeat Protein, Is Required for Stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell 2010, 22, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Tourasse, N.J.; Choquet, Y.; Vallon, O. PPR Proteins of Green Algae. RNA Biol. 2013, 10, 1526–1542. [Google Scholar] [CrossRef]
- Keeling, P.J. The Endosymbiotic Origin, Diversification and Fate of Plastids. Philos. Trans. R. Soc. B 2010, 365, 729–748. [Google Scholar] [CrossRef] [PubMed]
- Manna, S. An Overview of Pentatricopeptide Repeat Proteins and Their Applications. Biochimie 2015, 113, 93–99. [Google Scholar] [CrossRef]
- Wang, X.; An, Y.; Xu, P.; Xiao, J. Functioning of PPR Proteins in Organelle RNA Metabolism and Chloroplast Biogenesis. Front. Plant Sci. 2021, 12, 627501. [Google Scholar] [CrossRef]
- Sasaki, K.; Imai, R. Pleiotropic Roles of Cold Shock Domain Proteins in Plants. Front. Plant Sci. 2012, 2, 116. [Google Scholar] [CrossRef]
- Wang, H.; Kim, H.; Ki, J.S. Transcriptomic identification and expression analysis of cold shock domain protein (CSP) genes in the marine dinoflagellate Prorocentrum minimum. J. Appl. Phycol. 2021, 33, 843–854. [Google Scholar] [CrossRef]
- Anderson, G.H.; Hanson, M.R. The Arabidopsis Mei2 Homologue AML1 Binds AtRaptor1B, the Plant Homologue of a Major Regulator of Eukaryotic Cell Growth. BMC Plant Biol. 2005, 5, 2. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Phillips, M.J.; Moore, S.; Veit, B. A Description of the Mei2-like Protein Family; Structure, Phylogenetic Distribution and Biological Context. Dev. Genes Evol. 2004, 214, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Dankwa, D.A.; Davis, M.J.; Kappe, S.H.I.; Vaughan, A.M. A Plasmodium yoelii Mei2-Like RNA Binding Protein Is Essential for Completion of Liver Stage Schizogony. Infect. Immun. 2016, 84, 1336–1345. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Cheng, S.; Song, B.; Zhong, X.; Lin, X.; Li, W.; Li, L.; Zhang, Y.; Zhang, H.; Ji, Z.; et al. The Symbiodinium kawagutii Genome Illuminates Dinoflagellate Gene Expression and Coral Symbiosis. Science 2015, 350, 691–694. [Google Scholar] [CrossRef]
- Gong, S.; Li, G.; Jin, X.; Qiu, D.; Liang, J.; Yu, K.; Tan, Y.; Ma, X.; Xia, X. Photobleaching and Recovery of Symbiodiniaceae Effrenium voratum SCS01 Reveals Life form Transformation Under Thermal Stress. Front. Mar. Sci. 2021, 8, 740416. [Google Scholar] [CrossRef]
- Luo, H.; Wang, J.; Goes, J.I.; Gomes, H.d.R.; Al-Hashmi, K.; Tobias, C.; Koerting, C.; Lin, S. A Grazing-Driven Positive Nutrient Feedback Loop and Active Sexual Reproduction Underpin Widespread Noctiluca Green Tides. ISME Commun. 2022, 2, 103. [Google Scholar] [CrossRef]
- Akbar, M.A.; Mohd Yusof, N.Y.; Usup, G.; Ahmad, A.; Baharum, S.N.; Bunawan, H. Nutrient Deficiencies Impact on the Cellular and Metabolic Responses of Saxitoxin Producing Alexandrium minutum: A Transcriptomic Perspective. Mar. Drugs 2023, 21, 497. [Google Scholar] [CrossRef]
- Guillard, R.R.L.; Ryther, J.H. Studies of Marine Planktonic Diatoms: I. Cyclotella nana Hustedt, and Detonula confervacea (Cleve) Gran. Can. J. Microbiol. 1962, 8, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Krishan, A. Rapid Flow Cytofluorometric Analysis of Mammalian Cell Cycle by Propidium Iodide Staining. J. Cell Biol. 1975, 66, 188–193. [Google Scholar] [CrossRef]
- Saldarriaga, J.F.; ‘Max’ Taylor, F.J.R. Dinoflagellata. In Handbook of the Protists; Archibald, J.M., Simpson, A.G.B., Slamovits, C.H., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 625–678. ISBN 978-3-319-28147-6. [Google Scholar]
- Li, Z.; Trick, H.N. Rapid Method for High-Quality RNA Isolation from Seed Endosperm Containing High Levels of Starch. Biotechniques 2005, 38, 872–876. [Google Scholar] [CrossRef]
- Mundt, F.; Heinrich, S.; Hanelt, D. RNA Isolation from Taxonomically Diverse Photosynthetic Protists. Limnol. Oceanogr. Methods 2019, 17, 190–199. [Google Scholar] [CrossRef]
- Dutta, S.; Muthusamy, V.; Chhabra, R.; Zunjare, R.U.; Hossain, F. Two-Step Method for Isolation of High-Quality RNA from Stored Seeds of Maize Rich in Starch. 3 Biotech 2020, 10, 433. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.-J.; Pendi, F.H.; Hussain, H. Improved CTAB Method for RNA Extraction of Thick Waxy Leaf Tissues from Sago Palm (Metroxylon sagu Rottb.). Chem. Biol. Technol. Agric. 2022, 9, 63. [Google Scholar] [CrossRef]
- Jensen, T.; Saleh, L.; Bents, D.; Krohn, S.; Wu, Y.-C.; Mucke, M.; Boje, A.S.; Veltel, S.; Hennig, S.; Piker, L.; et al. Optimised Protocols for RNA Extraction from a Broad Taxonomic Range of Algae. J. Appl. Phycol. 2023, 35, 1743–1753. [Google Scholar] [CrossRef]
- Li, J.; Tang, C.; Song, C.; Chen, M.; Feng, Z.; Pan, Y. A Simple, Rapid and Effective Method for Total RNA Extraction from Lentinula edodes. Biotechnol. Lett. 2006, 28, 1193–1197. [Google Scholar] [CrossRef]
- Jiang, H.; Lei, R.; Ding, S.-W.; Zhu, S. Skewer: A Fast and Accurate Adapter Trimmer for next-Generation Sequencing Paired-End Reads. BMC Bioinform. 2014, 15, 182. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 February 2025).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024. [Google Scholar]
- BioBam. Bioinformatics OmicsBox—Bioinformatics Made Easy Version 3.4.5; BioBam: Valencia, Spain, 2019. [Google Scholar]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.-P.; Mi, H. PANTHER: Making Genome-Scale Phylogenetics Accessible to All. Prot. Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Warwick Vesztrocy, A.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python Library for Gene Ontology Analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Use R!; Springer: Cham, Switzerland, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Kolde, R. Pheatmap: Pretty Heatmaps; R Package Version 1.0.12; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Keeling, P.J.; Burki, F.; Wilcox, H.M.; Allam, B.; Allen, E.E.; Amaral-Zettler, L.A.; Armbrust, E.V.; Archibald, J.M.; Bharti, A.K.; Bell, C.J.; et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the Functional Diversity of Eukaryotic Life in the Oceans through Transcriptome Sequencing. PLoS Biol. 2014, 12, e1001889. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A Unified Bioinformatics Toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berdieva, M.; Safonov, P.; Palii, O.; Prilutsky, M.; Matantseva, O.; Skarlato, S. Transcriptomic Analysis of Cold-Induced Temporary Cysts in Marine Dinoflagellate Prorocentrum cordatum. Int. J. Mol. Sci. 2025, 26, 5432. https://doi.org/10.3390/ijms26125432
Berdieva M, Safonov P, Palii O, Prilutsky M, Matantseva O, Skarlato S. Transcriptomic Analysis of Cold-Induced Temporary Cysts in Marine Dinoflagellate Prorocentrum cordatum. International Journal of Molecular Sciences. 2025; 26(12):5432. https://doi.org/10.3390/ijms26125432
Chicago/Turabian StyleBerdieva, Mariia, Pavel Safonov, Olga Palii, Mikhail Prilutsky, Olga Matantseva, and Sergei Skarlato. 2025. "Transcriptomic Analysis of Cold-Induced Temporary Cysts in Marine Dinoflagellate Prorocentrum cordatum" International Journal of Molecular Sciences 26, no. 12: 5432. https://doi.org/10.3390/ijms26125432
APA StyleBerdieva, M., Safonov, P., Palii, O., Prilutsky, M., Matantseva, O., & Skarlato, S. (2025). Transcriptomic Analysis of Cold-Induced Temporary Cysts in Marine Dinoflagellate Prorocentrum cordatum. International Journal of Molecular Sciences, 26(12), 5432. https://doi.org/10.3390/ijms26125432




