1. Introduction
Rice (
Oryza sativa), serving as the staple food for more than half of the global population, plays a pivotal role in ensuring food security through yield stability. Although substantial efforts have been made and notable progress achieved in enhancing rice productivity, agricultural production remains severely threatened by adverse climatic conditions, with drought being the most prominent stressor [
1,
2,
3]. Climate change has led to increased frequency and intensity of drought events, posing significant challenges to rice cultivation. As a semi-aquatic crop particularly sensitive to water deficit, rice exhibits high vulnerability to drought stress, which has emerged as one of the predominant abiotic constraints limiting global rice production. The plant’s response to drought involves a complex regulatory network encompassing multifaceted physiological adaptations, biochemical adjustments, and molecular reprogramming [
4,
5,
6]. Therefore, elucidating the mechanistic basis of drought response and adaptive strategies in rice plants holds great promise for developing high drought-resistant cultivars, which is crucial for sustainable agricultural production and global food security under changing climate scenarios.
NAC (NAM, ATAF1/2, and CUC2) transcription factors (TFs) have emerged as central regulatory players in plant stress responses [
7]. Characterized by a conserved N-terminal DNA-binding domain and a highly divergent C-terminal transcriptional activation domain, this protein family exhibits distinctive structural features [
8]. Studies have demonstrated the pivotal roles of NAC TFs in developmental regulation, while accumulating evidence confirms their integral involvement in plant responses to abiotic stresses [
9]. In the context of drought resistance mechanisms in rice (
Oryza sativa), NAC TFs orchestrate a multifaceted regulatory network [
10,
11,
12]. Emerging evidence reveals distinct molecular strategies employed by specific NAC members to enhance drought tolerance: SNAC1 modulates stomatal movement to reduce water loss;
OsNAC066 activates antioxidant systems to alleviate oxidative damage;
OsXND1 and
OsNAC17 improve hydraulic conductivity through xylem development regulation and lignin biosynthesis, respectively; while
OsNAC15 coordinates stress signaling via ABA pathway mediation [
13,
14,
15,
16,
17]. Notably, the drought-responsive functions of
OsNAC67,
OsNAC45,
OsNAC006,
OsNAC14,
OsNAC95, and
OsNAC22 have been systematically documented, collectively establishing this TF family as a master regulator in plant stress adaptation [
16,
18,
19,
20,
21,
22,
23].
As a key member of the NAC transcription factor family,
OsNAC25 has been reported to regulate potassium deficiency responses and grain development. Furthermore, studies have demonstrated its role in coordinating starch biosynthesis homeostasis through physical interactions with
NAC20/26 proteins [
24,
25]. However, its potential involvement in drought resistance remained unexplored. In this investigation, the functional characterization of knockout and overexpression lines revealed that
OsNAC25 exhibits drought-inducible expression patterns and confers enhanced drought tolerance in rice (
Oryza sativa). Transgenic plants overexpressing
OsNAC25 demonstrated significantly improved survival rates under extreme drought conditions during both vegetative and reproductive stages. Transcriptomic profiling further identified enriched pathways associated with drought resistance mechanisms. Intriguingly, unlike most nuclear-localized NAC TFs,
OsNAC25 displays dual localization in both the cytoplasm and nucleus. This unique subcellular distribution suggests a potential novel post-translational regulatory mechanism operating under drought stress conditions.
2. Results
2.1. OsNAC25 Is a Drought-Induced Transcription Factor with High Expression
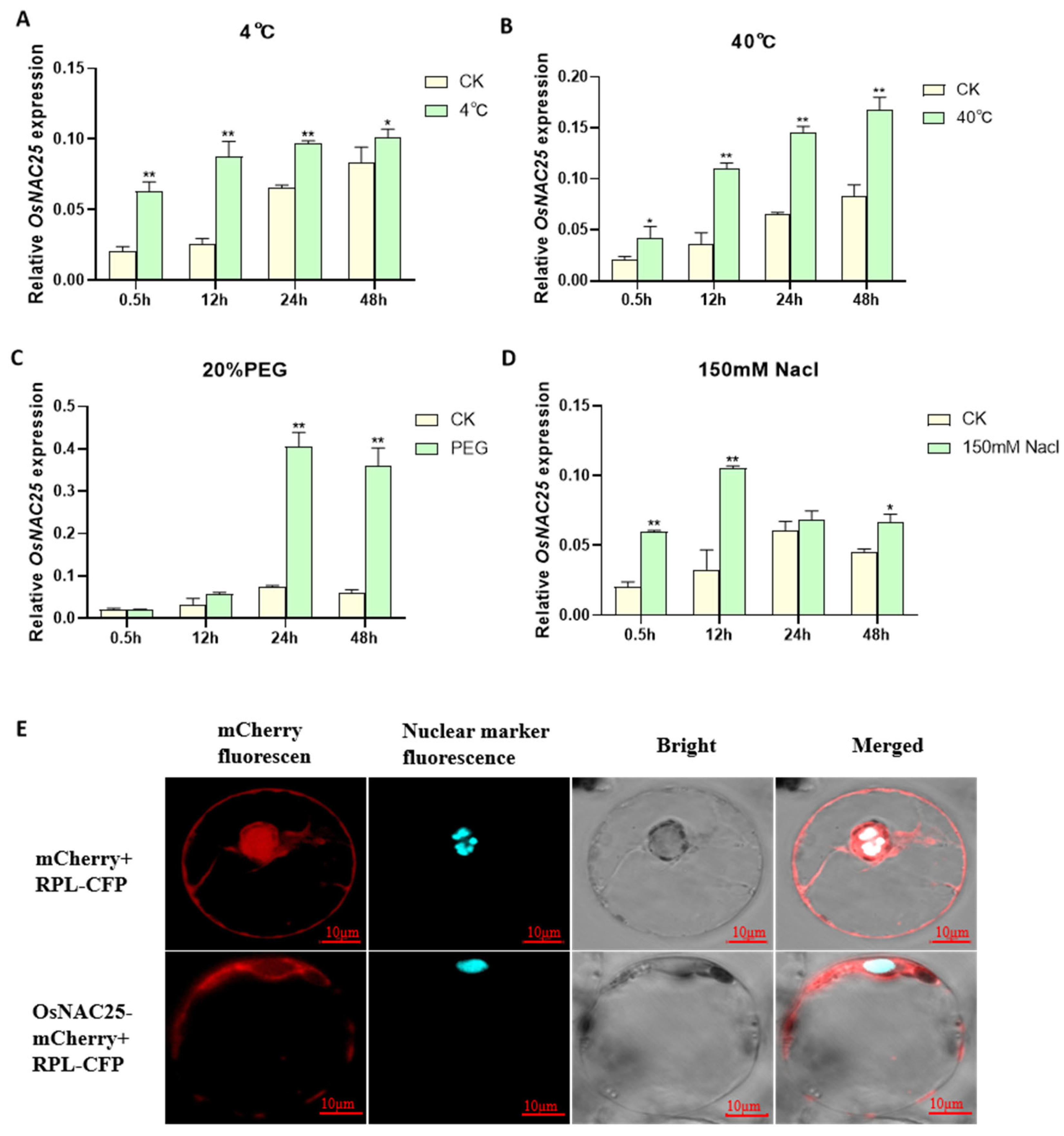
To elucidate the spatiotemporal expression dynamics of
OsNAC25 under abiotic stress, we systematically analyzed its transcriptional responses in rice seedlings exposed to low temperature (4 °C), high temperature (40 °C), salt stress (150 mM NaCl), and simulated drought (20% PEG-6000) over a 48-hour period using qRT-PCR. The results demonstrate distinct temporal and spatial heterogeneity in
OsNAC25 regulation across different stress conditions (
Figure 1). During the initial 12 h, low temperature, high temperature, and salt stress rapidly induced a significant upregulation of
OsNAC25, with expression levels increasing by 3.4-fold (4 °C), 3.1-fold (40 °C), and 3.2-fold (NaCl) compared to the control (CK) (
Figure 1A,B,D). In contrast, the PEG treatment showed no significant induction during this early phase (
Figure 1C). Beyond 12 h, the regulatory patterns diverged markedly: both low-temperature and salt stress exhibited diminishing effects, with the low-temperature group showing only a 1.2-fold difference by 48 h and the salt-treated group reverting to CK levels by 24 h. While high-temperature stress maintained a 2.0-fold elevation at 48 h, the induction magnitude decreased by 34% compared to earlier peaks. Strikingly, PEG-induced drought triggered a unique sustained response, with
OsNAC25 expression surging sharply to 5.4-fold at 24 h and further increasing to 5.6-fold by 48 h, without the decline observed under other stresses. Collectively,
OsNAC25 is responsive to multiple abiotic stresses but displays a delayed, high-amplitude, and persistent induction specifically under drought-like conditions, contrasting with the transient responses to temperature or salt stress. These findings strongly implicate
OsNAC25 as a drought-inducible transcription factor potentially central to rice drought adaptation mechanisms.
To clarify the subcellular localization of
OsNAC25, we constructed an
OsNAC25-mCherry fusion expression vector and performed co-localization analysis using a rice protoplast transient expression system. The co-transfection of
OsNAC25-mCherry with the nuclear marker RPL-CFP, alongside a negative control (empty mCherry vector co-transfected with RPL-CFP), revealed distinct localization patterns. Confocal microscopy showed that RPL-CFP signals were predominantly localized to the nucleus, while
OsNAC25-mCherry fluorescence was distributed in both the nucleus and cytoplasm, with significant co-localization observed between
OsNAC25-mCherry and RPL-CFP nuclear signals (
Figure 1E). These results demonstrate that
OsNAC25 is a nucleo-cytoplasmic transcription factor, suggesting its dual regulatory role: transcriptional regulation in the nucleus and potential involvement in cytoplasmic stress-responsive signaling pathways.
2.2. Overexpression of OsNAC25 Enhances Drought Tolerance During the Vegetative Growth Stage in Rice
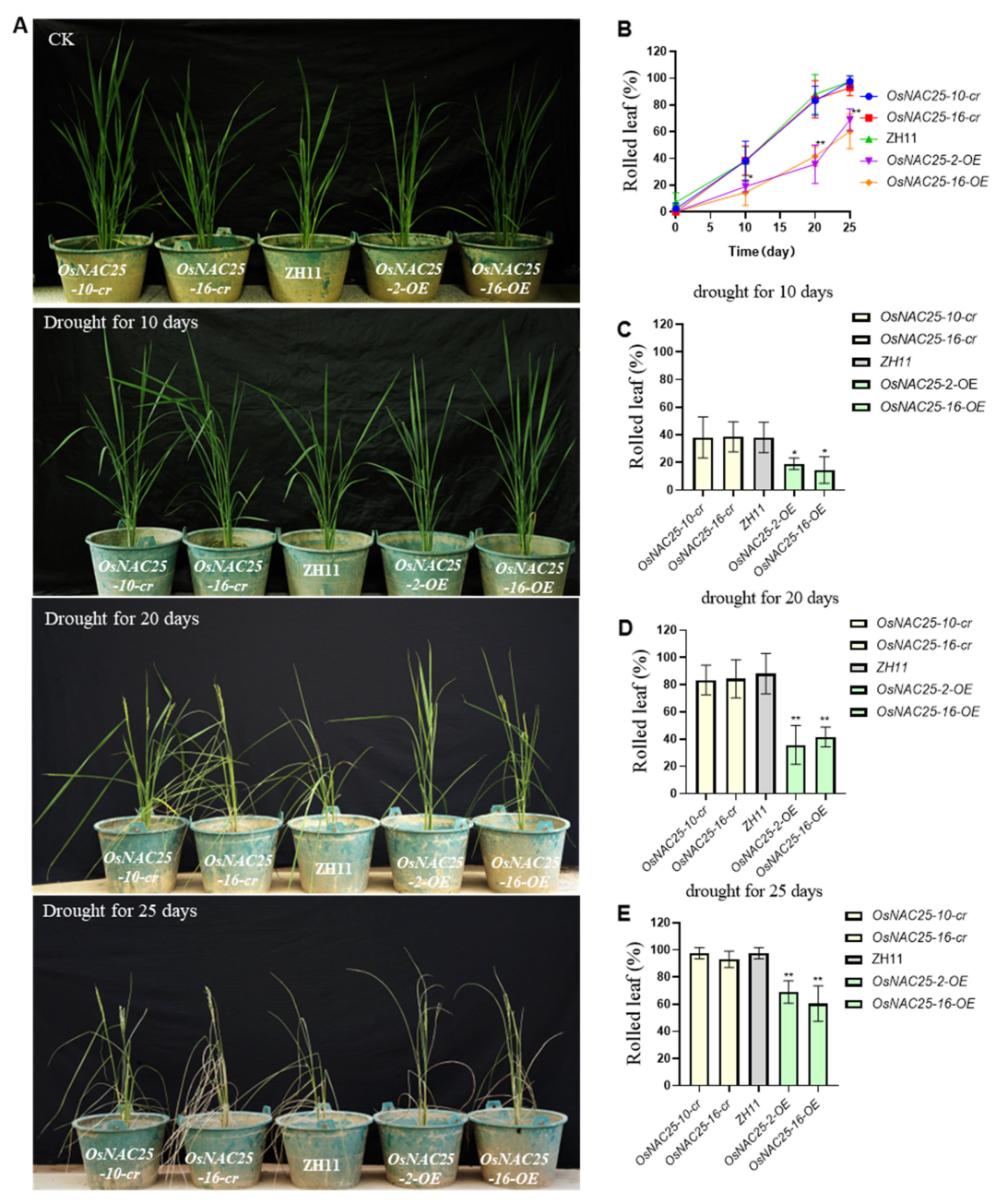
Based on qRT-PCR results indicating that OsNAC25 may regulate drought-responsive seedling development, we investigated its functional role in drought tolerance by subjecting two-leaf-one-heart stage seedlings of OsNAC25 overexpression lines (OsNAC25-OE), CRISPR/Cas9-generated knockout mutants (OsNAC25-cr), and ZH11plants to severe drought stress treatment.
Consequently, the seedlings at the two-leaf-one-heart stage were subjected to severe drought stress by withholding water. Within 12 h, visible symptoms of wilting and chlorosis emerged in leaves, and by 24 h, pronounced phenotypic divergence became evident: nearly all
OsNAC25-cr mutants and ZH11 plants exhibited severe wilting, whereas the
OsNAC25-OE lines remained upright (
Figure 2A). Following 24 h of drought treatment, the plants were re-watered and allowed to recover for one week. Survival rates were quantified, revealing that
OsNAC25-OE lines displayed significantly higher survival (86%) compared to
OsNAC25-cr (9.5%) and ZH11 (5.8%) plants (
Figure 2B). In summary, the overexpression of
OsNAC25 substantially enhances drought tolerance in rice.
Antioxidant enzymes play a critical role in scavenging reactive oxygen species (ROS) and enhancing plant stress tolerance. To investigate the role of OsNAC25 in mitigating oxidative stress under drought conditions, we analyzed the activities of key antioxidant enzymes—including catalase (CAT), peroxidase (POD), and superoxide dismutase (SOD)—as well as the malondialdehyde (MDA) content in the roots and leaves of ZH11, OsNAC25 mutants (OsNAC25-cr), and OsNAC25 overexpression lines (OsNAC25-OE).
Under drought stress,
OsNAC25-OE lines exhibited significantly higher CAT and POD activities compared to the ZH11 and
OsNAC25-cr mutants (
Figure 2D,E). CAT catalyzes the decomposition of hydrogen peroxide (H
2O
2) into water and oxygen. In
OsNAC25-OE plants, CAT activity increased by 1.5-fold in leaves and 778.1-fold in roots relative to ZH11. Similarly, POD, which scavenges peroxides, showed a 1.16-fold increase in leaf activity in the
OsNAC25-OE lines compared to ZH11, though root POD activity showed no statistically significant enhancement. Notably, SOD levels in
OsNAC25-OE lines were lower than those in the ZH11 and
OsNAC25-cr mutants (
Figure 2F), suggesting that
OsNAC25 may regulate antioxidant pathways through compensatory mechanisms or differential modulation. Despite reduced SOD levels,
OsNAC25-OE lines displayed a significantly lower MDA content than the ZH11 and
OsNAC25-cr plants, with leaf MDA levels reduced to 0.39-fold and root MDA to 0.73-fold of ZH11 values (
Figure 2C), indicating attenuated oxidative damage overall.
Therefore, OsNAC25 overexpression mitigates the toxic effects of reactive oxygen species (ROS) by enhancing antioxidant enzyme activities, thereby reducing oxidative damage in plant cells.
2.3. Overexpression of OsNAC25 Enhances Drought Tolerance During the Reproductive Growth Stage in Rice
OsNAC25 enhances drought tolerance during both the seedling and mature stages. To further investigate the role of
OsNAC25 in mature plants, we subjected
OsNAC25-OE,
OsNAC25-cr, and ZH11 plants to 25 days of drought stress starting at the post-booting stage and analyzed their phenotypes (
Figure 3A). Leaf rolling rates were quantified, revealing that
OsNAC25-OE lines exhibited a significantly lower rate (64.7%) compared to the
OsNAC25-cr mutants (95.3%) and ZH11 (97.6%) (
p < 0.05; three independent biological replicates). These results indicate that
OsNAC25-OE plants retain turgid leaves for a markedly extended duration under prolonged drought stress. Collectively, our findings demonstrate that the
OsNAC25 transcription factor functions in both seedling and mature stages to significantly enhance drought tolerance in rice.
2.4. Gene-Editing Knockout and Overexpression of OsNAC25 Alter the Rice Transcriptome Profile
To elucidate the regulatory network of
OsNAC25 under drought stress, RNA sequencing (RNA-seq) was performed on aerial tissues of 10-day-old seedlings from
OsNAC25-OE lines,
OsNAC25-cr mutants, and ZH11 (wild type) under both drought and control conditions. Sequencing was conducted using the Qsep400 high-throughput platform, generating 110.48 Gb of clean data, with each sample yielding ≥5.75 Gb of high-quality reads (Q30 base percentage ≥93.07%). Clean reads were aligned to the reference genome, achieving mapping efficiencies ranging from 93.69% to 98.50% (
Supplementary Table S1). Subsequent analyses included alternative splicing prediction, gene structure optimization, and novel gene discovery, identifying 3737 novel genes, of which 2483 were functionally annotated.
To validate RNA-seq results, eight differentially expressed genes (DEGs) were randomly selected from
OsNAC25-OE,
OsNAC25-cr, and ZH11 under drought and control conditions for qRT-PCR analysis. The expression levels of all eight genes were consistent with RNA-seq data (
Supplementary Figure S1), confirming the reliability of our transcriptomic findings.
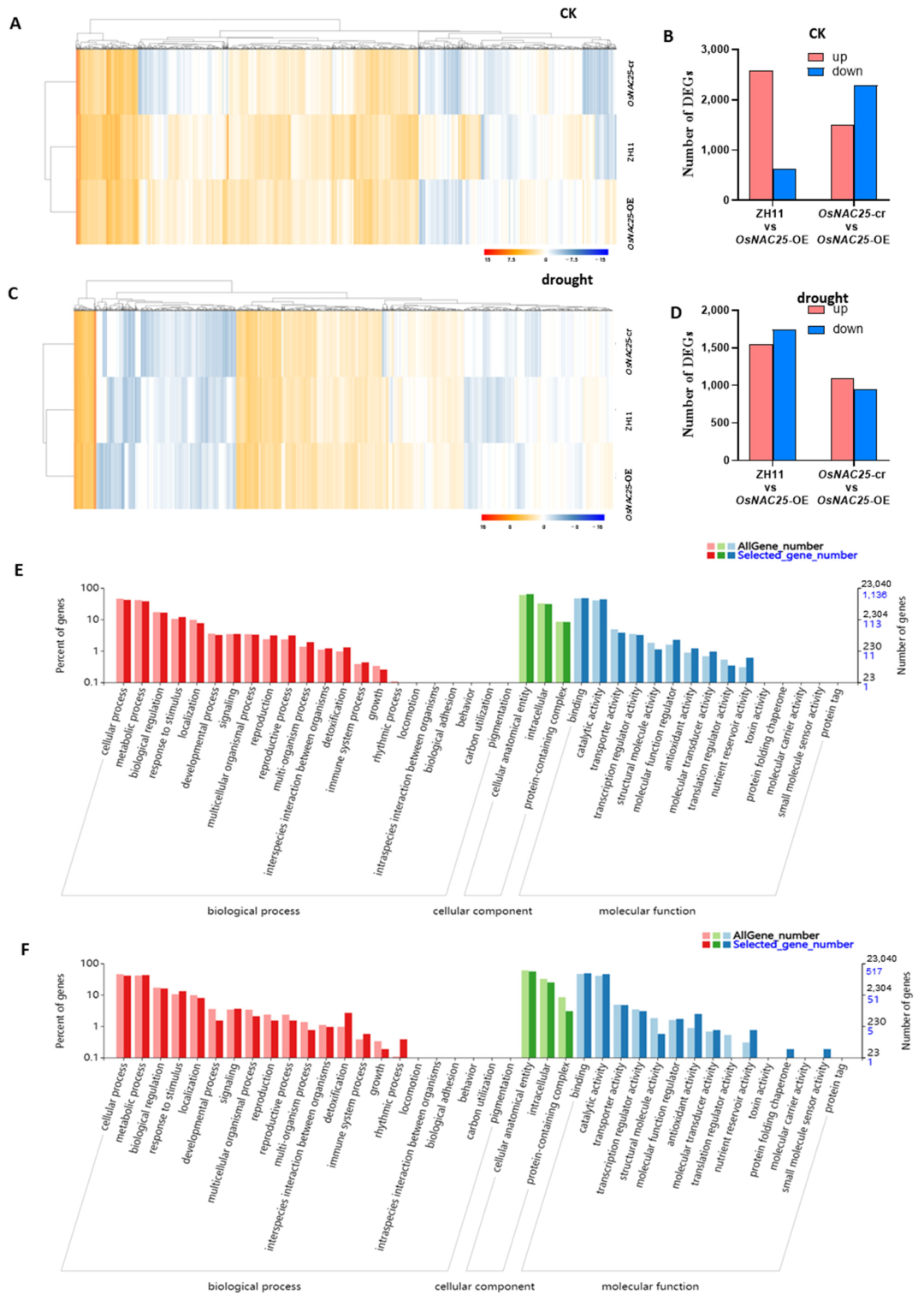
The RNA-seq results demonstrate significant alterations in gene expression under both experimental conditions (
Figure 4A,C). Under normal conditions, 2570 genes were upregulated and 615 genes downregulated in the
OsNAC25-OE lines compared to the wild-type ZH11, while 1506 genes were upregulated and 2285 genes downregulated relative to
OsNAC25-cr mutants (
Figure 4B). Following drought stress,
OsNAC25-OE exhibited 1548 upregulated and 1743 downregulated genes versus ZH11, and 1089 upregulated and 942 downregulated genes compared to the
OsNAC25-cr lines (
Figure 4D).
The Venn analysis identified 1601 constitutively differentially expressed genes (DEGs) shared between
OsNAC25-overexpressing (
OsNAC25-OE) transgenic lines and control groups (ZH11 wild-type +
OsNAC25-CRISPR) under well-watered conditions (
Supplemental Figure S2), indicating putative basal regulatory functions of
OsNAC25 before drought exposure. Post-drought comparative transcriptomics revealed 787 overlapping DEGs consistently regulated between
OsNAC25-OE and control genotypes (
Supplemental Figure S2).
The gene ontology (GO) enrichment analysis was performed on the 1601 (pre-drought) and 787 (post-drought) differentially expressed genes (DEGs) to identify associated biological processes (
Figure 4E,F). DEGs related to biological processes were primarily associated with cellular processes, metabolic processes, biological regulation, and response to stimuli. Cellular component-associated DEGs were predominantly linked to cellular anatomical entities and intracellular organelle functions. Molecular function-enriched DEGs mainly involved binding and catalytic activity. Among these, cellular/metabolic processes, biological regulation, response to stimuli, cellular anatomy/intracellular components, binding, and catalytic activity exhibited the most significant differential expression (
Figure 4E,F).
2.5. OsNAC25 Mediates Transcriptional Responses Under Drought Stress
The GO enrichment analysis identified 29 significantly enriched terms between
OsNAC25-cr mutants and
OsNAC25-OE lines. A further investigation of DEG biological functions via KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment highlighted the most impacted pathways: phenylpropanoid biosynthesis, cyanogenic amino acid metabolism, taurine and hypotaurine metabolism, plant hormone signal transduction, brassinosteroid biosynthesis, peroxisome activity, circadian rhythm in plants, carbon metabolism, carbon fixation in photosynthetic organisms, diterpenoid biosynthesis, and starch and sucrose metabolism (
Supplementary Figures S3–S6).
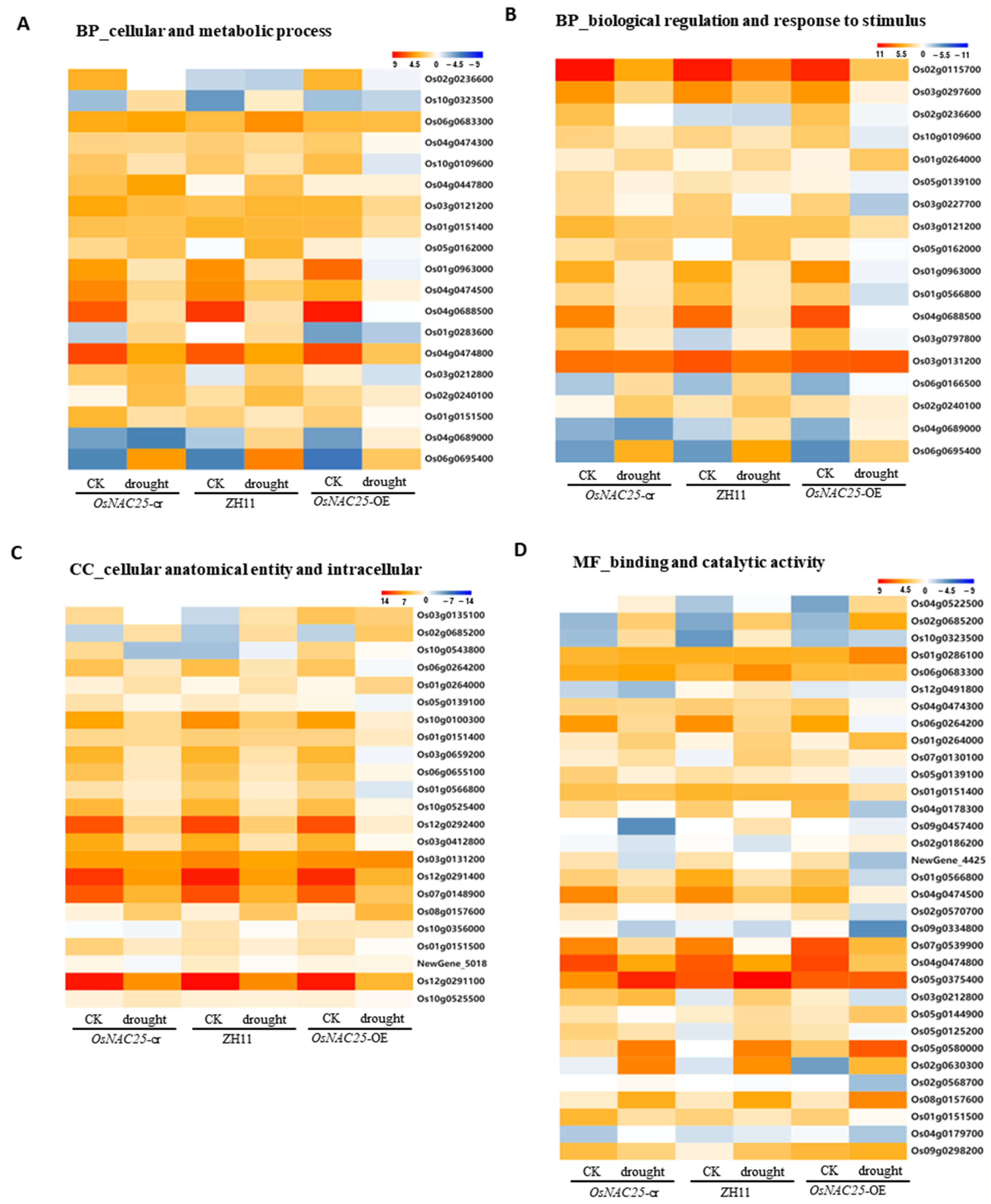
The KEGG pathway analysis identified eight prioritized pathways for further investigation, with the heatmap analysis focusing on key genes associated with cellular/metabolic processes, biological regulation, response to stimuli, cellular anatomy/intracellular components, binding, and catalytic activity. Among these, phenylpropanoid biosynthesis and taurine/hypotaurine metabolism emerged as the most significantly impacted pathways.
Key genes in phenylpropanoid biosynthesis included peroxidase
OsPrx30 (
Os02g0240100) and β-glucosidase
Os3BGlu6 (
Os03g0212800), both linked to drought resistance, while glutamate decarboxylase
OsGAD2 (
Os04g0447800) was central to taurine/hypotaurine metabolism (
Figure 5A). Plant hormone signaling transduction pathways exhibited marked transcriptional changes, notably in auxin transcriptional repressor
OsIAA14 (
Os03g0797800), PGL1 antagonist
OsPIL16 (
Os05g0139100), and ABA receptor
OsPYL4 (
Os03g0297600) (
Figure 4B). Enhanced ROS scavenging capacity was corroborated by the differential expression of catalase
OsCatC (
Os03g0131200) in peroxisome pathways (
Figure 4B). In the binding and catalytic activity pathways, diterpenoid biosynthesis gene
OsKSL12 (
Os02g0568700) and pyruvate phosphate dikinase
OsPPDKA (
Os03g0432100)—a carbon metabolism gene implicated in drought regulation—showed significant expression shifts (
Figure 5D). Intracellular pathways were further enriched for catalase
OsCatC (
Os03g0131200) and ferredoxin
OsFdC1 (
Os03g0659200), the latter maintaining redox homeostasis via photosynthetic electron transport under drought-induced ROS accumulation (
Figure 5D). The key genes identified in this study are systematically presented (
Table 1), which documents their differential expression profiles (upregulated/downregulated) under drought stress between: (i) wild-type ZH11 vs. overexpression lines, and (ii) knockout mutants vs. overexpression lines.
3. Discussion
As one of the largest transcription factor families in plants, the rice NAC family comprises 151 members [
10,
26]. This study focused on the functional characterization of
OsNAC25, revealing its dual expression activity in both the nucleus and cytoplasm, with its transcript levels being significantly induced by multiple abiotic stresses, particularly showing the most pronounced induction under drought stress. To elucidate its drought-responsive mechanism, we generated
OsNAC25 gene-edited materials (selecting two homozygous mutants,
OsNAC25-10-cr and
OsNAC25-16-cr) and overexpression lines (choosing those with highest expression levels,
OsNAC25-2-OE and
OsNAC25-16-OE) (
Supplementary Figure S7A,B). The phenotypic analysis demonstrated that overexpression lines exhibited markedly enhanced drought tolerance compared to ZH11under water deficit, whereas no significant phenotypic divergence was observed between gene-edited materials and ZH11 (
Figure 2A and
Figure 3A). A further investigation revealed a substantially higher expression abundance of
OsNAC25 in aerial tissues than in subterranean organs. The GUS histochemical staining analysis revealed that the gene exhibits a tissue-specific inducible expression pattern under drought stress conditions, with its promoter activity being specifically activated in the leaf and stem base regions, while no detectable activity was observed under normal growth conditions (
Supplementary Figure S7C,D). These findings indicate that
OsNAC25 primarily functions through aerial tissue-specific expression induced by drought stress. This contrasts with previously reported NACs from the SNAC subgroup that predominantly exhibit stronger root-specific than leaf-specific expression patterns when responding to abiotic stresses [
27,
28,
29]. Therefore, unlike SNACs,
OsNAC25 principally operates as a stress-inducible transcription factor in aboveground plant tissues.
The overexpression of
OsNAC25 enhances drought stress tolerance in transgenic materials, accompanied by elevated activities of peroxidase (POD) and catalase (CAT), reduced levels of malondialdehyde (MDA)—a harmful oxidative damage marker— and decreased superoxide dismutase (SOD) activity. Plants have evolved a sophisticated antioxidant system comprising both non-enzymatic and enzymatic components [
30,
31]. While sustained high levels of antioxidant enzymes (POD, CAT, SOD) are generally associated with ROS scavenging and stress tolerance, we observed a paradoxical reduction in SOD activity in overexpression lines. This suggests that the oxidative process involves more than just the connection between a single organelle and the nucleus. The cellular redox signaling hub may be seen as a key integrator of retrograde signals arising from different organelles, allowing communication between different cellular compartments and the nucleus [
32]. This divergence suggests that
OsNAC25 may not participate in SOD-mediated regulatory pathways.
RNA-seq analysis of
OsNAC25 material: The transcriptomic profiling of
OsNAC25 focused on cellular/metabolic processes, biological regulation, response to stimuli, cellular anatomical entities, intracellular organelle functions, molecular binding, and catalytic activity (
Figure 4E,F). The most significant pathways associated with cellular/metabolic processes were phenylpropanoid biosynthesis and glutathione metabolism. The phenylpropanoid biosynthesis pathway was predominantly enriched with peroxidase- and β-glucosidase-related genes. Plants possess antioxidant systems to mitigate oxidative damage, and these pathways directly correlate with enhanced antioxidant capacity, playing a pivotal role in improving drought tolerance [
33]. Glutathione is critically involved in drought stress responses and ROS scavenging [
34]. Under drought conditions, rice crops exhibit a well-documented reduction in glutathione levels, followed by a significant compensatory increase as an adaptive response [
31]. Multiple studies have proposed glutathione depletion as a potential biomarker for screening drought-resistant plant varieties [
34,
35]. Regarding biological regulation and stress responses, phytohormone signaling transduction—particularly pathways involving IAA (indole-3-acetic acid) and ABA (abscisic acid)—emerged as a crucial mechanism influencing plant growth and stress adaptation. Our KEGG enrichment analysis revealed substantial modifications in multiple phytohormone biosynthesis pathways.
Numerous binding pathways and catalytic activity-related processes were significantly influenced by
OsNAC25. Notably,
OsNAC25 modulated diterpenoid biosynthesis through pronounced effects on carbon metabolism and GA44-dioxygenase synthesis, establishing a direct linkage to drought stress adaptation [
36]. Photosynthesis, the primary driver of plant growth, provides essential energy for organic compound synthesis [
36,
37]. Extensive crop improvement research has prioritized the identification of quantitative trait loci (QTLs) associated with photosynthetic enhancement [
38]. Ferredoxin-encoding genes, integral to photosynthetic electron transport, maintain intracellular redox homeostasis. Intracellular organelles coordinate a complex network of thousands of genes governing metabolic, signaling, and biosynthetic functions [
39]. Hierarchical clustering analysis further elucidated drought-responsive molecular mechanisms, revealing
OsNAC25’s systemic regulatory role across these biological hierarchies.
Collectively, our integrated transcriptomic and physiological analyses established OsNAC25 as a nucleo-cytoplasmic transcription factor that regulates drought resilience in rice. OsNAC25 enhances drought tolerance by reducing oxidative damage, promoting ROS scavenging, modulating hormone signaling, and activating antioxidant-related pathways. This discovery offers a promising target for the molecular breeding of drought-resistant crops.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Wild-type rice (Oryza sativa cv. Zhonghua 11, ZH11) seeds were used in this study. All rice lines were propagated in paddy fields at Hunan Agricultural University. Rice seedlings were cultivated in a greenhouse under a 16-hour light/8-hour dark photoperiod at 30 °C. Transgenic overexpression lines were constructed using the OsNAC25 (LOC_Os11g31330) coding sequence (CDS) cloned into a modified T-DNA vector under the control of the ubiquitin promoter, with a 3 × FLAG tag fused to its C-terminus. OsNAC25 knockout mutants were generated via the pYLCRISPR/Cas9Pubi-H system. All plants were screened for homozygous lines at the T2 generation.
4.2. Subcellular Localization of OsNAC25 in Rice Protoplasts
To investigate the subcellular localization of OsNAC25, the primers mCherry-OsNAC25-F and mCherry-OsNAC25-R were designed using Vector NTI. The full-length coding sequence (CDS) of OsNAC25 was amplified from wild-type ZH11 cDNA and cloned into a fusion expression vector containing the mCherry fluorescent tag. The resulting mCherry-OsNAC25 construct was co-transfected with the RPL-CFP nuclear localization marker into rice protoplasts via PEG4000-mediated transformation. Transfected protoplasts were incubated in darkness at 28 °C for 16 h, and fluorescence signals were visualized using a Zeiss LSM780 confocal laser microscope (Carl Zeiss, Jena, Germany).
4.3. RNA Extraction and Quantitative Real-Time RT-PCR (qRT-PCR) Analysis
Total RNA was extracted from rice seedling leaves using the TransZol Up Plus RNA Kit (TransGen Biotech, Beijing, China). First-strand cDNA synthesis was performed with ≥1 μg of total RNA using the HiScript First Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). Gene expression levels were analyzed by qRT-PCR with Hieff qPCR SYBR Green Master Mix (YEASEN, Shanghai, China). Data were normalized to the reference gene Actin1 and quantified using the relative quantitative method (2−ΔΔCT method). Three biological replicates were conducted for each experiment. The primers used for qRT-PCR in this study are listed in
Table S2.
4.4. Enzyme Activity Assays
To determine the activities of MDA (malondialdehyde), CAT (catalase), SOD (superoxide dismutase), and POD (peroxidase), the corresponding assay kits (Solarbio Science & Technology Co., Ltd., Beijing, China) were employed according to the manufacturers’ protocols. Fresh rice seedling leaf samples were collected and immediately frozen in liquid nitrogen. For analysis, 0.1 g of tissue was homogenized in 1 mL of ice-cold extraction buffer. The homogenate was centrifuged at 8000× g for 10 min at 4 °C, and the supernatant was collected for enzymatic activity assays.
4.5. Transcriptome Analysis
To investigate the functional role of OsNAC25 in drought stress, RNA sequencing (RNA-seq) was conducted using wild-type (ZH11), OsNAC25-cr (CRISPR/Cas9 knockout mutants), and OsNAC25-OE (overexpression) lines. The experiment used 10-day-old seedlings grown under normal conditions as the controls. The treatment groups were grown under identical standard conditions for 9 days until reaching the two-leaf-one-heart stage, and then subjected to 24-hour drought stress treatment. For RNA extraction, a pooled sampling approach was adopted: each sample (e.g., OsNAC25-cr under control conditions) comprised a homogenate of tissues from eight individual plants, representing one biological replicate. Three biological replicates were established per treatment group, resulting in six experimental groups (ZH11, OsNAC25-cr, and OsNAC25-OE, each with control and drought-treated subgroups) and 18 total samples. Sequencing was performed by Baimaike Biotechnology (Beijing, China). The raw data were quality assessed using FastQC, followed by adapter and low-quality read removal (Q-score < 20) with Trimmomatic. Differential expression analysis was performed using the DESeqR package (1.10.1) with stringent thresholds (|log2(fold change)| > 1 and adjusted p-value < 0.01). The gene ontology (GO) enrichment analysis of differentially expressed genes was conducted using the clusterProfiler R (4.4.4) package, with KEGG pathway enrichment analysis also performed using the same package, using the whole-genome background as reference for statistical significance (adjusted p-value < 0.05). To validate RNA-seq reliability, eight randomly selected genes were analyzed via qRT-PCR using Actin as the internal reference gene. Relative expression levels were quantified before and after drought treatment. All assays included three biological replicates under rigorously standardized conditions.